Abstract
The morphological plasticity of root systems is critical for plant survival, and understanding the mechanisms underlying root adaptation to nitrogen (N) fluctuation is critical for sustainable agriculture; however, the molecular mechanism of N-dependent root growth in rice remains unclear. This study aimed to identify the role of the complementary high-affinity NO3− transport protein OsNAR2.1 in NO3−-regulated rice root growth. Comparisons with wild-type (WT) plants showed that knockdown of OsNAR2.1 inhibited lateral root (LR) formation under low NO3− concentrations, but not under low NH4+ concentrations. 15N-labelling NO3− supplies (provided at concentrations of 0–10 mM) demonstrated that (i) defects in LR formation in mutants subjected to low external NO3− concentrations resulted from impaired NO3− uptake, and (ii) the mutants had significantly fewer LRs than the WT plants when root N contents were similar between genotypes. LR formation in osnar2.1 mutants was less sensitive to localised NO3− supply than LR formation in WT plants, suggesting that OsNAR2.1 may be involved in a NO3−-signalling pathway that controls LR formation. Knockdown of OsNAR2.1 inhibited LR formation by decreasing auxin transport from shoots to roots. Thus, OsNAR2.1 probably functions in both NO3− uptake and NO3−-signalling.
Plants have diverse mechanisms for adapting to nutrient supply conditions. Among these mechanisms, the plasticity of root development is vital. Lateral roots (LRs) are generally more sensitive to variations in nutrient conditions compared to primary roots1,2. Lateral root development begins with the initiation of founder cells in the root pericycle just behind the primary root apex and continues with the formation of a cluster of cells constituting the LR primordium, followed by the formation of a radially symmetrical meristem3.
Nitrogen (N) is an essential macronutrient for plant growth and crop productivity. Changes in N availability in the nutrient medium induce plasticity in LR initiation and elongation4,5,6,7,8,9,10. Lateral root development in response to NO3− has been investigated extensively in the dicot model plant Arabidopsis5,10,11,12,13. A striking example of plasticity in LR development is seen in Arabidopsis responding to localised NO3− treatment through stimulation of LR elongation5. Studies of an Arabidopsis nitrate reductase double mutant suggested that the local stimulation of LR elongation is a consequence of the NO3− ion acting as a signal rather than as a nutrient. AtANR1 (a NO3−-inducible gene) and AtNRT1.1 (CHL1/NPF6.3) genes, which encode a transcription factor and a dual NO3− transporter, respectively, were proposed to consecutively control the stimulatory effect of NO3− on LR elongation5,6,11. The positive regulatory role of ANR1 in LR elongation was recently corroborated when overexpression in transgenic lines stimulated LR elongation whilst having no direct effect on LR density or primary root growth14. In addition to local NO3− responses, LR growth is stimulated in response to mild N deficiency and suppressed under excess N supply by systemic plant signals carrying information on the nutritional status of distant plant organs1,6,15,16,17,18. Recent studies have highlighted the role of AtNRT2.1 in root architecture response to low NO3− availability, especially in LR initiation16,19. Experiments using atnar2.1-1 identified the role of AtNAR2.1 in LR responses to low NO3− supply20; further work showed that the essential role of AtNAR2.1 for the localization of AtNRT2.1 to the plasma membrane20,21. miR393/AFB3 is a NO3−-responsive module that controls LR density in responses to external and internal N concentrations in Arabidopsis12,13. Interestingly, AGL7-clade MADS-box gene AtAGL21 plays a crucial role in both LR formation and elongation22. Thus, NO3−-dependent root development is apparently under the control of complex mechanisms, although its signalling components have remained largely unidentified.
Our understanding of NO3−-regulated LR development is limited in monocot plants, including rice, the model cereal plant (Oryza sativa L.)8,23. Elevated NO3− responsiveness in rice plants is strongly related to increased LR initiation regardless of whether the roots system was split or whole8,9. Recent work showed that the transcriptional levels of four ANR1-like genes (OsMADS25, OsMADS27, OsMADS57 and OsMADS61) were markedly modulated by NO3− availability24. Furthermore, in miR444a-overexpressing rice lines expression of the target ANR1-like genes was down-regulated and LR elongation was less responsive to localised NO3− 23. These studies indicated that ANR1-like genes might have a similar role in regulating root developmental response to NO3− in rice, despite the evolutionary distance between Arabidopsis and rice.
We previously reported that OsNAR2.1, a rice NAR2-like gene, has no known transport activity, but is required to complement high-affinity NO3− transport. Yeast two-hybridisation showed that OsNAR2.1 interacted with OsNRT2.1/2.2/2.3a, and knockdown of OsNAR2.1 suppressed expression of the three high-affinity-transport- system NO3− transporters, unlike OsNRT1.125,26,27,28. Knockdown of OsNAR2.1 impaired NO3− uptake; mutants had only ~65% of the N concentration measured in wild-type (WT) plants when the NO3− supply was limited. No such difference was found when a low concentration of NH4+ was provided as the sole inorganic N source27. In this study, we confirmed inhibition of LR formation in the osnar2.1 mutants (compared to WT plants) under low NO3− concentrations. 15N-labelling of NO3− supplies (provided at concentrations of 0–10 mM) and our localised-NO3− treatments showed that defective LR formation in OsNAR2.1 knockdown plants may be due (i) an impairment of NO3− uptake, and (ii) a NO3−-signalling function.
