Abstract
Candida albicans is a human opportunistic fungus and it is responsible for a wide variety of infections, either superficial or systemic. C. albicans is a polymorphic fungus and its ability to switch between yeast and hyphae is essential for its virulence. Once C. albicans obtains access to the human body, the host serum constitutes a complex environment of interaction with C. albicans cell surface in bloodstream. To draw a comprehensive picture of this relevant step in host-pathogen interaction during invasive candidiasis, we have optimized a gel-free shaving proteomic strategy to identify both, human serum proteins coating C. albicans cells and fungi surface proteins simultaneously. This approach was carried out with normal serum (NS) and heat inactivated serum (HIS). We identified 214 human and 372 C. albicans unique proteins. Proteins identified in C. albicans included 147 which were described as located at the cell surface and 52 that were described as immunogenic. Interestingly, among these C. albicans proteins, we identified 23 GPI-anchored proteins, Gpd2 and Pra1, which are involved in complement system evasion and 7 other proteins that are able to attach plasminogen to C. albicans surface (Adh1, Eno1, Fba1, Pgk1, Tdh3, Tef1, and Tsa1). Furthermore, 12 proteins identified at the C. albicans hyphae surface induced with 10% human serum were not detected in other hypha-induced conditions. The most abundant human proteins identified are involved in complement and coagulation pathways. Remarkably, with this strategy, all main proteins belonging to complement cascades were identified on the C. albicans surface. Moreover, we identified immunoglobulins, cytoskeletal proteins, metabolic proteins such as apolipoproteins and others. Additionally, we identified more inhibitors of complement and coagulation pathways, some of them serpin proteins (serine protease inhibitors), in HIS vs. NS. On the other hand, we detected a higher amount of C3 at the C. albicans surface in NS than in HIS, as validated by immunofluorescence.
Keywords: Candida albicans, shaving, surface proteins, human serum, complement pathways, coagulation pathways, GPI-anchored proteins, host-pathogen interaction
Introduction
The yeast Candida albicans is the most important opportunistic fungi, causing a wide variety of infections ranging from superficial to invasive candidiasis, and it is often found in the mucosal microbiota. C. albicans infections produce high morbidity and mortality in intensive care, post-surgery and cancer patients or other types of immunocompromised patients (Leleu et al., 2002; Almirante et al., 2005; Hube, 2006; Brown et al., 2012). The high mortality of these infections is determined by the diagnostic limitations, the scarcity of antifungal agents and the emergence of resistance to them (Viudes et al., 2002; Wilson et al., 2002). For these reasons, the incidence of invasive candidiasis is still high.
The cell wall of C. albicans is a dynamic and complex multi-layered structure located external to the plasma membrane. It is responsible for maintenance of the shape that characterizes each growth form (mainly yeast and hyphae) of the fungus (Klis et al., 2009). The cell wall mediates the first contact with the environment and integrates multiple cues into complex signaling networks that are coordinated by various transcription factors. Consequently, C. albicans differentially expresses cell surface proteins and virulence factors. The dimorphic transition that is the ability of C. albicans to reversibly switch from yeast to hyphal growth is essential for virulence; strains that are locked in either form are avirulent (Yan et al., 2013; Lu et al., 2014). The hyphal form allows the pathogen to penetrate into tissues to acquire nutrients or escape from the host defense, and yeast cells disseminate across the human body. There are many signals capable of inducing the dimorphic phenotypic switch: N-acetyl-D-glucosamine, physiological temperature and pH (37°C and neutral), high CO2 concentration, hypoxia and nutrient starvation; nonetheless, growing in human serum is the most physiological condition to study this process (Kumamoto and Vinces, 2005; Karkowska-Kuleta et al., 2009; Sudbery, 2011; Mayer et al., 2013; Ene et al., 2014).
C. albicans is able to adhere to host cells and tissues; for this reason, it exposes surface proteins such as adhesins and many other pathogenic factors (Chaffin, 2008). Adhesion to host tissue is a prerequisite for tissue invasion and infection. Among adhesins, there are the Als family (agglutinin-like sequence), the Hwp family and the Iff/Hyr family (de Groot et al., 2013). Pathogenic factors include tissue-digesting hydrolytic enzymes such as the Sap family (Secreted Aspartyl Proteinases) (Kretschmar et al., 2002; Buu and Chen, 2014), lipases and phosholipases. The Sap family is composed of 10 proteins with different expression patterns; they act optimally at acidic pH. Their main role is related to digesting molecules for nutrient acquisition and for digesting or distorting host cell membranes to avoid or resist antimicrobial attack by the host immune system (Naglik et al., 2003). Some adhesins are attached to the wall through a C-terminus glycosylphosphatidylinositol (GPI) anchorage sequence (De Groot et al., 2003; Pardo et al., 2004; Klis et al., 2009).
Human serum is a very complex body fluid; its qualitative and quantitative composition was a hot topic of study during years (Anderson and Anderson, 2002; Mitchell, 2010). In the Plasma Proteome Database, proteomic data of 10,546 proteins detected in human serum and plasma are collected (Nanjappa et al., 2014). The dynamic range of abundance of these proteins has more than 10 orders of magnitude of variation. The complement system is an efficient component of the innate immune response and establishes a link with the adaptive system. The three conventional pathways by which complement is activated (classical, alternative and lectin) are composed of around 30 proteins (Engleberg et al., 2012). It mediates several physiological processes such as tissue regeneration and clearance of immune complexes. C2 and factor B (FB) are the main heat-sensitive complement components (Soltis et al., 1979). C2 is the central component in the activation of classical and lectin pathways and FB is for alternative pathway; both are necessary for the formation of C3- and C5-convertases in the respective complement cascades aforementioned.
C. albicans develops several strategies to evade the complement system, delay the detection by the host immune systems and even capable of manipulate the host defenses for its own purpose (Collette and Lorenz, 2011). In this way, C. albicans acquires host-derived inhibitory proteins such as factor H (FH), factor H-like protein 1 (FHL-1) and C4b-binding protein (C4BP), to prevent activation of complement pathways on its surface. These three protein regulators are closely related molecules composed of variable numbers of short consensus repeat (SCR) domains. There are four C. albicans proteins described with the ability to fix FH on their surface: glycerol 3-phosphate-dehydrogenase 2 (Gpd2), high-affinity gluthathione transporter 1 (Hgt1), phosphoglycerate mutase 1 or complement regulator-acquiring surface protein 1 (Gpm1/CRASP1) and pH-regulated antigen 1 (Pra1/CRASP2) (Poltermann et al., 2007; Luo et al., 2009, 2013). With the exception of Hgt1, the other three are able also to bind plasminogen. Additionally, Pra1 and Hgt1 bind C4BP (Lesiak-Markowicz et al., 2011; Luo et al., 2011). Factor H mediates binding to polyanions (sialic acids and human heparin) present on C. albicans cell wall (Soares et al., 2000; Rodríguez de Córdoba et al., 2004; Green et al., 2013). Pra1 also binds fibrinogen and C3 directly (Zipfel et al., 2011).
The aim of this study is to know the human serum protein coat on C. albicans surface as well as which C. albicans proteins could be candidates for diagnostic markers, potential vaccines or therapeutic targets, as they are detected when human serum is present. The interaction between human serum proteins and other microorganism surface was tested by different proteomic approaches in Staphylococcus aureus and Streptococcus pyogenes (Dreisbach et al., 2011; Sjöholm et al., 2014). In this work, we carried out a gel-free proteomic approach by shaving live C. albicans cells with trypsin after serum interaction to obtain exposed peptides of C. albicans and human proteins. With this shaving strategy that was applied in our laboratory to C. albicans and S. cerevisiae cultured in synthetic media (Hernáez et al., 2010; Insenser et al., 2010; Gil-Bona et al., 2015), it is not necessary to perform sub-cellular fractionation. In the present work, we have optimized this protocol for the detection of C. albicans and human proteins together in conditions that mimic in vivo interaction during systemic infections; overall, 372 C. albicans and 214 human proteins were identified. Among C. albicans proteins, 147 are described as located at the cell surface. Interestingly Pra1 and Gpd2 are able to attach inhibitors of complement cascades and plasminogen; while a further 7 proteins are also capable of attaching plasminogen. Regarding human proteins, all main proteins of complement pathways were identified, and many proteins involved in coagulation and metabolism, as well as immunoglobulins or cytoskeletal proteins. Furthermore, in this work proteins of complement and coagulation pathways were observed surrounded the surface of C. albicans hyphal cells.
Materials and methods
Candida albicans strain and growth conditions
The C. albicans strain used in this work was SC5314, from a clinical isolate (Gillum et al., 1984). This strain was grown on YPD plates (2% D-glucose, 2% peptone, 1% yeast extract, and 2% agar) and incubated at 30°C. A colony was picked and grown in YPD medium at 30°C and 200 rpm during 6–8 h. The OD600nm was measured and diluted on minimum medium (MM) at 0.00003 OD600nm (2% D-glucose, 0.17% yeast nitrogen base, 0.5% (NH4)2SO4, 0.192% synthetic complete mixture (Kaiser) drop-out -ura and 0.01% uracil). Culture was grown at 30°C and 200 rpm overnight to obtain and collected at a concentration ~106 cells/ml. 1.5 × 107 cells were resuspended in 9 ml of Lee medium at pH 6.7 (Lee et al., 1975). One milliliter of human serum was added and the culture was grown during 5 h at 37°C and 200 rpm. Human sera were from healthy donor volunteers who did not have any clinical or microbiological evidence of infection. They were used in the experiments as normal serum (NS) or heat inactivated serum (HIS). Serum inactivation was done by heating at 56°C during 30 min in a water bath.
Surface shaving
Surface shaving procedure was adapted from previous experiment of our group (Hernáez et al., 2010; Gil-Bona et al., 2015). Briefly, after incubation of C. albicans with 10% human serum (normal serum-NS or heat inactivated serum-HIS), the cultures were centrifuged at 3500 rpm 5 min. Cell pellet was resuspended in 1 ml of phosphate buffer saline (PBS) with 0.1% Tween 20, centrifuged again and washed 6 times more with PBS. The last pellet was resuspended in 400 μl of ammonium bicarbonate (AMBI buffer; NH4HCO3) 25 mM pH 8.0 and 7.5 μl dithiothreitol (DTT) 1 M and 9 μg of recombinant sequencing grade trypsin (Roche) were added. DTT was added during the digestion as reducing agent to consent deep digestion of non-covalently or disulphide bridges associated protein. The samples were incubated at 37°C, 30 min and 600 rpm. After incubation, samples were centrifuged at 5000 rpm 5 min and the supernatant was filtered with a filter unit of 0.22 μm. The cell pellet was resuspended in 400 μl of fresh AMBI buffer and 100 μl of trifluoroacetic acid (TFA) 0.1% (v/v) were added to stop the proteolytic reaction. It was centrifuged and the supernatant was filtered again. Both peptide-supernatants were put together and processed for further proteomic analysis. Subsequently, originated peptides were cleaned up with a Poros R2 resin (ABSciex, Framingham, MA). Peptides were eluted with 80% acetonitrile in 0.1% TFA, dried in a Speed-Vac and resuspended in 0.1% formic acid. The samples were stored at −20°C before nano-LC-MS/MS analysis.
