Abstract
Nonlinear modulation of the dopamine signaling on brain functions can be estimated by the interaction effects of dopamine-related genetic variations. We aimed to explore the interaction effects of COMT rs4680 and DRD2 rs1076560 on intra-network connectivity using independent component analysis. In 250 young healthy adults, we identified 11 meaningful resting-state networks (RSNs), including the salience, visual, auditory, default-mode, sensorimotor, attention and frontoparietal networks. A two-way analysis of covariance was used to investigate COMT×DRD2 interactions on intra-network connectivity in each network, controlling for age, gender and education. Significant COMT×DRD2 interaction was found in intra-network connectivity in the left medial prefrontal cortex of the anterior default-mode network, in the right dorsolateral frontal cortex of the right dorsal attention network, and in the left dorsal anterior cingulate cortex of the salience network. Post hoc tests revealed that these interactions were driven by the differential effects of DRD2 genotypes on intra-network connectivity in different COMT genotypic subgroups. Moreover, even in the same COMT subgroup, the modulation effects of DRD2 on intra-network connectivity were different across RSNs. These findings suggest a network-dependent modulation of the DA-related genetic variations on intra-network connectivity.
Dopamine (DA), as a critical neurotransmitter, regulates movement, cognition and reward1,2,3. The modulation of the DA system is realized by impacting the structure and function of the brain. The DA signaling in brain tissue is regulated by the genetic variations in the DA pathway. For example, a single-nucleotide polymorphism (SNP; Val158Met) of the catechol-O-methyltransferase (COMT) can affect the degradation of synaptic dopamine4,5 and an SNP (rs1076560, G > T) of the dopamine D2 receptor (DRD2) can affect the function of the receptor6. Although the DA signaling in brain tissue cannot be measured in vivo, it can be roughly estimated by the carrying status of the DA-related genes. For example, the DA signaling is lower in COMT Val homozygotes than in Met-carriers and lower in DRD2 GG than in TT carriers. According to status of the COMT and DRD2, we can generate subgroups with different levels of the DA signaling. By investigating the interaction effects of the two SNPs, one can in vivo explore how the DA system modulates brain structure and function7,8,9.
The human brain consists of several functional independent networks, which serves different functions. However, to date, whether and how the DA signaling modulates these functional networks remains largely unknown. Using the interaction effects of COMT and DRD2, a previous study has revealed a functional system-dependent modulation of the DA signaling on functional connectivity density (FCD) in healthy young subjects. Brain regions (temporal pole and putamen) of the “control system” and those (medial prefrontal cortex and occipital cortex) of the “processing system” have been shown to exhibit a much different modulation by the DA-related genetic variations8. Because the nature of the previous study is voxel-wise connectivity analysis but not a typical network analysis, the specific modulation patterns of the DA signaling on functional networks are unclear. Independent component analysis (ICA) can identify multiple resting-state networks (RSNs), which can be used to directly investigate the intra-network connectivity in each RSN10.
In this study, we used the ICA approach to explore the specific modulation of the DA signaling on the intra-network connectivity in the RSNs in healthy young adults by analyzing the interaction effects between the COMT rs4680 and DRD2 rs1076560.
Materials and Methods
Subjects
The study has been approved by the Medical Research Ethics Committee of Tianjin Medical University, and written informed consent was obtained from each subject before the study. The method was carried out in accordance with the approved guidelines. A total of 250 healthy right-handed subjects (mean age: 22.7 ± 2.4 years; 115 males) were selected from 323 Chinese Han subjects who participated in this study after giving written informed consent. Seventy-three subjects were excluded from further analysis due to a lack of genetic data (29 subjects) or excessive head movement (14 subjects) during the functional magnetic resonance imaging (fMRI) scans or missing behavioral scales (30 subjects). Careful screening was performed to ensure that all participants had no any lifetime history of psychiatric or neurological illness and MR contraindications. Memory function was evaluated with the Chinese Revised Wechsler Memory Scale11, and executive function was tested using the Wisconsin Card Sorting Test12. Individual working memory capacity was assessed using the n-back task13. Depression levels were examined using the Beck Depression Inventory14, and anxiety levels were evaluated with the Self-Rating Anxiety Scale15. Temperamental characteristics were assessed using the Tridimensional Personality Questionnaire16. These above-mentioned behavioral scales reflect structural and functional characteristics of the brain and show genotypes differences17,18,19,20,21.
Genotyping
Genomic DNA was extracted from 3000 μl of whole blood using the EZgeneTM Blood gDNA Miniprep Kit (Biomiga). We determined the genotypes for COMT rs4680 and DRD2 rs1076560 of the subject using the PCR and ligation detection reaction (LDR) method22,23. The PCR primer sequences for COMT were as follows: forward: 5′ GGGCCTACTGTGGCTACTCA 3′, and reverse: 5′ CCCTTTTTCCAGGTCTGACA 3′. The PCR primer sequences for DRD2 were as follows: forward: 5′ AGCATCTCCATCTCCAGCTC 3′, and reverse: 5′ GAAAAAGGACAGGGGCAATC 3′. PCR was performed with a 20 μL reaction volume containing 1 μL genomic DNA, 0.4 μL primer mixture, 2 μL dNTPs, 0.6 μL Mg2+, 2 μL buffer, 4 μL Q-Solution, and 0.3 μL Taq DNA polymerase. The amplification protocol incorporates an initial denaturation and enzyme activation phase at 95°C for 15 min, followed by 35 cycles of denaturation at 94°C for 30 sec, annealing for 1 min and 30 sec at 59°C for COMT rs4680 and 56°C for DRD2 rs1076560, extension at 72°C for 1 min, and then a final extension at 72°C for 7 min. PCR products were verified in 3% agarose gels that had been stained with ethidium bromide to regulate the amount of DNA added to the LDR.
