Abstract
Tyrosine kinase inhibitors (TKIs) are anticancer drugs that may be co-administered with other drugs. The aims of this study are to investigate the inhibitory effects of TKIs on UDP-glucuronosyltransferase (UGT) activities, and to quantitatively evaluate their potential to cause drug-drug interactions (DDIs). Inhibition kinetic profiles of a panel of UGT enzymes (UGT1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B15, and 2B17) by four TKIs (axitinib, imatinib, lapatinib and vandetanib) were characterized by using hepatic microsomes and recombinant proteins. Lapatinib exhibited potent competitive inhibition against UGT1A1 activity with a Ki of 0.5 μM. Imatinib was found to exhibit broad inhibition on several UGTs, particularly potent competitive inhibition against UGT2B17 with a Ki of 0.4 μM. The TKIs also exerted intermediate inhibition against several UGTs (i.e., UGT1A7 by lapatinib; UGT1A1 by imatinib; UGT1A4, 1A7 and 1A9 by axitinib; and UGT1A9 by vandetanib). Results from modeling for the quantitative prediction of DDI risk indicated that the coadministration of lapatinib or imatinib at clinical doses could result in a significant increase in AUC of drugs primarily cleared by UGT1A1 or 2B17. Lapatinib and imatinib may cause clinically significant DDIs when co-administered UGT1A1 or 2B17 substrates.
Tyrosine-kinase inhibitors (TKIs) are anticancer drugs. Tyrosine kinases phosphorylate the tyrosine residues of proteins involved in the activation of signal transduction cascades that play key roles in biological processes including growth, differentiation and apoptosis in cancer cells1. Currently, more than 20 FDA-approved TKIs are used clinically. More than 80% of cancer cases are developed in patients older than 60 years old2 who typically have other medical conditions that require drug treatment3. As a result, TKIs have been commonly combined with other drugs in cancer patients4,5, and drug-drug interaction (DDI) involving TKIs is a potential clinical concern.
UDP-glucuronosyltransferases (UGT), a class of phase II enzymes, catalyze the conjugation of glucuronic acid to endogenous substances and exogenous compounds. UGT-catalyzed glucuronidation reactions account for approximately 35% of drugs eliminated by phase II enzymes (or one-seventh of the drugs prescribed in the United States in 2002)6. The human UGT superfamily involved in xenobiotics metabolism is comprised of 2 families: UGT1 and UGT27. UGT1A1, 1A3, 1A4, 1A6, 1A9, 2B7 and 2B15 are the main UGTs responsible for drug metabolism8 while UGT1A7, 1A8, 1A10 and 2B4 have also been found to metabolize drugs including mycophenolic acid and troglitazone9. Most UGT isoforms are expressed in liver except UGT1A7, 1A8 and 1A10 that are expressed mainly in intestines10,11.
Previous in vitro and in vivo studies indicate that TKIs may alter the hepatic elimination of co-administered drugs by inhibiting their metabolism. For example, nilotinib and erlotinib inhibit UGT1A1 activity, and gefitinib inhibits UGT1A1, 1A7, 1A9 and 2B7 activities12,13,14,15. A clinical study also showed that co-administration of lapatinib with irinotecan led to a ~40% increase in the AUC of SN-38 (an active metabolite of irinotecan and a UGT1A1 substrate)16, suggesting the possible inhibition of UGT1A1 activity by lapatinib. However, whether these TKIs affect activities of others UGT isoforms and whether other TKIs affect UGTs remain unknown.
In this study, four commonly used TKIs−axitinib, imatinib, lapatinib and vandetanib (Fig. 1)−were evaluated for their capabilities to inhibit UGT activities. The inhibition kinetics of each compound was further characterized, and the risks for clinically significant drug-drug interactions were estimated.
Figure 1. Chemical structures of axitinib, imatinib, lapatinib, and vandetanib.
Results
Inhibition of UGT Activity by TKIs
As a preliminary study, we first examined whether TKIs inhibit different UGTs. To this end, axitinib, imatinib, lapatinib, or vandetanib (or vehicle control) was incubated with a UGT substrate (4-methylumbelliferone (4-MU) for all UGTs except for UGT1A4; trifluoperazine (TFP) was used for UGT1A4) and one of recombinant UGT enzymes (UGT1A1, 1A3, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B15, and 2B17). Then, the extent of glucuronide metabolite production was examined. The results showed that at 100 μM concentration, TKIs inhibited the activity of UGT isoforms to varying extent (Table 1). For UGT isoforms whose activity is inhibited by >50% by individual TKIs, IC50 values of TKIs were further estimated. The summary of IC50 values is shown in Table 2.