Results
Knockdown of OsNAR2.1 inhibited root growth when N was supplied as NO3 −
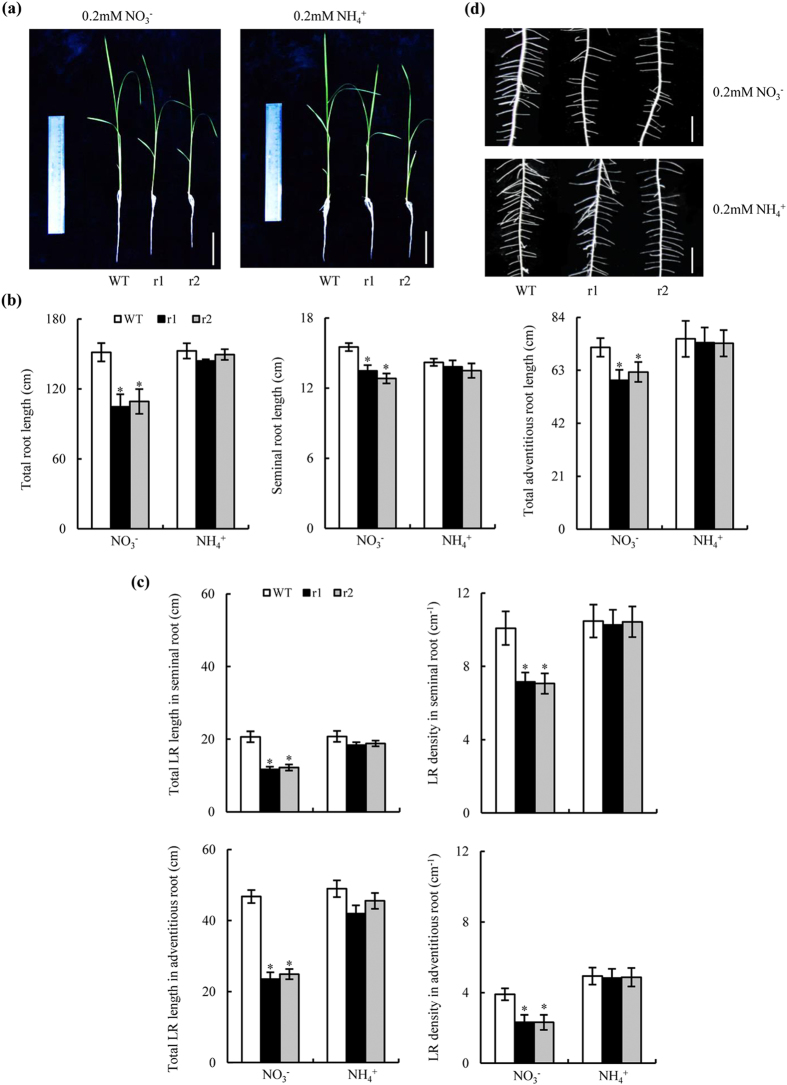
Total root lengths were shorter in osnar2.1 mutants than in WT plants at a NH4+:NO3− concentration ratio of 25:75. The difference in root length between the genotypes was greatest when the ionic ratio was 0:100 (Supplementary Fig. S1 online); mutant line total root lengths were ca. 30% of those in WT plants at this concentration ratio. However, total root lengths were similar between genotypes when NH4+-N was supplied (Fig. 1a,b). Reductions in total root lengths in the mutants were largely attributable to reduced total LR lengths (Fig. 1c). The total LR lengths of seminal and adventitious roots in the RNAi lines were ca. 64 and 53%, respectively, of those in WT plants when NO3−-N was supplied. The LR density in the RNAi lines was reduced to a greater extent than the mean LR length (Fig. 1c, Supplementary Fig. S2). The LR densities of the seminal and adventitious roots in the two osnar2.1 mutants were reduced by ca. 29% and 40%, respectively, in comparison with those in the WT plants (Fig. 1d). The lengths of the seminal and all adventitious roots in the osnar2.1 mutants were ca. 15% shorter than those of WT plants when NO3−-N was supplied (Fig. 1b). However, the numbers and mean lengths of adventitious roots were not different from those of WT plants (Supplementary Fig. S3 online).
Figure 1. Root morphology of wild-type (WT) and osnar2.1 knockdown lines (r1 and r2).
Rice seedlings were grown for 1 wk in hydroponic media containing 0.2 mM NO3− or NH4+. (a) Morphology of rice plants (bar = 5 cm); (b) Total root length (including the seminal root, adventitious roots and lateral roots [LR]); (c) LR morphology in seminal and adventitious roots (bar = 1 cm). (d) Morphology of LRs on the seminal root. Values are means ± SE (n = 6). *P < 0.05 (ANOVA) comparing WT plants and two mutant lines in the same treatment.
Knockdown of OsNAR2.1 repressed LR initiation when N was supplied as NO3 −
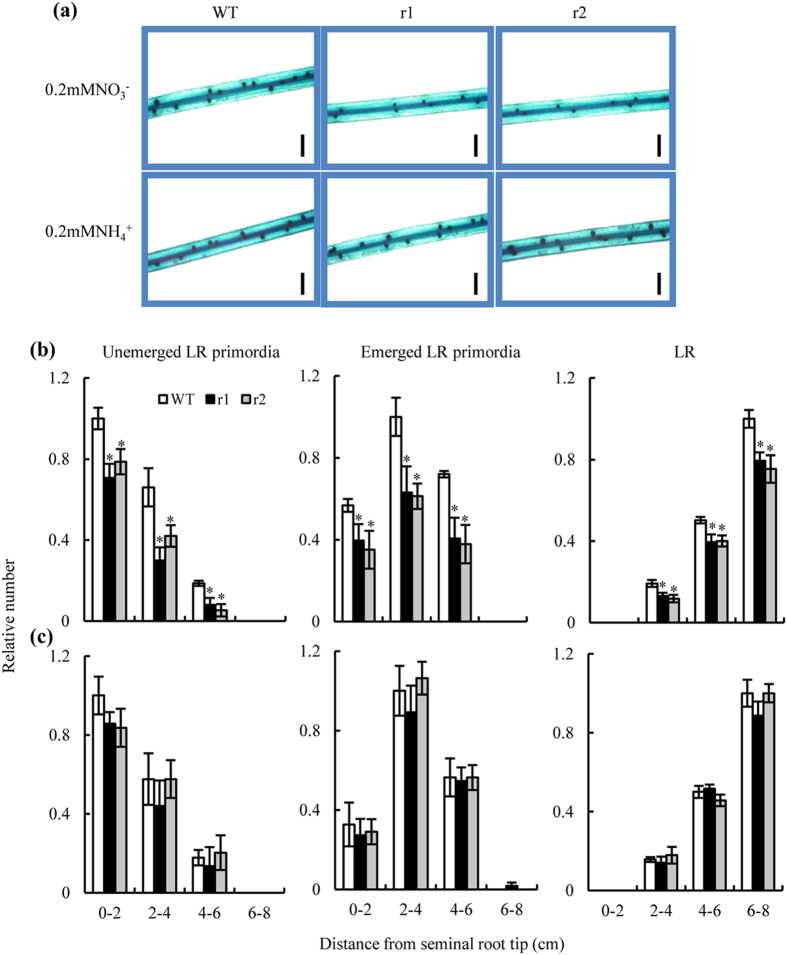
Two-week-old rice seedlings had seminal roots that were longer than the adventitious roots. Our preliminary experiment showed that the response of seminal root LRs to N treatment was similar to that of adventitious root LRs. We therefore chose seminal roots to represent root responses in our study of the effects of OsNAR2.1 on the rice root system. Compared to WT plants, the number of LR primordia was reduced markedly in the RNAi lines when NO3−-N was supplied (Fig. 2a). When NH4+-N was supplied, we observed no difference in LR development between the WT and the mutants (Fig. 2a,c). When NO3−- N was supplied, our microscopic observations detected significant differences between genotypes in (i) the numbers of unemerged and emerged LR primordia and (ii) the numbers of LRs at distances 0–8 cm behind the seminal root tips (Fig. 2b). A marked decrease in the number of LR primordia (number of primordia from the first division of the pericycle cells to emergence) resulted in a 23% reduction in the LR numbers in 6–8 cm-lengths of roots in the mutants (compared to roots in the WT plants) after incubation for 7 d with a NO3−-N supply.
Figure 2. Lateral root (LR) development on the seminal roots of wild-type (WT) and osnar2.1 knockdown lines (r1 and r2).
