Abstract
The long-term stability of protein therapeutics in the solid-state depends on the preservation of native structure during lyophilization and in the lyophilized powder. Proteins can reversibly or irreversibly unfold upon lyophilization, acquiring conformations susceptible to degradation during storage. Therefore, characterizing proteins in the dried state is crucial for the design of safe and efficacious formulations. This review summarizes the basic principles and applications of the analytical techniques that are commonly used to characterize protein structure, dynamics and conformation in lyophilized solids. The review also discusses the applications of recently developed mass spectrometry based methods (solid-state hydrogen deuterium exchange mass spectrometry (ssHDX-MS) and solid-state photolytic labeling mass spectrometry (ssPL-MS)) and their ability to study proteins in the solid-state at high resolution.
Keywords: Solid-state, lyophilization, protein structure, protein dynamics, protein conformations, protein stability, mass spectrometry, spectroscopy
INTRODUCTION
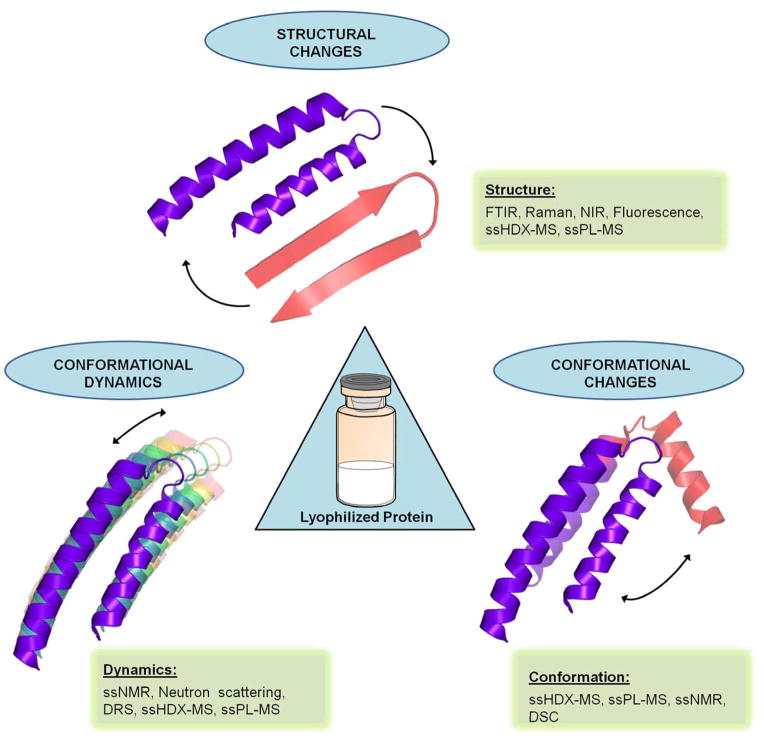
Recombinant therapeutic proteins offer promising treatments for many previously incurable diseases such as cancers, autoimmune diseases and hormone-related disorders. In 2013, the global biotherapeutic market reached $200.6 billion and is expected to reach $386.7 billion by the end of 2019 [1]. Because of the highly complex structure of proteins and their susceptibility to chemical and physical degradation, the successful production of therapeutic proteins can be challenging. To reduce degradation rates, protein drugs are often formulated as lyophilized (freeze-dried) solid-powders. However, degradation can still occur in the solid-state, particularly if the native structure is not retained during lyophilization (Figure 1).
Figure 1.
Structural properties of proteins in lyophilized solids and methods used for their characterization. Proteins can exhibit reversible or irreversible change in structural properties, which include change in secondary structure, tertiary structure, conformational dynamics and/or conformational changes upon lyophilization.
The development of a successful lyophilized protein drug includes careful selection of excipients and the lyophilization cycle. The lyophilization process involves freezing of the protein solution followed by the removal of water from the frozen solid under vacuum. These processes generate various stresses including ice crystal formation, increased local solute concentration and pH changes that can damage protein structure. Stabilizing agents such as low molecular weight disaccharides (e.g. sucrose and trehalose) are often included to protect protein structure during freezing and drying. Several studies have reported the mechanisms of molecular interactions by which protein stability is protected during lyophilization and in lyophilized powders [2–4]. The glassy nature of the solid matrix, typically an amorphous solids below its glass transition temperature (Tg), and the formation of hydrogen bonds between the protein molecule and excipients have been shown to be crucial for stability [5, 6].
Disaccharides with higher Tg have been shown to be better stabilizers in several studies [2, 7–10], while others have show that Tg alone does not determine the degree of protein stability in the solid-state [11–13]. Although any changes in the local environment can affect protein stability in the solid matrix, identifying the direct relationship between protein structural changes and stability is limited by the lack of high-resolution methods to analyze proteins in amorphous solids. Thus, there is a need for solid-state analytical techniques that can probe protein structure at high resolution. If available, such methods could be used to identify the most promising formulations, reducing our reliance on expensive and time consuming storage stability studies for formulation screening.
In this review, the basic principles of the techniques to characterize proteins in the solid-state are discussed briefly with selected examples. Fourier transform infrared (FTIR) spectroscopy, Raman spectroscopy and near-infrared (NIR) spectroscopy are useful for studying protein secondary structural changes in the solid-state, and fluorescence spectroscopy can monitor tertiary structural changes to a certain extent. Solid-state nuclear magnetic resonance (ssNMR) and neutron scattering are commonly employed to study protein dynamics in solid matrices. Differential scanning calorimetry (DSC) is used to characterize protein stability. Solid-state hydrogen-deuterium exchange mass spectrometry (ssHDX-MS) and photolytic labeling mass spectrometry (ssPL-MS) are relatively new techniques that are capable of mapping protein structure and conformations at high resolution in lyophilized solids (Table 1).
Table 1.
Methods used for the structural characterization of proteins in lyophilized powders.
| Method | Measurements | Structural Information | Level of Changes Measured |
|---|---|---|---|
| FTIR | C=O, N-H & C-N vibrations | Secondary structure | Global |
| Raman | C=O, N-H & C-N vibrations | Secondary structure | Global |
| NIR | C-H, N-H & O-H vibrations | Secondary structure | Global |
| Fluorescence | Fluorescence emission of aromatic amino acid residues (Trp, Tyr, Phe) | Tertiary structure | Local |
| DSC | Change in heat capacity at Tg | Conformation | Global |
| ssNMR | 1H, 13C, 15N chemical shifts | Dynamics & conformation | Global and local |
| Neutron Scattering | Neutron scattering by hydrogen atoms | Dynamics | Global and local |
| DRS | Rotational motions of dipole-bearing groups | Dynamics | Global and local |
| ssHDX-MS | Amide hydrogens exchanged with deuterium | Tertiary structure, dynamics & conformation | Global and local |
| ssPL-MS | Side-chains labeled with photolytic agent | Tertiary structure, dynamics & conformation | Global and local |
Tg = glass transition temperature
Global = domains of protein or whole protein molecule
Local = specific regions or amino acid residues of protein
FOURIER TRANSFORM INFRARED SPECTROSCOPY (FTIR)
Infrared (IR) vibrational spectroscopy is used to assess protein secondary structural changes in solution and in the solid-state. Proteins have several vibrational modes, of which the amide I region (1650 cm−1; mainly from stretching of the C=O carbonyl group of the peptide bond) and amide II region (1550 cm−1; mainly from N-H in-plane bending) are sensitive to changes in secondary structure [14]. The absorption of IR light by certain functional groups produces a unique peak in the ‘fingerprint region’ (~1500 to 500 cm−1). The hydrogen-bonding pattern of proteins differs based on the type and amount of secondary structural elements present (i.e. α-helices, β-sheets or β-turns). This results in differences in IR absorption by the polypeptide backbone, which are observed in the IR spectrum. IR signals typically consist of multiple overlapping bands that are poorly resolved. Hence, mathematical manipulations such as Fourier self-deconvolution and second derivative spectra are commonly used for band narrowing and enhanced signal resolution.