Table 1. Remaining activities (%) of UGTs inhibited by 100 μM TKIs.
| Lapatinib | Vandetanib | Imatinib | Axitinib | Pos. Ctrl. | |
|---|---|---|---|---|---|
| UGT1A1 | 30.8 ± 11.0 | 48.9 ± 4.9 | 34.1 ± 0.5 | 56.7 ± 3.4 | 37.0 ± 2.1 |
| UGT1A3 | 96.5 ± 7.5 | 104.1 ± 2.1 | 88.8 ± 5.1 | 47.8 ± 12.8 | 39.4 ± 6.8 |
| UGT1A4a | 40.7 ± 1.9 | 62.8 ± 0.0 | 59.1 ± 3.0 | 14.9 ± 1.8 | 20.3 ± 6.6 |
| UGT1A6 | 88.4 ± 5.9 | 55.1 ± 1.1 | 67.9 ± 7.1 | 85.8 ± 3.5 | 28.2 ± 10.0 |
| UGT1A7 | 37.7 ± 9.3 | 22.7 ± 2.7 | 8.5 ± 3.1 | 25.0 ± 2.5 | 25.6 ± 11.2 |
| UGT1A8 | 73.5 ± 10.6 | 56.0 ± 6.3 | 33.1 ± 9.0 | 54.6 ± 2.4 | 40.2 ± 5.3 |
| UGT1A9 | 51.1 ± 7.9 | 14.1 ± 4.7 | 49.2 ± 1.9 | 44.8 ± 0.2 | 15.2 ± 1.2 |
| UGT1A10 | 73.6 ± 4.8 | 49.0 ± 0.4 | 15.3 ± 9.5 | 55.5 ± 2.7 | 37.8 ± 8.0 |
| UGT2B4 | 105.1 ± 0.2 | 80.8 ± 6.6 | 71.0 ± 3.2 | 42.1 ± 1.6 | NDb |
| UGT2B7 | 90.5 ± 1.0 | 49.8 ± 3.7 | 29.8 ± 5.6 | 55.9 ± 9.7 | 16.4 ± 11.9 |
| UBT2B15 | 109.5 ± 3.1 | 79.2 ± 1.4 | 28.3 ± 3.3 | 49.3 ± 5.2 | 21.6 ± 11.9 |
| UBT2B17 | 94.8 ± 5.7 | 69.7 ± 3.1 | 14.7 ± 8.2 | 55.9 ± 0.3 | ND |
aTFP was used as a substrate, and for all other UGTs, 4-MU was used as substrate.
bno positive inhibitor available.
Table 2. The IC50 of TKIs for the inhibition of UGT activities.
| Axitinib (μM) | Imatinib (μM) | Lapatinib (μM) | Vandetanib (μM) | |
|---|---|---|---|---|
| UGT1A1 | NDa | 11.0 ± 2.8 | 0.5 ± 0.0 | ND |
| UGT1A3 | ND | ND | ND | ND |
| UGT1A4b | 14.2 ± 3.2 | ND | 3.8 ± 0.7 | ND |
| UGT1A7 | 3.3 ± 0.7 | 19.5 ± 4.1 | 2.7 ± 1.4 | 38.9 ± 4.3 |
| UGT1A8 | ND | 22.9 ± 3.8 | ND | ND |
| UGT1A9 | 3.6 ± 0.7 | ND | ND | 5.0 ± 0.4 |
| UGT1A10 | ND | 47.3 ± 7.6 | ND | ND |
| UGT2B7 | ND | 68.4 ± 10.8 | ND | ND |
| UGT2B15 | ND | 91.0 ± 18.3 | ND | ND |
| UGT2B17 | ND | 0.8 ± 0.1 | ND | ND |
anot determined; based on results from preliminary experiments showing that the extent of UGT inhibition at 100 μM of a respective TKI was <50%.
bTFP was used as a substrate.
UGT Inhibition Kinetics of TKIs
Except axitinib whose maximum plasma concentration (Cmax) is in the sub-micromolar range17, the Cmax values of the remaining TKIs tested in this study are at the micromolar level at typical oral doses. For TKIs that exhibit IC50 values lower or similar to the reported plasma Cmax (i.e., imatinib against UGT1A1 and 2B17; lapatinib against UGT1A1, 1A4, and 1A7; and vandetinib against UGT1A9), their UGT inhibitory kinetics profiles were further characterized.