LTQ-Orbitrap velos analysis, protein identification, and bioinformatics analysis
Peptides were analyzed by nano-LC-MS/MS analysis. Peptide samples were concentrated and desalted using C18-A1 ASY-column 2 cm pre-column (Thermo Scientific) and then eluted onto a Biosphere C18 column (Nano-Separations). Peptides were separated with a 140 min gradient (110 min from 0 to 40% Buffer B; Buffer A: 0.1% formic acid/2% acetonitrile; Buffer B: 0.1% formic acid in acetonitrile) at a flow-rate of 250 nl/min on a nano-Easy HPLC (Proxeon) coupled to a nano-electrospray ion source (Proxeon). Mass spectrometry experiments were performed using a LTQ-Orbitrap Velos (Thermo Scientific) in the positive ion mode. Full-scan MS spectra (m/z 400/1400) were acquired in the Orbitrap apparatus with a target value of 1,000,000 at a resolution of 60,000 at m/z 400 and the 15 most intense ions were selected for collision induced dissociation (CID) fragmentation in the LTQ with a target value of 10,000 and normalized collision energy of 38%. Precursor ion charge state screening and monoisotopic precursor selection were enabled. Singly charged ions and unassigned charge states were rejected. Dynamic exclusion was enabled with a repeat count of 1 and exclusion duration of 30 s.
Protein identification was carried out with mass spectra raw files using a licensed version of search engine MASCOT 2.3.0 with Proteome Discoverer software version 1.4.1.14 (Thermo Scientific). Searchers were made against C. albicans SC5314 (6221 sequences) and human database (20233 sequences) available on Candida Genome Database (Assembly 21 of CGD, http://www.candidagenome.org/) and Uniprot-SwissProt (http://www.uniprot.org) respectively, to identify peptides and proteins. Search parameters were oxidized methionine as variable modification, peptide mass tolerance 10 ppm, 2 missed trypsin cleavage sites and MS/MS fragment mass tolerance of 0.8 Da. In all protein identification, the FDR was <1%, using a Mascot Percolator, with a q-value of 0.01. Gene Ontology (GO) terms at CGD (http://www.candidagenome.org/cgi-bin/GO/goTermFinder) were used to classify C. albicans proteins.
As an estimation of the relative protein abundances the normalized spectral abundance factor (NSAF) was used (Zybailov et al., 2007), and the average of the normalized values was calculated. Mass spectrometry proteomics data have been deposited in C. albicans PeptideAtlas (Vialas et al., 2014) with the data set identifier PASS00446.
Cell permeability
Before and after trypsin treatment, C. albicans cell wall permeability was evaluated by staining with propidium iodide (PI). 106 cells/ml were incubated with 10 μl of PI 5 mM and the positive fluorescent cells (red staining) were checked, at least 200 cells of each sample were counted in a fluorescence microscope. Cells treated with 70% ethanol-PBS (v/v) were used as positive control.
Epifluorescence and confocal fluorescence microscopy
For detecting complement proteins on C. albicans surface after interaction with human serum, a solution of 106 cells/ml on Lee medium at pH 6.7 with 10% of human serum (NS or HIS) was prepared. 105 cells of this suspension were laid during 30 min, 1.5 or 5 h at 37°C on glass coverslips pre-coated with poly-L-lysine (1 mg/ml). Cells were fixed with formaldehyde 4% (w/v) (in PBS) for 20 min at room temperature (RT). Glass slides were washed twice with PBS. To stain cell membrane, PKH26 dye was added following technical instructions (Sigma Aldrich). Then, slides were blocked during 45 min at RT with buffer B (PBS plus bovine serum albumin (BSA) at 1 mg/ml). The slides were washed twice again with PBS and then incubated for 1.5 h at RT in the same buffer with an anti-C3, anti-factor B polyclonal serum (dilution 1:75 and 1:50, respectively) or only buffer. The slides were washed three times with PBS and further incubation for 1 h with an anti-rabbit IgG conjugated with Alexa-488 diluted at 1:500 in buffer B was done. Nuclei were stained with DAPI dye (5 μg/ml; 5 min at RT). Mounting medium Fluoromount-G (SouthernBiotech) was added to the preparations. Cells were then examined with epifluorescence and confocal microscopy images were collected using an Olympus FV1200 microscope.
Flow cytometry analysis
C. albicans cells were grown as previously described on Immunofluorescence assay section. In this case the incubation with human serum (NS or HIS) was done during 30 min to avoid larger hyphae formation. After that, cells were washed with PBS and fixed with formaldehyde 4% in PBS during 20 min at RT. Then, three washes with PBS were performed and samples were blocked with 0.1% BSA in PBS at 4°C overnight with gentle shaking. Samples were incubated with primary antibody or with buffer alone as negative control, for 2 h at RT. The primary antibodies were prepared in PBS with 0.1% BSA, anti-C3 and anti-FB at 1:75 and 1:50 dilutions, respectively. Four PBS washes were performed and incubated with secondary antibody below, an anti-rabbit conjugated with Alexa-488 diluted at 1:500, 2 h at RT with gently shaking. Then, samples were washed with PBS 4 times more. These cells were analyzed with a Guava easyCyte cytometer of Millipore.
Results
Gel-free proteomic approach to study protein interactions between C. albicans and human serum
In order to break through the broad spectrum of human serum proteins attached to C. albicans cells and the fungus surface proteins, we analyzed C. albicans cell surface after 5 h of incubation with 10% human serum by a gel-free proteomic strategy. Normal serum (NS) was used to mimic physiological conditions of C. albicans during invasive candidiasis and heat inactivated serum (HIS) to determine human proteins deposited on the C. albicans surface without conventional complement cascade activation. As human serum is the most important inductor of C. albicans dimorphic transition, after 5 h of incubation all C. albicans cells were in the hyphal form. To obtain the protein peptides, a shaving methodology was used, which consists of direct cell digestion with trypsin and MS/MS identification. Before digestion, cells were washed with PBS plus Tween-20 and several additional washes with PBS were done to remove human serum proteins weakly attached to C. albicans cells. We optimized parameters such as time and trypsin concentration during shaving of C. albicans cells, in order to control cell integrity and identify the highest number of proteins (from both, C. albicans and human). With short times of trypsin treatment, between 5 and 10 min, only human proteins were identified (data not shown). Otherwise, after 30 min, human and C. albicans proteins were detected; thus, this duration of trypsin incubation was selected. Before and after trypsin treatment, cells were treated with propidium iodide (PI) and the cell permeability state was analyzed under the microscope. Less than 1% positive staining was obtained in all conditions. Cell integrity was assessed with the purpose of avoiding cytoplasmic protein contamination after trypsin treatment.
Peptides obtained after C. albicans cells interaction with 10% human serum (NS or HIS) by shaving were recovered and identified by LC-MS/MS on a LTQ Orbitrap. Four biological replicates were made using NS and three using HIS. Mass spectra raw files were searched against C. albicans SC5314 and human databases (CGD and Uniprot-SwissProt, respectively) and proteins identified in at least two biological replicates with two or more peptides in one of them were selected. C. albicans-identified proteins are listed in Table 1 and Table S1, and human proteins in Table 2 and Table S2.
Table 1.
Selected C. albicans proteins identified after incubation with 10% NS or HIS related with cell surface and immunogenicity.
| Categories | Common in NS and HIS | Only identified in NS | Only in HIS | Total |
|---|---|---|---|---|
| GPI-anchored proteinsa | 11 | 12 | – | 23 |
| Als3, Cht2, Ecm33, Hyr1, Ihd1, Pga45, Phr1, Pir1, Rbt1, Rbt5, Sod5 | Als1, Crh11, Pga10, Pga4, Phr2, Plb3, Plb4.5, Rhd3, Sap9, Ssr1, Utr2, Ywp1 | – | ||
| Proteins involved in cell wall organization or biogenesisb(GO: 71554) | 8 | 18 | – | 26 |
| Act1, Bmh1, Gda1, Msb2, Sod1, Srb1, Tsa1, Yps7 | Agm1, Bgl2, Eng1, Frs1, Gfa1, Gsc1, Kex2, Mnn26, Mnt1, Pmi1, Pmt2, Ras1, Rho1, Rvs161, Sim1, Slk19, Sur7, Ypt31 | – | ||
| Cell surface proteinsb(GO: 9986) | 40 | 21 | 1 | 62 |
| Adh1, Cdc19, Cdc48, Cef3, Ddr48, Ece1, Efb1, Eft2, Egd2, Eno1, Fba1, Glx3, Hsp70, Hsp90, Ino1, Ipp1, Kar2, Met6, Mp65, Orf19.2478.1, Pdc11, Pdi1, Pgk1, Pma1, Pra1, Rpl10, Rpl17B, Rpl3, Rpl4B, Rps7A, Rps8A, Sam2, Ssa2, Ssb1, Tal1, Tdh3, Tef1, Tkl1, Tos1, Tpi1 | Ape2, Atp1, Atp2, Coi1, Csp37, Gpd2, Gph1, Hem13, Hsp104, Hsp21, Rpl13, Rpl14, Rpl19A, Rpl20B, Rpl6, Rps10, Rps6A, Ssc1, Ssz1, Ugp1, Xyl2 | Cht3 | ||
| Plasma membrane proteinsb (GO: 5886) | 8 | 28 | – | 36 |
| Ahp1, Orf19.6553, Cof1, Met15, Pfy1, Pgi1, Pil1, Tif | Ade17, Adh2, Bcy1, Bzz1, Cdc42, Chc1, Faa4, Gca1, Hgt6, Ist2, Kin2, Lsp1, Mdg1, Orf19.1564, Orf19.3003, Orf19.4216, Orf19.6160, Pet9, Pga63, Pst3, Rdi1, Sec26, Sec4, Shm2, Sso2, Tom70, Ypt1, Ykt6 | – | ||
| Others immunogenic proteins | 6 | 2 | – | 8 |
| Asc1, Gnd1, Grp2, Hxk2, Msi3, Sah1 | Acs2, Mdh1 | – | ||
| 155 |
The classification is hierarchical and exclusive. Proteins were included if they were detected in at least two replicates of one condition with at least 2 peptides in one replicate. There are four replicates of Normal Serum (NS) and three replicates of Heat Inactivated Serum (HIS). Protein names from Candida Genome Database (CGD) (Inglis et al., 2012).
GPI-cell wall proteins reviewed in De Groot et al. (2003), Pardo et al. (2004), Chaffin (2008), Klis et al. (2009).
Gene Ontology (GO) terms (in parentheses the GO identifier) at CGD.
Table 2.