Structural changes in lyophilized protein formulations have been studied using FTIR spectroscopy [15, 16]. Prestrelski et al demonstrated that amide I spectral features of freeze-dried granulocyte colony stimulating factor (G-CSF) were similar to those in solution, indicating retention of protein secondary structure [17]. Rehydration did not cause further spectral changes. In the same study, α-lactalbumin showed reversible conformational changes upon lyophilization and rehydration, whereas irreversible structural changes were detected for γ-interferon (γ-IFN), α-casein and basic fibroblast growth factor (bFGF) after lyophilization and rehydration. The FTIR spectra of proteins in the pre-lyophilized, lyophilized and rehydrated states were similar in the presence of disaccharides, suggesting the preservation of native protein structure by disaccharides in the solid-state. However, the FTIR spectra of γ-IFN lyophilized in the presence of mannitol or myoinositol differed from the spectra of the native form, consistent with perturbation of γ-IFN structure during lyophilization.
To a certain extent, increasing the amount of various carbohydrates in freeze-dried formulations generally results in increased intensity of the native protein FTIR band [6, 18]. A negative correlation between the degree of FTIR-derived native structure retention and aggregation propensity has been reported for lyophilized human growth hormone (hGH) [19], immunoglobulin G1 (IgG1)-sucrose formulation [20], lactate dehydrogenase [21] and γ-IFN [22]. FTIR band areas have also shown some correlation with moisture content in freeze-dried solids [6, 23]. In situ IR spectroscopy was used by Remmele et al to monitor lyophilization-induced changes in lysozyme [24]; however, the studies were carried out in D2O to avoid signal interference from water, limiting the use of IR spectroscopy for lyophilization process monitoring.
FTIR data are a low resolution, semi-quantitative measure of protein structure and are not always predictive of degradation rates in the solid-state. A limitation of FTIR is that it reflects global protein conformation and cannot detect subpopulations with local conformational changes that may affect stability. Aggregation due to changes in tertiary structure is also not detected by FTIR, highlighting the need for better resolution in the solid-state. FTIR measures such as peak height, bandwidth and spectral correlation coefficient (r) do not always correlate well with dehydrated protein secondary structure and degradation rate [4, 19, 25]. For example, studies with lyophilized myoglobin have shown that FTIR amide I band intensity and wavenumber correlated poorly with loss of monomer during storage [26]. Chang et al observed that native secondary structure retention, as measured by FTIR, correlated well with storage stability for sucrose-containing formulations, but did not in the presence of trehalose [20]. This implies that factors other than global secondary structure retention as measured by FTIR influence protein stability in the solid-state.
RAMAN SPECTROSCOPY
Raman spectroscopy is similar to IR spectroscopy in that both methods provide information about the nature of molecular vibrations. The difference between these techniques is that Raman spectroscopy measures the inelastic scattering of monochromatic light from polarizable molecules whereas IR spectroscopy measures light absorbed by molecules that undergo a change in dipole moment during vibration. This makes the two methods complementary. Added advantages of Raman spectroscopy are weak scattering and reduced noise due to water and ice, making it useful to study frozen solutions.
Raman spectroscopy has been used to examine the effects of freeze-thawing [27], presence of excipients [27, 28], lyophilization [29–31] and phase separation [32, 33] on protein backbone and side-chain conformation. This method has been used to quantify structural perturbation in a monoclonal antibody upon lyophilization or spray-drying [34]. Secondary structure content measured in the amide I region after lyophilization or spray-drying showed good correlation with storage stability. Reduced structural perturbation was observed for the solid-state protein with increasing amounts of carbohydrate excipients. Thus, Raman spectroscopy can be used as a tool to screen excipients in formulation design. Hedoux et al used Raman micro-spectroscopy to detect lyophilization-induced structural changes in β-lactoglobulin (βLg), bovine serum albumin and chymotrypsinogen in real time [35]. Spectral features related to protein structure were not altered significantly during freezing and rehydration. Peak broadening and frequency shifts were observed in the amide I and amide III regions in response to a faster cooling rate, as well as during primary drying, suggesting perturbed helices and increased solvent exposure of β-sheets. Spectra for lyophilized and thermally denatured proteins were not identical, indicating that spectral changes were not merely a result of increased temperature, but rather a consequence of vacuum-induced dehydration. In-line Raman spectroscopy has also been used to study the phase behavior of mannitol, a common bulking agent in lyophilized protein formulations [36]. Varying proportions of the amorphous form and α-, β-, δ- and hemihydrate polymorphs of mannitol were formed as a function of freezing rates and annealing. Interconversion between the forms was detected during secondary drying as well.
Like IR spectroscopy, Raman spectroscopy provides unique signals in the fingerprint region. Sample preparation is not required, making it simpler to use compared to other spectroscopic techniques. However, the inelastic Raman signal is inherently weak since most of the scattering is elastic Rayleigh scattering. Since heat generated by the laser light source may damage samples, spectra must be obtained quickly. Background fluorescence may also interfere with Raman signals.
NEAR INFRARED SPECTROSCOPY (NIR)
The measurement of C-H, N-H and O-H vibrations at frequencies in the combination (4000–5000 cm−1) and overtone (5000–13000 cm−1) spectral regions can be used to assess the conformation of proteins in the solid-state. The extent of intramolecular hydrogen bonding suggests the conformational stability of a protein in the solid-state. An unfolded protein contains fewer intramolecular hydrogen bonds than the properly folded molecule. In an NIR measurement, an increased or decreased extent of intramolecular hydrogen bonding is reflected by a decrease or increase in the A/II band (combination band) frequency, respectively [37, 38]. NIR spectra of freeze-dried proteins showing bands near 4369 and 4604 cm−1 have been associated with an α-helical structure, while bands at 4323, 4417, 4525–4535 cm−1 have been associated with a β-sheet structure [39, 40]. Using these band positions, NIR measurement of heat treated, lyophilized bovine serum albumin (BSA) showed an increase in β-sheet formation when compared to control [41].
NIR is non-destructive and non-invasive, and has several other advantages over FTIR and Raman spectroscopy for characterizing protein structure in the solid-state. Since the moisture content in the environment shows weak interference in the NIR spectra, the method does not require purging of the instrument with nitrogen gas, as required for FTIR. The data acquisition time for each sample in NIR is less than 2 min. Unlike FTIR, NIR does not require extensive data manipulation to obtain protein structural information. A comparison of secondary structural information from NIR and FTIR measurements showed strong correlation for lyophilized cytochrome c and α-chymotrypsinogen samples [18]. In another study, spectra from both NIR and FTIR indicated the presence of L-arginine and phosphate ion interactions in the solid-state [42]. NIR spectra of L-arginine co-lyophilized with phosphoric acid showed reduced intensity of the band for the amino or guanidyl group compared to L-arginine in the absence of phosphoric acid, consistent with the reduced intensity of signal for the NH3+ vibration absorbance in FTIR.
NIR has been shown to be valuable for monitoring protein conformation and stability during the entire lyophilization process, from the initial solution-state to the final solid product [42, 43]. Though NIR has gained acceptance in the biopharmaceutical industry, further research is required for the method to be routinely applied as an in-line tool for monitoring protein secondary structure during lyophilization.
SOLID-STATE FLUORESCENCE SPECTROSCOPY
Altered secondary structure in proteins is indicative of the loss of tertiary structure; however, observance of an unaltered secondary structure does not guarantee that the native tertiary structure has been retained. In a partially unfolded state, tertiary structure may be lost with little or no change in the secondary structure. In solution, when protein unfolding leads to the exposure of aromatic amino acids from a relatively hydrophobic core to the polar aqueous environment, a shift in the emission maximum (λmax) towards longer wavelength (i.e., a red shift) is observed. Conversely, the λmax shifts towards shorter wavelength (i.e., a blue shift) when aromatic residues are removed from an aqueous environment and buried in the hydrophobic core. Fluorescence spectroscopy has recently been used to study the tertiary structure of proteins in solid powders, in a manner similar to its use for proteins in aqueous solution [44, 45].