LR development in the seminal roots was measured after the seedlings had been grown for 1 wk in hydroponic media containing 0.2 mM NO3− or NH4+. (a) LR primordia in 2–4 cm seminal root from the root tip (bar = 1 mm); (b,c) Microscopic images of LR development at a concentration of 0.2 mM NO3− (b) or 0.2 mM NH4+ (c). Relative numbers for three LR stages were normalised to the largest number in WT plants. Values are means ± SE (n = 6). *P < 0.05 (ANOVA) comparing WT plants and two mutant lines in the same root zone.
N accumulation in the osnar2.1 mutants was inhibited across a wide range of NO3 − concentrations
We determined whether defective LR formation in the osnar2.1 mutants under NO3− supply was attributable to the inhibition of N uptake, by quantifying the level of N limitation experienced by rice seedlings through measurements of the cumulative uptake of 15N-NO3− during the entire 7 d period following transfer to media containing a range of labelled NO3− concentrations (Fig. 3a,b). Compared to WT plants, the mutants had reduced N accumulations in the shoots and the roots; reductions ranged between 61% and 18% at external NO3− concentrations of 0.05–10 mM.
Figure 3. Cumulative 15N-NO3− uptake in wild-type (WT) and osnar2.1 knockdown lines (r1 and r2).
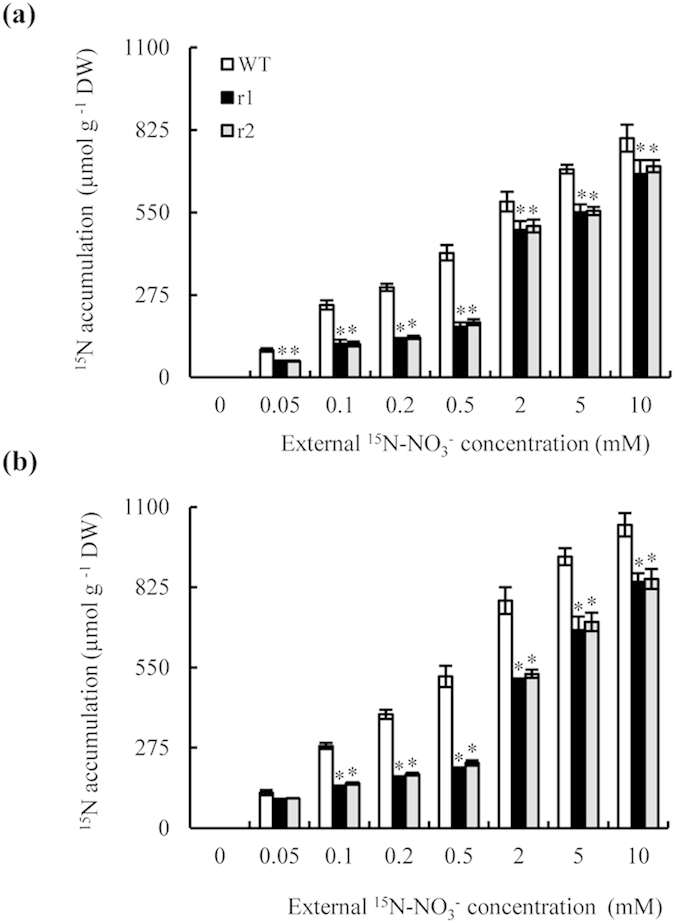
Rice seedlings were grown for 1 wk in hydroponic medium containing 10 mM NO3− and then transferred for a further week to media containing 0–10 mM 15N-NO3−. (a, b) Cumulative 15N-NO3− uptake in the shoots (a) and roots (b). Values are means ± SE (n = 6). *P < 0.05 (ANOVA) comparing WT plants and two mutant lines in the same treatment.
Lateral root formation responses to reduced root N accumulation
We investigated LR formation in rice plants after transfer from medium containing 10 mM NO3− to media containing a range of NO3− concentrations (0–10 mM). After 7 d, the elongation of seedling seminal and adventitious roots had responded little to the treatments (Fig. 1, Supplementary Fig. S3), but subsequent LR formation was strongly affected by external NO3− concentrations (Fig. 4a). Lateral root numbers on the seminal roots of WT plants increased with increasing external NO3− concentration (0.05–2 mM), but decreased with continued increases in external NO3− concentrations above 2 mM (Fig. 4a), in agreement with our previous findings29,30.
Figure 4. Changes in lateral root (LR) numbers in response to varying external and internal nitrate concentrations in wild-type (WT) and osnar2.1 knockdown lines (r1 and r2).
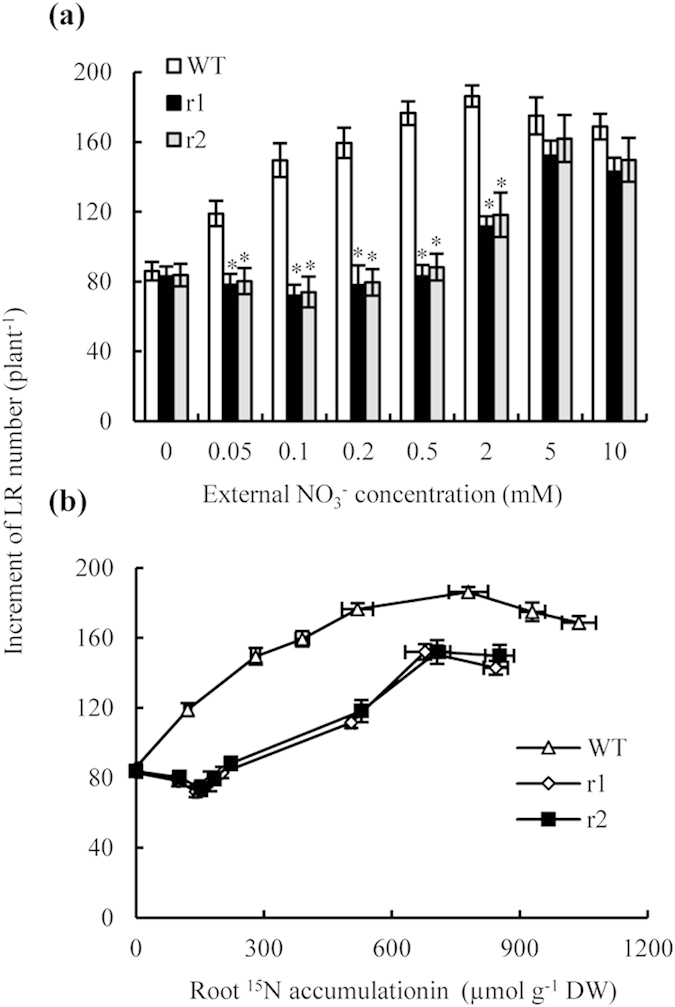
Rice seedlings were grown for 1 wk in hydroponic medium containing 10 mM NO3− and then transferred for a further week to media containing 0-10 mM 15N-NO3−. (a) LR number responding to varying external NO3− concentrations; (b) LR number responding to varying internal 15N accumulation levels in the roots. Values are means ± SE (n = 6). *P < 0.05 (ANOVA) comparing WT plants and two mutant lines in the same treatment.