Human proteins of complement and coagulation pathways identified on the surface of C. albicans after incubation with human serum.
| Accession numbera | Gene symbola | Description/Functiona | No. replicates (peptides on each replicate)b | |
|---|---|---|---|---|
| NS | HIS | |||
| COMPLEMENT PATHWAY | ||||
| P01023 | A2M | Alpha-2-macroglobulin | 2 (15,11) | 3 (10,9,70) |
| P04003 | C4BPA | C4b-binding protein alpha chain | 4 (5,3,4,9) | 3 (15,8,4) |
| P20851 | C4BPB | C4b-binding protein beta chain | 1 (2) | 2 (2,3) |
| P10909 | Clusterin | Extracellular chaperone or ApoJ | 4 (11,8,13,19) | 3 (11,13,18) |
| Q9BWP8 | COLEC11 | Collectin kidney protein 1 or collectin-11 | 4 (5,5,2,4) | 0 |
| P02745 | C1qA | Complement C1q subcomponent subunit A | 0 | 2 (1,4) |
| P02746 | C1qB | Complement C1q subcomponent subunit B | 3 (1,1,1) | 3 (5,5,4) |
| P02747 | C1qC | Complement C1q subcomponent subunit C | 3 (1,1,2) | 3 (7,5,6) |
| P00736 | C1r | Complement C1r subcomponent | 2 (2,7) | 3 (21,15,14) |
| Q9NZP8 | C1r-like | Complement C1r-like protein | 0 | 3 (6,5,5) |
| P09871 | C1s | Complement C1s subcomponent | 2 (1,10) | 3 (23,14,11) |
| P06681 | C2 | Complement component C2 | 1 (2) | 3 (19,15,10) |
| P01024 | C3 | Complement component C3 | 4 (103,106,141,147) | 3 (141,105,138) |
| P0C0L4 | C4-A | Complement component C4-A | 4 (32,32,94,98) | 3 (90,61,76) |
| P0C0L5 | C4-B | Complement component C4-B | 4 (32,33,95,98) | 2 (90,62) |
| P01031 | C5 | Complement component C5 | 3 (34,23,21) | 3 (68,46,70) |
| P13671 | C6 | Complement component C6 | 3 (20,18,4) | 3 (21,14,16) |
| P10643 | C7 | Complement component C7 | 4 (13,9,8,6) | 3 (6,11,16) |
| P07357 | C8-A | Complement component C8 alpha chain | 4 (13,11,6,7) | 3 (11,8,8) |
| P07358 | C8-B | Complement component C8 beta chain | 4 (5,6,16,27) | 3 (22,16,13) |
| P07360 | C8-G | Complement component C8 gamma chain | 4 (6,6,4,1) | 3 (6,4,9) |
| P02748 | C9 | Complement component C9 | 4 (17,15,17,22) | 3 (24,17,14) |
| P00751 | FB | Complement factor B | 4 (8,3,8,10) | 3 (47,31,28) |
| P00746 | FD | Complement factor D | 0 | 3 (2,2,4) |
| P08603 | FH | Complement factor H | 2 (5,3) | 3 (18,17,13) |
| P05156 | FI | Complement factor I | 1 (1) | 3 (3,3,6) |
| P27918 | FP | Complement factor P or Properdin | 4 (5,5,2,2) | 1 (3) |
| Q15485 | Ficolin-2 | Collagen/fibrinogen domain containing protein 2 | 0 | 2 (1,3) |
| O75636 | Ficolin-3 | Collagen/fibrinogen domain containing protein 3 | 1 (1) | 3 (5,2,7) |
| O00187 | MASP2 | Mannan-binding lectin serine protease 2 | 3 (2,2,5) | 1 (3) |
| P80108 | GPLD1 | Phosphatidylinositol-glycan-specific phospholipase D | 0 | 2 (4,15) |
| P05155 | C1INH | Plasma protease C1 inhibitor (Serpin G1) | 0 | 3 (20,14,24) |
| P04004 | VTN | Vitronectin or protein S | 4 (10,12,10,11) | 3 (14,11,13) |
| COAGULATION PATHWAY | ||||
| P01009 | SERPINA1 | Alpha-1-antitrypsin (AAT) | 4 (4,3,6,12) | 3 (37,35,47) |
| P08697 | SERPINF2 | Alpha-2-antiplasmin | 4 (2,2,9,13) | 3 (17,16,18) |
| P01008 | SERPINC1 | Antithrombin-III (AT-III) | 4 (2,2,5,10) | 3 (13,16,25) |
| P02749 | ApoH | Beta-2-glycoprotein 1; APC inhibitor; Apolipoprotein H | 2 (1,1) | 3 (8,3,4) |
| Q96IY4 | CPB2 | Carboxypeptidase B2; Thrombin-activable fibrinolysis inhibitor | 4 (2,1,12,10) | 3 (5,3,3) |
| P00740 | F9 | Coagulation factor IX | 4 (3,2,3,5) | 3 (5,4,4) |
| P12259 | F5 | Coagulation factor V | 4 (8,2,11,22) | 3 (11,4,8) |
| P08709 | F7 | Coagulation factor VII | 2 (2,3) | 2 (3,2) |
| P00742 | F10 | Coagulation factor X | 2 (2,2) | 3 (6,6,6) |
| P00748 | F12 | Coagulation factor XII | 1 (1) | 2 (4,8) |
| P00488 | F13A | Coagulation factor XIII A chain | 3 (1,2,1) | 2 (4,7) |
| P05160 | F13B | Coagulation factor XIII B chain | 0 | 3 (2,1,2) |
| P02671 | FGA | Fibrinogen alpha chain | 4 (13,12,3,7) | 3 (6,6,10) |
| P02675 | FGB | Fibrinogen beta chain | 4 (13,10,1,7) | 3 (6,11,14) |
| P02679 | FGG | Fibrinogen gamma chain | 3 (6,6,4) | 3 (2,5,10) |
| P02751 | FN1 | Fibronectin; Binds cell surfaces and various compounds (collagen, fibrin, heparin…) | 4 (60,62,15,16) | 3 (29,38,24) |
| P68871 | HBB | Hemoglobin subunit beta | 4 (7,9,3,10) | 3 (9,7,11) |
| Q9Y251 | HPA | Heparanase | 2 (1,3) | 0 |
| P05546 | HCF2 | Heparin cofactor 2 (Serpin D1) | 4 (8,11,34,28) | 3 (29,23,27) |
| P04196 | HRG | Histidine-rich glycoprotein | 4 (22,23,26,23) | 3 (21,24,17) |
| Q14520 | HABP2 | Hyaluronan-binding protein 2 | 0 | 3 (4,3,5) |
| P01042 | KNG1 | Kininogen-1 | 4 (6,2,11,15) | 3 (11,14,11) |
| P03952 | KLKB1 | Plasma kallikrein | 4 (7,8,2,1) | 3 (8,9,10) |
| P05154 | SERPINA5 | Plasma serine protease inhibitor | 0 | 2 (3,6) |
| P00747 | PLG | Plasminogen | 4 (5,6,21,34) | 3 (41,41,15) |
| P40197 | GP5 | Platelet glycoprotein V | 2 (2,2) | 3 (5,2,3) |
| Q9UK55 | SERPINA10 | Protein Z-dependent protease inhibitor | 4 (1,1,10,9) | 3 (6,5,7) |
| P00734 | F2 | Prothrombin | 4 (16,18,17,25) | 3 (21,17,10) |
| P04070 | PROC | Vitamin K-dependent protein C | 1 (3) | 2 (3,4) |
| P07225 | PROS1 | Vitamin K-dependent protein S | 3 (1,1,11) | 3 (6,3,14) |
| P04275 | VWF | von Willebrand factor | 4 (18,16,19,72) | 3 (52,36,9) |
Accession number, gene symbol and function from UniProtKB/Swiss-Prot database (Uniprot, 2014).
Proteins were included if they were identified in at least two replicates of one condition with at least 2 peptides in one replicate.
This analysis rendered the identification of 372 unique C. albicans proteins, 371 in NS and 134 in HIS (Table S1). There were 133 proteins identified in both conditions, accounting for 36% of the total C. albicans-identified proteins. The Orf19.8 (an ortholog of C. dubliniensis CD36) was the only protein identified with HIS and not with NS. Noteworthy, around ~33% of the C. albicans proteins were identified in at least six of the seven analyzed samples. Furthermore, of the 372 C. albicans-identified proteins, 60 proteins presented a signal peptide (SP) at CGD. On the other hand, 214 human proteins were identified; of these, 128 were common to both conditions (60%), 43 were present only after incubation with NS and the other 43 only with HIS (Table 2 and Table S2).
Analysis of C. albicans cell surface protein pattern after interaction with 10% human serum
Only those C. albicans identified proteins with a Gene Ontology (GO) annotation at CGD pointing either to surface localization, either to surface exposure or with the ability to induce human serum antibodies, were included in Table 1. We focused our study on these 155 C. albicans proteins classified into 5 categories: glycosylphosphatidylinositol (GPI)-anchored proteins; proteins involved in cell wall organization or biogenesis; cell surface proteins; plasma membrane proteins and other immunogenic proteins not present in the previous group. The groups are hierarchical and exclusive.
The first category includes 23 proteins attached to the plasma membrane or cell wall by a GPI-anchor; most are involved in biogenesis and maintenance of the C. albicans cell wall. All GPI-anchored proteins identified have a detected SP for entry into the secretory pathway at CGD. In the second category, there are 26 other proteins involved in cell wall organization or biogenesis. Among the 62 proteins included in the cell surface category, there are proteins located at the cell surface, although most of them do not have SP and could not reach this cell localization via the secretory pathway. Several of them have been described as “moonlighting” or multifunctional proteins. It is interesting that antibodies against a total of 52 C. albicans-identified proteins have been detected in human blood or serum from patients with invasive candidiasis (these immunogenic proteins are highlighted in Table 1). Many of these are abundant at the cell surface (as they were detected with more than 10 unique peptides) and were not secreted by the classical secretory pathway (not have SP), such as Eno1, Fba1, Hsp70, and 17 additional proteins.
It is also remarkable that two out of the four proteins described in C. albicans with the ability to fix the complement inhibitor factor H (FH) were identified. Pra1 was identified in both samples (NS and HIS), but Gpd2 was only in NS samples. Furthermore, we detected many C. albicans proteins described with the ability to bind plasminogen: Adh1, Eno1, Fba1, Gpd2, Pgk1, Pra1, Tdh3, Tef1, and Tsa1.
Analysis of human serum proteins that interact with C. albicans cell surface relevant to C. albicans-host interaction
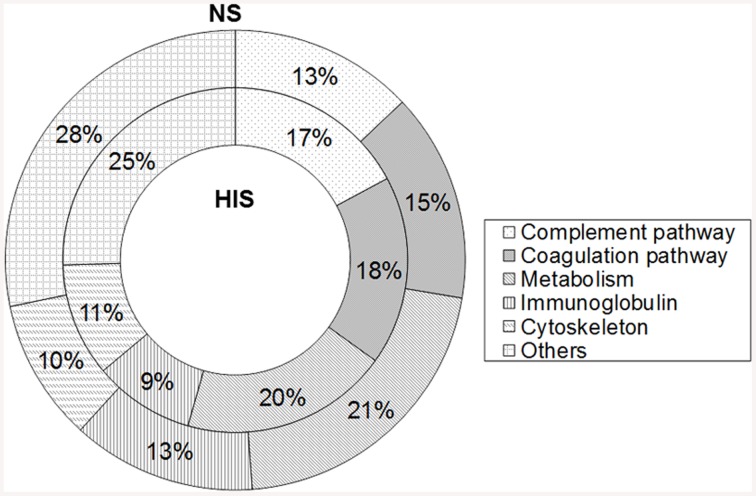
The 214 identified human proteins were classified into the following classes according to their function: complement pathway, coagulation pathway, metabolism, immunoglobulins, cytoskeleton, and others. Proteins belonging to complement and coagulation cascades are listed in Table 2 and the rest of the proteins are in Table S2. The percentages of proteins identified in each category in the two conditions tested are shown in Figure 1. The complement and coagulation pathways represent higher percentages of the total identified proteins when HIS was used and, on the other hand, the immunoglobulin group represents a higher percentage when NS was used.
Figure 1.
Percentage representation of human protein groups identified in normal serum (NS) and heat inactivated serum (HIS) samples by shaving C. albicans cells. Overall, 171 proteins were identified in both conditions and were classified into the complement pathway, coagulation pathway, metabolism, immunoglobulin, cytoskeleton, and others.