A limitation of solid-state fluorescence measurements is the strong background scattering of powders with high optical density. This problem can be largely overcome by analyzing the samples in the frontface mode. Few studies have used solid-state fluorescence spectroscopy to probe protein structure in amorphous powders. Steady-state Trp fluorescence spectroscopy has been used to study the tertiary structure of βLg and interferon alpha-2a (IFN-α2A) in lyophilized powders [44]. βLg lyophilized with guanidine hydrochloride was red shifted when compared to the pre-lyophilized sample. Similarly, emission spectra from polyethylene glycol (PEG)-induced IFN-α2A aggregates in freeze dried samples differed from those for the protein in solution. This shift in the λmax was associated with a change in the tertiary structure of βLg and IFN-α2A. In another study, a monoclonal antibody (mAb) lyophilized with sucrose and stored at higher temperature show decreased fluorescence intensity with greater aggregation [45]. Although fluorescence measurement offers the advantage over other spectroscopic techniques of providing tertiary structural information, the sensitivity of this method depends on the location of aromatic amino acids in the protein tertiary structure. An alternate approach may be co-lyophilization of proteins and low-molecular weight chromophores to probe the global structural changes. Freezing-induced structural perturbations in azurin have been monitored using 1-anilino-8-naphthalene sulfonate (ANS) as a fluorescent probe in the frozen state [46].
DIFFERENTIAL SCANNING CALORIMETRY (DSC)
Freeze-dried proteins are usually in a glassy state below their Tg, in which the rates of many chemical degradation reactions are reduced. The glassy state also restricts protein local motions and so may reduce physical degradation rates in the solid-state. DSC measures the change in heat capacity and produces a well-defined increase in signal at Tg, when the system changes from a glassy to rubbery state.
DSC is often used to supplement spectroscopic and chromatographic characterization of proteins in the solid-state. For example, Pikal et al used DSC in conjunction with FTIR and HPLC to study the degradation of lyophilized hGH at temperatures above Tg [47]. Aggregation was correlated with a loss of secondary structure and a decrease in area under the denaturation endotherm, and was reversible when trehalose was included in the formulation. In another study, Breen et al studied the chemical and physical stability of lyophilized monoclonal antibody formulations [48]. Increased moisture levels resulted in increased rates of Asp isomerization when stored above or below Tg, while aggregation rates increased with moisture levels when stored above, but not below Tg. Other studies have used DSC to investigate the mixing behavior of lyophilized protein-carbohydrate-polymer systems [38], to study the effect of protein on temperature-induced excipient crystallization [49] and to determine the physical form of carbohydrates in lyophilized protein formulations [50–52]. Modulated DSC (MDSC) is a related technique that uses sinusoidal changes in temperature to separate ‘non-reversing’ kinetic events (e.g. crystallization) from ‘reversing’ thermodynamic events (e.g. glass transition). ‘Reversing’ and ‘non-reversing’ heat flow components do not refer to the reversible or irreversible nature of events; they refer to the heat capacity component and time-dependent kinetic component of total heat flow, respectively. MDSC has been used to determine Tg values for systems with small changes in heat capacity [48], to correlate eutectic temperature events to cake collapse [53] and to study the effects of annealing on the thermal properties of frozen sucrose solutions [54].
DSC can be used to explore mixing behavior during lyophilization, since multiple glass transitions are detected in phase-separated systems. After ice crystallizes during lyophilization, increased effective solute concentration (freeze-concentration) may drive the formation of solute-rich and solute-poor phases. Several polymers as well as some small molecules are known to undergo phase separation upon freezing and show multiple Tg [55–57]. Proteins can partition into phase separated layers and subsequently degrade, as has been observed for hemoglobin and phase-separated PEG and dextran layers [58]. However, if the change in heat capacity associated with phase separation is small, it may not be detected by DSC.
Tg values are not always reliable predictors of protein stability. DSC was used to evaluate the stability of glucose-6-phosphate dehydrogenase (G6PD) during lyophilization and storage [13]. The pre-storage Tg value for G6PD co-lyophilized with 100 % sucrose was ~ 21 °C and increased with the inclusion of raffinose up to ~ 37 °C in the presence of 100 % raffinose. However, Tg values did not correlate with storage stability at temperatures above Tg. Greater G6PD activity (~ 80 %) was recovered with increasing sucrose ratios, while ~ 50 % activity was detected in the 100 % raffinose formulation after storage for 81 days.
Although formulations stored above their Tg are usually less stable than those stored below Tg, storage below Tg is not sufficient to ensure stability. Strickley and Anderson showed that lyophilized insulin stored at various humidity levels was degraded by deamidation and dimerization, despite remaining in the glassy state [59]. Although these degradation pathways are pH-mediated [60], there appeared to be sufficient mobility in the solid-state at < 12 % humidity for formation of the reactive intermediate. Similar deamidation and aggregation behavior below Tg has been reported for lyophilized rhIL1-ra, even in the presence of amorphous carbohydrate excipients [61]. Hence storage below Tg is necessary but not sufficient for storage stability, suggesting that DSC studies should be supplemented by other analytical methods when evaluating protein stability in lyophilized formulations.
SOLID-STATE NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY (ssNMR)
Measuring the radiofrequency radiation absorbed by the nuclei of 1H, 13C or 15N enables proteins to be characterized with atomic-level resolution. ssNMR has been widely used to study the structure of insoluble membrane proteins and amyloid fibrils. Unlike membrane proteins and amyloid fibrils, the orientations of proteins in amorphous solid are highly random, limiting the use of ssNMR for obtaining high resolution structural information. Nevertheless, ssNMR is an effective method for studying protein conformation and dynamics in lyophilized powders. ssNMR relaxation measurements have been used to monitor the mobility of protein side chains, surface groups, excipients and bound water molecules, and the results correlated to the stability of proteins in lyophilized solids [62–66]. Spin-lattice relaxation time (T1, ns-μs timescale), spin-lattice relaxation time in the rotating frame (T1ρ, μs-ms timescale) and spin-spin relaxation time (T2, ms-s timescale) can provide details of molecular motions in the solid-state. ssNMR spectroscopy can also be used to study protein-excipient interactions by measuring the differences in chemical shifts of functional groups of protein in different formulations.
T1 and T1ρ measurements were used to evaluate the molecular mobility of lysozyme lyophilized with different excipients [67, 68]. Lysozyme co-spray-dried with a high concentration of trehalose or sucrose and stored at 0% relative humidity showed decreased molecular mobility of the protein, suggesting a close association of protein and sugar [67]. In another study, an increase in T1 values was observed for lysozyme in the presence of lactose or trehalose, but gradually decreased with increasing hydration. These results correlated well with lysozyme aggregation and activity [68]. Yoshioka et al showed that the T1ρ of β-galactosidase (βGl) carbonyl carbon in the lyophilized βGl-sucrose formulation was less than in the βGl-stachyose system. The results correlated with the storage stability of βGl, with minimal aggregation observed for the sucrose containing formulation [66]. Similarly, the storage stability of insulin lyophilized with dextran and trehalose correlated with the T1ρ of the insulin carbonyl carbon, as measured by ssNMR. Formulation with dextran showed increased molecular motion and chemical degradation, whereas insulin lyophilized with trehalose showed minimal relaxation and greater stability [65].
Protein relaxation times have also been used to predict the storage stability in the solid-state. ssNMR data showed good correlation with the long-term storage stability of an IgG1 mAb. The spin-lattice relaxation time associated with protein (T1b) was similar in all the mAb formulation, whereas the spin-lattice relaxation time associated with mannitol (T1a) was lower in the mAb containing formulation when compared to mannitol alone. The decreased mannitol mobility was attributed to the inhibition of mannitol crystallization by the mAb through hydrogen bonding interactions. The lower T1a value correlated with greater protein stability during long-term storage [45]. Separovic et al studied the mobility of three different proteins (insulin, lysozyme and DNase) in lyophilized solids. A constant T1ρ and decreased T1 value with increase in the relative humidity was observed for these proteins. The change in the T1 value was associated with altered enzyme activity and aggregation [69].
NEUTRON SCATTERING SPECTROSCOPY
Protein motions in the solid-state are often influenced by extrinsic factors such as excipients, water content and temperature. Neutron scattering spectroscopy can be used to measure protein motions in lyophilized powders directly. The basic principles and types of neutron scattering have been explained in detail elsewhere [70–72]. In this method, the change in the intensity of neutron scattering by hydrogen atoms in the protein as a function of time is used to monitor protein dynamics. The effects of extrinsic factors on dynamics can be assessed by measuring the atomic mean-square displacements (MSD) in the protein molecule. Dynamic neutron scattering can also be used to probe protein-water interactions, in which the hydrogen bonds are formed and broken on the picosecond time scale [73].