For each SNP, three probes were designed for the LDR reactions: one common probe (rs4680: P-GCCAGCGAAATCCACCATCCGCTGGTTTTTTTTTTTTTTTTTTTT-FAM; rs1076560: P-GAAAGGGAGGGGCCAGTGAGATGGGTTTTTTTTTTTTTTTTTT-FAM) and two discriminating probes for the two alleles of each SNP (rs4680_A: TTTTTTTTTTTTTTTTTTTTCAGGCATGCACACCTTGTCCTTCAT; rs4680_G: TTTTTTTTTTTTTTTTTTTTTTCAGGCATGCACACCTTGTCCTTCAC; rs1076560_T: TTTTTTTTTTTTTTTTTTGTGTTTGCAGGAGTCTTCAGAGGGA; rs1076560_G: TTTTTTTTTTTTTTTTTTTTGTGTTTGCAGGAGTCTTCAGAGGGC). These reactions were conducted in a 10 μL mixture containing 1 μL buffer, 1 μL probe mix, 0.05 μL Taq DNA ligase, 1 μL PCR product, and 6.95 μL deionized water. The reaction program consisted of an initial heating at 95°C for 2 min, followed by 35 cycles of 30 sec at 94°C and 2 min at 50°C. Reactions were stopped by chilling the tubes in an ethanol–dry ice bath and adding 0.5 mL of 0.5 mM EDTA. Aliquots of the reaction products (1 μL) were mixed with 1 μL of loading buffer (83% formamide, 8.3 mM EDTA and 0.17% blue dextran) and 1 μL ABI GS-500 Rox-Fluorescent molecular weight marker and then denatured at 95°C for 2 min.
The samples were then chilled rapidly on ice prior to being loaded on a 5 Murea-5% polyacrylamide gel and electrophoresed on an ABI 3100 DNA sequencer at 3000 V. Finally, the fluorescent ligation products were analyzed and quantified using the ABI GeneMapper software.
Data acquisition
MRI data were acquired using a 3.0-Tesla MR system (Discovery MR750, General Electric, Milwaukee, WI, USA). Tight but comfortable foam padding was used to minimize head motion, and earplugs were used to reduce scanner noise. Resting-state fMRI data were collected using single-shot echo-planar imaging with the following imaging parameters: repetition time/echo time = 2000/30 ms; field of view = 240 mm ×240 mm; matrix = 64 × 64; flip angle = 90°, slice thickness = 4 mm; no gap; 40 interleaved transverse slices; and 180 volumes. All subjects were asked to keep their eyes closed, to think of nothing in particular and not to fall asleep, and as motionless as possible. Sagittal three-dimensional T1-weighted images were acquired by a brain volume sequence (repetition time/echo time = 8.1/3.1 ms; inversion time = 450 ms; field of view = 256 mm ×256 mm; matrix = 256 ×256; flip angle = 13°, slice thickness = 1 mm; no gap; 176 slices).
fMRI data preprocessing
The fMRI data were preprocessed using the Statistical Parametric Mapping (SPM8, http://www.fil.ion.ucl.ac.uk/spm) and Data Processing Assistant for Resting-State fMRI (DPARSF)24. We discarded the first 10 volumes of each functional time series because of the signal reaching equilibrium and the participants adapting to the scanning noise. The remaining 170 volumes were corrected for the acquisition time delay between different slices. Head movement parameters were tested by estimating the translation in each direction and the angular rotation on every axis for each volume. Fourteen subjects who had a maximum displacement >2 mm or a maximum rotation >2.0° were excluded from further analysis. The movement-corrected functional volumes were spatially normalized to the Montreal Neurological Institute (MNI) space and re-sampled to 3 × 3 × 3 mm3 voxels using the normalization parameters estimated during the unified segmentation. Then, the resulting images were smoothed using a Gaussian kernel of 6 × 6 × 6 mm3 full-width at half-maximum.
Identification of resting-state networks
We used group ICA (GICA) to decompose all the data into independent components (ICs) using the GIFT software (http://icatb.sourceforge.net/, version 1.3 h) with three steps: data reduction, ICA, and back reconstruction25. Data reduction was to reduce the size of the subject’s fMRI data using the principal components analysis. Two data reduction steps were adopted. After each subject’s fMRI data was reduced, the subjects were concatenated into one group and put through another data reduction step. ICA algorithm was then applied to the reduced dataset to identify ICs. The number (n = 31) of ICs was automatically estimated using the minimum description length (MDL) criterion26. ICASSO toolbox was used to determine the reliability of ICA algorithm. Specifically, ICA was run 100 times to obtain the final integrated output. The subject-specific time courses and spatial maps were back-reconstructed by a dual-regression method. A linear spatial regression was applied to the group-level spatial maps and individuals’ fMRI datasets to calculate matrices describing time courses for each component of each subject25. A linear temporal regression was then applied to these time-course matrices and individuals’ fMRI datasets to estimate subject-specific spatial maps27. For each subject-specific spatial component, the value of a voxel represents the relation of the time courses between this voxel and the subject-specific component. We defined this value as intra-network connectivity. To improve the normality of the data, we scaled the spatial maps to z-scores, which were used to voxel-wise compare the localized differences in the spatial component between different groups25,27. In this study, a total of 11 meaningful components were identified from the 31 ICs by visual inspection and a frequency analysis of the spectra of the estimated ICs28, potentially depicting functionally relevant RSNs.