Close to all components of the three complement pathways (33 proteins) were identified, most of them in the two conditions tested. However, there are some complement proteins that have been detected only in the NS sample (Collectin-11) or in the HIS sample [complement C1q subcomponent subunit A (C1qA), complement C1r-like protein, complement factor D (FD), ficolin-2, phosphatidylinositol-glycan-specific phospholipase D (GPLD1) and plasma protease C1 inhibitor (SERPING1 or C1INH)] (Table 3). The identification of many complement inhibitors such as alpha-2-macroglobulin (A2M), C1INH, FH, and factor I (FI) is also interesting. In relation to the coagulation pathway, 31 proteins were identified, with most of them also being detected in the two conditions. Although heparanase was only identified in NS sample and coagulation factor XIII B chain, hyaluronan-binding protein 2 and plasma serine protease inhibitor (SERPINA5) were only identified in the HIS one. Interestingly, antithrombin-III (SERPINC1) and plasminogen (PLG), two coagulation proteins that are relevant complement inhibitors, were identified. Focusing on human immunoglobulins IgG and IgM, they were identified in NS and HIS samples and with a similar number of peptides in both of them (Table S2). The category of metabolic proteins includes many apolipoproteins detected on the surface of C. albicans (Table S2).
Table 3.
Comparison of complement proteins identified on the cell surface of C. albicans by shaving after incubation with NS vs. HIS.
| Complement proteins | Normal serum (NS) | Heat inactivated serum (HIS) |
|---|---|---|
| Lectin pathway | 7 proteins | 7 proteins |
| Collectin-11, MASP2, Ficolin-3*, C2*, C3, C4-A, C4-B | MASP2*, Ficolin-2, Ficolin-3, C2*, C3, C4-A, C4-B | |
| Classical pathway | 8 proteins | 10 proteins |
| C1qB, C1qC, C1r, C1s, C2*, C3, C4-A, C4-B | C1qA, C1qB, C1qC, C1r, C1r-like, C1s, C2, C3, C4-A, C4-B | |
| Alternative pathway | 3 proteins | 4 proteins |
| C3, FB, FP | C3, FB, FD, FP* | |
| Membrane attack complex (MAC) | 7 proteins | 7 proteins |
| C5, C6, C7, C8-A, C8-B, C8-G, C9 | C5, C6, C7, C8-A, C8-B, C8-G, C9 | |
| Complement inhibitors | 7 proteins | 9 proteins |
| A2M, C4BPA, C4BPB*, clusterin, FH, FI*, vitronectin | A2M, C1INH, C4BPA, C4BPB, clusterin, FH, FI, GPLD1, vitronectin |
Proteins indicated in bold are only identified in one condition.
Proteins with
are identified only in one replicate of the indicated condition.
Interestingly, 13 proteins belonging to the serpin family were identified. Serpins are a relevant group of proteins with similar structures able to inhibit proteases. Some of them had only been observed in the HIS sample, such as SERPINA5, C1INH and cortisol-binding globulin (SERPINA6) (Table 4).
Table 4.
Identified proteins belonging to the SERPIN family.
| Accesion number | Gene name | Protein | Substrate | No. replicates (peptides on each replicate) | |
|---|---|---|---|---|---|
| NS | HIS | ||||
| P01009 | SERPINA1 | Alpha 1-antitrypsin | Human neutrophil elastase | 4 (4,3,6,12) | 3 (37,35,47) |
| P01011 | SERPINA3 | Alpha 1-antichymotrypsin | Cathepsin G | 2 (1,2) | 3 (27,19,30) |
| P29622 | SERPINA4 | Kallistatin | Kallikrein | 2 (5,10) | 3 (13,19,16) |
| P05154 | SERPINA5 | Plasma serine protease inhibitor | Protein C | 0 | 2 (3,6) |
| P08185 | SERPINA6 | Cortisol-binding globulin | Non-inhibitory; cortisol binding | 0 | 3 (11,8,12) |
| P05543 | SERPINA7 | Thyroxine-binding globulin | Non-inhibitory; thyroxine binding | 2 (2,1) | 3 (6,4,14) |
| P01019 | SERPINA8 | Angiotensionogen | Non-inhibitory; Precursor of angiotensin I | 4 (6,5,5,5) | 3 (11,13,19) |
| Q9UK55 | SERPINA10 | Protein Z-related protease inhibitor | Factor Z and XI | 4 (1,1,10,9) | 3 (6,5,7) |
| P01008 | SERPINC1 | Antithrombin-III | Thrombin and factors IXa, Xa, and XIa | 4 (2,2,5,10) | 3 (13,16,25) |
| P05546 | SERPIND1 | Heparin cofactor II | Thrombin | 4 (8,11,34,28) | 3 (29,23,27) |
| P36955 | SERPINF1 | Pigment epithelium derived factor | Non-inhibitory; Potent anti-angiogenic molecule | 1 (1) | 3 (11,10,19) |
| P08697 | SERPINF2 | Alpha 2-antiplasmin | Plasmin and trypsin | 4 (2,2,9,13) | 3 (17,16,18) |
| P05155 | SERPING1 | Plasma protease C1 inhibitor | C1r, C1s, F12a, kallikrein, F11a, plasmin, MASP1, MASP2 | 0 | 3 (20,14,24) |
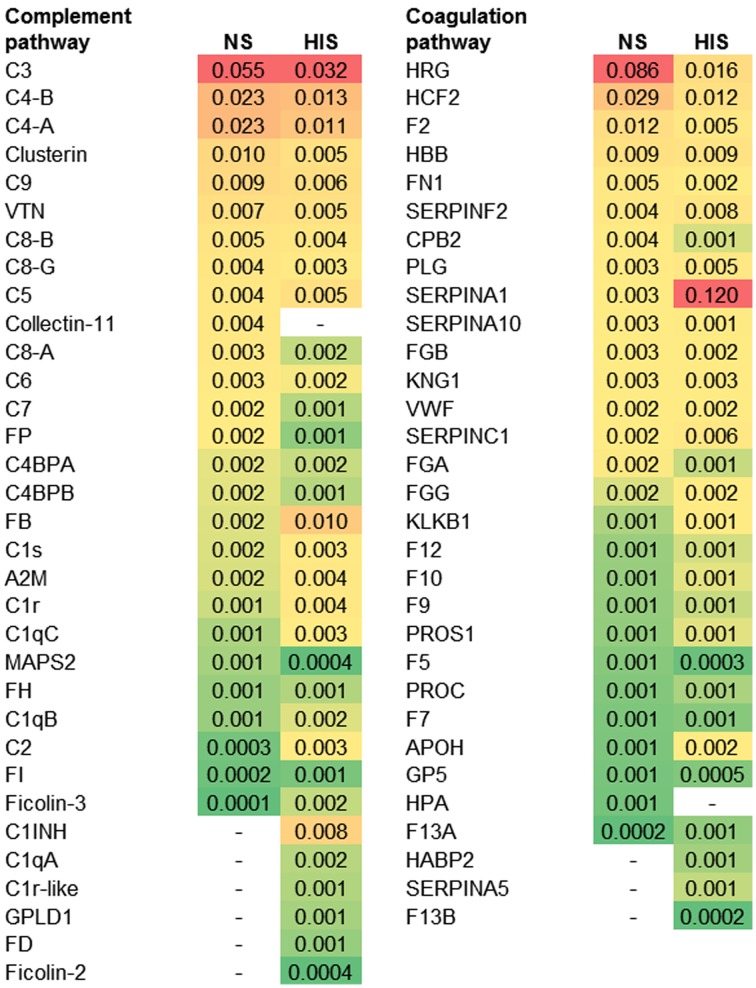
To estimate the relative abundance of human proteins identified on C. albicans surface, the normalized spectral abundance factor (NSAF) (Zybailov et al., 2007) of each protein was calculated and the average NSAF values among samples were assessed. We compared these NSAF values with the ranking of the original protein abundance in human plasma using data of the dynamic range of most abundant proteins adapted from Mitchell (2010). As shown in Figure S1, the relative abundance of proteins in the plasma is different to the relative abundance at the hypha surface after interaction with human serum. For example, C3 protein of complement pathways was identified as one of the most abundant proteins on the surface of C. albicans in both conditions and it was not among the most abundant proteins in plasma. Also, in the apolipoprotein family, the relative abundance of some proteins is different in the plasma vs. the fungal surface. ApoB100 and ApoE are less abundant in human plasma than ApoCIII and both proteins showed higher NSAF than ApoCIII.
The abundance ranking of the proteins involved in the complement and coagulation pathways identified on C. albicans surface is shown in Figure 2. C3, C4-B, and C4-A had the highest NSAF results among complement proteins in both serum conditions. Complement components of Membrane Attack Complex (MAC: C6, C7, C8-A, C8-B, C8-G, and C9) had higher NSAF in NS than in HIS samples and only C5 had higher NSAF in HIS than in NS samples. In the coagulation pathway, it is notable that SERPINA1 (alpha-1-antitrypsin) is much more abundant in the HIS sample than other proteins such as HRG (histidine-rich glycoprotein), HCF2 (SERPIND1) or F2 (prothrombin), which are more abundant in the NS sample than SERPINA1.
Figure 2.
Protein ranking of complement and coagulation proteins identified on normal serum (NS) and heat inactivated serum (HIS) samples according to the average of NSAF. C. albicans was incubated with 10% human serum at 37°C (normal serum-NS or heat inactivated serum-HIS) and NSAF was calculated for proteins belonging to complement and coagulation proteins. The proteins were ordered according to their NSAF obtained in NS samples, from more to less abundant. Gradient color ranges from red (more abundant) to green (less abundant).
During complement activation, C3, C4, and C5 are proteolyzed by cleavage at a specific site and small fragments are produced; these fragments are called anaphylotoxins or complement peptides, and are indicated with the letter “a” (Walport, 2001a,b). The analysis of the identified peptides belonging to: (i) anaphylotoxin regions (C3a, C4a, and C5a), (ii) released domains during proteolysis activation of complement pathways (C2b and FBa), and (iii) of the MAC-interaction regions (MACPF) is shown in Table 5. The tendency was the identification of more peptides belonging to these regions in HIS samples than in NS samples. It is remarkable that peptides from C2b and C5a were not identified in NS samples.
Table 5.
Analysis of peptides corresponding to cleavage fragments produced during complement activation and to contact regions in MAC formation.
| Region | No. peptides | |||
|---|---|---|---|---|
| Normal Serum-NS | Heat Inactivated Serum-HIS | |||
| Total | Only in NS | Total | Only in HIS | |
| C2b | 0 | 0 | 4 | 4 |
| C3a | 8 | 2 | 10 | 3 |
| C4a | 5 | 1 | 4 | 1 |
| C5a | 0 | 0 | 5 | 5 |
| Ba | 1 | 0 | 5 | 4 |
| C6-MACPF | 13 | 0 | 19 | 6 |
| C7-MACPF | 4 | 0 | 15 | 12 |
| C8-A-MACPF | 4 | 0 | 8 | 4 |
| C8-B-MACPF | 19 | 1 | 19 | 0 |
Identified peptides belong to the indicated released region or remain part of the full-length protein.
Gray background highlights identification in HIS condition.