The effects of temperature and moisture content on protein dynamics in lyophilized formulations have been studied using elastic neutron scattering [74, 75]. Measurement of lysozyme dynamics at various temperatures in a glucose-water matrix showed that protein motion is driven by the relaxation of the external matrix [74]. The dynamics of green fluorescent protein (GFP) at different hydration levels and temperatures showed that the water of hydration suppresses GFP dynamics at temperatures less than ~200K and increases protein motions at higher temperatures. When compared to other globular proteins, the suppression of GFP dynamics by water at lower temperatures may be due to its beta barrel structure [75].
Several studies have shown that the internal protein dynamics observed in neutron scattering varies with buffer composition in the lyophilized powder [76, 77]. For example, human butyrylcholinesterase lyophilized in Tris-HCl buffer showed no difference in protein internal motions when compared to a sample lyophilized in the salt-free state (control), while protein lyophilized in sodium phosphate buffer showed altered dynamics relative to control [76]. In another study, lysozyme lyophilized with trehalose at a 1:1 ratio show increased MSD values on the nanosecond time scale at low hydration (h = 0.075), suggesting that lysozyme preferentially interacted with water rather than with trehalose in the hydrated matrix. The relaxation of hydrogen bonds between lysozyme and water was slowed significantly in the presence of trehalose [77].
Neutron scattering results have also been shown to correlate well with the storage stability of lyophilized proteins [78–80]. Neutron scattering studies of lyophilized hGH showed greater suppression of fast dynamics in formulations containing sucrose than in those containing trehalose. The greater storage stability observed for the sucrose-containing formulations suggested that, well below the Tg, fast dynamics is directly related to protein instability [78]. Neutron scattering of deuterated protein powders allowed specific monitoring of motions of non-exchangeable hydrogen atoms in methyl groups and amino acid side chains [81].
Although neutron scattering can be complementary to the vibrational spectroscopy methods, one of its main disadvantages is that it requires a reactor for the neutron source, which limits accessibility and makes the method expensive and impractical for routine use. In addition, as the incident neutron flux is not high, the technique requires large samples for good quality data.
DIELECTRIC RELAXATION SPECTROSCOPY (DRS)
Dielectric relaxation spectroscopy (DRS) is a noninvasive method used to probe protein motions over a wide frequency range (from 10−6 to 105 s) both above and below the Tg. The theory behind DRS and its application to the characterization of pharmaceuticals have been discussed elsewhere [82–84]. In dielectric spectroscopy measurements, an electrical perturbation is applied across the sample and the resultant current is measured as the response. The response can be expressed in terms of the real and imaginary permittivity. The relaxation time is obtained as the inverse of the frequency maximum in the imaginary permittivity. Dielectric techniques essentially measure the rotational motions of dipole-bearing groups in the molecule. As the molecular motions are probed by stimulating different electrical dipole groups, the method allows clear differentiation between groups involved in global and local dynamics.
DRS can be used to evaluate the temperature dependence of the relaxation times by measuring the dielectric behavior of samples over a range of temperatures. DRS measurements have been used to probe the molecular mobility of a lyophilized mAb below the Tg and correlated with the stability of the mAb during storage [10]. Larger relaxation times were correlated with greater stability of the mAb in sucrose and trehalose containing formulations under refrigerated conditions. The effect of hydration on the dynamics of a protein in lyophilized powders has been studied using DRS measurements [85, 86]. The temperature-dependent relaxation time of lysozyme determined by neutron scattering spectroscopy has been shown to agree with the relaxation times observed by DRS [86]. Since the relaxation processes of water and protein overlap in hydrated lyophilized solids, the correct assignment of spectra in DRS can be challenging, and additional research is required before DRS is widely adopted in formulation development.
HYDROGEN-DEUTERIUM EXCHANGE MASS SPECTROMETRY (ssHDX-MS)
Hydrogen-deuterium exchange (HDX) has been widely used to study protein structure, protein-protein interactions and protein-ligand interactions in aqueous solution. When a protein is exposed to deuterium oxide (D2O) buffer, the hydrogen atoms of the backbone amide groups exchange with deuterons based on their solvent accessibility. Amide hydrogens exposed to solvent and not involved in intramolecular hydrogen bonding exchange with deuterons faster than those involved in the secondary structure. Measuring the rate and extent of deuterium uptake by the protein can provide information on its conformations under different conditions. HDX was traditionally coupled with NMR analysis to determine protein structure and dynamics at high resolution [87–90]. However, HDX-NMR requires large sample sizes and is typically limited to the analysis of smaller proteins. In HDX-MS, proteins with deuterated amide groups have a heavier mass that can be measured in any high resolution mass spectrometer (MS). HDX-MS requires very small sample sizes (picomoles of protein) and the size of the protein analyzed is limited only by the mass range of the instrument. Proteolysis of deuterated protein prior to MS analysis allows structural measurements at peptide level resolution. Over the last two decades, HDX-MS has been used extensively to characterize proteins from several thousand daltons to large macromolecular assemblies, including ribosomes and viral particles [91–93].
Recently, our group has adapted HDX-MS to characterize proteins in lyophilized formulations (ssHDX-MS). In ssHDX-MS, a protein in a lyophilized powder is exposed to D2O vapor under controlled relative humidity and temperature. The deuteration reaction (“in-exchange”) is quenched by reconstituting the sample in ice cold acidic buffer (pH ~2.7) and the sample subjected to online proteolytic digestion before MS analysis. In the solid-state, the level of deuteration may reflect the spatial distribution of conformational states and protein-excipient interactions as well as the conformational states of the protein molecule itself. Calmodulin lyophilized with trehalose and calcium chloride showed decreased deuterium uptake in different regions of the protein [94–96]. Formulations containing calcium chloride showed decreased deuteration in the calcium binding loops, whereas trehalose containing formulations showed protection mainly in the α-helix regions. ssHDX-MS of myoglobin lyophilized with sucrose showed decreased deuterium uptake when compared to a mannitol containing formulation [97, 98]. These studies suggest that proteins or regions with higher retention of native structure in the solid-state show greater protection from deuterium uptake.
Comparing mass spectral peak widths for deuterated protein at similar deuteration level provides information on the spatial and/or conformational heterogeneity of the protein in the lyophilized powders. Myoglobin freeze-dried with mannitol showed peak broadening at an intermediate level of deuterium uptake, suggesting greater heterogeneity when compared to a sucrose containing formulation [98]. In another study, myoglobin lyophilized alone or in the presence of sodium chloride showed ~2-fold greater peak width at 15% deuterium uptake than formulations containing sucrose or mannitol and showed greater aggregation during long-term storage [26].
A recent study has shown that deuterium incorporation in ssHDX-MS for five different myoglobin formulations was strongly correlated with the long-term storage stability of protein in the solid-state [26]. Myoglobin lyophilized with sucrose showed decreased deuterium uptake and greater storage stability than formulations with mannitol or sodium chloride. Myoglobin formations with higher sucrose content showed greater protection from HDX and improved stability. In contrast, storage stability correlated weakly with the FTIR band position or band intensity. Results such as these show the promise of ssHDX-MS as a screening tool for developing solid-state protein formulations, and support its further development.
PHOTOLYTIC LABELING MASS SPECTROMETRY (ssPL-MS)
Methods such as FTIR and DSC are limited in that they provide information at the bulk level. Although relatively simple and frequently used, they fail to provide high-resolution information about local protein structure and environment and are also not always predictive of stability, as described above. Photolytic labeling-mass spectrometry is a new technique being developed by our group that is complementary to ssHDX-MS and provides peptide- to residue-level information about the protein side-chain environment in the solid state. In ssPL-MS, a small photoactive labeling reagent is co-lyophilized with the protein and other formulation excipients. The resulting solid is irradiated with UV light to activate the photoactive label, which forms a stable covalent bond with matrix-accessible side-chains.
