Abstract
Genetic and epigenetic alterations have been identified as to contribute directly or indirectly to the generation of transitional cell carcinoma of the urinary bladder (TCC-UB). We have previously found that amplification of chromosome 6p22 is significantly associated with the muscle-invasive rather than superficial TCC-UB. Here, we demonstrated that Sox4, one of the candidate oncogenes located within the chromosome 6p22 amplicon, confers bladder cancer stem cell (CSC) properties. Down-regulation of Sox4 led to the inhibition of cell migration, colony formation as well as mesenchymal-to-epithelial transition (MET). Interestingly, knockdown of Sox4 also reduced the sphere formation, enriched cell population with high levels of aldehyde dehydrogenase (ALDH high) and tumor formation potential. Using gene expression profiling, we further identified novel Sox4 target genes. Last, immunohistochemistry analysis of human bladder tumor tissue microarrays (TMAs) indicated that high Sox4 expression was correlated with advanced cancer stages and poor survival rate. In summary, our data show that Sox4 is an important regulator of the bladder CSC properties and it may serve as a biomarker of the aggressive phenotype in bladder cancer.
Keywords: bladder cancer, chromosome 6p22 amplification, Sox4, bladder cancer stem cell
Introduction
Bladder cancer is the fifth most common malignancies worldwide 1. Many genetic and epigenetic alterations have been identified as to contribute directly or indirectly to the generation of transitional cell carcinoma of the urinary bladder (TCC-UB) 2. In general, bladder cancer can be divided into two classes, majority of them (~80%) are non-muscle-invasive bladder cancer representing low-grade bladder cancer, as well as the muscle-invasive bladder cancer (~20%) that is of high-grade and has less favorable prognosis 2. Genetic alterations occurring in low-grade superficial TCC-UB are most frequently caused by activating mutations of proto-oncogenes, of which fibroblast growth factor receptor 3 (FGFR3) and HRAS are most prevalent, with mutations in up to 75% and 30% of the papillary tumors, respectively 3 and 4. Since both these oncogenes activate the RAS/MEK/ERK signaling pathway, they appear to be mutually exclusive 5. In contrast, the majority of muscle-invasive TCC-UB arises through inactivation of the tumor suppressor pathways of TP53, RB1 or PTEN 2, 3 and 6. These mutations result in genomic instability and an anti-apoptotic phenotype, which enables tumor progression through accumulation of mutations. We and others found that amplification of chromosome 6p22 was significantly associated with muscle-invasive TCC-UB in contrast to superficial TCC-UB 7, 8, 9 and 10. In addition, the rate of 6p22.3 amplification in pN>1 patients (37%) was more than twice that of pN1 (17%) patients, indicating that there are potential oncogenes residing in this region that may contribute to muscle-invasive bladder cancer progression and invasion 10.
One of the candidate oncogenes at the chromosome 6p22 locus is Sox4. Sox4 is a member of the Sox (SRY-related HMG-box) family of transcription factors with a critical role in the embryonic development and in cell-fate determination during organogenesis of the heart, pancreas, central nerve system and T lymphocyte differentiation 11, 12, 13 and 14. Elevated Sox4 expression has been documented in all major human cancers, including breast, lung, brain, prostate, colorectal, bladder, pancreatic, ovarian cancer and leukemia 15. Through the oncogenic-retrovirus-induced insertional mutagenesis analysis, Sox4 locus has been identified as one of the common integration sites, providing the first evidence that Sox4 may act as an oncogene 16 and 17. Sox4 has been reported to function as an oncogene in more advanced stages of hepatocellular carcinomas (HCC) by promoting intrahepatic metastasis 18. In breast cancer, sox4 induced epithelial-to-mesenchymal transition (EMT) and cooperated with the RAS oncogene in cancer progression 19. More recently, it has been reported that sox4 acts as a master regulator of EMT by controlling the epigenetic program 20. Last, using a conditional knockout of Sox4 in stratified epithelia, Foronda et al. demonstrated that loss of functional Sox4 increased the skin stem cell quiescence and resistance to chemical carcinogenesis 21.
The cancer stem cell (CSC) hypothesis is based on the observation that many cancers, including bladder cancer, are driven by a subpopulation of tumor-initiating cells (popularly known as CSCs) and promote tumor growth 22 and 23. In addition, CSCs are resistant to chemotherapy and radiation treatments, which potentially explains the limitations of these agents in curing human malignancies 24, 25 and 26. The normal human urothelium comprises a layer of basal cells that are in contact with the basement membrane, several layers of intermediate cells and a single layer of large, superficial 'umbrella' cells with a specialized apical membrane to accommodate bladder expansion and contraction 2. The basal cells residing at the tumor-stroma interface have been indicated possessing tumor-initiating ability and may represent the origin cells of bladder cancer 27 and 28. Recent study suggests that such cancer stem cells may contribute to therapeutic resistance by repopulating residual tumors between chemotherapy cycles 29.
Here we demonstrated that Sox4, one of the candidate oncogenes located in the chromosome 6p22 amplicon, confers bladder cancer stem cell (CSC) properties. Down-regulation of Sox4 led to mesenchymal-to-epithelial transition (MET) and inhibition of cell migration. Of note, knockdown of sox4 reduced sphere formation, aldehyde-dehydrogenase-high (ALDHhigh) cell population and potential of tumor formation. Furthermore, immunohistochemistry (IHC) analysis of human bladder tumor tissue microarrays (TMAs) indicated that high sox4 expression correlated with advanced tumor stages and poorer survival of urinary bladder cancer patients.
Methods
Cell Culture
Human bladder cancer cell lines, SCaBER, HT-1197, HT-1376, 5637, J82, T24,TCCSUP, SW-780 and RT-4 were purchased from the American Type Culture Collection (ATCC, VA). CLS-439, RT-112, RT-112-D21 were purchased from Cell Line Service (CLS, Germany). SV-HUC, UROTSA, 5637, RT-112, RT-112-D21, 253J, 253J-BV cells were cultivated in RPMI-1640 medium with 10% fetal bovine serum (FBS). T24, CLS-439, RT4 cells were cultivated in McCoy's 5A medium with 10% fetal bovine serum (FBS). SCaBER, SW-780, HT-1197, HT-1376, J82, TCCSUP cells were cultivated in EMEM medium with 10% fetal bovine serum (FBS). All media were supplemented with 100 units/ml penicillin, 100 µg/ml streptomycin and 2mM glutamine. All cells were cultured in a humidified atmosphere of 95% air and 5% CO2 at 37 ºC. Transfection was performed using X-tremeGENE 9 DNA Transfection Reagent following the manufacturer's protocol (Roche). Packaging of lentivirus, cell transduction and drug selection were performed following standard protocols. For knockdown experiments, control shRNA was designed to target green fluorescent protein (GFP), a gene not expressed endogenously. shRNA hairpins targeting human Sox4 were cloned into pLKO.1 shRNA vector. The target sequences are listed (in the 5'-3' direction): shSOX4-I: 5'-AGCGACAAGATCCCTTTCATT-3', shSOX4-II: 5'-CCTTTCTACTTGTCGCTAAAT-3'. SMART pool: ON-Target plus Sox4 siRNA were purchased from GE Dharmacon (L-011779-00-0005). Transfection was performed using DharmaFECT transfection reagent 1 following the manufacturer's protocol (GE Dharmacon).