Intra-network connectivity analysis
The eleven meaningful independent components representing RSNs were entered into a random-effect one-sample t-test. We used t > 20 and a cluster size of >100 voxels to improve the representation of each brain network. A sample-specific spatial map was generated for each component (Fig. 1). A two-way analysis of covariance (ANCOVA) was used to investigate the interaction effects between the COMT and DRD2 on intra-network connectivity in each RSN mask with gender, age and years of education as nuisance covariates. A correction for multiple comparisons was performed using a Monte Carlo simulation, resulting in a corrected threshold of p < 0.05 (AlphaSim program in AFNI software (http://afni.nimh.nih.gov/). Parameters: single voxel p= 0.05, 5000 simulations, cluster connection radius = 5 mm; with a gray matter mask and a resolution of 3 mm ×3 mm ×3 mm).
Figure 1. Cortical representation of the resting state networks (RSNs) identified by independent component analysis.
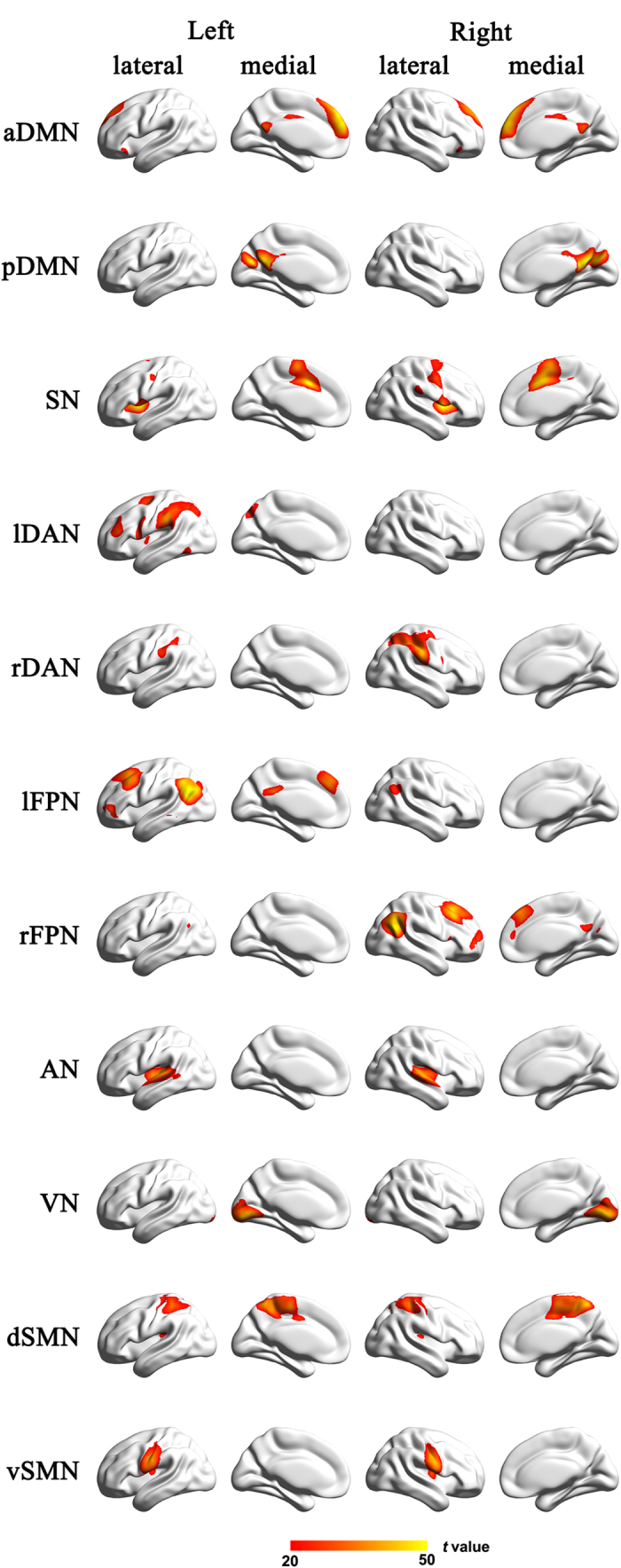
Data are displayed on the lateral and medial surfaces of the left and right hemispheres of a brain surface map. aDMN, anterior default mode network; AN, auditory network; dSMN, dorsal sensorimotor network; lDAN, left dorsal attention network; lFPN, left frontoparietal network; pDMN, posterior default mode network; rDAN, right dorsal attention network; rFPN, right frontoparietal network; SN, salience nerwork; vSMN, ventral sensorimotor; VN, visual network. The color scale represents the t values in each RSN.
Results
Demographic and genetic characteristics
The demographic data are summarized in Table 1. The group of distributions in both COMT rs4680 genotypes (120 Val/Val, 106 Met/Val, and 24 Met/Met) and DRD2 rs1076560 genotypes (39 TT, 115 GT, and 96 GG) were in Hardy–Weinberg equilibrium (p > 0.05). Subjects with either homozygous or heterozygous for the Met-allele of COMT were merged into a group of Met-allele carriers according to previous method to address skewed genotypic distributions19,29,30. There was no significant interaction effect or main effect (p > 0.05) of COMT and DRD2 on any of the demographic, cognitive (memory and execution) and psychological (depression, anxiety, and personality) variables.
Table 1. Demographic data of subjects in fMRI analysis (n = 250).