Lateral root development is modulated by internal plant N status11,16,18,31. A plot of the number of new LRs formed during the experimental period against the internal root N content in WT plants (Fig. 4b) showed clearly that LR numbers increased with internal root N content. N accumulation increased when the concentration in the root was less than ca. 780 μmol g−1 (external NO3− ≤ 2 mM); however, LR formation was inhibited when the internal N content in the root was greater than ca. 780 μmol g−1 (external NO3− ≥ 2 mM). A similar trend was detected when we plotted the number of newly formed LRs in the osnar2.1 knockdown mutants against a range of internal root N contents. These results suggest that the inhibition of LR initiation in osnar2.1 mutants at a low NO3− concentrations is due to impaired NO3− uptake. However, defects in LR formation of osnar2.1 mutants were not completely explainable by reduced NO3− uptake because when internal N concentrations in the roots of WT plants and mutants were similar, the mutants still had fewer visible LRs than the WT plants.
LR formation was less sensitive to localised NO3 − supply in the osnar2.1 mutants than in WT plants
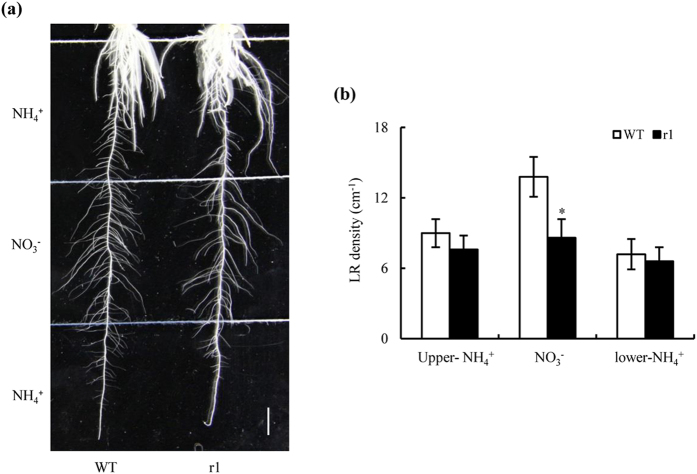
Our data to this point suggested that OsNAR2.1 may be involved in a signalling function; we therefore examined the responses of seminal root LRs to localised NO3− supply in WT plants and osnar2.1 mutants. LR densities on seminal roots provided with a localised NO3− supply were 33% lower in osnar2.1 mutants than in WT plants (Fig. 5a,b). However, LR densities were similar on seminal roots of mutants and WT plants when we provided a localised NH4+ supply. These data suggest that a knockdown of OsNAR2.1 inhibited LR formation via a NO3−-signalling pathway when NO3−-N was supplied.
Figure 5. Effect of a localised NO3− supply on lateral root (LR) development in wild-type (WT) and osnar2.1 knockdown line (r1).
Rice seeds were germinated in trays over a period of 2 d and then transferred to plant culture dishes containing 0.6% Phytagel™ media. Culture dishes were divided into three 5-cm segments; the upper and lower segments were supplemented with 0.2 mM NH4+, and the middle segment was supplemented with 0.2 mM NO3−. (a) Image of LRs responding to localised NO3− supply after a 3 wk growth period. Bar = 1 cm; (b) LR density in three segments. Values are means ± SE (n = 8). *P < 0.05 (ANOVA) comparing WT and r1 plants in the same treatment.
Knockdown of OsNAR2.1 inhibited auxin transport from the shoot to the root when N was supplied as NO3 −
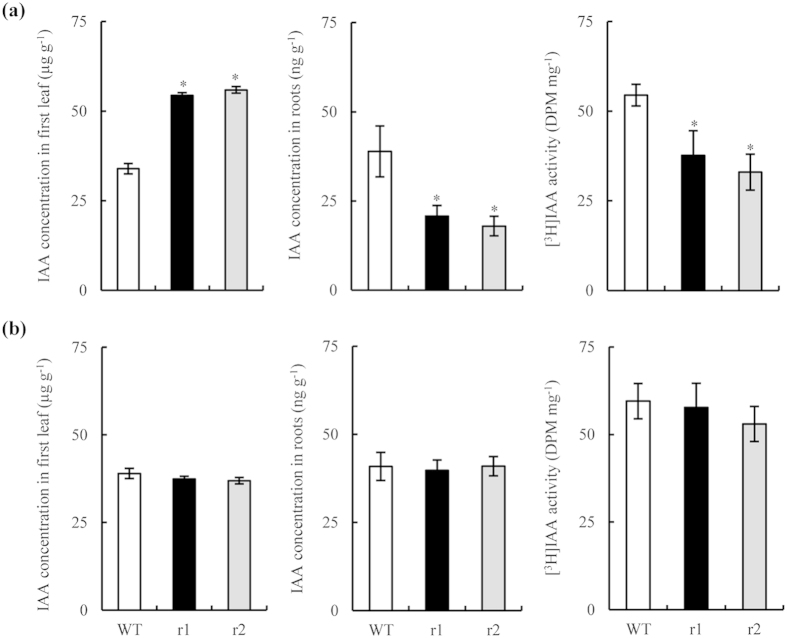
Several plant hormones control LR formation; auxin plays a pivotal role32,33. To determine whether auxin affected LR formation in osnar2.1 mutants, we analysed endogenous IAA concentrations in the first leaf and in the root. IAA concentrations in the first leaf of the two osnar2.1 mutants supplied with NO3− were elevated by ca. 62% in comparison with WT seedlings; conversely, IAA concentrations in the roots of two osnar2.1 mutants were ca. 50% lower than those in WT plants (Fig. 6a). When NH4+-N was supplied, auxin distributions were similar between genotypes (Fig. 6b). Thus, knockdown of OsNAR2.1 probably inhibited auxin polar transport from shoots to roots when NO3−-N was supplied. We therefore conducted [3H]IAA transport assays. When NO3−-N was supplied to the two mutant seedlings, [3H]IAA transport from their shoots to their roots was reduced significantly, and the [3H]IAA activity in the roots was reduced in consequence. These data confirm that auxin polar transport from shoots to roots was inhibited in osnar2.1 mutants supplied with NO3− as an N source.
Figure 6. IAA concentration in the first leaf and roots and [3H]IAA transport in wild-type (WT) and osnar2.1 knockdown lines (r1 and r2).
Seedlings were grown hydroponically for 1 wk in nutrient solution containing 0.2 mM NO3− (a) or NH4+ (b). Values are means ± SE (n = 6). *P < 0.05 (ANOVA) comparing WT plants and two mutant lines.