Antibodies and immunoblot analysis
Sox4 antibody was purchased from Diagenode; β-actin antibody from Upstate. For protein extraction, cells were washed with phosphate-buffered saline (PBS) and collected with IP buffer: 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 20% glycerol, 0.5% NP-40, plus 1x CompleteTM EDTA-free Protease Inhibitor Cocktail (Roche) or 1x HaltTM EDTA-free Protease and Phosphotase Inhibitor (Thermo Scientific). Cell lysate was cleared by centrifugation at 14,000 rpm for 20 min at 4 ºC. Lysate was loaded onto 4-15% MINI-PROTEAN TGX gel (Bio-Rad) with 4X SDS sample buffer. For immunoblot, proteins were transferred onto Immobilon-P membrane (Millipore), detected by various antibodies and visualized with ECL Plus Western Blotting Detection Reagents (GE Healthcare).
Real-time RT-PCR
For RNA preparation and qRT-PCR, RNA was extracted using the Trizol reagent (Invitrogen). cDNA synthesis was performed using the First-Strand cDNA Synthesis Kit (GE Healthcare) and quantitative real-time RT-PCR was performed using Power SYBR Green PCR Master Mix (Invitrogen). Sequences of the qPCR primer pairs (in the 5'-3' direction) are in Table A.
Table A.
Sequences of the qPCR primer
| FORWARD | REVERSE | |
|---|---|---|
| NEDD9 | GGAACTCATCCTCCACAACAA | GGACCAGCTGCACTCATTTA |
| FERMT2 | TCCTGATTCCAGTTGCAGAAG | TCTTGCCTTTGGAGGCTAATC |
| TJP1 | GCAGCCACAACCAATTCATAG | GAAAGGTAAGGGACTGGAGATG |
| TJP3 | GGCGGGAAAGTTCAGTAGATT | GGCACTCTGTAGATGTCATAGC |
| SHANK2 | GTCTCCTGGTACCCAAAGAAAT | CCTTGTCCCATGTGTGATAGAA |
| PARD6B | GAGGATGAAGACAGCGAAGAA | TGACTCCAGGCTCTCAGTATTA |
| CRB3 | CCCTCTCCTTTCTTTCAGTTCTC | CAGCAATGAAGGGAGACAGAT |
| SOX4 | ACCGGGACCTGGATTTTAAC | AAACCAGGTTGGAGATGCTG |
| β-ACTIN | CCAACCGCGAGAAGATGA | CCAGAGGCGTACAGGGATAG |
| GAPDH | GTGAAGGTCGGAGTCAACGG | GAGGTCAATGAAGGGGTCATTG |
| CCND1 | CCTCGGTGTCCTACTTCAAATGTG | ACAGGAAGCGGTCCAGGTA |
| CDK1 | CAGGTCAAGTGGTAGCCATGAAA | CCTGGAATCCTGCATAAGCACAT |
| FGFR3 | GGGAAACACAAAAACATCATCAACCT | GCAGGTGTCGAAGGAGTAGTC |
| MYB | CATATATAGCAGTGACGAGGTGATG | TCTGTTCCACCAGCTTCTTCAG |
| MYC | GCTTCTCTGAAAGGCTCTCCTT | AAATACGGCTGCACCGAGT |
| TUBA1A | GTGTTGGACCGAATTCGCAA | AGCTTGGACTTCTTGCCATAATCA |
| TUBA1C | CGCAGACCCCTTCAAGTTCT | AATGGTCTTGTCACTTGGCATCT |
| TUBB3 | GGG CCT TTG GAC ATC TCT TCA | AGTCGCAGTTTTCACACTCCTT |
| TUBB4B | TCTTCTGCTGCTGTT TGTCTACTT | GGTCGATGCCGTGCTCAT |
| S100A8 | AGACCGAGTGTCCTCAGTATATC | TGCCACGCCCATCTTTATC |
| TWIST1 | CCGGAGACCTAGATGTCATTG | CCACGCCCTGTTTCTTTG |
| SNAI1 | CTCTTTCCTCGTCAGGAAGC | GGCTGCTGGAAGGTAACTC |
| SNAI2 | CCAAACTACAGCGAACTGGA | GTGGTATGACAGGCATGGAG |
| ZEB1 | AACCCAACTTGAACGTCACA | ATTACACCCAGACTGCGTCA |
| ZEB2 | CGATCCAGACCGCAATTAAC | TGCTGACTGCATGACCATC |
| E47 | GGACGAGGAGAACACGTCAG | CACCTCGTCCGTACTGCTG |
Measurements were performed in triplicate and standardized to the levels of β-actin and GAPDH.
Clonogenic assay and colony formation in soft agar
To evaluate the difference in cell survival and proliferation under the condition of Sox4 knockdown, cells were plated at a density of 200 per well in a 6-well plate. Clones with >50 cells were fixed, stained and scored at 12 days.
Colony formation in soft agar
Cells (1X104 or 5X104) were added to 1.5ml of 0.4% agar and layered onto 2ml of 0.5% agar beds in six-wells plates. Cells were fed with 1ml of medium with 0.4% agar every 7 days for 3 weeks, after which colonies were stained with 0.02% iodonitrotetrazolium chloride (Sigma-Aldrich) and photographed. Colonies larger than 50 µm in diameter were counted as positive for growth. Assays were conducted in duplicate in three independent experiments.
Immunofluorescence microscopy analysis
Bladder cancer cells were cultured on coverslips to appropriate density. Cells were fixed with 4% paraformaldehyde for 15 min and then permeabilized with 0.1% Triton X-100 for 15 min. After blocking in 3% BSA for 30 min, slides were incubated with the primary antibody against CDH1 (BD Bioscience, San Joes, CA). After washing with PBS, slides were incubated with Alexa Fluor 594-conjugated secondary antibodies (Life Technologies) and examined under a Leica microscope (Leica Microsystems, Inc. Buffalo, NY). Each batch of slides contained a positive and negative control.
Isolation of ALDH1A1+ cell population by Aldefluor assay and fluorescence-activated cell sorting (FACS)
An Aldefluor kit (STEMCELL Technologies, Vancouver, Canada) was used to detect ALDH1A1 positive populations according to the manufacturer's protocol. Briefly, the brightly fluorescent ALDH1A1-expressing cells were detected using an Arial cell sorter (BD Biosciences, San Jose, CA). Side-scattered and forward-scattered profiles were used to reduce cell doublets. Specific ALDH1A1 activity was based on the difference between the presence/absence of the Aldefluor inhibitor diethylaminobenzaldehyde (DEAB).
Bladder sphere formation assay
Bladder sphere formation assay was performed by plating 5X103 cells in serum-free DMEM media (Gibco) supplemented with EGF (20 ng/mL), FGF (20 ng/ml) and B27 (2%) into ultra-low attachment 6-well plates (Corning). Spheres were allowed to grow for 7 days. Total spheres greater than 100 μm in diameter were counted. Each experimental group was done in triplicate and same experiments were repeated at least three times.
In vivo tumor growth assay
The tumor formation assay performed as described 30. Briefly, 1×106 shControl or shSox4 transduced RT-112 were subcutaneously injected into the female NOD/SCID mice of 6-8 weeks old. For serial dilution experiments, shControl or shSox4 transduced RT-112 cells in exponential growth phase were harvested and suspended in PBS (50% matrigel), and 1×104, 1×103, 1×102 shControl or shSox4 transduced RT-112 cells were subcutaneously injected into the female NOD/SCID mice of 6-8 weeks old. The SCID mice were generated at the Roswell Park Cancer Institute. Tumor sizes were measured twice a week using calipers. The care and use of animals was approved by the Institutional Animal Care and Use Committee of the Roswell Park Cancer Institute (Buffalo, NY).