| Genotypic groups | n | Age (years) | Years of education | Gender |
|---|---|---|---|---|
| COMT | ||||
| Val/Val | 120 | 22.7 (2.4) | 15.8 (2.0) | 54:56 |
| Met carriers | 130 | 22.6 (2.4) | 15.5 (2.1) | 61:67 |
| F (P) | 250 | 0.11 (0.74) | 0.14 (0.71) | 0.49(0.49) |
| DRD2 | ||||
| TT | 39 | 22.8 (2.6) | 15.7 (2.0) | 15:13 |
| GT | 115 | 22.6 (2.3) | 15.6 (2.2) | 56:59 |
| GG | 96 | 22.7 (2.4) | 15.8 (2.0) | 44:51 |
| F (P) | 250 | 0.27 (0.76) | 0.41 (0.66) | 0.41(0.68) |
| COMT×DRD2 | ||||
| Val/Val-TT | 21 | 22.4 (2.4) | 15.4 (1.9) | 7:14 |
| Val/Val-GT | 56 | 22.3 (2.4) | 15.7 (2.1) | 28:28 |
| Val/Val-GG | 43 | 23.3 (2.3) | 16.2 (2.0) | 19:24 |
| Met carriers-TT | 18 | 23.4 (3.0) | 16.1 (2.1) | 8:9 |
| Met carriers-GT | 59 | 22.8 (2.3) | 15.4 (2.3) | 28:31 |
| Met carriers-GG | 53 | 22.1 (2.4) | 15.4 (1.9) | 25:27 |
| F (P) | 250 | 3.93 (0.02) | 1.63 (0.20) | 0.39 (0.68) |
Components of the resting-state networks
As shown in Fig. 1, eleven meaningful components were identified by visual inspection, including anterior and posterior default mode networks (aDMN and pDMN), salience network (SN), left (lDAN) and right (rDAN) dorsal attention networks, ventral (vSMN) and dorsal (dSMN) sensorimotor networks, left (lFPN) and right (rFPN) frontoparietal networks, visual network (VN), and auditory network (AN). The components and locations of these RSNs were consistent with previous studies31,32,33,34.
Interaction effects of COMT and DRD2 on the intra-network connectivity
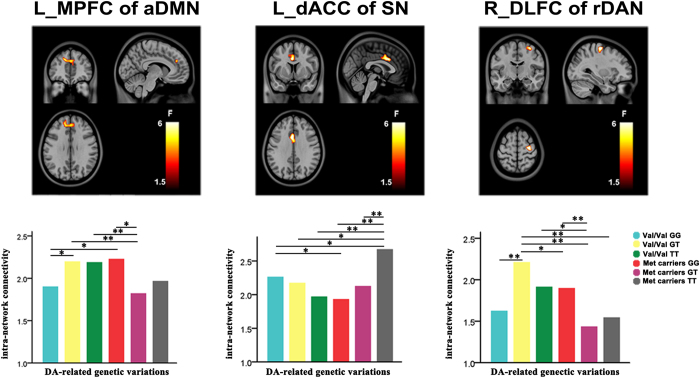
Although we focused on the interaction effects, we also found significant main effects of the two SNPs on intra-network connectivity (Supplementary information). The COMT × DRD2 interaction effects on intra-network connectivity was found in the left medial prefrontal cortex (MPFC) (peak MNI coordinates: x = −15, y = 45, z = 33) of the aDMN, in the left dorsal anterior cingulate cortex (dACC) (x = −3, y = 12, z = 33) of the SN, and in the right dorsolateral frontal cortex (x = 33, y = −15, z = 63) of the rDAN (Fig. 2 and Table 2). However, there were no significant interaction effects (p < 0.05, Alphasim correction) on the intra-network connectivity between these two SNPs.
Figure 2. The interaction effects of COMT and DRD2 on intra-network connectivity in the brain regions (top row) of brain networks, and the intergroup differences in intra-network connectivity in each cluster of brain networks (bottom row).
Color bar shows the F values. The horizontal axes of the bar plots (bottom row) represent six genotypic subgroups interacted by COMT and DRD2. The vertical axes represent the z-value of the intra-network connectivity. aDMN, anterior default mode network; dACC, dorsal anterior cingulate cortex; DLFC, dorsolateral frontal cortex; L, left; MPFC, medial prefrontal cortex; R, right; rDAN, right dorsal attention network; SN salience network. *represents p < 0.05, uncorrected for multiple comparisons; **represents p < 0.05/15 = 0.0033, Bonferroni corrected for multiple comparisons.
Table 2. Brain areas showed significant COMT×DRD2 interaction effects on brain networks.
| Brain regions | RSN | Cluster size (voxels) | Peak intensity | MNI Coordinates (x, y, z) |
|---|---|---|---|---|
| Left medial prefrontal cortex | aDMN | 75 | 6.5649 | −15, 45, 33 |
| Left dorsal anterior cingulated cortex | SN | 93 | 7.3553 | −3, 12, 33 |
| Right dorsolateral frontal cortex | rDAN | 60 | 10.9014 | 33, −15, 63 |
Post hoc tests revealed that the DA-related genetic variations exhibited different nonlinear modulation patterns on intra-network connectivity in different RSNs. The intergroup differences in intra-network connectivity in the MPFC of the DMN are shown in the lower row of the left column in Fig. 2 and Table 3. In the COMT Val/Val carriers, the DRD2 GG subgroup exhibited weaker intra-network connectivity than the DRD2 GT subgroup (p = 0.013, uncorrected). In contrast, in the COMT Met carriers, the DRD2 GG subgroup exhibited stronger intra-network connectivity than the DRD2 GT subgroup (p < 0.05, Bonferroni corrected). The intergroup differences in intra-network connectivity in the DLFC of the rDAN are shown in the lower row of the middle column in Fig. 2 and Table 4. In the COMT Val/Val carriers, the DRD2 GG subgroup had weaker intra-network connectivity than the DRD2 GT subgroup (p < 0.05, Bonferroni corrected). In contrast, in the COMT Met carriers, the DRD2 GG subgroup showed stronger intra-network connectivity than the DRD2 GT subgroup (p < 0.05, Bonferroni corrected). The intergroup differences in intra-network connectivity in the dACC of the SN are shown in the lower row of the right column in Fig. 2 and Table 5. In the COMT Val/Val carriers, intra-network connectivity did not exhibit any significant differences among the DRD2 genotypic subgroups. However, in the COMT Met carriers, both the DRD2 GG and GT subgroups exhibited weaker intra-network connectivity than the DRD2 TT subgroup (p < 0.05, Bonferroni corrected).