Labeling reagents used in solution are usually pH-dependent and amino acid-specific. For example, NHS esters hydrolyze at neutral to alkaline pH and predominantly label primary amine-containing residues. In contrast, UV-sensitive labeling reagents, such as photoactive amino acid analogs (PAAs; e.g. L-2-amino-4,4′ azipentanoic acid) are not pH sensitive and can be activated even in the solid-state. PAAs contain a photoreactive diazirine group that is activated at 350–365 nm and produces a reactive carbene that labels proteins by addition or insertion reactions. Diazirine-containing PAAs are thought to be non-selective and can label the entire protein surface, although there is some indication of carbene preference for acidic residues in solution [99].
In a recent study, the matrix exposure of lyophilized apomyoglobin side-chains was determined with peptide-level resolution using ssPL-MS [100]. Excipient effects on side-chain accessibility in the solid-state were also observed. Similar ssPL-MS studies, in conjunction with ssHDX-MS, have been performed for holomyoglobin lyophilized with trehalose or sorbitol [101]. The two methods provided complementary information, in that better structure retention coincided with lower deuterium uptake but greater PAA labeling, whereas structure perturbation resulted in greater deuterium uptake and lower PAA labeling. ssPL-MS is a qualitative method and allows the solid-state protein structure and environment to be probed with high resolution. However the stochastic nature of labeling and the promiscuity of the carbene result in populations of heterogeneously labeled protein [99, 100]. This characteristic, combined with mass spectrometric differences in ionization efficiency between labeled and unlabeled peptides, make quantification difficult. The limitation can be addressed by using an internal standard peptide to normalize abundance (peak height or area). However, one must be cautious when interpreting data, taking into account biasing factors such as possible sequestration of the labeling reagent, affinity of the reagent for certain residues, solid-state reactivity and concentration of the reagent. Nevertheless, the method shows considerable promise for mapping local protein-protein, protein-water and protein-excipient interactions in lyophilized solids with high resolution.
CONCLUSIONS
The increasing numbers of lyophilized protein therapeutics demand high resolution analytical methods for their characterization. FTIR and Raman spectroscopy are routinely used to measure secondary structure and batch-to-batch variations in biopharmaceutical products. Non-invasive and non-destructive techniques such as NIR can be further developed as in-line tools to monitor protein structure during the lyophilization process. Though solid-state fluorescence spectroscopy is not widely used in the biopharmaceutical industry, recent studies have demonstrated its capability to probe protein tertiary structure in amorphous solids. Protein dynamics monitored using ssNMR, DSC, neutron scattering and DRS have shown good correlation with the stability of proteins in the solid-state, but are not capable of localizing the observed effects to particular regions in proteins. Both ssHDX-MS and ssPL-MS are relatively new techniques that can provide complementary information about protein conformations and structural changes with peptide level resolution.
Acknowledgments
The authors gratefully acknowledge financial support from NIH RO1 GM085293.
Footnotes
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
References
- 1.Biotechnology. BCC Research. Feb, 2015. Biologic Therapeutic Drugs: Technologies and Global Markets. Report Code: BIO079C. [Google Scholar]
- 2.Wang W. Lyophilization and development of solid protein pharmaceuticals. Int J Pharm. 2000:1–60. doi: 10.1016/s0378-5173(00)00423-3. [DOI] [PubMed] [Google Scholar]
- 3.Carpenter JF, Chang BS, Garzon-Rodriguez W, Randolph TW. Rational design of stable lyophilized protein formulations: theory and practice. Pharm Biotechnol. 2002;13:109–33. doi: 10.1007/978-1-4615-0557-0_5. [DOI] [PubMed] [Google Scholar]
- 4.Chang L, Shepherd D, Sun J, Ouellette D, Grant KL, Tang XC, Pikal MJ. Mechanism of protein stabilization by sugars during freeze-drying and storage: native structure preservation, specific interaction, and/or immobilization in a glassy matrix? J Pharm Sci. 2005;94:1427–44. doi: 10.1002/jps.20364. [DOI] [PubMed] [Google Scholar]
- 5.Carpenter JF, Crowe JH. The mechanism of cryoprotection of proteins by solutes. Cryobiology. 1988:244–55. doi: 10.1016/0011-2240(88)90032-6. [DOI] [PubMed] [Google Scholar]
- 6.Allison SD, Chang B, Randolph TW, Carpenter JF. Hydrogen bonding between sugar and protein is responsible for inhibition of dehydration-induced protein unfolding. Arch Biochem Biophys. 1999;365:289–98. doi: 10.1006/abbi.1999.1175. [DOI] [PubMed] [Google Scholar]
- 7.Green JL, Angell CA. Phase relations and vitrification in saccharide-water solutions and the trehalose anomaly. The Journal of Physical Chemistry. 1989;93:2880–2882. [Google Scholar]
- 8.Bell LN, Hageman MJ, Muraoka LM. Thermally induced denaturation of lyophilized bovine somatotropin and lysozyme as impacted by moisture and excipients. J Pharm Sci. 1995;84:707–12. doi: 10.1002/jps.2600840608. [DOI] [PubMed] [Google Scholar]
- 9.Buitink J, van den Dries IJ, Hoekstra FA, Alberda M, Hemminga MA. High critical temperature above T(g) may contribute to the stability of biological systems. Biophys J. 2000:1119–28. doi: 10.1016/S0006-3495(00)76365-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duddu SP, Zhang G, Dal Monte PR. The relationship between protein aggregation and molecular mobility below the glass transition temperature of lyophilized formulations containing a monoclonal antibody. Pharm Res. 1997;14:596–600. doi: 10.1023/a:1012196826905. [DOI] [PubMed] [Google Scholar]
- 11.Cicerone MT, Soles CL. Fast dynamics and stabilization of proteins: binary glasses of trehalose and glycerol. Biophys J. 2004:3836–45. doi: 10.1529/biophysj.103.035519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buera MP, Rossi S, Moreno S, Chirife J. DSC confirmation that vitrification is not necessary for stabilization of the restriction enzyme EcoRI dried with saccharides. Biotechnol Prog. 1999:577–9. doi: 10.1021/bp990032u. [DOI] [PubMed] [Google Scholar]
- 13.Davidson P, Sun WQ. Effect of sucrose/raffinose mass ratios on the stability of co-lyophilized protein during storage above the Tg. Pharm Res. 2001;18:474–9. doi: 10.1023/a:1011002326825. [DOI] [PubMed] [Google Scholar]
- 14.Barth A, Zscherp C. What vibrations tell us about proteins. Q Rev Biophys. 2002;35:369–430. doi: 10.1017/s0033583502003815. [DOI] [PubMed] [Google Scholar]