Exogenous application of NAA recovered LR initiation in osnar2.1 mutants
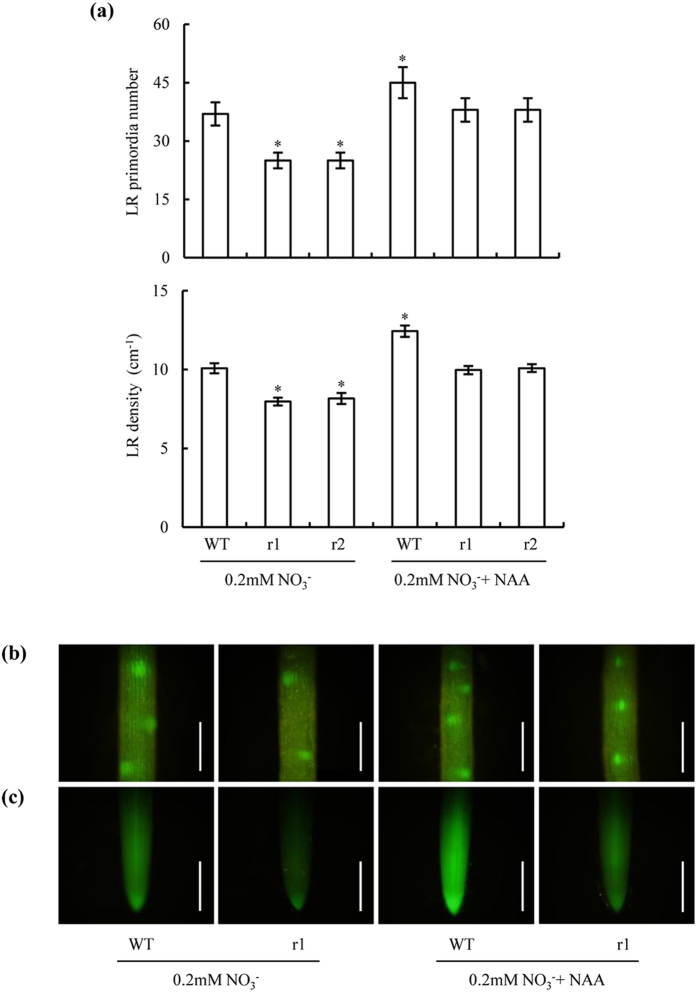
Application of NAA (1 nM) counteracted the effects of OsNAR2.1 knockdown on LR primordium numbers and LR density when N was supplied as NO3− (Fig. 7a,b). LR initiation in osnar2.1 mutants supplied with NO3− and treated with NAA was similar to LR initiation in WT plants supplied with NO3− as the N source. Furthermore, exogenous NAA application enhanced LR initiation in WT rice seedlings supplied with NO3−. These findings were concordant with changes in DR5::GFP expression levels in seminal root tips (Fig. 7c). Thus, reduced IAA concentrations in the two osnar2.1 mutants played a key role in the inhibition of LR initiation.
Figure 7. Lateral root (LR) primordium numbers and LR density in seminal roots of wild-type (WT) and osnar2.1 knockdown lines (r1 and r2).
Seedlings were grown for 1 wk in nutrient solution containing 0.2 mM NO3− with or without NAA application (1 nM) in agar media. (a) LR primordium numbers and LR density in seminal roots; (b) DR5::GFP expression in the LR primordium; (c) DR5::GFP expression in the seminal root tips. Bar = 1 mm. Values are means ± SE (n = 6). *P < 0.05 (ANOVA) comparing WT plants subjected to 0.2 mM NO3− and five other treatments.
Knockdown of OsNAR2.1 reduced expression levels of PINs in the roots
Most auxin transport occurs via the polar transport stream, which is facilitated by proteins of the PIN family34. Our qRT-PCR analysis showed that the expression levels of PIN1c, PIN2, PIN9, and PIN10a-b in the roots subjected to NO3− treatments were significantly lower in the two osnar2.1 mutants than in the WT rice seedlings (Fig. 8a). When NH4+-N was supplied, PIN expression levels were similar between the two osnar2.1 mutants and the WT (Fig. 8b).
Figure 8. qRT-PCR analysis of PIN family genes in wild-type (WT) and osnar2.1 knockdown lines (r1 and r2).
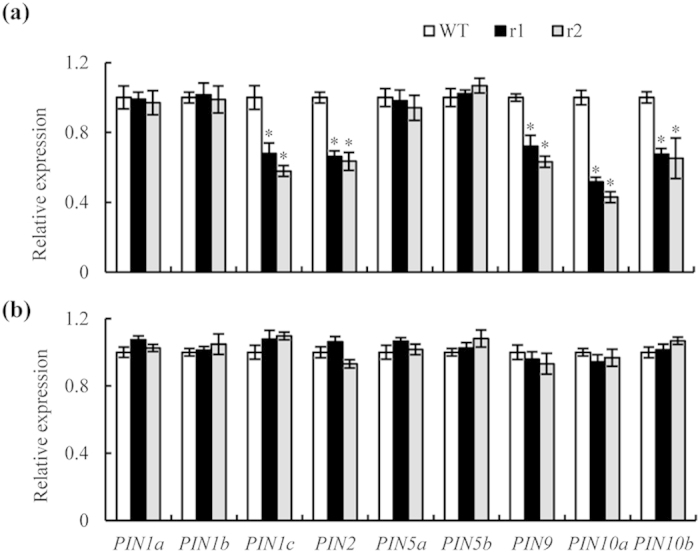
Seedlings were grown hydroponically for 1 wk in nutrient solution containing 0.2 mM NO3− or NH4+. (a,b) Expression levels in medium containing (a) 0.2 mM NO3− or (b) 0.2 mM NH4+. Relative mRNA levels for individual genes were normalised relative to OsACT. Values are means ± SE (n = 4). *P < 0.05 (ANOVA) comparing expression levels of the same gene in WT plants and two mutant lines.
Discussion
Mounting evidence shows that LR development is highly plastic and responsive to N treatments1,4,11,17,20,29,30,35. The striking effects of localised-NO3− and high NO3− media on LR growth have been investigated extensively in the model plant Arabidopsis5,10,11,15; however, the mechanism of N-dependent LR growth in rice remains unclear.
We observed inhibition of LR formation in the osnar2.1 mutants (compared to WT plants) subjected to low NO3− concentrations, but not in those subjected to low NH4+ concentrations. Knockdown of OsNAR2.1 reportedly impairs NO3− uptake at a NO3− concentration of 0.2 mM; the concentration of N in the mutants was only ca. 65% of that in the WT plants, but no differences were observed among genotypes at an NH4+ concentration of 0.2 mM27. Our study is the first to determine whether the inhibition of LR initiation in osnar2.1 mutants at low NO3− concentrations is due to impaired NO3− uptake. When we plotted LR number under varying external NO3− concentrations against root N content, we found that trends in the relationship were similar between mutants and WT plants: LR numbers increased with root N content up to a concentration of 700–780 μmol g−1; above this concentration, LR numbers decreased (Fig. 4b). Under external NO3− (15N-NO3−) concentrations of 5 and 10 mM, impaired NO3− uptake in the osnar2.1 mutants resulted in root N accumulations of 700–780 μmol g−1. Therefore, similar LR formations in WT plants and the two mutants were not unexpected at high NO3− concentrations (Fig. 4a). Reduced root N accumulation in osnar2.1 mutants subjected to low external NO3− concentrations inhibited LR formation (in comparison with WT rice plants). Overall, these results suggest that the knockdown of OsNAR2.1 probably reduced LR formation under low external NO3− concentrations via impairment of NO3− uptake.