RNAseq analyses
The sequencing libraries are prepared with the TruSeq Stranded Total RNA kit (Illumina Inc), from 1ug total RNA. Following manufacturer's instructions, the first step depletes rRNA from total RNA. After ribosomal depletion, the remaining RNA is purified, fragmented and primed for cDNA synthesis. Fragmented RNA is then reverse transcribed into first strand cDNA using random primers. The next step removes the RNA template and synthesizes a replacement strand, incorporating dUTP in place of dTTP to generate ds cDNA. AMPure XP beads are used to separate the ds cDNA from the second strand reaction mix resulting in blunt-ended cDNA. A single 'A' nucleotide is then added to the 3' ends of the blunt fragments. Multiple indexing adapters, containing a single 'T' nucleotide on the 3' end of the adapter, are ligated to the ends of the ds cDNA, preparing them for hybridization onto a flow cell. Libraries are purified and validated for appropriate size on a 2100 Bioanalyzer High Sensitivity DNA chip (Agilent Technologies, Inc.). The DNA library is quantitated using KAPA Biosystems qPCR kit, and then normalized to 2nM prior to pooling. Libraries are pooled together in an equimolar fashion, following experimental design criteria. Each pool is denatured, diluted to 10pM, and clustered to individual lanes of a HiSeq Flow Cell using an Illumina cBot and the corresponding single-read or paired-end read TruSeq cluster kit. Pooled, clustered samples are then run on a HiSeq2500 sequencer according to the manufacturer's recommended protocol (Illumina Inc.).
Raw reads passed quality filter from Illumina RTA have been first pre-processed by using FASTQC for sequencing base quality control. Then the reads were mapped to the human (hg19) and ENSEMBLE annotation database using STAR 31. Second-round QC using RSeQC was applied to mapped bam files to identify potential RNAseq library preparation problems. From the mapping results, reads that matched a single unique location in the genome were kept, allowing up to two mismatches for further analysis. The number of reads aligning to each gene was calculated using HTSeq 32. Differentially expressed genes were identified using DESeq2, a variance-analysis package developed to infer the statically significant difference in RNAseq data. In addition, biological hypothesis was tested using generalized linear model implemented in DESeq2 by construct corresponding contrasts. List of the differentially expressed genes (DEGs) was analyzed for enriched Gene Ontology and/or KEGG pathway term with the GAGE Bioconductor package 33. Finally, the GSAASeqSP was employed for genome-wide gene set association in contrast to DEGs alone 34.
Patient validation cohort
Patients included in this study (total 309) were diagnosed and treated for transitional cell carcinoma of the urinary bladder (TCC-UB) between July 1970 and April 2011 at the Roswell Park Cancer Institute. Selection of patients included all in this time period with adequate material in the RPCI archival bank, i.e., adequate material for tissue microarray (TMA) construction and follow-up in the RPCI Tumor Registry. The RPCI Institutional Review Board gave approval for this study. The median patient age at first diagnosis was 69 years (average 68; range 36-91) with 242 males and 67 females. The majority (255 of 309; 82%) were either a current smoker at the time of diagnosis or reported a history of smoking. To more specifically describe this cohort of 309 patients, they were then divided into two general cohorts as either superficial TCC-UB (pTa, Tis, T1) or muscle-invasive TCC-UB (>pT2).
Tissue microarrays
Three 1-mm tissue cores from formalin-fixed paraffin embedded donor blocks were precisely arrayed into a new recipient paraffin block, including tumor specimens as well as controls. Specimens for controls within the TMA consisted of multiple cores of normal tissue from 10 different organs, including heart, colon, kidney, adrenal, ovary, myometrium, brain, thyroid, lung, and prostate, representing slightly more than 20% of all the cores in a TMA.
Immunohistochemistry (IHC)
IHC were carried out on Formalin-Fixed Paraffin Embedded (FFPE) tumor blocks (procured by RPCI Pathology Network Shared Resource) from bladder cancer patients. Polyclonal primary anti-Sox4 antibody was purchased from Abcam. Slides were de-paraffinized and rehydrated by washing with xylene and ethanol, brought to up to near boiling temperatures in 10 μM buffered citrate and immersed in 0.03% hydrogen peroxide in order to cease peroxidase activity. Slides were incubated in 0.03% casein in phosphate buffered saline (PBS), incubated in the primary antibody diluted in PBS Tween-20 (PBST), washed with 0.03% casein in PBS and then incubated with a biotinylated secondary antibody. Slides were incubated with DakoCytomation Envisioning staining kit as directed by manufacturer for a color reaction. The slides had been counterstained with hematoxylin to allow for analysis of nuclear morphology in the tumor and stromal cells.
Statistical Analysis
Association between clinical/histological covariates and sox4 expression was evaluated using fisher's exact test. Survival time associations were tested with a log-rank test. Other statistical analysis of data was performed using the SPSS statistics software package (SPSS, IL). All results are expressed as mean ± SD.
Results
Sox4 is required for bladder cancer cell colony formation
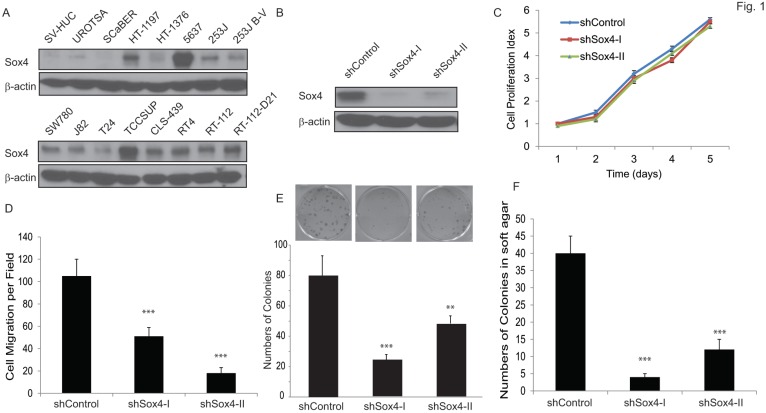
We first examined the Sox4 protein level in a panel of bladder cancer cell lines (Figure 1A). Of note, Sox4 expression was not detectable in the immortalized non-tumorigenic urothelial epithelial SV-HUC and UROTSA cells, whereas it was highly expressed in the majority of muscle-invasive bladder cancer cells, especially in the 6p22-amplicon containing 5637 and TCCSUP cells. To further investigate the role of Sox4 in cell proliferation, we designed lentiviral shRNA constructs that specifically targeted Sox4 and transduced them into the RT-112 bladder cancer cells. The knockdown efficiency was confirmed by immunoblotting (Figure 1B). Knockdown of Sox4 has no effects on cell proliferation in 2D culture (Figure 1C). To our great interest, knockdown of Sox4 significantly reduced the cell migration (Figure 1D). In addition, ablation of Sox4 dramatically reduced the potential for clonogenicity as well as the colony formation in soft agar (Figure 1E, F). Together, these results strongly suggested that Sox4 may play an important role in the bladder cancer cell migration and transformation.
Figure 1.