- 15.Andya JD, Hsu CC, Shire SJ. Mechanisms of aggregate formation and carbohydrate excipient stabilization of lyophilized humanized monoclonal antibody formulations. AAPS PharmSci. 2003;5:E10. doi: 10.1208/ps050210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sinha S, Li Y, Williams TD, Topp EM. Protein conformation in amorphous solids by FTIR and by hydrogen/deuterium exchange with mass spectrometry. Biophys J. 2008;95:5951–61. doi: 10.1529/biophysj.108.139899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prestrelski SJ, Tedeschi N, Arakawa T, Carpenter JF. Dehydration-induced conformational transitions in proteins and their inhibition by stabilizers. Biophys J. 1993;65:661–71. doi: 10.1016/S0006-3495(93)81120-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bai S, Nayar R, Carpenter JF, Manning MC. Noninvasive determination of protein conformation in the solid state using near infrared (NIR) spectroscopy. J Pharm Sci. 2005;94:2030–8. doi: 10.1002/jps.20416. [DOI] [PubMed] [Google Scholar]
- 19.Costantino HR, Carrasquillo KG, Cordero RA, Mumenthaler M, Hsu CC, Griebenow K. Effect of excipients on the stability and structure of lyophilized recombinant human growth hormone. J Pharm Sci. 1998;87:1412–20. doi: 10.1021/js980069t. [DOI] [PubMed] [Google Scholar]
- 20.Chang LL, Shepherd D, Sun J, Tang XC, Pikal MJ. Effect of sorbitol and residual moisture on the stability of lyophilized antibodies: Implications for the mechanism of protein stabilization in the solid state. J Pharm Sci. 2005;94:1445–55. doi: 10.1002/jps.20363. [DOI] [PubMed] [Google Scholar]
- 21.Prestrelski SJ, Arakawa T, Carpenter JF. Separation of freezing- and drying-induced denaturation of lyophilized proteins using stress-specific stabilization. II. Structural studies using infrared spectroscopy. Arch Biochem Biophys. 1993;303:465–73. doi: 10.1006/abbi.1993.1310. [DOI] [PubMed] [Google Scholar]
- 22.Dong A, Prestrelski SJ, Allison SD, Carpenter JF. Infrared spectroscopic studies of lyophilization- and temperature-induced protein aggregation. J Pharm Sci. 1995;84:415–24. doi: 10.1002/jps.2600840407. [DOI] [PubMed] [Google Scholar]
- 23.Careri G, Gratton E, Yang PH, Rupley JA. Correlation of IR spectroscopic, heat capacity, diamagnetic susceptibility and enzymatic measurements on lysozyme powder. Nature. 1980;284:572–3. doi: 10.1038/284572a0. [DOI] [PubMed] [Google Scholar]
- 24.Remmele RL, Jr, Stushnoff C, Carpenter JF. Real-time in situ monitoring of lysozyme during lyophilization using infrared spectroscopy: dehydration stress in the presence of sucrose. Pharm Res. 1997;14:1548–55. doi: 10.1023/a:1012170116311. [DOI] [PubMed] [Google Scholar]
- 25.Wang B, Tchessalov S, Warne NW, Pikal MJ. Impact of sucrose level on storage stability of proteins in freeze-dried solids: I. Correlation of protein-sugar interaction with native structure preservation. J Pharm Sci. 2009;98:3131–44. doi: 10.1002/jps.21621. [DOI] [PubMed] [Google Scholar]
- 26.Moorthy BS, Schultz SG, Kim SG, Topp EM. Predicting Protein Aggregation during Storage in Lyophilized Solids Using Solid State Amide Hydrogen/Deuterium Exchange with Mass Spectrometric Analysis (ssHDX-MS) Mol Pharm. 2014;11:1869–79. doi: 10.1021/mp500005v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roessl U, Leitgeb S, Pieters S, De Beer T, Nidetzky B. In situ protein secondary structure determination in ice: Raman spectroscopy-based process analytical tool for frozen storage of biopharmaceuticals. J Pharm Sci. 2014;103:2287–95. doi: 10.1002/jps.24072. [DOI] [PubMed] [Google Scholar]
- 28.Hedoux A, Paccou L, Achir S, Guinet Y. Mechanism of protein stabilization by trehalose during freeze-drying analyzed by in situ micro-raman spectroscopy. J Pharm Sci. 2013;102:2484–94. doi: 10.1002/jps.23638. [DOI] [PubMed] [Google Scholar]
- 29.Pieters S, Vander Heyden Y, Roger JM, D’Hondt M, Hansen L, Palagos B, De Spiegeleer B, Remon JP, Vervaet C, De Beer T. Raman spectroscopy and multivariate analysis for the rapid discrimination between native-like and non-native states in freeze-dried protein formulations. Eur J Pharm Biopharm. 2013;85:263–71. doi: 10.1016/j.ejpb.2013.03.035. [DOI] [PubMed] [Google Scholar]
- 30.Yu NT, Jo BH. Comparison of protein structure in crystals and in solution by laser raman scattering. I. Lysozyme. Arch Biochem Biophys. 1973;156:469–74. doi: 10.1016/0003-9861(73)90296-8. [DOI] [PubMed] [Google Scholar]
- 31.Yu NT. Comparison of protein structure in crystals, in lyophilized state, and in solution by laser Raman scattering. 3. Alpha-Lactalbumin. J Am Chem Soc. 1974;96:4664–8. doi: 10.1021/ja00821a049. [DOI] [PubMed] [Google Scholar]
- 32.Padilla AM, Ivanisevic I, Yang Y, Engers D, Bogner RH, Pikal MJ. The study of phase separation in amorphous freeze-dried systems. Part I: Raman mapping and computational analysis of XRPD data in model polymer systems. J Pharm Sci. 2011;100:206–22. doi: 10.1002/jps.22269. [DOI] [PubMed] [Google Scholar]
- 33.Padilla AM, Pikal MJ. The study of phase separation in amorphous freeze-dried systems, part 2: investigation of Raman mapping as a tool for studying amorphous phase separation in freeze-dried protein formulations. J Pharm Sci. 2011;100:1467–74. doi: 10.1002/jps.22380. [DOI] [PubMed] [Google Scholar]
- 34.Sane SU, Wong R, Hsu CC. Raman spectroscopic characterization of drying-induced structural changes in a therapeutic antibody: correlating structural changes with long-term stability. J Pharm Sci. 2004;93:1005–18. doi: 10.1002/jps.20014. [DOI] [PubMed] [Google Scholar]
- 35.Hedoux A, Paccou L, Achir S, Guinet Y. In situ monitoring of proteins during lyophilization using micro-Raman spectroscopy: a description of structural changes induced by dehydration. J Pharm Sci. 2012;101:2316–26. doi: 10.1002/jps.23172. [DOI] [PubMed] [Google Scholar]
- 36.Kauppinen A, Toiviainen M, Aaltonen J, Korhonen O, Jarvinen K, Juuti M, Pellinen R, Ketolainen J. Microscale freeze-drying with Raman spectroscopy as a tool for process development. Anal Chem. 2013;85:2109–16. doi: 10.1021/ac3027349. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y, Cho R-K, Sakurai K, Miura T, Ozaki Y. Studies on Spectra/Structure Correlations in Near-Infrared Spectra of Proteins and Polypeptides. Applied Spectroscopy. 1994;48:6. [Google Scholar]
- 38.Katayama DS, Carpenter JF, Menard KP, Manning MC, Randolph TW. Mixing properties of lyophilized protein systems: a spectroscopic and calorimetric study. J Pharm Sci. 2009;98:2954–69. doi: 10.1002/jps.21467. [DOI] [PubMed] [Google Scholar]
- 39.Robert P, Devaux MF, Mouhous N, Dufour E. Monitoring the Secondary Structure of Proteins by Near-Infrared Spectroscopy. Applied Spectroscopy. 1999;53:226–232. [Google Scholar]
- 40.Miyazawa M, Sonoyama M. Second derivative near infrared studies on the structural characterisation of proteins. Journal of Near Infrared Spectroscopy. 1998;6:253–257. [Google Scholar]
- 41.Izutsu K, Fujimaki Y, Kuwabara A, Hiyama Y, Yomota C, Aoyagi N. Near-infrared analysis of protein secondary structure in aqueous solutions and freeze-dried solids. J Pharm Sci. 2006;95:781–9. doi: 10.1002/jps.20580. [DOI] [PubMed] [Google Scholar]
- 42.Izutsu K, Fujimaki Y, Kuwabara A, Aoyagi N. Effect of counterions on the physical properties of l-arginine in frozen solutions and freeze-dried solids. Int J Pharm. 2005:161–9. doi: 10.1016/j.ijpharm.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 43.Pieters S, Beer TD, Heyden YV. Near-infrared and Raman Spectroscopy: Potential Tools for Monitoring of Protein Conformational Instability during Freeze-drying Processes. The Review of American Pharmaceutical Business & Technology. 2012 [Google Scholar]