Interestingly, inhibition of LR growth by OsNAR2.1 knockdown cannot be fully explained by reduced N uptake in the mutants. When internal root N contents in the roots of the WT and mutant plants were similar across the entire range of N contents (i.e. under external 15N-NO3− concentrations ranging from 0.05 to 10 mM), the osnar2.1 mutant plants always had significantly fewer LRs than the WT plants; e.g. when internal root N contents reached 520 μmol g−1, LR numbers in the two mutants were 33% fewer than those in the WT plants. Furthermore, the LR formation response to localised NO3− supply was weaker in the osnar2.1 mutants than in WT plants (Fig. 5). Thus, the NO3−-responsive OsNAR2.1 gene may be involved in a signalling pathway controlling LR formation in environments supplied with NO3−-N.
The expression levels of OsNRT2.1, OsNRT2.2, and OsNRT2.3a in the roots of two osnar2.1 mutants are reportedly much reduced in comparison with the abundant expression levels in WT plants27. Recent evidence suggests that in the model plant Arabidopsis, AtNRT1.1 (CHL1) is a NO3− sensor that activates the ANR1-mediated NO3−-signalling pathway to regulate LR proliferation5,11,35,36,37. Furthermore, AtNRT2.1 may have a direct stimulatory role in LR initiation under low NO3− concentrations19. Interestingly, AtNAR2.1 may also participate in the signalling pathway that integrates nutritional cues for LR proliferation by targeting AtNRT2.1 at the plasma membrane20,21. Therefore, it is reasonable to postulate that the inhibition of LR initiation in the osnar2.1 mutants is also related to the interaction of proteins affected by the knockdown of OsNAR2.1. The transcript levels of OsNAR2.1 and the three NO3− transporters are rapidly induced by NO3− supply, with peak levels occurring 1–2 h after the initiation of treatments25,27,38,39. Furthermore, OsNAR2.1, OsNRT2.1, and OsNRT2.2 are expressed abundantly throughout the primary and lateral roots25,26,27, and OsNRT2.3a is expressed abundantly in the root stelar cells, particularly in the xylem parenchyma tissue40. More experiments are required to elucidate the participation of OsNAR2.1 regulatory networks in rice LR formation under low NO3− concentrations.
Plants adjust their growth and development in response to changing environmental conditions through the perception and integration of external signals into the signalling pathways of plant hormones, such as auxin12,13,33,41,42,43,44,45. Auxin plays dominant roles in the specification of the founder cells that initiate LR formation and in the later stages of LR development32,33. Diverse environmental and endogenous signals may be integrated to mediate changes in auxin distribution via effects on polar transport46,47. Lateral root growth is not stimulated by localised NO3− supply in the auxin-insensitive mutant axr4, which suggests an overlap between the auxin and NO3− signalling pathways6. These two signalling pathways are further linked through an effect on auxin transport via AtNRT1.137.
In our study, (i) elevated IAA concentrations in mutant leaves, (ii) reduced IAA concentrations in mutant roots, and (iii) repressed mutant [3H]IAA transport from the shoot to root (in comparison with WT plants) suggested that auxin polar transport was inhibited by the knockdown of OsNAR2.1. These effects were correlated with a decrease in the transcript levels of five PIN genes in mutant roots (relative to WT plants). Exogenous NAA application restored LR initiation and DR5::GFP expression levels in seminal root tips of osnar2.1 mutants to levels similar to those in WT plants, further demonstrating that a reduced auxin concentration contributed to repressed LR initiation in the osnar2.1 mutants. OsNAR2.1 expression is dominant in roots and minimal in shoots25,38. Furthermore, in comparison with normal nutrient supply conditions, IAA transport is reduced in rice when N supply is limiting29,30. Thus, the repression of [3H]IAA transport from the shoot to the root in the osnar2.1 mutants (relative to WT plants) under low external NO3− concentrations can probably be attributed to N starvation system signals that inhibited auxin polar transport from the shoot to the root.
In conclusion, OsNAR2.1 had a key function in coordinating LR formation at low external NO3− concentrations. Its effects on LR formation most likely operated through a combination of roles in both NO3− uptake and NO3− signalling.
Methods
Plant material and growth conditions
The Nipponbare rice ecotype was used as the WT. The osnar2.1 mutant lines (T2 generation) obtained from RNA interference were reported in previous work27. Plants were grown in a greenhouse under natural light at day/night temperatures of 30 °C/18 °C. Seven-day-old seedlings of uniform size and vigour were transplanted into holes in a lid placed over the tops of 7-L pots (four holes per lid and three seedlings per hole). Nutrient solutions varying from one-quarter to half strength were applied for 2 d, and then full-strength nutrient solution was applied for a further week. Pots receiving NH4+ or NO3− were filled with 0.2 mM N solutions. The treatment protocol for providing NO3− concentrations of 0–10 mM to the plants followed a previously reported methodology16. Seven-day-old WT and mutant seedlings were transplanted into nutrient solutions varying from one-quarter to half strength for 2 d and then into nutrient solution containing 10 mM NO3− for 1 additional wk. Rice plants were transferred to nutrient solutions containing various NO3− (15N-NO3−, atom% 15N: 99%) concentrations (0–10 mM) for 1 wk before harvest.
The full chemical composition of the International Rice Research Institute (IRRI) nutrient solution was (mM): 0.3 KH2PO4, 0.35 K2SO4, 1.0 CaCl2, 1.0 MgSO4·7H2O, 0.5 Na2SiO3, and (μM) 20.0 Fe-EDTA, 9.0 MnCl2, 0.39 (NH4)6Mo7O24, 20.0 H3BO3, 0.77 ZnSO4 and 0.32 CuSO4; pH 5.5. The nutrient solution was replaced with fresh solution daily. NO3− and NH4+ were supplied in the nutrient medium as Ca(NO3)2 and (NH4)2SO4. To exclude the potential effects of calcium (Ca2+) on the treatments, we supplemented the solutions in the same experimental system with Ca2+ (as CaCl2) to the levels experienced by plants under the highest NO3− concentrations. The nitrification inhibitor dicyandiamide (7.0 μM) was added to each pot to prevent NH4+ oxidation.
Measurement of root system architecture
The rice root system comprises seminal and adventitious roots, each bearing LRs48. We used previously-described LR developmental stage categories3; stages I–XII were grouped here into the “unemerged primordia” category. The primordia of LR were classified into unemerged and emerged primordia. An emerged LR primordium >0.5 mm long (visible to the unaided eye) was classified as a LR, and the primordium was considered to have been activated9.
To visualise the development of LRs, we stained the seminal roots with methylene blue49. These roots were fixed in formalin/acetic acid/alcohol (FAA) solution (10:5:85 v/v/v) at 4 °C for at least 24 h. After fixation, LR primordia were rinsed for 10 min in water and then stained with a 0.01% w/v methylene blue solution. After the roots had been stained, counting the numbers of LR primordia and LRs was straightforward. The scaleplate in the stereomicroscope we used (Olympus Optical Co. Ltd, Tokyo, Japan) simplified determinations of the lengths of emerged primordia and LRs. The lengths of seminal and adventitious roots were measured using a ruler, and LR density was calculated by dividing the LR number by root length. Total root length and LR length were measured using the WinRhizo scanner-based image analysis system (Regent Instruments, Montreal, QC, Canada).