Validation of proteomic results
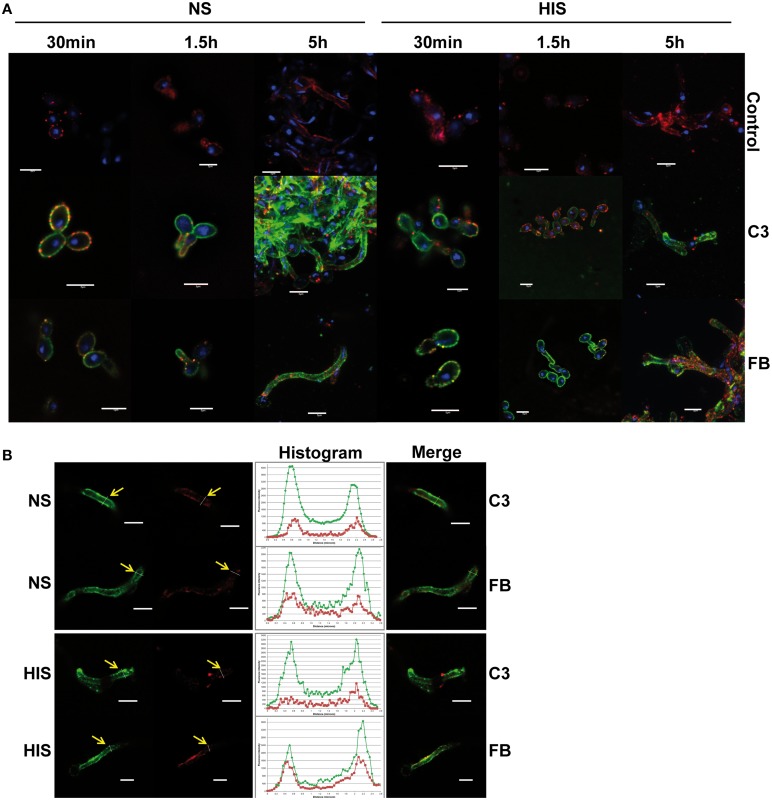
To validate the proteomic results, firstly the deposition level of C3 and FB on the C. albicans surface was analyzed by immunofluorescence microscopy and flow cytometry assays. C3 is a common component of the three complement cascades and together with A2M, are the two highest abundant complement proteins in human serum. Factor B (FB) is an abundant complement protein specific of the alternative pathway. C. albicans cells were incubated with NS and HIS and treated with anti-C3 and anti-FB antibodies (Figure 3). We observed the deposition of C3 and FB along the surface of all C. albicans cells when incubated with human serum (NS or HIS) and some areas with higher intensity in the case of C3 after 5 h of incubation with NS (Figure 3A, left panels). These differences between the fluorescence intensity of FB and C3 can be correlated with the number of peptides of these proteins in the samples at 5 h. When cells were incubated with HIS, the intensity of FB was slightly higher than in the case of NS incubation (it is well observed in short incubation times) (Figure 3A, right panels). To achieve flow cytometry assays, the incubation time of C. albicans cells with human serum was decreased to avoid the formation of large filaments. Within 30 min of incubation, the deposition of C3 was higher among the population in NS samples than in HIS (Figure S3). The intensity of FB was lower than that of C3, as expected, and less homogenous in both samples (NS and HIS) (upper panels). Mean fluorescence intensity (MFI) of two independent experiments were collected and showed that, in all cases, the MFI for C3 and FB was higher when cells were incubated with NS than with HIS at short times, although the differences are not significant in the case of FB (bottom panel).
Figure 3.
Detection of C3 and factor B on C. albicans surface after incubation with human serum. (A) Immunofluorescence assay to detect C3 and factor B (FB) on C. albicans surface incubated 30 min, 1.5 and 5 h with normal serum (NS; left panels) or heat inactivated serum (HIS; right panels). Control images showed the background of secondary antibody (anti-rabbit-A488). (B) Co-localization of C3 and FB with C. albicans plasmatic membrane (staining with PKH26) was examined by the analysis of confocal microscopy images, after 5 h of incubation with NS and HIS. Representative images are shown. Yellow arrows indicate position of transversal sections of illustrative histograms. Co-localization analysis was performed using FiJi software. Red staining of plasmatic membrane was done with PKH26, blue staining of DNA with DAPI and green staining with anti-rabbit-A488. Line in bottom right corner indicated in all figures corresponds to a 5 μm.
All of these data together showed that C3 and FB deposition was clearly observed at short incubation times, and C3 enhanced its accumulation on the surface with longer incubation time and more dramatically when the incubation was with NS. However, FB accumulation on C. albicans surface did not appear to increase with time, maybe because it can be masked by the rest of the complement proteins (Figure 3A).
Furthermore, to address whether C3 and FB are close to the cytoplasmic membrane, fluorescence co-localization was evaluated with confocal microscopy images by the analysis of transversal sections across cells (Figure 3B). As observed, both C3 and FB proteins are localized around the plasmatic membrane, staining when C. albicans cells were incubated with NS and also with HIS.
Afterwards, in order to evaluate the deposition of human IgGs on C. albicans surface after 5 h of incubation with 10% human serum, IgGs were detected in NS and HIS samples, with higher intensity on the yeast surface than on the hyphal surface in both samples (Figure S2).
Discussion
Our work shows, for the first time to our knowledge, the identification by a shaving approach of 147 C. albicans surface proteins and 214 human serum proteins attached to the surface of the fungus in the same report. The more relevant C. albicans proteins identified are GPI-anchored proteins and proteins involved in cell wall organization or biogenesis. The most relevant human proteins identified belong to complement and coagulation pathways. Indeed, all main human complement proteins were identified on the surface of C. albicans. The identified proteins were reproducible among the experiments and would represent the physiological mode of action of the immune system against C. albicans or components of the immune system used for C. albicans to evade, disturb or use on its own purpose the host defenses.
Analysis of C. albicans surface proteins
With the objective of unraveling the protein surface pattern of C. albicans hyphal cells, we analyzed our data in comparison to the data obtained by our group using the same methodology of cell shaving over yeast and hyphal cells (growth also in Lee's medium, but without human serum) (Gil-Bona et al., 2015). This analysis rendered 304 proteins identified in all C. albicans assays (yeasts and hyphae with and without human serum) that we consider the C. albicans common core of cell surface proteins and not morphotype specific (Table S3). Also, a hyphal protein pattern of 55 proteins identified on C. albicans hypha forms, but not identified in C. albicans yeasts, was obtained. Among them, Als3, Hyr1, and Sod5 were associated with hyphae in different works (Heilmann et al., 2011; Sosinska et al., 2011; Sudbery, 2011). They are proteins that are expected to be detected, indicating that the other 52 proteins are at least more abundant or accessible in hypha than in yeast cells. Another example is Ece1, which is a hyphal-specific protein with unknown function (Fan et al., 2013) (Table S3).
Other proteins were associated with hyphae in some works and described as morphotype-independent in others, for example, Phr1 and Rbt1 (Heilmann et al., 2011; Ragni et al., 2011; Monniot et al., 2013). We identified both proteins in all shaving C. albicans experiments. Furthermore, Phr2 (a homolog of Phr1) was also identified in all conditions. The differences observed at the level of detection of Phr1 and Phr2 among the samples were possibly related to the optimum pH for expression of each one (Sosinska et al., 2011; Dühring et al., 2015) (Table S1). On the other hand, Rhd3 and Ywp1 were reported as yeast-specific and were found to be part of the common core pattern in our study (Heilmann et al., 2011). A relevant protein found in C. albicans common core pattern is Msb2; this stabilizes the fungal cell wall and inactivates human antimicrobial peptides (Dühring et al., 2015).
It is interesting to know that GPI-anchored proteins identified in this work are part of the common core of C. albicans surface proteins or of the hyphal-form specific ones. There are 17 proteins belonging to the common core in our analysis, and also Ece1 and Sod5, which are part of the hyphal-specific group, whose genes were up-regulated under blood growth (Fradin et al., 2003).
Remarkably, 12 proteins were only identified on the C. albicans hyphal surface when it was grown with 10% human serum and we highlighted Pra1, Sap5, Tef1, and 5 more Orfs with unknown function (Table S3). In this work, two members of the Sap family, Sap9 and Sap5, were identified. Sap9 is part of C. albicans common core while Sap5 shows up in hyphae induced with the human serum group. Both genes were up-regulated in C. albicans biofilm associated with bloodstream infections (Joo et al., 2013). Ten out of these twelve proteins are identified for the first time on the surface of C. albicans; Pra1 and Tef1 have been previously identified in the cell wall of C. albicans.
Many proteins described as “moonlighting” or multifunctional proteins in C. albicans were identified in this work (Nombela et al., 2006; Chaffin, 2008) (indicated with * in Table S1). Among them, Gpd2 has been described as a virulence factor on microorganism surface because it mediates complement evasion; meanwhile, there is a glycerol 3-phosphate-dehydrogenase in the cytoplasm that is involved in the intracellular accumulation of glycerol to control osmotic pressure (Luo et al., 2013). Tsa1 is involved in oxidative stress response in the cytoplasm and in hyphal cell wall biogenesis (Urban et al., 2005). Tdh3 is a glycolytic enzyme (glyceraldehyde-3-phosphate dehydrogenase) in the cytoplasm and able to attach fibronectin and laminin at the cell surface (Gozalbo et al., 1998).
Regarding C. albicans proteins that interact with complement inhibitor proteins (FH and PLG), the expression of Gpd2 and Pra1 is more prominent at the hyphal tip (Zipfel et al., 2011; Luo et al., 2013). Pra1 is also able to bind C4BP and it is also known that the hyphal tip is a prominent binding site for C4BP (Meri et al., 2004; Luo et al., 2011). For these reasons and because it facilitates host tissue penetration, the hyphal tip is considered an important factor of pathogenesis. We identified these two proteins on the C. albicans surface grown with 10% human serum that activate hyphal growth. Focusing on the mechanism to evade complement activation mediated by plasminogen coated surfaces, as plasminogen is an important complement cascade inhibitor in the fluid phase and on the human surface (Barthel et al., 2012), the identification of nine out of the eleven C. albicans proteins with the ability to bind PLG (Adh1, Eno1, Fba1, Gpd2, Pgk1, Pra1, Tdh3, Tef1, and Tsa1) is another highlight of this study (Crowe et al., 2003; Jong et al., 2003; Barthel et al., 2012).
Human serum proteins with the ability to interact with C. albicans surface
We detected human plasma proteins from different abundance strata on C. albicans surface; most of them are from the most abundant proteins or from intermediate stratum (Anderson and Anderson, 2002; Mitchell, 2010). Due to the heat treatment of HIS serum, FB and C2 become non-functional; in this situation, other human serum proteins could interact further with the surface of C. albicans. Remarkably, C. albicans surface could be less accessible to trypsin digestion when it was grown with 10% HIS as the number of coated of human proteins is greater. This could explain the observation of higher sum of peptide-spectrum match (PSMs) of human serum proteins in the HIS condition in comparison with the NS condition. In any case, it is not possible to discard that some proteins differentially observed in HIS are a result of changes in their binding properties due to protein denaturation.
Albumin is the most abundant protein in human plasma with close to two orders of magnitude more than the rest of proteins (Mitchell, 2010), but it was not detected as the highest abundant protein on C. albicans surface incubated with human serum (either NS or HIS) (Figure S1). Also, there are some proteins only detected on C. albicans surface in HIS samples that have intermediate abundance in human plasma such as C1INH, C1qA, and RBP4. Serotransferrin and fibrinogen (FGG, FGA and FGB) were more abundant in NS condition and in the other side apolipoprotein A-I and transthyretin in HIS condition. All of these data indicate that the interaction between human proteins and C. albicans surface did not respond solely to their protein abundance in human serum. The study of C3 and FB deposition on C. albicans surface after incubation with 10% human serum was carried out by immunofluorescence with specific antibodies. The clearly deposition of C3 and FB was observed at short times of interaction in Figure 3A.