- 44.Sharma VK, Kalonia DS. Steady-state tryptophan fluorescence spectroscopy study to probe tertiary structure of proteins in solid powders. J Pharm Sci. 2003;92:890–9. doi: 10.1002/jps.10354. [DOI] [PubMed] [Google Scholar]
- 45.Park J, Nagapudi K, Vergara C, Ramachander R, Laurence JS, Krishnan S. Effect of pH and excipients on structure, dynamics, and long-term stability of a model IgG1 monoclonal antibody upon freeze-drying. Pharm Res. 2013;30:968–84. doi: 10.1007/s11095-012-0933-z. [DOI] [PubMed] [Google Scholar]
- 46.Gabellieri E, Strambini GB. Perturbation of protein tertiary structure in frozen solutions revealed by 1-anilino-8-naphthalene sulfonate fluorescence. Biophys J. 2003:3214–20. doi: 10.1016/S0006-3495(03)74739-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pikal MJ, Rigsbee D, Roy ML. Solid state stability of proteins III: calorimetric (DSC) and spectroscopic (FTIR) characterization of thermal denaturation in freeze dried human growth hormone (hGH) J Pharm Sci. 2008;97:5122–31. doi: 10.1002/jps.21386. [DOI] [PubMed] [Google Scholar]
- 48.Breen ED, Curley JG, Overcashier DE, Hsu CC, Shire SJ. Effect of moisture on the stability of a lyophilized humanized monoclonal antibody formulation. Pharm Res. 2001;18:1345–53. doi: 10.1023/a:1013054431517. [DOI] [PubMed] [Google Scholar]
- 49.Costantino HR, Curley JG, Wu S, Hsu CC. Water sorption behavior of lyophilized protein–sugar systems and implications for solid-state interactions. International Journal of Pharmaceutics. 1998;166:211–221. [Google Scholar]
- 50.Han Y, Jin B-S, Lee S-B, Sohn Y, Joung J-W, Lee J-H. Effects of sugar additives on protein stability of recombinant human serum albumin during lyophilization and storage. Archives of Pharmacal Research. 2007;30:1124–1131. doi: 10.1007/BF02980247. [DOI] [PubMed] [Google Scholar]
- 51.Carpenter JF, Prestrelski SJ, Arakawa T. Separation of freezing- and drying-induced denaturation of lyophilized proteins using stress-specific stabilization. I. Enzyme activity and calorimetric studies. Arch Biochem Biophys. 1993;303:456–64. doi: 10.1006/abbi.1993.1309. [DOI] [PubMed] [Google Scholar]
- 52.Cleland JL, Lam X, Kendrick B, Yang J, Yang TH, Overcashier D, Brooks D, Hsu C, Carpenter JF. A specific molar ratio of stabilizer to protein is required for storage stability of a lyophilized monoclonal antibody. J Pharm Sci. 2001;90:310–21. doi: 10.1002/1520-6017(200103)90:3<310::aid-jps6>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 53.Duru C, Swann C, Dunleavy U, Mulloy B, Matejtschuk P. The importance of formulation in the successful lyophilization of influenza reference materials. Biologicals. 2015;43:110–6. doi: 10.1016/j.biologicals.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 54.Chang L, Milton N, Rigsbee D, Mishra DS, Tang X, Thomas LC, Pikal MJ. Using modulated DSC to investigate the origin of multiple thermal transitions in frozen 10% sucrose solutions. Thermochimica Acta. 2006;444:141–147. [Google Scholar]
- 55.Izutsu K, Aoyagi N, Kojima S. Effect of polymer size and cosolutes on phase separation of poly(vinylpyrrolidone) (PVP) and dextran in frozen solutions. J Pharm Sci. 2005;94:709–17. doi: 10.1002/jps.20292. [DOI] [PubMed] [Google Scholar]
- 56.Akers MJ, Milton N, Byrn SR, Nail SL. Glycine crystallization during freezing: the effects of salt form, pH, and ionic strength. Pharm Res. 1995;12:1457–61. doi: 10.1023/a:1016223101872. [DOI] [PubMed] [Google Scholar]
- 57.Izutsu K-i, Shigeo K. Phase separation of polyelectrolytes and non-ionic polymers in frozen solutions. Physical Chemistry Chemical Physics. 2000;2:123–127. [Google Scholar]
- 58.Heller MC, Carpenter JF, Randolph TW. Manipulation of lyophilization-induced phase separation: implications for pharmaceutical proteins. Biotechnol Prog. 1997;13:590–6. doi: 10.1021/bp970081b. [DOI] [PubMed] [Google Scholar]
- 59.Strickley RG, Anderson BD. Solid-state stability of human insulin. II. Effect of water on reactive intermediate partitioning in lyophiles from pH 2–5 solutions: stabilization against covalent dimer formation. J Pharm Sci. 1997;86:645–53. doi: 10.1021/js9700311. [DOI] [PubMed] [Google Scholar]
- 60.Darrington RT, Anderson BD. The role of intramolecular nucleophilic catalysis and the effects of self-association on the deamidation of human insulin at low pH. Pharm Res. 1994;11:784–93. doi: 10.1023/a:1018909220255. [DOI] [PubMed] [Google Scholar]
- 61.Chang BS, Beauvais RM, Dong A, Carpenter JF. Physical factors affecting the storage stability of freeze-dried interleukin-1 receptor antagonist: glass transition and protein conformation. Arch Biochem Biophys. 1996;331:249–58. doi: 10.1006/abbi.1996.0305. [DOI] [PubMed] [Google Scholar]
- 62.Yoshioka S, Aso Y, Kojima S, Sakurai S, Fujiwara T, Akutsu H. Molecular mobility of protein in lyophilized formulations linked to the molecular mobility of polymer excipients, as determined by high resolution 13C solid-state NMR. Pharm Res. 1999;16:1621–5. doi: 10.1023/a:1018973125010. [DOI] [PubMed] [Google Scholar]
- 63.Yoshioka S, Aso Y, Izutsu K, Terao T. Stability of beta-galactosidase, a model protein drug, is related to water mobility as measured by 17O nuclear magnetic resonance (NMR) Pharm Res. 1993;10:103–8. doi: 10.1023/a:1018933315538. [DOI] [PubMed] [Google Scholar]
- 64.Yoshioka S, Aso Y, Kojima S. The effect of excipients on the molecular mobility of lyophilized formulations, as measured by glass transition temperature and NMR relaxation-based critical mobility temperature. Pharm Res. 1999;16:135–40. doi: 10.1023/a:1018891317006. [DOI] [PubMed] [Google Scholar]
- 65.Yoshioka S, Miyazaki T, Aso Y. Beta-relaxation of insulin molecule in lyophilized formulations containing trehalose or dextran as a determinant of chemical reactivity. Pharm Res. 2006;23:961–6. doi: 10.1007/s11095-006-9907-3. [DOI] [PubMed] [Google Scholar]
- 66.Yoshioka S, Miyazaki T, Aso Y, Kawanishi T. Significance of local mobility in aggregation of beta-galactosidase lyophilized with trehalose, sucrose or stachyose. Pharm Res. 2007;24:1660–7. doi: 10.1007/s11095-007-9296-2. [DOI] [PubMed] [Google Scholar]
- 67.Suihko EJ, Forbes RT, Apperley DC. A solid-state NMR study of molecular mobility and phase separation in co-spray-dried protein-sugar particles. Eur J Pharm Sci. 2005:105–12. doi: 10.1016/j.ejps.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 68.Lam YH, Bustami R, Phan T, Chan HK, Separovic F. A solid-state NMR study of protein mobility in lyophilized protein-sugar powders. J Pharm Sci. 2002:943–51. doi: 10.1002/jps.10089. [DOI] [PubMed] [Google Scholar]
- 69.Separovic F, Lam YH, Ke X, Chan HK. A solid-state NMR study of protein hydration and stability. Pharm Res. 1998;15:1816–21. doi: 10.1023/a:1011993620177. [DOI] [PubMed] [Google Scholar]
- 70.Smith JC. Protein dynamics: comparison of simulations with inelastic neutron scattering experiments. Q Rev Biophys. 1991;24:227–91. doi: 10.1017/s0033583500003723. [DOI] [PubMed] [Google Scholar]
- 71.Pynn R. Neutron Scattering-A Non-destructive Microscope for Seeing Inside Matter. In: Liang L, Rinaldi R, Schober H, editors. Neutron Applications in Earth, Energy and Environmental Sciences. Springer; US: 2009. pp. 15–36. [Google Scholar]