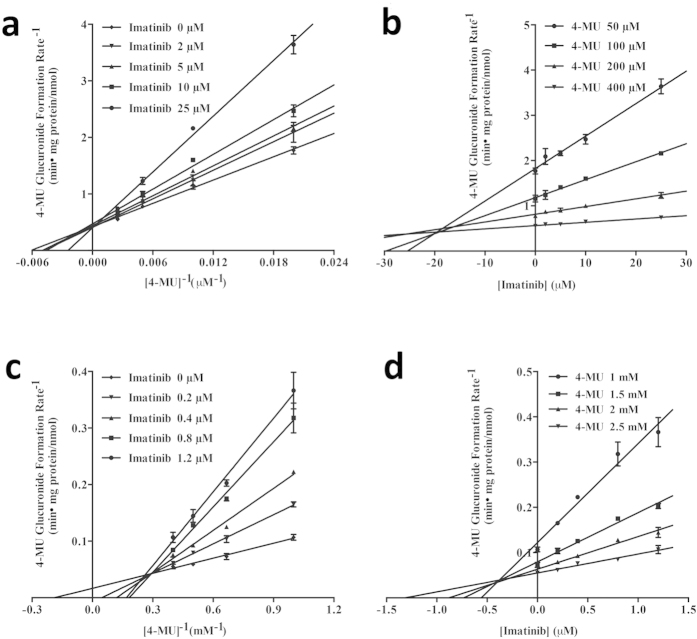
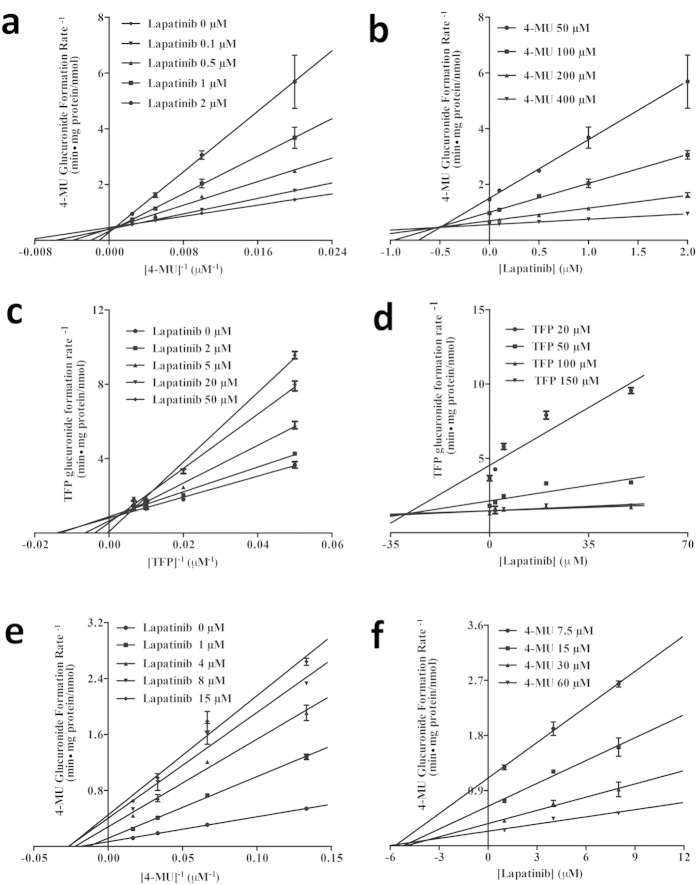
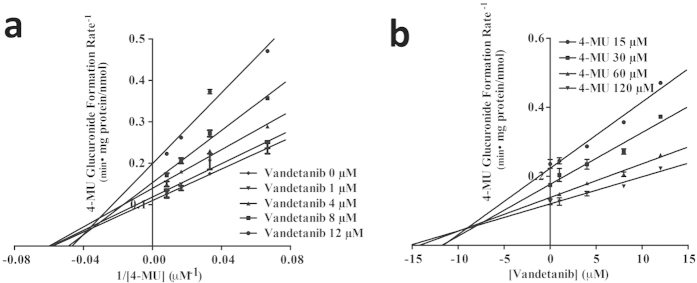
The kinetic results were shown in Table 3. Imatinib inhibited UGT1A1 and 2B17 activities (Fig. 2) by a competitive inhibitory mechanism. Ki values of imatinib were estimated to be 19 μM and 0.4 μM against the UGT1A1 and UGT2B17, respectively. Lapatinib (Fig. 3) exhibited a potent competitive inhibition against UGT1A1 with a Ki of 0.5 μM, a mixed inhibition against UGT1A4 with a Ki of 36.4 μM, a non-competitive inhibition with a Ki of 4.5 μM against UGT1A7. Vandetanib showed a mixed inhibition against UGT1A9 with a Ki of 9.0 μM (Fig. 4).