Table 3. Intergroup differences in intra-network connectivity in the MPFC of the DMN.
| P values | Val/Val-GT | Val/Val-TT | Met carriers-GG | Met carriers-GT | Met carriers-TT |
|---|---|---|---|---|---|
| Val/Val-GG | 0.013* | 0.066 | 0.007* | 0.494 | 0.693 |
| Val/Val-GT | 0.952 | 0.788 | 0.001** | 0.145 | |
| Val/Val-TT | 0.795 | 0.014* | 0.238 | ||
| Met carriers-GG | <0.001** | 0.102 | |||
| Met carriers-GT | 0.357 |
*p < 0.05, uncorrected for multiple comparisons; **p < 0.05/15 = 0.0033, Bonferroni corrected for multiple comparisons.
Table 4. Intergroup differences in intra-network connectivity in the DLFC of the rDAN.
| P values | Val/Val-GT | Val/Val-TT | Met carriers-GG | Met carriers-GT | Met carriers-TT |
|---|---|---|---|---|---|
| Val/Val-GG | <0.001** | 0.175 | 0.097 | 0.242 | 0.724 |
| Val/Val-GT | 0.151 | 0.043* | <0.001** | 0.002** | |
| Val/Val-TT | 0.936 | 0.020* | 0.152 | ||
| Met carriers-GG | 0.0025** | 0.107 | |||
| Met carriers-GT | 0.614 |
*p < 0.05, uncorrected for multiple comparisons; **p < 0.05/15 = 0.0033, Bonferroni corrected for multiple comparisons.
Table 5. Intergroup differences in intra-network connectivity in the dACC of the SN.
| P values | Val/Val-GT | Val/Val-TT | Met carriers-GG | Met carriers-GT | Met carriers-TT |
|---|---|---|---|---|---|
| Val/Val-GG | 0.499 | 0.092 | 0.014* | 0.297 | 0.0266* |
| Val/Val-GT | 0.224 | 0.053 | 0.699 | 0.005* | |
| Val/Val-TT | 0.817 | 0.346 | 0.001** | ||
| Met carriers-GG | 0.114 | <0.001** | |||
| Met carriers-GT | 0.002** |
*p < 0.05, uncorrected for multiple comparisons; **p < 0.05/15 = 0.0033, Bonferroni corrected for multiple comparisons.
Discussion
In the present study, we investigated COMT ×DRD2 interactions on intra-network connectivity in healthy young adults. The SN, aDMN and rDAN exhibited significant COMT×DRD2 interactions that were driven by the differential effects of DRD2 genotypes on intra-network connectivity in different COMT genotypic subgroups. Moreover, even in the same COMT subgroup, the genetic modulation of the DRD2 on intra-network connectivity was different across RSNs. Because the different genotypic combinations of COMT and DRD2 may represent different presumed dopamine signaling, our findings indicate that the nonlinear modulation of the DA system on intra-network connectivity is RSN-dependent.
COMT plays a critical role in the degradation of the catecholamine neurotransmitters (noradrenaline, adrenaline and dopamine), accounting for more than 60% of the DA degradation in the prefrontal cortex due to the lack of DA transporter4. The COMT gene contains a functional polymorphism (Val158Met; rs4680). Because the Met alleles leads to a fourfold decrease in enzyme activity at body temperature, in the Val homozygotes individuals show the greater COMT activity and the lower dopamine level than the Met carriers35. There was two splice isoforms of DRD2: the short (D2S) isoform locates presynaptically and the long (D2L) isoform distributes postsynaptically36. SNP rs1076560 (G > T) within the DRD2 gene at intron 6 was associated with decreased expression of D2S relative to D2L37. In contrast to the association between GG genotype and more expression of D2S38, T-allele carriers are related to more expression of D2L39. Because the D2S isoform inhibits dopamine release40, subjects with DRD2 GG genotype predict lower DA signaling than T-allele carriers. Thus, subjects with Val/Val and GG genotypes may exhibit the lowest DA signaling and subjects with Met and TT genotypes may have the highest DA signaling.
The DMN consists of the posterior cingulate cortex, the lateral parietal cortex and the ventromedial prefrontal cortex, which plays an important role in human cognitive functions41,42,43. The structural and functional impairments of the DMN have been reported in several neuropsychiatric diseases, and these impairments have been associated with cognitive deficits44. In consistent with respective modulation effect of COMT or DRD2 on the MPFC connectivity45,46, we found a COMT×DRD2 interaction effect on intra-network connectivity in the left MPFC of the aDMN in healthy young subjects. The interaction was driven by different effects of DRD2 on connectivity between COMT genotypic subgroups. The DRD2 GG subgroup exhibited weaker intra-network connectivity than the GT subgroup in the COMT Val/Val carriers; however, this modulation is reversed in the COMT Met carriers. Our findings are also consistent with previously reported modulation effect of the dopamine-related genetic variations on DMN connectivity8,47. That is, Val/Val-TT or Met-GG carriers with optimal DA levels showed higher intra-DMN connectivity than Val/Val-GG with lower DA levels and Met-TT carriers with higher DA levels.
The SN mainly consists of the fronto-insular cortex (FIC) and the dACC48. The SN serves to identify salient stimuli around environment and to initiate control signals in time to coordinate functions of other cognitive-related networks49. In consistent with a previous imaging genetics study that reported a COMT×DRD2 interaction on the FCD of left dACC in healthy young adults8, we found an interaction effect on intra-network connectivity in the left dACC of the SN, which was driven by the lack of DRD2 modulation in the COMT Val/Val carriers and the strong modulation in the COMT Met carriers. As a critical component of the SN, the dACC was associated with decision-making and reward system50,51. This modulation effect of the DA-related genetic variations on the intra-SN connectivity may help for understanding the relationship between DA system and cognitive performance52,53.