Knockdown of Sox4 inhibits cell migration and colony formation. (A) Expression of Sox4 protein in a panel of bladder cancer cell lines. Increased Sox4 level is revealed by immunoblot in the 6p22-amplicon-containing 5637 and TCC-SUP cells. (B) Knockdown of Sox4 in RT-112 cells as confirmed by immunoblot (β-actin was used as a loading control). (C) Knockdown of Sox4 has no effect on RT-112 cell proliferation examined by MTT assay. (D) Knockdown of Sox4 inhibits cell migration in the transwell cell migration assay. (E) Knockdown of Sox4 inhibits the clonogenicity potential. (F) Loss-of-function Sox4 reduces the colony formation in soft agar.
Depletion of Sox4 induces mesenchymal-to-epithelial transition (MET)
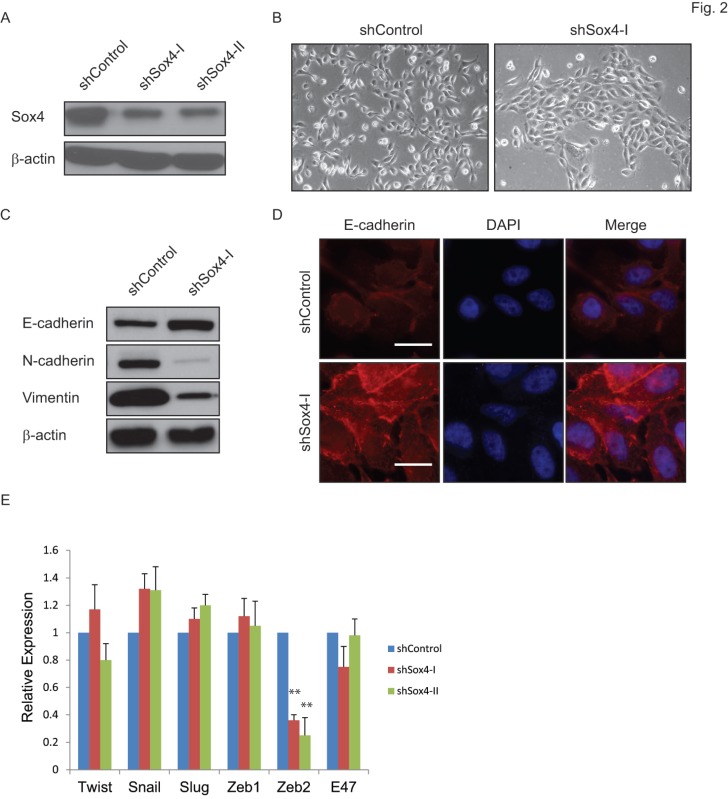
Sox4 has been indicated as a regulator of the TGF-β-induced epithelial-to-mesenchymal transition (EMT) 19. To test whether knockdown of Sox4 might convert the EMT phenotype to MET in bladder cancer, we transduced the bladder cancer 5637 cells that containing the 6p22 amplicon with the shSox4 lentiviral constructs (Figure 2A). As expected, knockdown of Sox4 induced morphological changes of cells from the scattered to the epithelial-like phenotype (Figure 2B). Meanwhile, down-regulation of Sox4 increased the level of epithelial marker CDH-1 and reduced the mesenchymal markers CDH-2 and vimentin as revealed by immunoblotting (Figure 2C). Immunofluorescence staining further showed that Sox4 depletion preserved the expression of epithelial marker E-cadherin on the cell membrane (Figure 2D). Interestingly, we found knockdown of Sox4 induced dramatically repression of EMT inducer ZEB2 expression (Figure 2E). All these results demonstrated that loss-of-function of Sox4 was capable of reversing bladder cancer cells from the malignant-associated mesenchymal back to more differentiated epithelial-like phenotype and it may through down-regulation of ZEB2.
Figure 2.
Ablation of Sox4 induces mesenchymal-to-epithelial transition (MET). (A) Knockdown of Sox4 in 5637 cells as confirmed by immunoblot (β-actin was used as a loading control). (B) Images of control and Sox4-targeted 5637 cells. Knockdown of Sox4 reverses the mesenchymal state of cells back to an epithelial-like state. (C) Increased epithelial marker (E-cadherin) and decreased mesenchymal markers (vimentin and N-Cadherin) in Sox4-knockdown cells as revealed by immunoblot. (D) Increased and plasma membrane-localized E-cadherin in Sox4-knockdown cells as revealed by immunofluorescence microscopy (scale bar=20µm). (E) Knockdown of Sox4 in 5637 cells induced dramatically repression of ZEB2 expression as revealed by real time RT-PCR. (** p<0.01)
Sox4 is required for bladder sphere and tumor formation
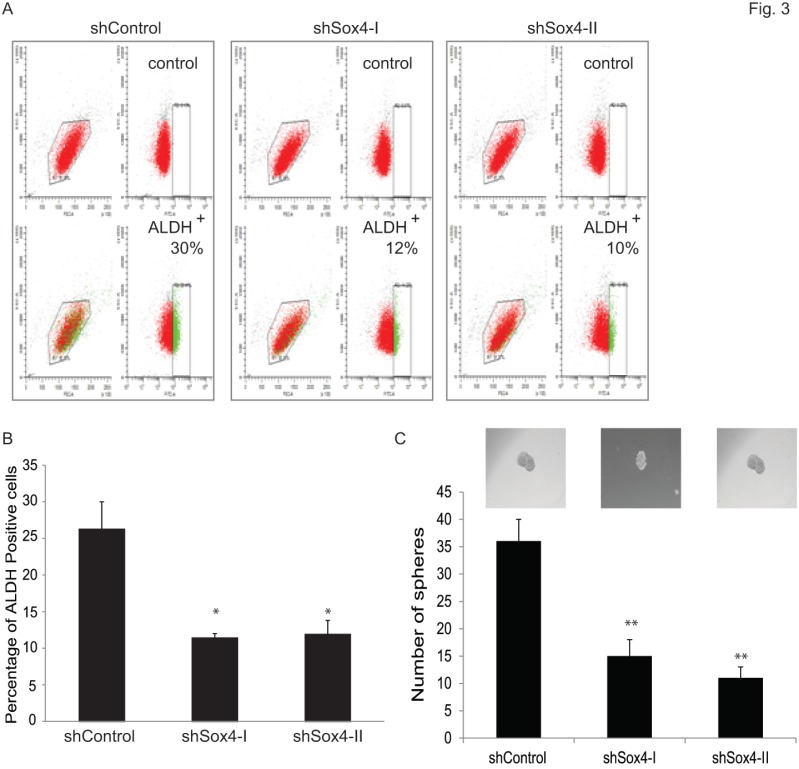
Accumulating evidence points to the existence and contribution of tumor-initiating cells (TICs) and/or cancer stem cells (CSCs) in tumor progression and relapse 22. Recently, some reliable assays have been reported to identify and analyze such cells. For example, Aldehyde dehydrogenase 1 A1 (ALDH1A1) has been demonstrated as a marker for cancer stem or stem-like cells in human malignancy including bladder cancer 35 and 36. Also, sphere culture is being used increasingly as a method to enrich stem cells based on their property of anchorage-independent growth 29 and 30. It is generally agreed that the tumor-sphere-forming cells have higher capacity of proliferation, self-renewal and tumorigenicity. Therefore, to test whether ablation of Sox4 would alter the bladder cancer stem properties, we first examined the ALDH1A1-positive population at different Sox4 status in the RT-112 bladder cells by FACS (Figure 3A). It was found that knockdown of Sox4 significantly decreased the ALDH1A1-positive cell population (Figure 3B). To further test whether Sox4 contributes to the self-renewal properties, we next performed the bladder sphere formation. Of great interest, knockdown of Sox4 also dramatically reduced the sphere formation (Figure 3C). Taken together, our in vitro data so far indicated that Sox4 may contribute to the bladder cancer stem cell traits.