Table 3. The inhibition kinetic parameters and types of TKIs on UGTs.
| enzymesresource | Substrate |
Ki (μM)/inhibition type |
||
|---|---|---|---|---|
| Imatinib | Lapatinib | Vandetanib | ||
| UGT1A1 | 4-MU | 19.1 ± 2.8/competitive | 0.5 ± 0.1/competitive | – |
| UGT1A1 | SN-38 | – | 0.6 ± 0.1/competitive | – |
| UGT1A4 | TFP | – | 36.4 ± 5.0/mixed | – |
| UGT1A7 | 4-MU | – | 4.5 ± 0.5/noncompetitive | – |
| UGT1A9 | 4-MU | – | – | 9.0 ± 1.2/mixed |
| UGT2B17 | 4-MU | 0.4 ± 0.1/competitive | – | – |
| HLMs | SN-38 | – | 1.6 ± 0.2/noncompetitive | – |
Figure 2.
Lineweaver-Burk and Dixon plots for imatinib inhibition of 4-MU glucuronidation by UGT1A1 (a and b) and UGT2B17 (c and d) proteins.Data shown are the mean ± standard deviation of triplicate experiment.
Figure 3.
Lineweaver-Burk and Dixon plots for lapatinib inhibition of 4-MU (for UGT1A1, (a) and (b); and UGT1A7, (e) and (f)) or TFP (for UGT1A4, (c) and (d)) glucuronidation. Data shown are the mean ± standard deviation of triplicate experiment.
Figure 4.
Lineweaver-Burk (a) and Dixon plot (b) for vandetinib inhibition of 4-MU glucuronidation by UGT1A9 proteins. Data shown are the mean ± standard deviation of triplicate experiment.
Inhibition SN-38 Glucuronidation by Lapatinib
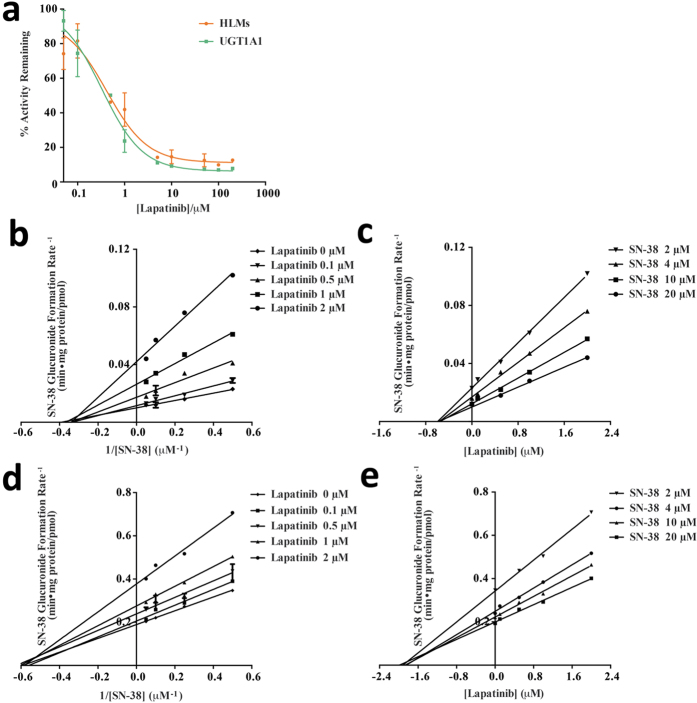
Because UGT1A1 inhibition can exhibit substrate-dependency18, the inhibition of UGT1A1 activity by lapatinib was further examined using SN-38, a drug mainly metabolized by UGT1A119. IC50 values were estimated for the inhibition of SN-38 glucuronidation by lapatinib in human microsomes and recombinant UGT1A1 proteins, and their kinetic parameters were estimated. Lapatinib showed strong inhibition against SN-38 glucuronidation, the IC50 of which was 0.33 μM in UGT1A1 and 0.42 μM in human liver microsomes (HLMs) (Fig. 5a). The estimated Ki values were 0.6 μM and 1.6 μM for UGT1A1 and HLMs, respectively, which were similar to the Ki values for the inhibition of 4-MU glucuronidation by lapatinib. Interestingly, however, the kinetic profile of inhibition of SN-38 glucuronidation was different from that of 4-MU glucuronidation; while inhibition of 4-MU glucuronidation by lapatinib was competitive, the inhibition of SN-38 glucuronidation by lapatinib (Fig. 5b–e) was non-competitive in both UGT1A1 and HLMs (Table 3).
Figure 5.
(a) Inhibition of SN-38 glucuronidation by lapatinib in human liver microsomes (HLM) or by recombinant UGT1A1 proteins. (b–e) Lineweaver-Burk and Dixon plots for lapatinib inhibition of SN-38 glucuronidation by UGT1A1 (b,c) or in human microsomes (d,e). Data shown are the mean ± standard deviation of triplicate experiment.