The DAN including parts of the intraparietal cortex and superior frontal cortex, is involved in preparing and applying top-down selection for stimuli and responses54,55. Genetic variations in COMT and DRD2 have been associated with attention performance56,57 and attention bias58. In this study, we provided a possible pathway that DA-related genetic variation may affect human attention via modulating the intra-network connectivity of the DAN. Specifically, we found a COMT×DRD2 interaction on intra-network connectivity in the right dorsolateral frontal cortex of the rDAN, which was driven by the inversed modulation of DRD2 on connectivity in the COMT Val/Val and Met carriers.
The functional networks of the human brain have been divided into two functional systems: the “processing system” includes the visual, sensorimotor, and DMN; and the “control system” consists of the fronto-parietal, attention, and SN34. By investigating the interaction effects of COMT and DRD2, a previous study has revealed a functional system-dependent modulation of the DA signaling on FCD in healthy young subjects. Brain regions of the SN of the “control system” exhibit the highest FCD in DRD2 GG in the COMT Val/Val carriers and the lowest FCD in GG subgroup in the COMT Met carriers; however, brain regions of the visual network and DMN of the “processing system” showed an inverse modulation: the lowest FCD in DRD2 GG in the COMT Val/Val carriers and the highest FCD in GG in the COMT Met carriers8. In the present study, the modulation patterns on the SN and DMN connectivity are consistent with the hypothesis of functional system-dependent modulation. Because the DAN is thought to be a typical functional network of the “control system”34, the completely different modulation effects of dopamine-related genetic variations on intra-network connectivity in the DAN and SN of the “control system” may support a network-dependent modulation of the DA signaling on functionality of the brain. In contrast to significant differences on RSN connectivity, we did not found any significant differences in the cognitive, emotional, or personality scores among different genotypic groups. These findings may indicate that imaging measures are more sensitive than behavioral measures in detecting potential genetic effects in healthy young subjects. The lack of significant differences in behavioral performance across genotypic groups may also exclude the possibility that our imaging genetics findings are resulted from behavioral differences across genotypes.
Several limitations should be noted when one interprets our results. Although separating the subjects into different groups before applying GICA can remain the maximal differences across groups, this approach may bring mismatched components for different groups, which will create difficulty for intergroup comparisons. To avoid the need for matching components between groups, we performed GICA on combined data from both subject groups and then reconstructed subject-specific maps and time courses for group comparisons. However, the resulting ICA components tend to be similar among subjects, reducing the statistical power of the following ANCOVA. Another limitation of this study is the use of a relatively higher voxel-level threshold (p < 0.05) in the cluster-level correction for multiple comparisons. Thus, our results should be treated as preliminary and need to be further validated.
In conclusion, with a relatively large sample of healthy young adults and an ICA analysis, we explored the interaction effects of COMT and DRD2 on intra-network connectivity in resting-state networks. We found that a different modulation of DRD2 on intra-network connectivity in different COMT genotypic subgroups drove the interactions. Moreover, the effects DRD2 on connectivity in COMT subgroups were different across resting-state networks, suggesting that the nonlinear modulation of the dopamine-related genes on functionality of the brain is network-dependent.
Additional Information
How to cite this article: Zhao, F. et al. Network-Dependent Modulation of COMT and DRD2 Polymorphisms in Healthy Young Adults. Sci. Rep. 5, 17996; doi: 10.1038/srep17996 (2015).
Supplementary Material
Acknowledgments
The authors thank Drs. Zhenyu Zhou and Ziheng Zhang of the GE Healthcare MR Research China for their support and assistance. This work was supported by the National Basic Research Program of China (973 program, Grant 2011CB707800), the Natural Science Foundation of China (Grants 81271551, 91132301, 81501451 and 81301201), and the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant XDB02030300).
Footnotes
Author Contributions F.S.Z., X.J.Z. and C.S.Y. designed the experiment and wrote the protocol and the draft of manuscript text. W.Q. and F.L. performed image processing and statistical analyses. Q.X. and J.P.W. collected the magnetic resonance imaging (MRI) data and genetic data. Q.H.W. prepared Figures 1 and 2. All authors contributed to and have approved the final manuscript.
References
- Brozoski T. J., Brown R. M., Rosvold H. E. & Goldman P. S. Cognitive deficit caused by regional depletion of dopamine in prefrontal cortex of rhesus monkey. Science 205, 929–932 (1979). [DOI] [PubMed] [Google Scholar]
- Schultz W. Getting formal with dopamine and reward. Neuron 36, 241–263 (2002). [DOI] [PubMed] [Google Scholar]
- Witte A. V. & Floel A. Effects of COMT polymorphisms on brain function and behavior in health and disease, NeuroImage 55, 225–232, doi: 10.1016/j.brainresbull.2011.11.012 (2012). [DOI] [PubMed] [Google Scholar]
- Mannisto P. T. & Kaakkola S. Catechol-O-methyltransferase (COMT): biochemistry, molecular biology, pharmacology, and clinical efficacy of the new selective COMT inhibitors. Pharmacol Rev 51, 593–628 (1999). [PubMed] [Google Scholar]
- Seamans J. K. & Yang C. R. The principal features and mechanisms of dopamine modulation in the prefrontal cortex. Prog Neurobiol 74, 1–58, doi: 10.1016/j.pneurobio.2004.05.006 (2004). [DOI] [PubMed] [Google Scholar]
- Khan Z. U., Mrzljak L., Gutierrez A., de la Calle A. & Goldman-Rakic P. S. Prominence of the dopamine D2 short isoform in dopaminergic pathways, Proc Natl Acad Sci USA 95, 7731–6 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M., Schmitz A., Corr P. & Hennig J. Molecular genetics support Gray’s personality theory: the interaction of COMT and DRD2 polymorphisms predicts the behavioural approach system. Int J Neuropsychopharmacol 9, 155–166, doi: 10.1017/s1461145705005419 (2006). [DOI] [PubMed] [Google Scholar]
- Tian T., Qin W., Liu B., Jiang T. & Yu C. Functional connectivity in healthy subjects is nonlinearly modulated by the COMT and DRD2 polymorphisms in a functional system-dependent manner. J Neurosci 33, 17519–17526, doi: 10.1523/jneurosci.2163-13.2013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler D. A. et al. Motor impulsivity in Parkinson disease: associations with COMT and DRD2 polymorphisms. Scand J Psychol 55, 278–286, doi: 10.1111/sjop.12113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Ven V. G., Formisano E., Prvulovic D., Roeder C. H. & Linden D. E. Functional connectivity as revealed by spatial independent component analysis of fMRI measurements during rest. Hum Brain Mapp 22, 165–178, doi: 10.1002/hbm.20022 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y. Manual of Wechsler Memory Scale-China Revised. Changsha, China: Hunan Mapping (1989). [Google Scholar]
- Heaton R. K. Wisconsin Card Sorting Test: Computer Version 3 For Windows, Research Edition. Psychological Assessment Resources: Odessa, FL (1999). [Google Scholar]
- Owen A. M., McMillan K. M., Laird A. R. & Bullmore E. N-back working memory paradigm: a meta-analysis of normative functional neuroimaging studies. Hum Brain Mapp 25, 46–59, doi: 10.1002/hbm.20131 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck A. T., Steer R. A. & Brown G. K. Manual for the beck depression inventory-II. (1996).