Figure 3.
Knockdown of Sox4 reduces the ADLH-positive population and bladder sphere formation. (A) FACS analysis for ALDH1A+ population of shControl or shSox4 transduced RT-112 cells using the Aldefluor assay. Cells incubated with DEAB were used to establish the baseline fluorescence (top panel); and the brightly fluorescent ALDHA1+ cells were detected in the green fluorescence channel (lower panel). (B) Quantifications of ALDH1A+ population of shControl or shSox4 transduced RT-112 cells using the Aldefluor assay. (* p<0.05). (C) Quantifications of bladder sphere formation of shControl or shSox4 transduced RT-112 cells. (** p<0.01)
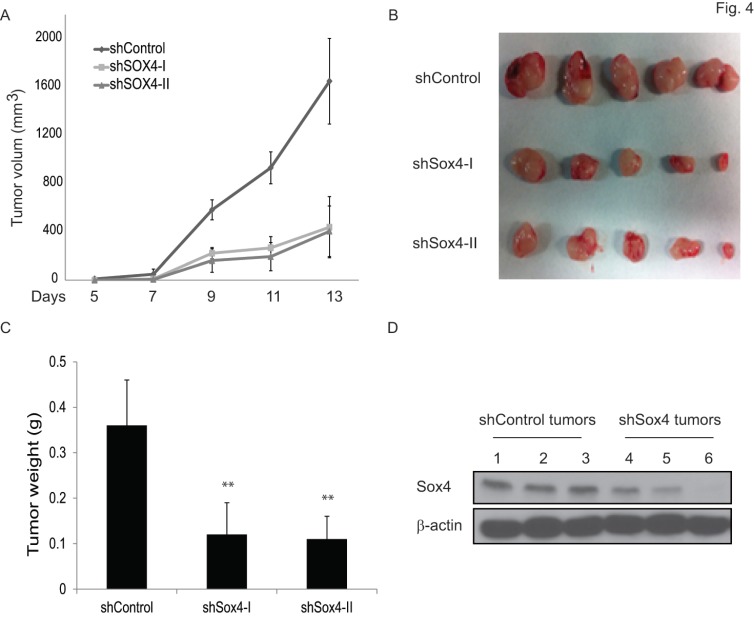
To test the functional contribution of Sox4 to primary tumor formation in vivo, we subcutaneously transplanted the sh-Control or sh-Sox4 transduced RT-112 cells into the SCID mice and followed up the tumor growth. As expected, knockdown of Sox4 in RT-112 cells resulted in significantly reduced primary tumor size (Figure 4A, B) and weight (Figure 4C). Immunoblotting analysis further documented an efficient depletion of Sox4 expression in these harvested primary tumors (Figure 4D). Last, to further characterize the tumor initiation properties of Sox4, we performed limited dilution analysis and found that the tumorigenicity potential was significantly reduced in the Sox4-knockdown cells (Table 1). Together, these results showed that Sox4 confers bladder cancer stem cell traits and plays an important role in the bladder tumorigenesis.
Figure 4.
Knockdown of Sox4 inhibits tumor growth in vivo. (A) Tumor growth from subcutaneously implanted shControl or shSox4 transduced RT-112 cells. (B) Images of primary tumors induced from shControl or shSox4 transduced RT-112 cells. (C) Weight of induced tumors from subcutaneously implanted shControl or shSox4 transduced RT-112 cells. (** p<0.01). (D) Expression of Sox4 protein in shControl or shSox4 tumors as revealed by immunoblot.
Table 1.
Limiting dilution analysis of tumorigenesis of RT112 cells expressing scrambled control (siControl) or siSox4.
| Groups | Dosage of cells | No. of mice with tumors |
|---|---|---|
| shControl | 1X104 | 8/8 |
| 1X103 | 8/8 | |
| 1X102 | 6/8 | |
| shSox4-I | 1X104 | 5/8 |
| 1X103 | 2/8 | |
| 1X102 | 0/8 | |
| shSox4-II | 1X104 | 4/8 |
| 1X103 | 2/8 | |
| 1X102 | 0/8 |
Identification of Sox4 target genes
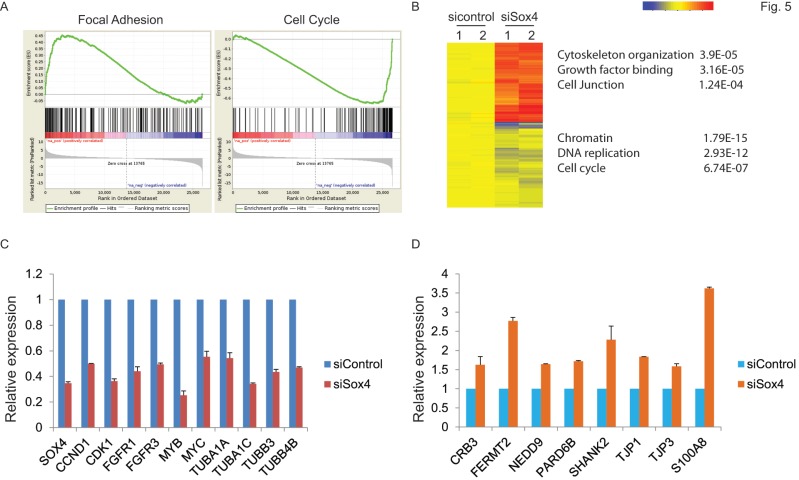
Next, to extensively examine the Sox4-mediated pro-tumorigenic transcriptional program, we performed RNA-seq differential gene and transcript analysis on the siControl or siSox4 transduced RT-112 cells after 48 hours. Genes found to be differentially expressed according to the Sox4 status were further analyzed for Sox4-binding motifs within one-kilobase of their transcription start site and altogether 1117 genes were identified in siSox4 cells (fold change log2>1.5; p<0.05) (Figure S1). Gene Set Enrichment Analysis (GSEA) of differentially regulated transcripts in the siSox4 gene cluster/signature revealed an overrepresentation of up-regulated genes associated with focal adhesion and down-regulated genes associated with cell cycle (Figure 5A). Furthermore, using DAVID (database for annotation, visualization and discovery), we wanted to assess whether any GO biological or molecular processes were statistically over- or under-represented. Indeed, some over-represented pathways (p < 0.001) were identified, including those involved in cytoskeleton organization, growth factor binding, cell junction, chromatin remodeling, DNA replication and cell cycle (Figure 5B; Figure S1).
Figure 5.
Gene expression profiling reveals Sox4 regulated genes. (A) GSEA enrichment plot shows that focal adhesion components are highly enriched and cell cycle regulated genes significantly reduced in response to siSox4 in RT-112 cells. (B) Heat map of hierarchically clustered genes in RT-112 cells expressing scrambled control (sicontrol) or siSox4 (siSox4). Expression levels are from duplicate samples. Significantly up-regulated (red) and down-regulated (blue) GO terms and enrichment p values by DAVID analysis are shown. (C) Real-time RT-PCR of Sox4, CCND1, CDK1, FGFR1, FGFR3, MYB, MYC, TUBA1A, TUBA1C, TUBB3 and TUBB4B mRNA followed by treatment with siControl or siSox4 in the RT-112 cells. GAPDH and β-actin were used as internal controls. (D) Real-time RT-PCR of CRB3, FERMT2, NEDD9, PARD6B, SHANK2, TJP1, TJP3 and S100A8 mRNA followed by treatment with siControl or siSox4 in the RT-112 cells. GAPDH and β-actin were used as internal controls.