Quantitative Prediction of DDI Potential
The risk of DDIs due to the inhibition of UGTs by TKIs was evaluated by predicting the ratio of area under the plasma drug concentration versus time curve (AUC) of a victim drug in the presence and absence of the TKI (i.e., AUCi/AUC). To this end, the hepatic or systemic concentrations of each TKI were first estimated using previously reported clinical pharmacokinetic data (Table 4). For the inhibition of UGT1A1, 1A9, and 2B17, the maximum unbound hepatic input concentrations of TKIs were estimated. For the inhibition of UGT1A7, which is mainly expressed in intestines, systemic drug concentration was estimated. The AUC ratios of each inhibitor were estimated based on Ki values obtained in this study and the calculated hepatic or plasma concentrations of TKIs. The AUC changes of the UGT2B17 substrate by imatinib (at the time for Cmax) were predicted to be significant. For example, UGT2B17 inhibition by imatinib was predicted to increase the AUC of 4-MU by 140% when the fraction of the UGT substrate metabolized by the inhibited enzyme (fm) of a victim drug was 90%, which was much greater than the cut-off value for potential clinical significance (i.e., 25%)20. Also, lapatinib was predicted to cause ~25% increase in AUC of UGT1A1 substrates (4-MU and SN-38) when the fm is 1, indicating a potential DDI risk with clinical significance. On the other hand, axitinib could not cause the increase in AUC of UGT1A4, 1A7, or 1A9 substrates when the fm is 1, and the AUCi/AUC ratios were close to 1. UGT1A9 inhibition by Vandetanib, UGT1A1 inhibition by imatinib, and UGT1A7 inhibition by lapatinib were predicted to increase the AUC of substrate by about 2%, 3%, and 1%, indicating minor clinical impacts of the TKIs on the pharmacokinetics of other UGT substrates.
Table 4. Pharmacokinetic parameters of TKIs used in the estimation of AUC changes.
| Drugs | Axitinib | Imatinib | Lapatinib | Vandetanib | |
|---|---|---|---|---|---|
| Dose (mg) | 5 | 400 | 1250 | 300 | |
| Dosing Interval (hr) | 12 | 24 | 24 | 24 | |
| Cmax (μM) | 0.16 | 4.41 | 4.2 | 3.32 | |
| Absorption Rate Constant (ka, h-1) | 0.523 | 1.64 | 0.7 | 0.3 | |
| Fraction absorbed (Fa) | 0.88 | 1 | 0.73 | 0.935 | |
| Plasma Unbound Fraction (fu,p) | 0.01 | 0.05 | 0.01 | 0.06 | |
| Oral CL (L/h) | 45 | 11.2 | 114 | 13.2 | |
| t1/2 (h) | 3.2 | 22 | 24 | 456 | |
| Calculated Concentrations | Iav (μM) | 0.024 | 2.52 | 0.79 | 1.99 |
| Imax,u (μM) | 0.00067 | 0.18 | 0.011 | 0.12 | |
| Iin,u (μM) | 0.00086 | 0.7 | 0.12 | 0.23 | |
| References | 17,40,45 | 38,39,46 | 41,42 | 43,44 | |
Discussion
In recent years, a number of TKIs of different classes have been approved for cancer treatment. Among these TKIs, several TKIs of the 4-anilinoquinazoline class (erlotinib and gefitinib) and pyridine class (sorafenib) have been shown to inhibit UGTs12,13,15,21. In the present study, we selected four commonly used TKIs, including two 4-anilinoquinazolines (lapatinib and vandetanib), one pyrimidine (imatinib) and one indazol (axitinib) to investigate whether these TKIs could inhibit a panel of UGTs activities. To this end, 4-MU was used as a global probe substrate for UGT enzymes (except for UGT1A4) based on the following. The kinetic behavior of 4-MU glucuronidation mediated by different UGT enzymes has been fully characterized22. Also, specific probe substrates for most individual UGTs have not been identified yet23 while 4-MU has been demonstrated to be a robust probe substrate in assessing the activity of most recombinant human UGTs in many studies12,24,25. Thus, characterization of UGT inhibition using 4-MU as a probe compound should provide solid preliminary data to estimate UGT inhibitory potentials of perpetrator drugs.
Important inhibition is uncommon for compounds with IC50 values greater than about 75–100 μM because sufficiently high levels are not clinically achieved26. So in the current study, for UGT isoforms whose activity is inhibited by >50% by individual TKIs at 100 μM, IC50 and Ki values of TKIs were further estimated. Most notable finding in our study was UGT inhibition by imatinib and lapatinib. Our study has revealed that imatinib is a broad inhibitor of several UGTs. Imatinib is a potent inhibitor of UGT2B17; an intermediate inhibitor of UGT1A1; and a weak inhibitor of UGT1A7, 1A8, 1A9, 1A10, 2B7, and 2B15. The human UGT2B enzymes are involved in the metabolism of steroid hormones as well as bile acids, acidic steroids, and fatty acids27. Among UGT2Bs, UGT2B17 exhibits high levels of activity against androgens and xenobiotics. UGT2B17 substrate drugs include carcinogens, coumarins, anthraquinones, flavonoids, nonsteroidal anti-inflammatory drugs, monoterpenoids, phenols, suberoylanilide hydroxamic acid (SAHA), and exemestane28,29,30. Therefore, the potent inhibition of UGT2B17 by imatinib could potentially have a significant clinical effect on the metabolism of UGT2B17 substrates. In fact, our results from quantitative prediction of DDI risk indicate that the co-administration of imatinib at clinical doses can result in over 2-fold increases in AUC of a UGT2B17 substrate drug.