- Zung W. W. A rating instrument for anxiety disorders. Psychosomatics 12, 371–379 (1971). [DOI] [PubMed] [Google Scholar]
- Cloninger C. R., Svrakic D. M. & Przybeck T. R. A psychobiological model of temperament and character. Archives of general psychiatry 50, 975–990 (1993). [DOI] [PubMed] [Google Scholar]
- Enoch M. A., Xu K., Ferro E., Harris C. R. & Goldman D. Genetic origins of anxiety in women: a role for a functional catechol-O-methyltransferase polymorphism. Psychiatr Genet 13, 33–41, doi: 10.1097/01.ypg.0000054709.85338.c3 (2003). [DOI] [PubMed] [Google Scholar]
- Reuter M. & Hennig J. Association of the functional catechol-O-methyltransferase VAL158MET polymorphism with the personality trait of extraversion. Neuroreport 16, 1135–1138 (2005). [DOI] [PubMed] [Google Scholar]
- Li J. et al. COMT val158met modulates association between brain white matter architecture and IQ. Am J Med Genet B Neuropsychiatr Genet 150B, 375–380, doi: 10.1002/ajmg.b.30825 (2009). [DOI] [PubMed] [Google Scholar]
- Chen C. et al. Sex modulates the associations between the COMT gene and personality traits. Neuropsychopharmacology 36, 1593–1598, doi: 10.1038/npp.2011.39 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green A. E., Kraemer D. J., Deyoung C. G., Fossella J. A. & Gray J. R. A gene-brain-cognition pathway: prefrontal activity mediates the effect of COMT on cognitive control and IQ. Cereb Cortex 23, 552–559, doi: 10.1093/cercor/bhs035 (2013). [DOI] [PubMed] [Google Scholar]
- Thomas G. et al. Capillary and microelectrophoretic separations of ligase detection reaction products produced from low-abundant point mutations in genomic DNA. Electrophoresis 25, 1668–1677, doi: 10.1002/elps.200405886 (2004). [DOI] [PubMed] [Google Scholar]
- Yi P. et al. PCR/LDR/capillary electrophoresis for detection of single-nucleotide differences between fetal and maternal DNA in maternal plasma. Prenat Diagn 29, 217–222, doi: 10.1002/pd.2072 (2009). [DOI] [PubMed] [Google Scholar]
- Chao-Gan Y. & Yu-Feng Z. DPARSF: A MATLAB Toolbox for “Pipeline” Data Analysis of Resting-State fMRI. Front Syst Neurosci 4, 13, doi: 10.3389/fnsys.2010.00013 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun V., Adali T., Pearlson G. & Pekar J. A method for making group inferences from functional MRI data using independent component analysis. Hum Brain Mapp 14, 140–151, (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. O., Adalı T. & Calhoun V. D. Estimating the number of independent components for functional magnetic resonance imaging data. Hum Brain Mapp 28, 1251–1266, doi: 10.1002/hbm.20359 (2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantini D. et al. Electrophysiological signatures of resting state networks in the human brain. Proc Natl Acad Sci USA 104, doi: 10.1073/pnas.0700668104 13170–13175 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W. et al. Evaluating the effective connectivity of resting state networks using conditional Granger causality. Biological cybernetics 102, 57–69, doi: 10.1007/s00422-009-0350-5 (2010). [DOI] [PubMed] [Google Scholar]
- Taylor W. D. et al. The COMT Val158Met polymorphism and temporal lobe morphometry in healthy adults. Psychiatry Res 155, 173–177, doi: 10.1016/j.pscychresns.2007.01.005 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera M. et al. Putative role of the COMT gene polymorphism (Val158Met) on verbal working memory functioning in a healthy population. Am J Med Genet B Neuropsychiatr Genet 147B, 898–902, doi: 10.1002/ajmg.b.30705 (2008). [DOI] [PubMed] [Google Scholar]
- Damoiseaux J. S. et al. Consistent resting-state networks across healthy subjects. Proc Natl Acad Sci USA 103, 13848–13853, doi: 10.1073/pnas.0601417103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca M., Beckmann C. F., De Stefano N., Matthews P. M. & Smith S. M. fMRI resting state networks define distinct modes of long-distance interactions in the human brain. Neuroimage 29, 1359–1367, doi: 10.1016/j.neuroimage.2005.08.035 (2006). [DOI] [PubMed] [Google Scholar]
- Mantini D., Perrucci M. G., Del G. C., Romani G. L. & Corbetta M. Electrophysiological signatures of resting state networks in the human brain. Proc Natl Acad Sci USA 104, 13170–13175, doi: 10.1073/pnas.0700668104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J. D. et al. Functional network organization of the human brain. Neuron 72, 665–678, doi: 10.1016/j.neuron.2011.09.006 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M. et al. Catechol O-methyltransferase mRNA expression in human and rat brain: evidence for a role in cortical neuronal function. Neuroscience 116, 127–137 (2003). [DOI] [PubMed] [Google Scholar]
- Usiello A. et al. Distinct functions of the two isoforms of dopamine D2 receptors. Nature 408, 199–203, doi: 10.1038/35041572 (2000). [DOI] [PubMed] [Google Scholar]