Among these candidate genes, we further validated by qRT-PCR cell cycle regulated genes, such as CCND1, CDK1, FGFR1, FGFR3, MYB and MYC, as specifically down-regulated in response to siSox4 (Figure 5C). In addition, we found that tight junction proteins, e.g., CRB3, TJP1, TJP3, were specifically up-regulated in response to knock down of Sox4 (Figure 5D). Our gene expression profiling data demonstrated that Sox4 is not only involved in the classic cell cycle regulation, but also in other signaling pathways and cellular processes that regulate cell migration and invasion.
High Sox4 expression is present in the muscle-invasive bladder cancer patient samples and correlated with poor survival
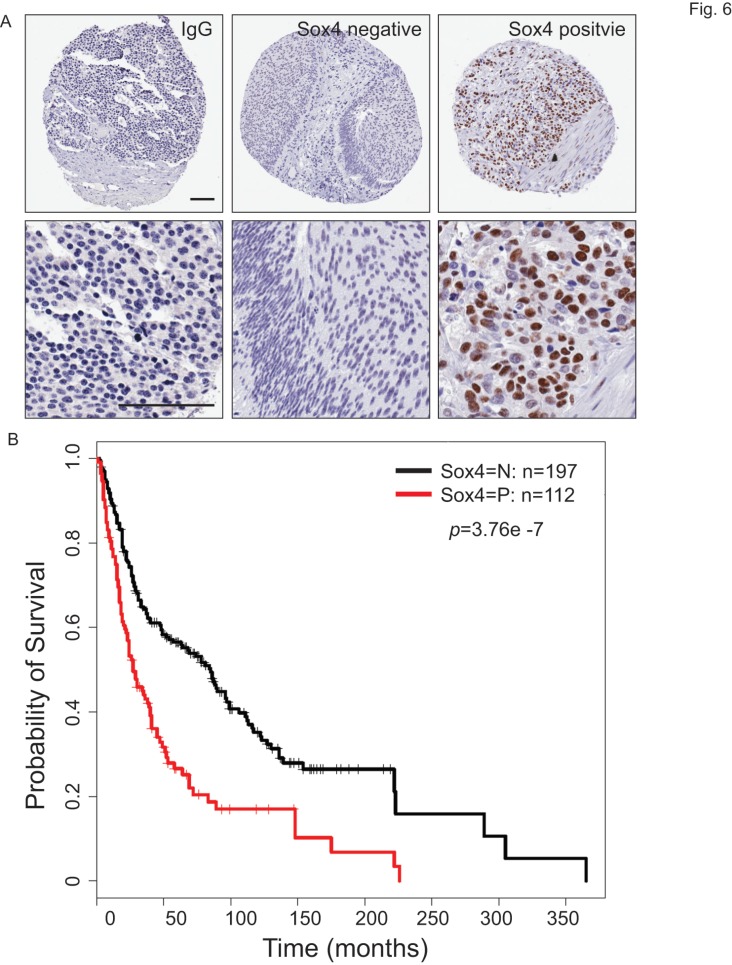
To understand the clinical relevance of Sox4 expression in bladder cancer patients, we carried out the immunohistochemistry staining analysis using bladder cancer tissue microarrays (TMAs) (Figure 6A). In our cohort of TCC-UB, the Sox4 expression was detected in both non-muscle-invasive and muscle-invasive tumors, but with a strikingly higher rate in the muscle-invasive samples (p=7.00E-04) (Table 2). Comparison of Sox4-positivity in females (34%; 23/67) versus males (37%; 89/242) (t-test p=0.77), age <60 (35%; 23/64) versus age ≥60 (36%; 9/245) (p=1) and smokers (37%; 96/255) versus non-smokers (31%; 16/52) (p=0.43) revealed no significant difference. Intensive examination of Sox4 within the standard pathological settings such as tumor grade, depth of invasion (pT) and nodal metastasis (pN) proposed for its association with a more aggressive phenotype. For example, in regard to tumor invasion, significantly different Sox4 positivity was observed between superficial (23%; 20/87) and muscle-invasive TCC-UB (46%; 65/142) (p=7.00E-04). Similarly, a strong association between Sox4 and more advanced tumor stage was also noted, as only 5% (3/59) of low-stage TCC-UB expressed Sox4 in contrast to 46% (109/203) (p=3.71E-10) of high-stage TCC-UB. Such significance of Sox4 as a marker for aggressiveness was accentuated in the evaluation of nodal metastatic disease for the rate of Sox4 expression in pN>1 (61%; 20/33) was significantly higher than that in pN0 patients (34%; 55/160) (p=0.0061). Of particular note, there was a remarkable association between Sox4 and poor survival (p=3.76E-07) (Figure 6B). Together, our IHC analysis of bladder cancer TMAs revealed that high Sox4 expression was significantly associated with higher tumor grade, invasiveness and unfavorable patient outcome.
Figure 6.
Sox4 expression in bladder cancer patient tumor microarrays (TMAs). (A) Imaging examples of IHC staining of IgG control, negative and positive Sox4 expression in bladder cancer TMAs. (scale bar=100µm). (B) Kaplan-Meier survival curve of Sox4-positive and -negative bladder cancer patients. Statistical significance was determined by the log-rank test (p=3.76E-07).
Table 2.
Clinicopathological features of patients with Sox4 expression.
| Total | Sox4 positive | |||||
|---|---|---|---|---|---|---|
| # | % | # | % | p | ||
| Total Patients | 309 | 100% | 112 | 36% | ||
| Sex | Female | 67 | 21% | 23 | 34% | 0.77 |
| Male | 242 | 79% | 89 | 37% | ||
| Age | <60 | 64 | 21% | 23 | 35% | 1 |
| >60 | 245 | 79% | 89 | 36% | ||
| Smoking History | Current or previous smoker | 255 | 82% | 96 | 37% | 0.43 |
| Never smoked | 52 | 17% | 16 | 31% | ||
| Bladder Cancer | Non-muscle invasive | 87 | 28% | 20 | 23% | 7.00E-04 |
| Muscle invasive | 142 | 46% | 65 | 46% | ||
| Bladder Cancer Grade | Low | 59 | 19% | 3 | 5% | 3.71E-10 |
| High | 238 | 77% | 109 | 46% | ||
| Nodal Status MIBC | pN0 | 160 | 52% | 55 | 34% | 0.0061 |
| N>1 | 33 | 11% | 20 | 61% | ||
| Outcome MIBC | Evidence Of This Cancer | 114 | 37% | 50 | 44% | 0.33 |
| No Evidence Of This Cancer | 62 | 20% | 22 | 35% | ||
Discussion
In the present study, we found that the transcription factor Sox4 is upregulated in muscle-invasive bladder cancer cells. Up-regulation of Sox4 is partially through the gain of DNA copy numbers. On the other hand, knockdown of Sox4 inhibited cell migration and colony formation of bladder cancer cells and reversed the tumor cells from a mesenchymal state to epithelial-like state. Of note, loss-of-function of Sox4 reduced the ALDHhigh population and sphere formation, which manifest cancer stem cell (CSC) properties, as well as decreased dramatically the potential for primary tumor formation. Furthermore, our RNAseq gene expression profiling identified Sox4-regulated pathways involved in cytoskeleton regulation, chromatin remodeling, cell cycle regulation and so on. Last, we found that high level of Sox4 expression was frequently observed in the muscle-invasive subset and had a significant correlation with more advanced tumor stage, lymph-node metastasis and poor survival. As a potential biomarker for TCC-UB, examination of Sox4 level can greatly add to the risk assessment of advanced pT or pN stage in the pre-cystectomy biopsy setting as typical surgical pathology can only provide information about the presence or absence of muscle invasion.