Our data offer in vitro evidence that lapatinib is a potent inhibitor of UGT1A1. UGT1A1 is broadly expressed in human organs including liver, intestines, and kidney31,32,33; its expression levels in the intestines and kidney are one third as high as that in liver11. About 15% of top 200 prescribed drugs in the United States in 2002 are eliminated mainly via glucuronidation by UGT1A16, and the inhibition of UGT1A1 can have clinically significant impacts on drug therapy with a narrow therapeutic index drug such as irinotecan. Irinotecan is a chemotherapeutic agent commonly used for the treatment of colorectal cancer. Irinotecan requires metabolic activation by carboxylesterase 2 to the active metabolite SN-38. SN-38 is mainly eliminated via glucuronidation by UGT1A1, with minor contribution by UGT1A3, 1A6, and 1A919. Because UGT1A1 inhibition can exhibit substrate-dependency18, two UGT1A1 substrates, 4-MU and SN-38, were used in the kinetic study of lapatinib. Interestingly, the kinetic profile of inhibition of SN-38 glucuronidation was different from that of 4-MU glucuronidation. This finding offers new experimental evidence that the reduction of UGT1A1 activity might vary with the substrate, and also for the opinion that UGT1A1 has two or more binding sites for xenobiotics and endobiotics18. Our results from the quantitative prediction of DDI risk indicate that the coadministration of lapatinib at clinical doses could result in around 25% increase in AUC of SN-38. Because the extent of UGT inhibition in in vivo systems can be underestimated when the prediction is based solely on in vitro parameters34, our prediction of SN-38 and lapatinib interaction appears consistent with the clinical observation that co-administration of lapatinib increased the AUC of SN-38 by an average of 41%16.
Different from above two TKIs, vandetanib was found as an intermediate inhibitor of UGT1A9, which is also involved in the glucuronidation of a number of drugs9. UGT1A9 is expressed in both human liver and some extrahepatic tissues including the gastrointestinal tract9. Significant evidence exists supporting a role for gastrointestinal UGTs as modifiers of pharmacokinetics and biological responses to drugs and xenobiotics35. Our data show that vandetanib might influence the first-pass effect and bioavailability of more orally administered drugs. This issue requires additional attentions, especially when modern medicinal chemistry tends to synthesize polar chemicals to avoid the metabolism by CYPs, which makes Phase II enzymes become the main metabolizing enzymes such as UGTs.
It is noteworthy that the extrapolation from in vitro data to in vivo drug interactions should be taken with caution. Various factors may affect the accuracy of prediction, including enzyme resources, incubation systems, protein binding, active transporters, the selection of probe substrates, and the accumulation of products in the reaction medium so on. In addition, UGTs in the gastrointestinal tract may contribute significantly to the first-pass metabolism of orally administered drugs that undergo glucuronidation. The concentrations of substrates in the intestine may be different from the concentrations used here to predict the AUC ratio. Therefore, further systemic studies are needed to clarify the in vivo effects.
In summary, the present findings highlight potential DDIs involving specific TKIs and UGT substrates, namely by inhibition of UGT1A1 and 2B17 by lapatinib and imatinib, respectively. Our study provides a basis for design of clinical studies for investigation of DDIs involving TKIs.
Methods
Chemicals and Reagents
Axitinib, imatinib, lapatinib, vandetanib, SN-38 and SN-38 glucuronide (SN-38G) were purchased from Toronto Research Chemicals, Inc (Toronto, Ontario, Canada). Tris-HCl, 4-methylumbelliferone (4-MU), 4-methylumbelliferone-β-D-glucuronide (4-MUG), alamethicin and uridine 5-diphosphoglucuronic acid trisodium salt (UDPGA) were purchased from Sigma-Aldrich (St. Louis, MO). Trifluoperazine (TFP) was purchased from Enzo Life Science (Farmingdale, NY). All other reagents were of high-performance liquid chromatography (HPLC) grade or the highest grade commercially available. A panel of recombinant human UGT supersomes (UGT1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B15, and 2B17) expressed in baculovirusinfected insect cells was purchased from BD Gentest (Woburn, MA). Pooled human liver microsomes (HLMs) (n = 50 donors) were purchased from Invitrogen (Carlsbad, CA).