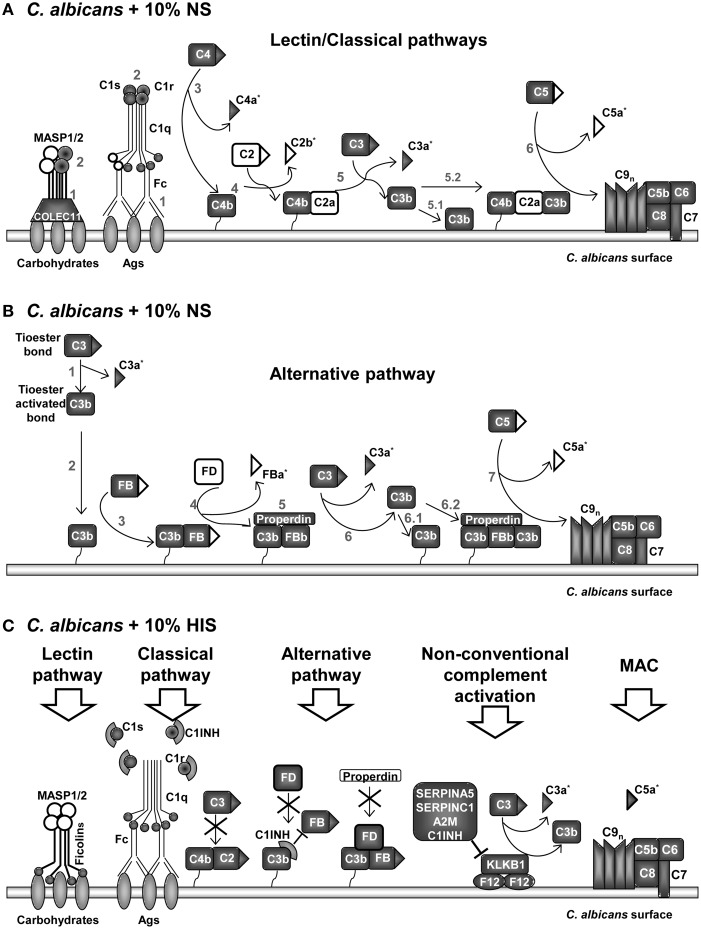
A graphical summary of the identified proteins at C. albicans surface from the different complement pathways in NS and HIS conditions is presented in Figure 4. The enrichment on the surface of C. albicans in proteins belonging to the lectin, classical and alternative complement pathways is visibly observed in normal conditions (NS); almost all of the main proteins of complement pathways were identified (Figures 4A,B). Peptides belonging to anaphylotoxin region are less frequently identified in NS than in HIS condition; this could be related to that C2b, C3a, FBa, and C5a fragments were released to the medium under this condition (Table 5). Furthermore, two peptides (CCEDGMR and CCEDGMRENPMR), which are part of the LRK26 peptide derived from C3a and with antimicrobial effects on C. albicans surface (Sonesson et al., 2007), were only identified in NS. Peptides belonging to the C5a region were only identified in HIS samples, just as more peptides belonging to the MACPF region of C6, C7, and C8-A. Furthermore, lower NSAF for all components of MAC, except C5, were obtained with HIS (Figure 2), which could be correlated with the enhanced identification of C5a peptides in this condition. In contrast, no identification of peptides belonging of C5a region and higher NSAF for the rest of components of MAC in NS samples may indicate MAC formation. MACPF-domains would not be accessible to trypsin digestion because they are points of interaction for MAC formation and would be protected in this condition. Furthermore, we identified clusterin and vitronectin (VTN) with higher NSAF in NS condition and both are able to interact with MAC in plasma, and thus prevent the cytolysis of host cells (Chauhan and Moore, 2006). Further experiments are required to determine the formation and the functionality of MAC on C. albicans surface.
Figure 4.
Model of interactions among proteins of the complement pathways identified on C. albicans surface after incubation with human serum. Proteins identified in normal serum (NS) samples in (A,B). Proteins belonging to lectin and classical pathways are represented in (A) and to alternative pathway in (B). Proteins mainly identified on heat inactivated serum (HIS) in (C). Identified proteins on samples are indicated with gray background and not-identified in white. Complement fragments released during complement activation are indicated with triangles. Peptides belonging to complement fragments that can be released or be part of a full-length protein are indicated with *. Numbers in (A,B) correspond to sequential steps during complement pathways activation. (A) Collectin-11 was only identified in NS samples. MASP2 was identified in NS and MASP1 was not identified in any sample. Fc (constant region of Ig) of IgGs or IgMs start classical complement pathway. (B) Properdin was identified in NS and Factor D (FD) was not. (C) Ficolin-2, Ficolin-3, C1qA, C1INH, and FD were identified only in HIS samples. C1INH dissociates the C1qrs complex, blocks alternative C3 convertase and inhibits C3 activation by kallikrein. C2 was identified in HIS, including 4 peptides of C2b. Five peptides belonging to FBa were identified in HIS condition. Signally, peptides belonging to C5a fragment were only identified in HIS samples.
In the same line of thought, in NS samples, properdin (FP) and not factor D (FD) were identified, while the opposite happened with HIS samples (Figure 4B vs. Figure 4C). The results are in agreement with a functional alternative complement pathway in NS condition on the surface of C. albicans. On the other hand, in HIS samples, FD could still be attached to the C3bB complex and we suppose that C3 convertase is not formed. In this situation, FP could not stabilize the complex and would be not identified on HIS samples. C1INH is able to bind to immobilized C3b and block progression of the complement cascade (Jiang et al., 2001). Serpin role of C1INH produces dissociation of C1qrs complex and the identification of these proteins in the HIS condition was enhanced, possibly for this reason. C3 and C4 (C4-A and C4-B) were identified with less NSAF and C2 and FB with higher NSAF in HIS samples (Figure 4C). In a current study about Streptococcus pyogenes interaction with human plasma, C2, C1qA, and FD were not detected on the surface of the microorganism (Sjöholm et al., 2014). In our experiments, when C. albicans cells were incubated with HIS, we detected not only these proteins, but also C1INH, and the enrichment of FB, C1qB, and C1qC. This is an interesting observation that may be due to the absence of functional convertases; FB and C2 might be more accessible to trypsin digestion. We assume that the unsuitable formation of complement convertases, or at least in a less extended manner, is responsible for the enhanced peptide identification of C2, FB, FD, C5, and the C1qrs complex in HIS samples.
Coagulation and complement pathways are highly connected, with many cross-talks (Kazatchkine and Jouvin, 1984; Amara et al., 2008, 2010; Peerschke et al., 2008; Diamond, 2013; Verschoor and Langer, 2013). Many complement inhibitors were identified on the surface of C. albicans and in general more abundant in HIS samples. The innate immune system strives to respond and activates complement cascades by non-conventional mechanisms (thrombin (F2), PLG, Factor XII (F12), kallikrein (KLKB1), ficolin-2 and -3) and bypass locking points in absence of functional FB and C2 (Nielsen et al., 1996; Rittirsch et al., 2008) (Figures 2, 4C). In HIS samples, KLKB1 inhibitors were enriched, such as SERPINA5, SERPINC1, A2M, and C1INH (Gadjeva et al., 1998; Paréj et al., 2013).
A relevant group of proteins is serpin family (serine protease inhibitors), which are broadly distributed among eukaryote organisms. Serpins are able to inhibit proteases by a specific mechanism where covalent complexes are formed with target proteases (Law et al., 2006). Since serpin has to be cleaved to inhibit the target substrates, inhibition also consumes the serpin; serpins are therefore irreversible enzyme inhibitors. For these reasons, the identification of many serpins on C. albicans surface and more interestingly in HIS samples is an interesting observation because they may be inactivating their substrates attached to the surface of C. albicans or themselves coating C. albicans surface in the sense to avoid interactions between their substrates and C. albicans (Table 4).
Interestingly, the apolipoproteins classified in the metabolism group are involved in lipid transport and also play a relevant role in host defense as part of the innate immune system (Grunfeld and Feingold, 2008). Furthermore, C. albicans grows better in lipoprotein-depleted plasma due to two factors: greater availability of lipids and reduced neutralizing candidacidal factor (Vonk et al., 2004). ApoE has a relevant role in systemic candidiasis infection and showed higher NSAF in NS samples (Figure S1). ApoE-/- mice showed an increased mortality in comparison with wild type (85 vs. 52%) (Vonk et al., 2004), and also an impaired immune response in Klebsiella pneumonia infection (de Bont et al., 2000).
In summary, the mechanisms developed by C. albicans to evade, interfere or use for its benefit host defenses are very wide and part of its pathogenic evolution. The analysis of C. albicans surface after incubation with human serum to obtain a partial breakdown of the immunological crosstalk between C. albicans and human serum proteins is a relevant target of study. C. albicans mimic human immune evasion model using many surface proteins to fix plasminogen and complement inhibitors (FH and C4BP) to protect against complement activation. Furthermore, the identification of Pra1, Sap5, Tef1, and 9 more proteins only identified in C. albicans hyphal surface induced with 10% human serum and not in other published conditions with an equivalent approach is a relevant highlight of this analysis.