Lateral root responses to localised NO3 − supply
Rice seeds were germinated in trays over 2 d and then transferred into plant culture dishes (23 × 23 cm) containing 0.6% Phytagel™ media. Culture dishes were divided into three 5-cm segments; the upper and lower segments were supplemented with 0.2 mM NH4+, and the middle segment was supplemented with 0.2 mM NO3−. We photographed the representative morphologies of LRs in seminal roots after a 3 wk growth period. Each of the LR densities was calculated by dividing the LR number by the length of the seminal root segment.
The full chemical composition in the procedures using Phytagel medium was similar to that used in the hydroponic medium. To exclude the potential effects of calcium (Ca2+) on the treatments, the solutions in the three segments were supplemented with Ca2+ (as CaCl2) up to concentrations matching those in the NO3− treatments.
Measurement of 15N concentration
15N concentration was assayed in plants grown hydroponically27. After grinding in liquid N2, one aliquot of powder was dried to a constant weight at 70 °C. Approximately 6 mg powder from each sample were analysed using an Isotope Ratio Mass Spectrometer system (Thermo Fisher Scientific). To analyse the change in LR growth in response to the nutrient solution containing varying NO3− concentrations, the same experiment without NO3− labelling was performed at the same time. After harvest, LR number in the seminal root was analysed through recording of the LR number before and after the transfer of rice plants to nutrient solutions containing various NO3− concentrations.
Determination of IAA
We measured the concentrations of IAA in the first leaf and in the root9. Fresh weights of the samples were measured, after which specimens were immediately frozen in liquid N2. We performed sample measurement of free IAA by high-performance liquid chromatography. A standard IAA sample was obtained from Sigma-Aldrich (St. Louis, MO, USA).
The pDR5::GFP construct was transformed into WT plants and osnar2.1 mutants using Agrobacterium tumefaciens (strain EHA105) to determine the patterns of IAA distribution in rice plants. To construct the pDR5::GFP vector, we amplified a 720-bp cDNA fragment of green fluorescent protein (GFP) from the cloning vector pSAT6A-EGFP-N1 using the following primer set: FP, GGATCCATGGTGAGCAA GGGCGAGGAGCT; RP, GAGCTCTCACTTGTACAGCTCGTCCATG. This fragment was inserted into the vector of pDR5::GUS at the BamHI and SacI sites. The pDR5::GUS construct was kindly provided by the Ping Wu laboratory at Zhejiang University, Hangzhou, China. We analysed the fluorescence of GFP in the cells using 543-nm helium-neon and 488-nm argon lasers using a confocal laser scanning microscope (LSM410; Carl Zeiss, Oberkochen, Germany).
[3H]IAA-transport assay
Shoot-to-root auxin transport in intact plants was assayed9. WT plants and osnar2.1 mutants were pre-cultured for 1 wk in 0.2 mM NO3− and NH4+ solutions. We applied 20 μL [3H] IAA solution to the cut surfaces after excision of rice shoots 4 cm above the root-shoot junction; plants were then kept in darkness for 18 h. The [3H] IAA solution applied contained 0.5 μM [3H] (20 Ci mmol−1) in 2% dimethylsulphoxide (DMSO), 25 mM 2-(N-morpholino) ethanesulphonic acid (MES) buffer (pH 5.2) and 0.25% agar. The root-shoot junction was dissected out and weighed before incubation in the scintillation solution for >18 h. [3H] IAA radioactivity was detected using a multipurpose scintillation counter (LS6500, Beckman-Coulter, Fullerton, CA, USA).
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis
Total RNA was isolated from the roots of rice seedlings. RNA extraction, reverse transcription, and the qRT-PCR procedures followed a previously-reported procedure50. The primer sets targeting the PIN genes are listed in Supplementary Table S1.
Data analysis
Data from experiments were pooled for calculation of means and standard errors (SE) and analysed by one-way ANOVA followed by the LSD test at P ≤ 0.05 to determine the statistical significance of the differences between individual treatments. All statistical evaluations were conducted using the SPSS (version 11.0) statistical software (SPSS Inc., Chicago, IL).
Additional Information
How to cite this article: Huang, S. et al. Knockdown of the partner protein OsNAR2.1 for high-affinity nitrate transport represses lateral root formation in a nitrate-dependent manner. Sci. Rep. 5, 18192; doi: 10.1038/srep18192 (2015).
Supplementary Material
Acknowledgments
This work was funded by the Ministry of Science and Technology of China (No. 2011CB100302), the National Nature Science Foundation of China (No. 31071846, 31172022, 31372122 and 31471936), Innovative Research Team Development Plan of the Ministry of Education of China (No. IRT1256), the 111 Project (No. 12009), PAPD in Jiangsu Province of China, China Scholarship Council (CSC), and Innovative Plan of Jiangsu Province of China (CXLX13_280 and 568).
Footnotes
Author Contributions Y.L.Z., S.J.H. and S.C. designed the experiments and wrote the manuscript. S.J.H., S.C., Z.H.L., C.M.Z. and M.Y. conducted the measurements, data analysis. J.G.C. assisted with the experiment. X.R.F. and G.H.X. assisted with the manuscript. These authors reviewed the manuscript before the submission.