Inhibition of UGT Glucuronidation Assay
The nonselective substrate of UGTs, 4-MU, was used as a probe substrate for all UGT isoforms except for UGT1A4. A typical incubation mixture with a total volume of 100 μl contained recombinant UGT isoform (final concentration: 0.1, 0.15, 0.05, 0.01, 0.01, 0.01, 0.01, 0.5, 0.05, 0.15 and 0.1 mg/ml for recombinant UGT1A1, 1A3, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B15, and 2B17, respectively), 10 mM UDPGA, 5 mM MgCl2, 100 mM Tris-HCl buffer (pH 7.4), and 4-MU in the absence or presence of a TKI (100 μM). Incubations with 4-MU were performed at the final concentration corresponding to the apparent Km or S50 value reported for each isoform (110, 1200, 110, 15, 750, 30, 80, 1200, 350, 250, and 2000 μM 4-MU for UGT1A1, 1A3, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B15, and 2B17, respectively)12. For UGT1A4, TFP was selected as the substrate. TFP (final concentration: 40 μM) was incubated with one of four TKIs and UGT1A4 (0.1 mg/ml). The following known UGT inhibitors were used as positive controls (at 100 μM): diclofenac for UGT1A1, 1A6, 1A7, and 1A9; hecogenin for UGT1A4, androsterone for UGT1A3, 2B7, and 2B15; and phenylbutazone for UGT1A8 and 1A10, respectively12,22,34. As there are no known inhibitors of UGT2B4 and 2B17, positive controls for these enzymes were not included. All the inhibitors and 4-MU or TFP were dissolved in DMSO. The final concentration of DMSO in the incubation system was 1% (v/v). The UGT reaction mixtures were pre-incubated at 37 °C for 5 min, and reactions were initiated by adding UDPGA (final concentration 5 mM). The following incubation times were used for different UGT isoforms: 120 min for UGT1A1, 1A10, 2B4, 2B7, 2B15, and 2B17; 75 min for UGT1A3; and 30 min for UGT1A4, UGT1A6, 1A7, 1A8, and 1A9. Rate of product formation for each isoform was linear with respect to protein concentration and incubation time. All the reactions were quenched by adding chilled acetonitrile (100 μl) containing phenytoin (1 μM) as an internal standard. The mixtures were centrifuged at 16,000 g for 16 min to obtain the supernatant. Samples were then analyzed in LC-MS/MS. The relative rate of 4-MUG or trifluoperazine glucuronide (TFPG) formation for each isoform was linear with respect to protein concentration and incubation time.
4-MUG concentrations were determined by using an Agilent 1200 HPLC interfaced with Applied Biosystems Qtrap 3200 equipped with an electrospray ion source. Chromatographic separation was carried out with a Waters XTerra MS C18 column (2.1 × 50 mm, 3.5 μm; Waters Corporation, Milford, MA). The mobile phase consisted of 0.1% formic acid in water (A) and acetonitrile (B) and was delivered at 250 μl/min. The gradient was initiated at 85% mobile phase A to 90% mobile phase B over 1 min, held constant for 1.5 min, and then restored to the initial composition until 10 min. The MRM parameters for 4-MUG and phenytoin as the internal standard were 353.05/177.20 and 253.30/182.20 in positive ion mode.
Signals for TFPG were detected by using Applied Biosystems Qtrap 4000 with an electrospray ion source interfaced with a Shimazu Prominence Modular HPLC. The mobile phase consisted of 0.1% formic acid in water (A) and acetonitrile (B). The column was first equilibrated at 25% mobile phase B for 1.5 min at a 200 μl/min flow rate. The elution was then ramped linearly to 90% mobile phase B over 1.5 min, maintained for 5 min, and followed by a return to initial conditions. TFPG was detected using MRM pair of 584/408 in positive ionization mode36. Phenytoin was used as the internal standard.
Inhibition of SN-38 Glucuronidation Assay
SN-38 glucuronidation activity was determined with a slight modification of a previously published method13. SN-38 was incubated in the absence or presence of different concentrations of lapatinib (0–200 μM). Reactions were performed using recombinant UGT1A1 protein (0.1 mg/ml) or pooled HLMs (0.4 mg/ml). When HLMs were used, 50 μg/mg protein alamethicin was added into incubation systems for 15 min preincubation on ice. Alamethicin was dissolved in 90/10 incubation buffer/ethanol. The final concentration of ethanol was less than 0.1%. After preincubation for 5 min, the reaction was started by adding UDPGA (final concentration 5 mM). Incubation was performed at 37 °C for 30 min and stopped by adding chilled acetonitrile. The samples were processed as described above for analysis using LC-MS/MS.