Author contributions
EM designed and performed the experiments, analysis of results, and writing of the manuscript. CP designed and performed the experiments. CH performed the experiments. MH performed the experiments and analysis of results. CN research support. LM designed the experiments, analysis of results and critically revised the manuscript. CG designed the experiments, analysis of results, and critically revised the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by BIO2012-31767 from the Ministerio de Economía y Competitividad, PROMPT (S2010/BMD-2414) from the Comunidad Autónoma de Madrid, REIPI-Spanish Network for the Research in Infectious Diseases (RD12/0015/0004) and PRB2 (PT13/0001/0038) funded by the ISCIII and FEDER. CP was the recipient of a fellowship from the program Continuous training for teachers of the Pontificia Javeriana University (Colombia) in conjunction with COLCIENCIAS-LASPAU University of Harvard. The proteomic analyses were carried out in the Proteomic Units of Complutense University of Madrid and of Centro de Investigaciones Biológicas (CSIC), members of ProteoRed network. The Immunofluorescence Microscopy images were carried out in Centro Nacional de Microscopía Electrónica (ICTS)-UCM. The authors would like to thank Dr. Fernando Vivanco [Fundación Jimenez Diaz, Universidad Complutense, Madrid (Spain)] and Dra. Pilar Sánchez-Corral Gómez [Unidad de Investigación de Inmunopatología, Hospital Universitario La Paz, Madrid (Spain)] for the gift of C3 and FB antibodies, respectively. These results are lined up with the Spanish Initiative on the Infectious Diseases-Human Proteome Project (B/D-HPP).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01343
References
- Almirante B., Rodríguez D., Park B. J., Cuenca-Estrella M., Planes A. M., Almela M., et al. (2005). Epidemiology and predictors of mortality in cases of Candida bloodstream infection: results from population-based surveillance, Barcelona, Spain, from 2002 to 2003. J. Clin. Microbiol. 43, 1829–1835. 10.1128/JCM.43.4.1829-1835.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amara U., Flierl M. A., Rittirsch D., Klos A., Chen H., Acker B., et al. (2010). Molecular intercommunication between the complement and coagulation systems. J. Immunol. 185, 5628–5636. 10.4049/jimmunol.0903678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amara U., Rittirsch D., Flierl M., Bruckner U., Klos A., Gebhard F., et al. (2008). Interaction between the coagulation and complement system. Adv. Exp. Med. Biol. 632, 71–79. 10.1007/978-0-387-78952-1_6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson N. L., Anderson N. G. (2002). The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics 1, 845–867. 10.1074/mcp.R200007-MCP200 [DOI] [PubMed] [Google Scholar]
- Barthel D., Schindler S., Zipfel P. F. (2012). Plasminogen is a complement inhibitor. J. Biol. Chem. 287, 18831–18842. 10.1074/jbc.M111.323287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G. D., Denning D. W., Gow N. A., Levitz S. M., Netea M. G., White T. C. (2012). Hidden killers: human fungal infections. Sci. Transl. Med. 4, 165rv113. 10.1126/scitranslmed.3004404 [DOI] [PubMed] [Google Scholar]
- Buu L. M., Chen Y. C. (2014). Impact of glucose levels on expression of hypha-associated secreted aspartyl proteinases in Candida albicans. J. Biomed. Sci. 21:22. 10.1186/1423-0127-21-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collette J. R., Lorenz M. C. (2011). Mechanisms of immune evasion in fungal pathogens. Curr. Opin. Microbiol. 14, 668–675. 10.1016/j.mib.2011.09.007 [DOI] [PubMed] [Google Scholar]
- Crowe J. D., Sievwright I. K., Auld G. C., Moore N. R., Gow N. A., Booth N. A. (2003). Candida albicans binds human plasminogen: identification of eight plasminogen-binding proteins. Mol. Microbiol. 47, 1637–1651. 10.1046/j.1365-2958.2003.03390.x [DOI] [PubMed] [Google Scholar]
- Chaffin W. L. (2008). Candida albicans cell wall proteins. Microbiol. Mol. Biol. Rev. 72, 495–544. 10.1128/MMBR.00032-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan A. K., Moore T. L. (2006). Presence of plasma complement regulatory proteins clusterin (Apo J) and vitronectin (S40) on circulating immune complexes (CIC). Clin. Exp. Immunol. 145, 398–406. 10.1111/j.1365-2249.2006.03135.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bont N., Netea M. G., Demacker P. N., Kullberg B. J., van der Meer J. W., Stalenhoef A. F. (2000). Apolipoprotein E-deficient mice have an impaired immune response to Klebsiella pneumoniae. Eur. J. Clin. Invest. 30, 818–822. 10.1046/j.1365-2362.2000.00715.x [DOI] [PubMed] [Google Scholar]
- de Groot P. W., Bader O., de Boer A. D., Weig M., Chauhan N. (2013). Adhesins in human fungal pathogens: glue with plenty of stick. Eukaryotic Cell 12, 470–481. 10.1128/EC.00364-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groot P. W., Hellingwerf K. J., Klis F. M. (2003). Genome-wide identification of fungal GPI proteins. Yeast 20, 781–796. 10.1002/yea.1007 [DOI] [PubMed] [Google Scholar]
- Diamond S. L. (2013). Systems biology of coagulation. J. Thromb. Haemost 11 (Suppl. 1), 224–232. 10.1111/jth.12220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreisbach A., van der Kooi-Pol M. M., Otto A., Gronau K., Bonarius H. P., Westra H., et al. (2011). Surface shaving as a versatile tool to profile global interactions between human serum proteins and the Staphylococcus aureus cell surface. Proteomics 11, 2921–2930. 10.1002/pmic.201100134 [DOI] [PubMed] [Google Scholar]
- Dühring S., Germerodt S., Skerka C., Zipfel P. F., Dandekar T., Schuster S. (2015). Host-pathogen interactions between the human innate immune system and Candida albicans—Understanding and modeling defense and evasion strategies. Front. Microbiol. 6:625. 10.3389/fmicb.2015.00625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ene I. V., Brunke S., Brown A. J., Hube B. (2014). Metabolism in Fungal Pathogenesis. Cold Spring Harb. Perspect. Med. 4. 10.1101/cshperspect.a019695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engleberg N. C., Dermody T. S., DiRita V. (eds.). (2012). Innate immunity in Schaechter's Mechanisms of Microbial Disease (Philadelphia, PA: Lippincott Williams & Wilkins; ), 66–90. [Google Scholar]
- Fan Y., He H., Dong Y., Pan H. (2013). Hyphae-specific genes HGC1, ALS3, HWP1, and ECE1 and relevant signaling pathways in Candida albicans. Mycopathologia 176, 329–335. 10.1007/s11046-013-9684-6 [DOI] [PubMed] [Google Scholar]
- Fradin C., Kretschmar M., Nichterlein T., Gaillardin C., d'enfert C., Hube B. (2003). Stage-specific gene expression of Candida albicans in human blood. Mol. Microbiol. 47, 1523–1543. 10.1046/j.1365-2958.2003.03396.x [DOI] [PubMed] [Google Scholar]
- Gadjeva M., Dodds A. W., Taniguchi-Sidle A., Willis A. C., Isenman D. E., Law S. K. (1998). The covalent binding reaction of complement component C3. J. Immunol. 161, 985–990. [PubMed] [Google Scholar]
- Gil-Bona A., Parra-Giraldo C. M., Hernáez M. L., Reales-Calderon J. A., Solis N. V., Filler S. G., et al. (2015). Candida albicans cell shaving uncovers new proteins involved in cell wall integrity, yeast to hypha transition, stress response and host-pathogen interaction. J. Proteomics 127(Pt B), 340–351. 10.1016/j.jprot.2015.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillum A. M., Tsay E. Y., Kirsch D. R. (1984). Isolation of the Candida albicans gene for orotidine-5'-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198, 179–182. 10.1007/BF00328721 [DOI] [PubMed] [Google Scholar]
- Gozalbo D., Gil-Navarro I., Azorín I., Renau-Piqueras J., Martínez J. P., Gil M. L. (1998). The cell wall-associated glyceraldehyde-3-phosphate dehydrogenase of Candida albicans is also a fibronectin and laminin binding protein. Infect. Immun. 66, 2052–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J. V., Orsborn K. I., Zhang M., Tan Q. K., Greis K. D., Porollo A., et al. (2013). Heparin-binding motifs and biofilm formation by Candida albicans. J. Infect. Dis. 208, 1695–1704. 10.1093/infdis/jit391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunfeld C., Feingold K. R. (2008). HDL and innate immunity: a tale of two apolipoproteins. J. Lipid Res. 49, 1605–1606. 10.1194/jlr.E800011-JLR200 [DOI] [PubMed] [Google Scholar]
- Heilmann C. J., Sorgo A. G., Siliakus A. R., Dekker H. L., Brul S., de Koster C. G., et al. (2011). Hyphal induction in the human fungal pathogen Candida albicans reveals a characteristic wall protein profile. Microbiology 157, 2297–2307. 10.1099/mic.0.049395-0 [DOI] [PubMed] [Google Scholar]
- Hernáez M. L., Ximénez-Embún P., Martínez-Gomariz M., Gutiérrez-Blázquez M. D., Nombela C., Gil C. (2010). Identification of Candida albicans exposed surface proteins in vivo by a rapid proteomic approach. J. Proteomics 73, 1404–1409. 10.1016/j.jprot.2010.02.008 [DOI] [PubMed] [Google Scholar]
- Hube B. (2006). Infection-associated genes of Candida albicans. Future Microbiol. 1, 209–218. 10.2217/17460913.1.2.209 [DOI] [PubMed] [Google Scholar]
- Inglis D. O., Arnaud M. B., Binkley J., Shah P., Skrzypek M. S., Wymore F., et al. (2012). The Candida genome database incorporates multiple Candida species: multispecies search and analysis tools with curated gene and protein information for Candida albicans and Candida glabrata. Nucleic Acids Res. 40, D667–D674. 10.1093/nar/gkr945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insenser M. R., Hernáez M. L., Nombela C., Molina M., Molero G., Gil C. (2010). Gel and gel-free proteomics to identify Saccharomyces cerevisiae cell surface proteins. J. Proteomics 73, 1183–1195. 10.1016/j.jprot.2010.02.005 [DOI] [PubMed] [Google Scholar]
- Jiang H., Wagner E., Zhang H., Frank M. M. (2001). Complement 1 inhibitor is a regulator of the alternative complement pathway. J. Exp. Med. 194, 1609–1616. 10.1084/jem.194.11.1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jong A. Y., Chen S. H., Stins M. F., Kim K. S., Tuan T. L., Huang S. H. (2003). Binding of Candida albicans enolase to plasmin(ogen) results in enhanced invasion of human brain microvascular endothelial cells. J. Med. Microbiol. 52, 615–622. 10.1099/jmm.0.05060-0 [DOI] [PubMed] [Google Scholar]
- Joo M. Y., Shin J. H., Jang H. C., Song E. S., Kee S. J., Shin M. G., et al. (2013). Expression of SAP5 and SAP9 in Candida albicans biofilms: comparison of bloodstream isolates with isolates from other sources. Med. Mycol. 51, 892–896. 10.3109/13693786.2013.824623 [DOI] [PubMed] [Google Scholar]
- Karkowska-Kuleta J., Rapala-Kozik M., Kozik A. (2009). Fungi pathogenic to humans: molecular bases of virulence of Candida albicans, Cryptococcus neoformans and Aspergillus fumigatus. Acta Biochim. Pol. 56, 211–224. [PubMed] [Google Scholar]
- Kazatchkine M. D., Jouvin M. H. (1984). Interactions between the alternative complement pathway and proteases of the coagulation system. Adv. Exp. Med. Biol. 167, 235–239. 10.1007/978-1-4615-9355-3_19 [DOI] [PubMed] [Google Scholar]
- Klis F. M., Sosinska G. J., de Groot P. W., Brul S. (2009). Covalently linked cell wall proteins of Candida albicans and their role in fitness and virulence. FEMS Yeast Res. 9, 1013–1028. 10.1111/j.1567-1364.2009.00541.x [DOI] [PubMed] [Google Scholar]
- Kretschmar M., Felk A., Staib P., Schaller M., Hess D., Callapina M., et al. (2002). Individual acid aspartic proteinases (Saps) 1-6 of Candida albicans are not essential for invasion and colonization of the gastrointestinal tract in mice. Microb. Pathog. 32, 61–70. 10.1006/mpat.2001.0478 [DOI] [PubMed] [Google Scholar]
- Kumamoto C. A., Vinces M. D. (2005). Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell. Microbiol. 7, 1546–1554. 10.1111/j.1462-5822.2005.00616.x [DOI] [PubMed] [Google Scholar]