References
- Gruber B. D., Giehl R. F. H., Friedel S. & von Wirén N. Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 163, 161–179 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H., De Smet I. & Ding Z. Shaping a root system: regulating lateral versus primary root growth. Trends Plant Sci. 19, 426–431 (2014). [DOI] [PubMed] [Google Scholar]
- Malamy J. E. & Benfey P. N. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development. 124, 33–44 (1997). [DOI] [PubMed] [Google Scholar]
- Drew M. C. & Saker L. R. Nutrient Supply and the Growth of the Seminal Root System in Barley II. Localized, compensatory increases in lateral root growth and rates of nitrate uptake when nitrate supply is restricted to only part of the root system. J. Exp. Bot. 26, 79–90 (1975). [Google Scholar]
- Zhang H. M. & Forde B. G. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science. 279, 407–409 (1998). [DOI] [PubMed] [Google Scholar]
- Zhang H. M., Jennings A., Barlow P. W. & Forde B. G. Dual pathways for regulation of root branching by nitrate. Proc. Nalt. Acad. Sci. USA. 96, 6529–6534 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojon A., Krouk G., Perrine-Walker F. & Laugier E. Nitrate transceptor(s) in plants. J. Exp. Bot. 62, 2299–2308 (2011). [DOI] [PubMed] [Google Scholar]
- Song W. et al. Nitrate supply affects root growth differentially in two rice cultivars differing in nitrogen use efficiency. Plant Soil. 343, 357–368 (2011). [Google Scholar]
- Song W. et al. Auxin distribution is differentially affected by nitrate in roots of two rice cultivars differing in responsiveness to nitrogen. Ann. Bot. 112, 1383–1393 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde B. G. Nitrogen signalling pathways shaping root system architecture: an update. Curr. Opin. Plant Biol. 21, 30–36 (2014). [DOI] [PubMed] [Google Scholar]
- Remans T. et al. The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc. Nalt. Acad. Sci. USA. 103, 19206–19211 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal E. A. et al. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Nalt. Acad. Sci. USA. 107, 4477–4482 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal E. A., Moyano T. C., Riveras E., Contreras-Lopez O. & Gutierrez R. A. Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. Proc. Nalt. Acad. Sci. USA. 110, 12840–12845 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y., Bernreiter A., Filleur S., Abram B. & Forde B. G. Overexpressing the ANR1 MADS-box gene in transgenic plants provides new insights into its role in the nitrate regulation of root development. Plant Cell Physiol. 53, 1003–1016 (2012). [DOI] [PubMed] [Google Scholar]
- Zhang H. M. & Forde B. G. Regulation of Arabidopsis root development by nitrate availability. J. Exp. Bot. 51, 51–59 (2000). [PubMed] [Google Scholar]
- Remans T. et al. A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol. 140, 909–921 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffel S. et al. Nitrogen economics of root foraging: Transitive closure of the nitrate-cytokinin relay and distinct systemic signaling for N supply vs. demand. Proc. Nalt. Acad. Sci. USA. 108, 18524–18529 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya T. et al. CLE-CLAVATA1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc. Nalt. Acad. Sci. USA. 111, 2029–2034 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little D. Y. et al. The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc. Nalt. Acad. Sci. USA. 102, 13693–13698 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsel M. et al. Nitrate signaling and the two component high affinity uptake system in Arabidopsis. Plant Signal. Behav. 2, 260–262 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsel M. et al. Characterization of a two-component high-affinity nitrate uptake system in Arabidopsis. Physiology and protein-protein interaction. Plant Physiol. 142, 1304–1307 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L. et al. MADS-Box transcription factor AGL21 regulates lateral root development and responds to multiple external and physiological signals. Mol. Plant. 7, 1653–1669 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Wang H., Hamera S., Chen X. & Fang R. miR444a has multiple functions in rice nitrate-signaling pathway. Plant J. 78, 44–55 (2014). [DOI] [PubMed] [Google Scholar]
- Yu C. et al. The effects of fluctuations in the nutrient supply on the expression of five members of the AGL17 clade of MADS-box genes in rice. PLoS One. 9, e105597 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H. et al. Spatial expression and regulation of rice high-affinity nitrate transporters by nitrogen and carbon status. J. Exp. Bot. 62, 2319–2332 (2011). [DOI] [PubMed] [Google Scholar]
- Feng H. et al. Multiple roles of nitrate transport accessory protein NAR2 in plants. Plant Signaling Behavior. 6, 1–4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M. et al. Rice OsNAR2.1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a nitrate transporters to provide uptake over high and low concentration ranges. Plant Cell Environ. 34, 1360–1372 (2011). [DOI] [PubMed] [Google Scholar]
- Liu X. et al. Identification and functional assay of the interaction motifs in the partner protein OsNAR2.1 of the two-component system for high-affinity nitrate transport. New Phytol. 1, 74–84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H. et al. Strigolactones are involved in phosphate-and nitrate-deficiency-induced root development and auxin transport in rice. J. Exp. Bot. 65, 6735–6746 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H. et al. Formation of rice root regulated by nitrogen deficiency. Acta Pedologica Sinica. 51, 183–189 (2014). [Google Scholar]
- Ma W. et al. Auxin biosynthetic gene TAR2 is involved in low nitrogen mediated reprogramming of root architecture in Arabidopsis. Plant J. 78, 70–79 (2014). [DOI] [PubMed] [Google Scholar]
- Forde B. G. Local and long-range signaling pathways regulating plant responses to nitrate. Annu. Rev. Plant Biol. 53, 203–224 (2002). [DOI] [PubMed] [Google Scholar]
- De Smet I., Vanneste S., Inzé D. & Beeckman T. Lateral root initiation or the birth of a new meristem. Plant Mol. Biol. 60, 871–887 (2006). [DOI] [PubMed] [Google Scholar]
- Friml J. et al. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426, 147–153 (2003). [DOI] [PubMed] [Google Scholar]
- Ho C. H., Lin S. H., Hu H. C. & Tsay Y. F. CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194(2009). [DOI] [PubMed] [Google Scholar]
- Mounier E., Pervent M., Ljung K., Gojon A. & Nacry P. Auxin-mediated nitrate signalling by NRT1.1 participates in the adaptive response of Arabidopsis root architecture to the spatial heterogeneity of nitrate availability. Plant Cell Environ. 37, 162–174 (2014). [DOI] [PubMed] [Google Scholar]
- Krouk G. et al. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell. 18, 927–937 (2010). [DOI] [PubMed] [Google Scholar]
- Araki R. & Hasegawa, H. Expression of rice (Oryza sativa L.) genes involved in high-affinity nitrate transport during the period of nitrate induction. Breeding Sci. 56, 295–302 (2006). [Google Scholar]
- Cai C. H. et al. Gene structure and expression of high-affinity nitrate transport system in rice roots. J. Integr. Plant Biol. 50, 443–451 (2008). [DOI] [PubMed] [Google Scholar]
- Tang T. et al. Knockdown of a rice stelar nitrate transporter alters long-distance translocation but not root influx. Plant Physiol. 160, 2052–2063 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bucio J., Cruz-Ramírez A. & Herrera-Estrella L. The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 6, 280–287 (2003). [DOI] [PubMed] [Google Scholar]
- Malamy J. E. Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ. 28, 67–77 (2005). [DOI] [PubMed] [Google Scholar]
- Rubio V. et al. Plant hormones and nutrient signaling. Plant Mol. Biol. 69, 361–373 (2009). [DOI] [PubMed] [Google Scholar]
- Krouk G. et al. A framework integrating plant growth with hormones and nutrients. Trends Plant Sci. 16, 178–182 (2011). [DOI] [PubMed] [Google Scholar]
- Kazan K. Auxin and the integration of environmental signals into plant root development. Ann. Bot. 112, 1655–1665 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S. & Friml J. Auxin: a trigger for change in plant development. Cell. 136, 1005–1016 (2009). [DOI] [PubMed] [Google Scholar]
- Wang J. R. et al. Expression of PIN genes in rice (Oryza sativa L.): tissue specificity and regulation by hormones. Mol. Plant. 2, 823–831 (2009). [DOI] [PubMed] [Google Scholar]
- Coudert Y., Périn C., Courtois B., Khong N. G. & Gantet P. Genetic control of root development in rice, the model cereal. Trends Plant Sci. 15, 219–226 (2010). [DOI] [PubMed] [Google Scholar]
- Johnson J. F., Vance C. P. & Allan D. L. Phosphorus deficiency in Lupinus albus. (Altered lateral root development and enhanced expression of phosphoenolpyruvate carboxylase). Plant Physiol. 112, 31–41 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Fan X., Song W., Zhang Y. & Xu G. Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnol. J. 10, 139–149 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.