- Moyer R. A. et al. Intronic polymorphisms affecting alternative splicing of human dopamine D2 receptor are associated with cocaine abuse. Neuropsychopharmacology 36, 753–762, doi: 10.1038/npp.2010.208 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. et al. Polymorphisms in human dopamine D2 receptor gene affect gene expression, splicing, and neuronal activity during working memory. Proc Natl Acad Sci USA 104, 20552–20557, doi: 10.1073/pnas.0707106104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolino A. et al. Functional variants of the dopamine receptor D2 gene modulate prefronto-striatal phenotypes in schizophrenia. Brain 132, 417–425, doi: 10.1093/brain/awn248 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouge-Pont F. et al. Changes in extracellular dopamine induced by morphine and cocaine: crucial control by D2 receptors. J Neurosci 22, 3293–3301, doi: 20026322 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler M. E. & Buckner R. L. Functional dissociation among components of remembering: control, perceived oldness, and content. J Neurosci 23, 3869–3880 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampson M., Driesen N. R., Skudlarski P., Gore J. C. & Constable R. T. Brain connectivity related to working memory performance. J Neurosci 26, 13338–13343, doi: 10.1523/jneurosci.3408-06.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield-Gabrieli S. et al. Associations and dissociations between default and self-reference networks in the human brain. Neuroimage 55, 225–232, doi: 10.1016/j.neuroimage.2010.11.048 (2011). [DOI] [PubMed] [Google Scholar]
- Broyd S. J. et al. Default-mode brain dysfunction in mental disorders: a systematic review. Neurosci Biobehav Rev 33, 279–296, doi: 10.1016/j.neubiorev.2008.09.002 (2009). [DOI] [PubMed] [Google Scholar]
- Liu B. et al. Prefrontal-related functional connectivities within the default network are modulated by COMT val158met in healthy young adults, J Neurosci 30, 64–69, doi: 10.1523/JNEUROSCI.3941-09.2010 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambataro F. et al. DRD2 genotype-based variation of default mode network activity and of its relationship with striatal DAT binding. Schizophr Bull 39, 206–216, doi: 10.1093/schbul/sbr128 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L. C., O’Neil J. P. & Jagust W. J. Genetic effects on behavior are mediated by neurotransmitters and large-scale neural networks. Neuroimage 66, 203–214, doi: 10.1016/j.neuroimage.2012.10.090 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley W. W. et al. Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci 27, 2349–2356, doi: 10.1523/jneurosci.5587-06.2007 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon V. & Uddin L. Q. Saliency, switching, attention and control: a network model of insula function. Brain Struct Funct 214, 655–667, doi: 10.1007/s00429-010-0262-0 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima K. & Tanji J. Role for cingulate motor area cells in voluntary movement selection based on reward. Science 282, 1335–1338 (1998). [DOI] [PubMed] [Google Scholar]
- Montag C., Weber B., Jentgens E., Elger C. & Reuter M. An epistasis effect of functional variants on the BDNF and DRD2 genes modulates gray matter volume of the anterior cingulate cortex in healthy humans. Neuropsychologia 48, 1016–1021, doi: 10.1016/j.neuropsychologia.2009.11.027 (2010). [DOI] [PubMed] [Google Scholar]
- Stice E., Yokum S., Burger K., Epstein L. & Smolen A. Multilocus genetic composite reflecting dopamine signaling capacity predicts reward circuitry responsivity. J Neurosci 32, 10093–10100, doi: 10.1523/jneurosci.1506-12.2012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig M. V., Antzoulatos E. G. & Miller E. K. Prefrontal dopamine in associative learning and memory. Neuroscience 282C, 217–229, doi: 10.1016/j.neuroscience.2014.09.026 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbetta M. & Shulman G. L. Control of goal-directed and stimulus-driven attention in the brain. Nat Rev Neurosci 3, 201–215, doi: 10.1038/nrn755 (2002). [DOI] [PubMed] [Google Scholar]
- Corbetta M., Patel G. & Shulman G. L. The reorienting system of the human brain: from environment to theory of mind. Neuron 58, 306–324, doi: 10.1016/j.neuron.2008.04.017 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opgen-Rhein C. et al. Executive attention in schizophrenic males and the impact of COMT Val108/158Met genotype on performance on the attention network test. Schizophr Bull 34, 1231–1239, doi: 10.1093/schbul/sbm155 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K. K., Schote A. B., Meyer J. & Frings C. Genes of the dopaminergic system selectively modulate top-down but not bottom-up attention. Cogn Affect Behav Neurosci 15, 104–116, doi: 10.3758/s13415-014-0320-9 (2015). [DOI] [PubMed] [Google Scholar]
- Gong P. et al. Genetic variations in COMT and DRD2 modulate attentional bias for affective facial expressions. PLoS One 8, e81446, doi: 10.1371/journal.pone.0081446 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.