The SOX family members are critical in many developmental processes involving both the maintenance of stem cells and control of the terminal differentiation of a wide variety of cell types 37. Sox4 was reported to play an important role in the cardiac outflow tract development and differentiation of pro-B and T cells 14. Of interest, elevated expression of Sox4 has been reported in various solid tumors 15, yet disparate observation that Sox4 overexpression in the bladder HU609 cell line decreased cell viability and induced apoptosis also exists 38. Here we demonstrated that ablation of Sox4 inhibited colony formation and reduced the bladder CSC potential as well as tumor formation in vivo. Such discrepancy may result from the different genetic background of cell lines. Consistently with our observation, it has been reported that Sox4 potentiated RAS-induced breast tumorigenesis 19 and reduction of whole-body Sox4 expression in a transgenic mouse model displayed lower cancer incidence 21. Taken together, these studies indicate that Sox4 may cooperate with other genes to promote solid tumorigenesis and metastasis.
As a transcription factor, Sox4 most likely functions in malignant processes through its direct regulation of target genes. As revealed by our gene expression profiling analysis, Sox4 regulates a wide range of cell cycle related genes, such as CCND1, CDK1, FGFR1, FGFR3, MYB and MYC, which is consistent with other study results on hepatocellular carcinoma (HCC) 18, prostate cancer 39 and 40, adenoid cystic carcinoma (ACC) 41, lung cancer 42 and endometrial cancer 43. In addition, we also identified decreased Ezh2 level in response to knockdown of Sox4 (Figure S1); and Ezh2 expression has been reported in a recent study to be directly regulated by Sox4 20. Of particular note, our data also showed possibility of Sox4 as a transcriptional repressor, as we found that knockdown of Sox4 activated some cell junction proteins, including CRB3, TJP1 and TJP3. Further characterization of these Sox4 target genes is in order and may contribute greatly to a better understanding of the CSC regulation, tumorigenesis and metastasis.
In summary, we provided here evidence that Sox4 in 6p22-amplified bladder tumors is a potential oncogene of importance. Specifically, the correlation between Sox4 expression and poor patient survival strongly suggest for its role as a prognostic biomarker for bladder cancer patients. Future directions may include investigation of the interaction between Sox4 and other genes in the 6p22 amplicon, e.g., E2F3. Meanwhile, other oncogenes, such as CCND1 and MYC, that are not in the 6p22 amplicon but frequently altered in bladder cancer may also interact with Sox4 cooperatively or synergistically in cancer development.
Supplementary Material
Supplemental Figure 1. List of up-regulated and down-regulated genes in sicontrol compared to siSox4 in RT-112 cells; significantly up-regulated and down-regulated GO terms by DAVID analysis in sicon compared to siSox4 in RT-112 cells.
Acknowledgments
We thank Dr. Yuesheng Zhang (RPCI) kindly share the UROTSA cells with us. We thank RPCI-PRN for providing us the bladder tumor microarrays (TMAs) and performing the immunohistochemistry staining. We thank Dr. Sean Glenn and RPCI-GSR for performing the RNAseq. L. Zhang was supported by the National Natural Science Fund of China (no. 81202055). This work was supported by the Roswell Park Cancer Institute and National Cancer Institute (NCI) grant #P30 CA016056, Roswell Park Alliance Foundation and in part by the National Cancer Institute (NCI) R21 CA179693, the American Cancer Society Research Scholar Grant RSG-14-214-01-TBE (to J.Z).
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. International journal of cancer Journal international du cancer. 2010;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Knowles MA, Hurst CD. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nature reviews Cancer. 2015;15:25–41. doi: 10.1038/nrc3817. [DOI] [PubMed] [Google Scholar]
- 3.Bakkar AA, Wallerand H, Radvanyi F, Lahaye JB, Pissard S, Lecerf L. et al. FGFR3 and TP53 gene mutations define two distinct pathways in urothelial cell carcinoma of the bladder. Cancer research. 2003;63:8108–12. [PubMed] [Google Scholar]
- 4.Jebar AH, Hurst CD, Tomlinson DC, Johnston C, Taylor CF, Knowles MA. FGFR3 and Ras gene mutations are mutually exclusive genetic events in urothelial cell carcinoma. Oncogene. 2005;24:5218–25. doi: 10.1038/sj.onc.1208705. [DOI] [PubMed] [Google Scholar]
- 5.Castillo-Martin M, Domingo-Domenech J, Karni-Schmidt O, Matos T, Cordon-Cardo C. Molecular pathways of urothelial development and bladder tumorigenesis. Urologic oncology. 2010;28:401–8. doi: 10.1016/j.urolonc.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 6.Cordon-Cardo C, Wartinger D, Petrylak D, Dalbagni G, Fair WR, Fuks Z. et al. Altered expression of the retinoblastoma gene product: prognostic indicator in bladder cancer. Journal of the National Cancer Institute. 1992;84:1251–6. doi: 10.1093/jnci/84.16.1251. [DOI] [PubMed] [Google Scholar]
- 7.Nord H, Segersten U, Sandgren J, Wester K, Busch C, Menzel U. et al. Focal amplifications are associated with high grade and recurrences in stage Ta bladder carcinoma. International journal of cancer Journal international du cancer. 2010;126:1390–402. doi: 10.1002/ijc.24954. [DOI] [PubMed] [Google Scholar]
- 8.Hurst CD, Tomlinson DC, Williams SV, Platt FM, Knowles MA. Inactivation of the Rb pathway and overexpression of both isoforms of E2F3 are obligate events in bladder tumours with 6p22 amplification. Oncogene. 2008;27:2716–27. doi: 10.1038/sj.onc.1210934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oeggerli M, Tomovska S, Schraml P, Calvano-Forte D, Schafroth S, Simon R. et al. E2F3 amplification and overexpression is associated with invasive tumor growth and rapid tumor cell proliferation in urinary bladder cancer. Oncogene. 2004;23:5616–23. doi: 10.1038/sj.onc.1207749. [DOI] [PubMed] [Google Scholar]
- 10.Shen H, Morrison CD, Zhang J, Underwood W 3rd, Yang N, Frangou C. et al. 6p22.3 amplification as a biomarker and potential therapeutic target of advanced stage bladder cancer. Oncotarget. 2013;4:2124–34. doi: 10.18632/oncotarget.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung M, Abu-Elmagd M, Clevers H, Scotting PJ. Roles of Sox4 in central nervous system development. Brain research Molecular brain research. 2000;79:180–91. doi: 10.1016/s0169-328x(00)00109-1. [DOI] [PubMed] [Google Scholar]
- 12.Lioubinski O, Muller M, Wegner M, Sander M. Expression of Sox transcription factors in the developing mouse pancreas. Developmental dynamics: an official publication of the American Association of Anatomists. 2003;227:402–8. doi: 10.1002/dvdy.10311. [DOI] [PubMed] [Google Scholar]
- 13.Schilham MW, Moerer P, Cumano A, Clevers HC. Sox-4 facilitates thymocyte differentiation. European journal of immunology. 1997;27:1292–5. doi: 10.1002/eji.1830270534. [DOI] [PubMed] [Google Scholar]