- 72.Harroun T, Wignall G, Katsaras J. Neutron Scattering for Biology. In: Fitter J, Gutberlet T, Katsaras J, editors. Neutron Scattering in Biology - Techniques and Applications. Springer; US: 2006. pp. 1–18. [Google Scholar]
- 73.Doster W, Settles M. Protein-water displacement distributions. Biochim Biophys Acta. 2005:173–86. doi: 10.1016/j.bbapap.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 74.Cornicchi E, Marconi M, Onori G, Paciaroni A. Controlling the protein dynamical transition with sugar-based bioprotectant matrices: a neutron scattering study. Biophys J. 2006:289–97. doi: 10.1529/biophysj.106.081752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nickels JD, O’Neill H, Hong L, Tyagi M, Ehlers G, Weiss KL, Zhang Q, Yi Z, Mamontov E, Smith JC, Sokolov AP. Dynamics of protein and its hydration water: neutron scattering studies on fully deuterated GFP. Biophys J. 2012;103:1566–75. doi: 10.1016/j.bpj.2012.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gabel F, Weik M, Doctor BP, Saxena A, Fournier D, Brochier L, Renault F, Masson P, Silman I, Zaccai G. The influence of solvent composition on global dynamics of human butyrylcholinesterase powders: a neutron-scattering study. Biophys J. 2004:3152–65. doi: 10.1016/S0006-3495(04)74363-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lerbret A, Affouard F, Hedoux A, Krenzlin S, Siepmann J, Bellissent-Funel MC, Descamps M. How strongly does trehalose interact with lysozyme in the solid state? Insights from molecular dynamics simulation and inelastic neutron scattering. J Phys Chem B. 2012;116:11103–16. doi: 10.1021/jp3058096. [DOI] [PubMed] [Google Scholar]
- 78.Pikal MJ, Rigsbee D, Roy ML, Galreath D, Kovach KJ, Wang B, Carpenter JF, Cicerone MT. Solid state chemistry of proteins: II. The correlation of storage stability of freeze-dried human growth hormone (hGH) with structure and dynamics in the glassy solid. J Pharm Sci. 2008;97:5106–21. doi: 10.1002/jps.21374. [DOI] [PubMed] [Google Scholar]
- 79.Wang B, Pikal MJ. The impact of thermal treatment on the stability of freeze dried amorphous pharmaceuticals: I. Dimer formation in sodium ethacrynate. J Pharm Sci. 2010;99:663–82. doi: 10.1002/jps.21959. [DOI] [PubMed] [Google Scholar]
- 80.Wang B, Cicerone MT, Aso Y, Pikal MJ. The impact of thermal treatment on the stability of freeze-dried amorphous pharmaceuticals: II. Aggregation in an IgG1 fusion protein. J Pharm Sci. 2010;99:683–700. doi: 10.1002/jps.21960. [DOI] [PubMed] [Google Scholar]
- 81.Fomina M, Schiro G, Cupane A. Hydration dependence of myoglobin dynamics studied with elastic neutron scattering, differential scanning calorimetry and broadband dielectric spectroscopy. Biophys Chem. 2014;185:25–31. doi: 10.1016/j.bpc.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 82.Smith G, Duffy AP, Shen J, Olliff CJ. Dielectric relaxation spectroscopy and some applications in the pharmaceutical sciences. J Pharm Sci. 1995;84:1029–44. doi: 10.1002/jps.2600840902. [DOI] [PubMed] [Google Scholar]
- 83.Pearson DS, Smith G. Dielectric analysis as a tool for investigating the lyophilization of proteins. 1998;1:108–117. [Google Scholar]
- 84.Raju GG. Dielectrics in Electric Fields. Marcel Dekker; New York: 2003. [Google Scholar]
- 85.Schiro G, Cupane A, Vitrano E, Bruni F. Dielectric relaxations in confined hydrated myoglobin. J Phys Chem B. 2009;113:9606–13. doi: 10.1021/jp901420r. [DOI] [PubMed] [Google Scholar]
- 86.Khodadadi S, Pawlus S, Sokolov AP. Influence of hydration on protein dynamics: combining dielectric and neutron scattering spectroscopy data. J Phys Chem B. 2008;112:14273–80. doi: 10.1021/jp8059807. [DOI] [PubMed] [Google Scholar]
- 87.Fridkin M, Wilchek M, Sheinblatt M. NMR studies of H-D exchange of alpha-CH group of amino acid residues in peptides. Biochem Biophys Res Commun. 1970:458–64. doi: 10.1016/0006-291x(70)90735-7. [DOI] [PubMed] [Google Scholar]
- 88.Thiery C, Nabedryk-Viala E, Menez A, Fromageot P, Thiery JM. Hydrogen exchange kinetics and dynamic structure of erabutoxin B from 1H NMR and infrared spectrometry. Biochem Biophys Res Commun. 1980:889–97. doi: 10.1016/0006-291x(80)91159-6. [DOI] [PubMed] [Google Scholar]
- 89.Clark AF, Gerken TA, Hogg RW. Proton nuclear magnetic resonance spectroscopy and ligand binding dynamics of the Escherichia coli L-arabinose binding protein. Biochemistry. 1982;21:2227–33. doi: 10.1021/bi00538a035. [DOI] [PubMed] [Google Scholar]
- 90.Desai UR, Osterhout JJ, Klibanov AM. Protein Structure in the Lyophilized State: A Hydrogen Isotope Exchange/NMR Study with Bovine Pancreatic Trypsin Inhibitor. Journal of the American Chemical Society. 1994;116:9420–9422. [Google Scholar]
- 91.Yamamoto T, Izumi S, Gekko K. Mass spectrometry of hydrogen/deuterium exchange in 70S ribosomal proteins from E. coli. FEBS Lett. 2006:3638–42. doi: 10.1016/j.febslet.2006.05.049. [DOI] [PubMed] [Google Scholar]
- 92.Tuma R, Coward LU, Kirk MC, Barnes S, Prevelige PE., Jr Hydrogen-deuterium exchange as a probe of folding and assembly in viral capsids. J Mol Biol. 2001:389–96. doi: 10.1006/jmbi.2000.4383. [DOI] [PubMed] [Google Scholar]
- 93.Suchanova B, Tuma R. Folding and assembly of large macromolecular complexes monitored by hydrogen-deuterium exchange and mass spectrometry. Microb Cell Fact. 2008:12. doi: 10.1186/1475-2859-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li Y, Williams TD, Schowen RL, Topp EM. Characterizing protein structure in amorphous solids using hydrogen/deuterium exchange with mass spectrometry. Anal Biochem. 2007;366:18–28. doi: 10.1016/j.ab.2007.03.041. [DOI] [PubMed] [Google Scholar]
- 95.Li Y, Williams TD, Schowen RL, Topp EM. Trehalose and calcium exert site-specific effects on calmodulin conformation in amorphous solids. Biotechnol Bioeng. 2007;97:1650–3. doi: 10.1002/bit.21362. [DOI] [PubMed] [Google Scholar]
- 96.Li Y, Williams TD, Topp EM. Effects of excipients on protein conformation in lyophilized solids by hydrogen/deuterium exchange mass spectrometry. Pharm Res. 2008;25:259–67. doi: 10.1007/s11095-007-9365-6. [DOI] [PubMed] [Google Scholar]
- 97.Sophocleous AM, Zhang J, Topp EM. Localized hydration in lyophilized myoglobin by hydrogen-deuterium exchange mass spectrometry. 1. Exchange mapping. Mol Pharm. 2012;9:718–26. doi: 10.1021/mp3000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sophocleous AM, Topp EM. Localized hydration in lyophilized myoglobin by hydrogen-deuterium exchange mass spectrometry. 2. Exchange kinetics. Mol Pharm. 2012;9:727–33. doi: 10.1021/mp2004093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jumper CC, Bomgarden R, Rogers J, Etienne C, Schriemer DC. High-resolution mapping of carbene-based protein footprints. Anal Chem. 2012;84:4411–8. doi: 10.1021/ac300120z. [DOI] [PubMed] [Google Scholar]
- 100.Iyer LK, Moorthy BS, Topp EM. Photolytic labeling to probe molecular interactions in lyophilized powders. Mol Pharm. 2013;10:4629–39. doi: 10.1021/mp4004332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Moorthy BS, Iyer LK, Topp EM. Mass spectrometric approaches to study protein structure and interactions in lyophilized powders. J Vis Exp. 2015 doi: 10.3791/52503. [DOI] [PMC free article] [PubMed] [Google Scholar]