- Law R. H., Zhang Q., McGowan S., Buckle A. M., Silverman G. A., Wong W., et al. (2006). An overview of the serpin superfamily. Genome Biol. 7, 216. 10.1186/gb-2006-7-5-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. L., Buckley H. R., Campbell C. C. (1975). An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia 13, 148–153. 10.1080/00362177585190271 [DOI] [PubMed] [Google Scholar]
- Leleu G., Aegerter P., Guidet B. (2002). Systemic candidiasis in intensive care units: a multicenter, matched-cohort study. J. Crit. Care 17, 168–175. 10.1053/jcrc.2002.35815 [DOI] [PubMed] [Google Scholar]
- Lesiak-Markowicz I., Vogl G., Schwarzmuller T., Speth C., Lass-Flörl C., Dierich M. P., et al. (2011). Candida albicans Hgt1p, a multifunctional evasion molecule: complement inhibitor, CR3 analogue, and human immunodeficiency virus-binding molecule. J. Infect. Dis. 204, 802–809. 10.1093/infdis/jir455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Su C., Liu H. (2014). Candida albicans hyphal initiation and elongation. Trends Microbiol. 22, 707–714. 10.1016/j.tim.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S., Blom A. M., Rupp S., Hipler U. C., Hube B., Skerka C., et al. (2011). The pH-regulated antigen 1 of Candida albicans binds the human complement inhibitor C4b-binding protein and mediates fungal complement evasion. J. Biol. Chem. 286, 8021–8029. 10.1074/jbc.M110.130138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S., Hoffmann R., Skerka C., Zipfel P. F. (2013). Glycerol-3-phosphate dehydrogenase 2 is a novel factor H-, factor H-like protein 1-, and plasminogen-binding surface protein of Candida albicans. J. Infect. Dis. 207, 594–603. 10.1093/infdis/jis718 [DOI] [PubMed] [Google Scholar]
- Luo S., Poltermann S., Kunert A., Rupp S., Zipfel P. F. (2009). Immune evasion of the human pathogenic yeast Candida albicans: Pra1 is a Factor H, FHL-1 and plasminogen binding surface protein. Mol. Immunol. 47, 541–550. 10.1016/j.molimm.2009.07.017 [DOI] [PubMed] [Google Scholar]
- Mayer F. L., Wilson D., Hube B. (2013). Candida albicans pathogenicity mechanisms. Virulence 4, 119–128. 10.4161/viru.22913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meri T., Blom A. M., Hartmann A., Lenk D., Meri S., Zipfel P. F. (2004). The hyphal and yeast forms of Candida albicans bind the complement regulator C4b-binding protein. Infect. Immun. 72, 6633–6641. 10.1128/IAI.72.11.6633-6641.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. (2010). Proteomics retrenches. Nat. Biotechnol. 28, 665–670. 10.1038/nbt0710-665 [DOI] [PubMed] [Google Scholar]
- Mochon A. B., Jin Y., Kayala M. A., Wingard J. R., Clancy C. J., Nguyen M. H., et al. (2010). Serological profiling of a Candida albicans protein microarray reveals permanent host-pathogen interplay and stage-specific responses during candidemia. PLoS Pathog. 6:e1000827. 10.1371/annotation/eff399e1-51e0-43b1-bdf4-1a11e9ada9bd [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monniot C., Boisramé A., Da Costa G., Chauvel M., Sautour M., Bougnoux M. E., et al. (2013). Rbt1 protein domains analysis in Candida albicans brings insights into hyphal surface modifications and Rbt1 potential role during adhesion and biofilm formation. PLoS ONE 8:e82395. 10.1371/journal.pone.0082395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naglik J. R., Challacombe S. J., Hube B. (2003). Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol. Mol. Biol. Rev. 67, 400–428. 10.1128/MMBR.67.3.400-428.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanjappa V., Thomas J. K., Marimuthu A., Muthusamy B., Radhakrishnan A., Sharma R., et al. (2014). Plasma Proteome Database as a resource for proteomics research: 2014 update. Nucleic Acids Res. 42, D959–D965. 10.1093/nar/gkt1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen E. W., Johansen H. T., Høgåsen K., Wuillemin W., Hack C. E., Mollnes T. E. (1996). Activation of the complement, coagulation, fibrinolytic and kallikrein-kinin systems during attacks of hereditary angioedema. Scand. J. Immunol. 44, 185–192. 10.1046/j.1365-3083.1996.d01-298.x [DOI] [PubMed] [Google Scholar]
- Nombela C., Gil C., Chaffin W. L. (2006). Non-conventional protein secretion in yeast. Trends Microbiol. 14, 15–21. 10.1016/j.tim.2005.11.009 [DOI] [PubMed] [Google Scholar]
- Pardo M., Monteoliva L., Vázquez P., Martínez R., Molero G., Nombela C., et al. (2004). PST1 and ECM33 encode two yeast cell surface GPI proteins important for cell wall integrity. Microbiology 150, 4157–4170. 10.1099/mic.0.26924-0 [DOI] [PubMed] [Google Scholar]
- Paréj K., Dobó J., Závodszky P., Gál P. (2013). The control of the complement lectin pathway activation revisited: both C1-inhibitor and antithrombin are likely physiological inhibitors, while alpha2-macroglobulin is not. Mol. Immunol. 54, 415–422. 10.1016/j.molimm.2013.01.009 [DOI] [PubMed] [Google Scholar]
- Peerschke E. I., Yin W., Ghebrehiwet B. (2008). Platelet mediated complement activation. Adv. Exp. Med. Biol. 632, 81–91. 10.1007/978-0-387-78952-1_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitarch A., Abian J., Carrascal M., Sánchez M., Nombela C., Gil C. (2004). Proteomics-based identification of novel Candida albicans antigens for diagnosis of systemic candidiasis in patients with underlying hematological malignancies. Proteomics 4, 3084–3106. 10.1002/pmic.200400903 [DOI] [PubMed] [Google Scholar]
- Pitarch A., Jiménez A., Nombela C., Gil C. (2006). Decoding serological response to Candida cell wall immunome into novel diagnostic, prognostic, and therapeutic candidates for systemic candidiasis by proteomic and bioinformatic analyses. Mol. Cell. Proteomics 5, 79–96. 10.1074/mcp.M500243-MCP200 [DOI] [PubMed] [Google Scholar]
- Pitarch A., Jiménez A., Nombela C., Gil C. (2008). Serological proteome analysis to identify systemic candidiasis patients in the intensive care unit: analytical, diagnostic and prognostic validation of anti-Candida enolase antibodies on quantitative clinical platforms. Proteomics Clin. Appl. 2, 596–618. 10.1002/prca.200780039 [DOI] [PubMed] [Google Scholar]
- Pitarch A., Nombela C., Gil C. (2011). Prediction of the clinical outcome in invasive candidiasis patients based on molecular fingerprints of five anti-Candida antibodies in serum. Mol. Cell. Proteomics 10:M110.004010. 10.1074/mcp.M110.004010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poltermann S., Kunert A., von der Heide M., Eck R., Hartmann A., Zipfel P. F. (2007). Gpm1p is a factor H-, FHL-1-, and plasminogen-binding surface protein of Candida albicans. J. Biol. Chem. 282, 37537–37544. 10.1074/jbc.M707280200 [DOI] [PubMed] [Google Scholar]
- Ragni E., Calderon J., Fascio U., Sipiczki M., Fonzi W. A., Popolo L. (2011). Phr1p, a glycosylphosphatidylinsitol-anchored beta(1,3)-glucanosyltransferase critical for hyphal wall formation, localizes to the apical growth sites and septa in Candida albicans. Fungal Genet. Biol. 48, 793–805. 10.1016/j.fgb.2011.05.003 [DOI] [PubMed] [Google Scholar]
- Rittirsch D., Flierl M. A., Ward P. A. (2008). Harmful molecular mechanisms in sepsis. Nat. Rev. Immunol. 8, 776–787. 10.1038/nri2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez de Córdoba S., Esparza-Gordillo J., Goicoechea de Jorge E., Lopez-Trascasa M., Sánchez-Corral P. (2004). The human complement factor H: functional roles, genetic variations and disease associations. Mol. Immunol. 41, 355–367. 10.1016/j.molimm.2004.02.005 [DOI] [PubMed] [Google Scholar]
- Sjöholm K., Karlsson C., Linder A., Malmström J. (2014). A comprehensive analysis of the Streptococcus pyogenes and human plasma protein interaction network. Mol. Biosyst. 10, 1698–1708. 10.1039/C3MB70555B [DOI] [PubMed] [Google Scholar]
- Soares R. M., de A Soares R. M., Alviano D. S., Angluster J., Alviano C. S., Travassos L. R. (2000). Identification of sialic acids on the cell surface of Candida albicans. Biochim. Biophys. Acta 1474, 262–268. 10.1016/S0304-4165(00)00003-9 [DOI] [PubMed] [Google Scholar]
- Soltis R. D., Hasz D., Morris M. J., Wilson I. D. (1979). Studies on the nature of heat-labile anti-complementary activity in normal human serum. Clin. Exp. Immunol. 37, 310–322. [PMC free article] [PubMed] [Google Scholar]
- Sonesson A., Ringstad L., Nordahl E. A., Malmsten M., Mörgelin M., Schmidtchen A. (2007). Antifungal activity of C3a and C3a-derived peptides against Candida. Biochim. Biophys. Acta 1768, 346–353. 10.1016/j.bbamem.2006.10.017 [DOI] [PubMed] [Google Scholar]
- Sosinska G. J., de Koning L. J., de Groot P. W., Manders E. M., Dekker H. L., Hellingwerf K. J., et al. (2011). Mass spectrometric quantification of the adaptations in the wall proteome of Candida albicans in response to ambient pH. Microbiology 157, 136–146. 10.1099/mic.0.044206-0 [DOI] [PubMed] [Google Scholar]
- Sudbery P. E. (2011). Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 9, 737–748. 10.1038/nrmicro2636 [DOI] [PubMed] [Google Scholar]
- Uniprot C. (2014). Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res. 42, D191–D198. 10.1093/nar/gkt1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban C., Xiong X., Sohn K., Schröppel K., Brunner H., Rupp S. (2005). The moonlighting protein Tsa1p is implicated in oxidative stress response and in cell wall biogenesis in Candida albicans. Mol. Microbiol. 57, 1318–1341. 10.1111/j.1365-2958.2005.04771.x [DOI] [PubMed] [Google Scholar]
- Verschoor A., Langer H. F. (2013). Crosstalk between platelets and the complement system in immune protection and disease. Thromb. Haemost. 110, 910–919. 10.1160/TH13-02-0102 [DOI] [PubMed] [Google Scholar]
- Vialas V., Sun Z., Loureiro Y Penha C. V., Carrascal M., Abián J., Monteoliva L., et al. (2014). A Candida albicans PeptideAtlas. J. Proteomics 97, 62–68. 10.1016/j.jprot.2013.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viudes A., Pemán J., Cantón E., Ubeda P., López-Ribot J. L., Gobernado M. (2002). Candidemia at a tertiary-care hospital: epidemiology, treatment, clinical outcome and risk factors for death. Eur. J. Clin. Microbiol. Infect. Dis. 21, 767–774. 10.1007/s10096-002-0822-1 [DOI] [PubMed] [Google Scholar]
- Vonk A. G., De Bont N., Netea M. G., Demacker P. N., van der Meer J. W., Stalenhoef A. F., et al. (2004). Apolipoprotein-E-deficient mice exhibit an increased susceptibility to disseminated candidiasis. Med. Mycol. 42, 341–348. 10.1080/13693780410001657135 [DOI] [PubMed] [Google Scholar]
- Walport M. J. (2001a). Complement. First of two parts. N. Engl. J. Med. 344, 1058–1066. 10.1056/NEJM200104053441406 [DOI] [PubMed] [Google Scholar]
- Walport M. J. (2001b). Complement. Second of two parts. N. Engl. J. Med. 344, 1140–1144. 10.1056/NEJM200104123441506 [DOI] [PubMed] [Google Scholar]
- Wilson L. S., Reyes C. M., Stolpman M., Speckman J., Allen K., Beney J. (2002). The direct cost and incidence of systemic fungal infections. Value Health 5, 26–34. 10.1046/j.1524-4733.2002.51108.x [DOI] [PubMed] [Google Scholar]
- Yan L., Yang C., Tang J. (2013). Disruption of the intestinal mucosal barrier in Candida albicans infections. Microbiol. Res. 168, 389–395. 10.1016/j.micres.2013.02.008 [DOI] [PubMed] [Google Scholar]
- Zipfel P. F., Skerka C., Kupka D., Luo S. (2011). Immune escape of the human facultative pathogenic yeast Candida albicans: the many faces of the Candida Pra1 protein. Int. J. Med. Microbiol. 301, 423–430. 10.1016/j.ijmm.2011.04.010 [DOI] [PubMed] [Google Scholar]
- Zybailov B. L., Florens L., Washburn M. P. (2007). Quantitative shotgun proteomics using a protease with broad specificity and normalized spectral abundance factors. Mol. Biosyst. 3, 354–360. 10.1039/b701483j [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.