- 14.Schilham MW, Oosterwegel MA, Moerer P, Ya J, de Boer PA, van de Wetering M. et al. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380:711–4. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- 15.Vervoort SJ, van Boxtel R, Coffer PJ. The role of SRY-related HMG box transcription factor 4 (SOX4) in tumorigenesis and metastasis: friend or foe? Oncogene. 2013;32:3397–409. doi: 10.1038/onc.2012.506. [DOI] [PubMed] [Google Scholar]
- 16.Du Y, Spence SE, Jenkins NA, Copeland NG. Cooperating cancer-gene identification through oncogenic-retrovirus-induced insertional mutagenesis. Blood. 2005;106:2498–505. doi: 10.1182/blood-2004-12-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin MS, Fredrickson TN, Hartley JW, Suzuki T, Akagi K, Morse HC 3rd. High-throughput retroviral tagging for identification of genes involved in initiation and progression of mouse splenic marginal zone lymphomas. Cancer research. 2004;64:4419–27. doi: 10.1158/0008-5472.CAN-03-3885. [DOI] [PubMed] [Google Scholar]
- 18.Liao YL, Sun YM, Chau GY, Chau YP, Lai TC, Wang JL. et al. Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene. 2008;27:5578–89. doi: 10.1038/onc.2008.168. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Liang Q, Lei Y, Yao M, Li L, Gao X. et al. SOX4 induces epithelial-mesenchymal transition and contributes to breast cancer progression. Cancer research. 2012;72:4597–608. doi: 10.1158/0008-5472.CAN-12-1045. [DOI] [PubMed] [Google Scholar]
- 20.Tiwari N, Tiwari VK, Waldmeier L, Balwierz PJ, Arnold P, Pachkov M. et al. Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer cell. 2013;23:768–83. doi: 10.1016/j.ccr.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 21.Foronda M, Martinez P, Schoeftner S, Gomez-Lopez G, Schneider R, Flores JM. et al. Sox4 links tumor suppression to accelerated aging in mice by modulating stem cell activation. Cell reports. 2014;8:487–500. doi: 10.1016/j.celrep.2014.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ailles LE, Weissman IL. Cancer stem cells in solid tumors. Curr Opin Biotechnol. 2007;18:460–6. doi: 10.1016/j.copbio.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 23.Chan KS, Espinosa I, Chao M, Wong D, Ailles L, Diehn M. et al. Identification, molecular characterization, clinical prognosis, and therapeutic targeting of human bladder tumor-initiating cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14016–21. doi: 10.1073/pnas.0906549106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frame FM, Maitland NJ. Cancer stem cells, models of study and implications of therapy resistance mechanisms. Advances in experimental medicine and biology. 2011;720:105–18. doi: 10.1007/978-1-4614-0254-1_9. [DOI] [PubMed] [Google Scholar]
- 25.Kakarala M, Wicha MS. Implications of the cancer stem-cell hypothesis for breast cancer prevention and therapy. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2008;26:2813–20. doi: 10.1200/JCO.2008.16.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou BB, Zhang H, Damelin M, Geles KG, Grindley JC, Dirks PB. Tumour-initiating cells: challenges and opportunities for anticancer drug discovery. Nature reviews Drug discovery. 2009;8:806–23. doi: 10.1038/nrd2137. [DOI] [PubMed] [Google Scholar]
- 27.He X, Marchionni L, Hansel DE, Yu W, Sood A, Yang J. et al. Differentiation of a highly tumorigenic basal cell compartment in urothelial carcinoma. Stem cells. 2009;27:1487–95. doi: 10.1002/stem.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho PL, Kurtova A, Chan KS. Normal and neoplastic urothelial stem cells: getting to the root of the problem. Nature reviews Urology. 2012;9:583–94. doi: 10.1038/nrurol.2012.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurtova AV, Xiao J, Mo Q, Pazhanisamy S, Krasnow R, Lerner SP. et al. Blocking PGE2-induced tumour repopulation abrogates bladder cancer chemoresistance. Nature. 2015;517:209–13. doi: 10.1038/nature14034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frangou C, Li YW, Shen H, Yang N, Wilson KE, Blijlevens M, Molecular profiling and computational network analysis of TAZ-mediated mammary tumorigenesis identifies actionable therapeutic targets. Oncotarget; 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: generally applicable gene set enrichment for pathway analysis. BMC bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiong Q, Mukherjee S, Furey TS. GSAASeqSP: a toolset for gene set association analysis of RNA-Seq data. Scientific reports. 2014;4:6347. doi: 10.1038/srep06347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moreb JS. Aldehyde dehydrogenase as a marker for stem cells. Current stem cell research & therapy. 2008;3:237–46. doi: 10.2174/157488808786734006. [DOI] [PubMed] [Google Scholar]
- 36.Su Y, Qiu Q, Zhang X, Jiang Z, Leng Q, Liu Z. et al. Aldehyde dehydrogenase 1 A1-positive cell population is enriched in tumor-initiating cells and associated with progression of bladder cancer. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2010;19:327–37. doi: 10.1158/1055-9965.EPI-09-0865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harley V, Lefebvre V. Twenty Sox, twenty years. The international journal of biochemistry & cell biology. 2010;42:376–7. doi: 10.1016/j.biocel.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 38.Aaboe M, Birkenkamp-Demtroder K, Wiuf C, Sorensen FB, Thykjaer T, Sauter G. et al. SOX4 expression in bladder carcinoma: clinical aspects and in vitro functional characterization. Cancer research. 2006;66:3434–42. doi: 10.1158/0008-5472.CAN-05-3456. [DOI] [PubMed] [Google Scholar]
- 39.Liu P, Ramachandran S, Ali Seyed M, Scharer CD, Laycock N, Dalton WB. et al. Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer research. 2006;66:4011–9. doi: 10.1158/0008-5472.CAN-05-3055. [DOI] [PubMed] [Google Scholar]
- 40.Scharer CD, McCabe CD, Ali-Seyed M, Berger MF, Bulyk ML, Moreno CS. Genome-wide promoter analysis of the SOX4 transcriptional network in prostate cancer cells. Cancer research. 2009;69:709–17. doi: 10.1158/0008-5472.CAN-08-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pramoonjago P, Baras AS, Moskaluk CA. Knockdown of Sox4 expression by RNAi induces apoptosis in ACC3 cells. Oncogene. 2006;25:5626–39. doi: 10.1038/sj.onc.1209566. [DOI] [PubMed] [Google Scholar]
- 42.Medina PP, Castillo SD, Blanco S, Sanz-Garcia M, Largo C, Alvarez S. et al. The SRY-HMG box gene, SOX4, is a target of gene amplification at chromosome 6p in lung cancer. Human molecular genetics. 2009;18:1343–52. doi: 10.1093/hmg/ddp034. [DOI] [PubMed] [Google Scholar]
- 43.Huang YW, Liu JC, Deatherage DE, Luo J, Mutch DG, Goodfellow PJ. et al. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer research. 2009;69:9038–46. doi: 10.1158/0008-5472.CAN-09-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. List of up-regulated and down-regulated genes in sicontrol compared to siSox4 in RT-112 cells; significantly up-regulated and down-regulated GO terms by DAVID analysis in sicon compared to siSox4 in RT-112 cells.