The concentration of SN-38G was determined by using an Agilent 1200 HPLC interfaced with Applied Biosystems Qtrap 5500. The MRM setting for SN-38G was 569.2/393.2 in the positive mode. The same internal standard was used for quantification. The mobile phase consisted of 0.1% formic acid in water (A) and acetonitrile (B). The column was first equilibrated at 10% mobile phase B for 1 min at 250 μl/min. The elution was then ramped linearly to 90% mobile phase B over 2 min, maintained for 2 min, and followed by a return to initial conditions.
Determination of IC50
The same incubation condition as for the preliminary inhibition study was used for each UGT isoform, except that the concentration of each inhibitor was ranged from 0 to 200 μM. The relative formation rate of 4-MUG, TFPG or SN-38G in the presence vs. absence of a TKI was estimated, and the half maximal inhibitory concentration (IC50) value for each TKI was estimated by using GraphPad Prism 5 software (La Jolla, CA).
Determination of K i
4-MU at four concentrations (ranging from 0.5 × Km to 4 × Km) was incubated with recombinant UGT1A1, 1A3, 1A7, 1A8, 1A9, 1A10, 2B7, 2B15 or 2B17 at a final protein concentration as described in the UGT glucuronidation inhibition assay in the presence or absence of TKI at four different concentrations (ranging from IC50/4 to 4 × IC50). The incubation time for each UGT isoform was the same as that used to determine IC50. SN-38 (2, 4, 10 or 20 μM) was incubated with UGT1A1 (0.1 mg/ml) or HLMs (0.4 mg/ml for) in the presence of lapatinib (0, 0.1, 0.5, 1 or 2 μM). Ki values were calculated using nonlinear regression according to the models for competitive inhibition, noncompetitive inhibition, or mixed inhibition. Global nonlinear regression analysis in the kinetics module of GraphPad Prism 5 software was used to simultaneously fit all data to the equations for competitive inhibition, non-competitive inhibition, and uncompetitive inhibition. The type of inhibition was determined based on Lineweaver-Burk plot and the best fitting of data to the equations. Goodness of fit to kinetic and inhibition models was assessed from the F statistic, r2 values, parameter standard error estimates and 95% confidence intervals. Kinetic constants are reported as the mean value ± standard error of the parameter estimate. Lineweaver-Burk plot and Dixon plot were also used as graphical representation of the type of inhibition and Ki.
Estimation of in vivo TKI Concentrations
The average systemic plasma concentration ([I]av), the maximum systemic plasma concentration ([I]max), the maximum unbound systemic plasma concentration ([I]max,u), the maximum hepatic input concentration ([I]in) and the maximum unbound hepatic input concentration ([I]in,u) of TKIs after repeated oral administration were estimated based on the previously reported equations37. The values of parameters needed were obtained from the literature37,38,39,40,41,42,43,44,45.
Prediction of DDI Risk from In Vitro Data
The magnitude of the inhibition of TKIs was estimated as the ratio of the area under the curve (AUCi/AUC) of UGT substrates in the presence and absence of the inhibitor. The ratio was calculated based on the eq. 1 for drugs orally administered37:
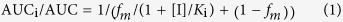 |
where AUCi and AUC are the AUC in the presence and absence of inhibitor, respectively; Ki is inhibition constant from the in vitro inhibition experiment; fm is the fraction of the UGT substrate metabolized by the inhibited enzyme; and [I] is the TKI concentration at the enzyme active site.
In view of the general assumption that only unbound drug is available for interaction with the enzyme active site, and the aim of DDI research is to prevent the highest potential risk, the maximum unbound hepatic input concentration ([I]in,u) was used for [I] in this study. Of note, clinically observed maximum unbound systemic plasma or blood concentration (Cmax,u) was not used because the Cmax,u is known to be much lower than the [I]in,u37. Also, for the UGTs expressed in gastrointestinal track (such as UGT1A7), the maximum unbound systemic plasma concentration ([I]max,u) was used to represent [I]. The prediction of AUC ratio is for any potential UGT substrates with undetermined fm, a range of fm between 0.1 and 1 was included for calculating AUCi/AUC ratio.
Additional Information
How to cite this article: Zhang, N. et al. Drug-Drug Interaction Potentials of Tyrosine Kinase Inhibitors via Inhibition of UDP-Glucuronosyltransferases. Sci. Rep. 5, 17778; doi: 10.1038/srep17778 (2015).
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81072698), the Fundamental Research Funds for the Central Universities, and the National Institutes of Health Eunice Kennedy Shriver National Institute of Child Health and Human Development [Grant HD065532].
Footnotes
Author Contributions N.Z., Y.L. and H.J. conceived and designed the experiments. N.Z. and Y.L. performed the experiments. All authors had roles in the collection, management, analysis and interpretation of the data, and preparation, review and approval of the article.
References
- Paul M. K. & Mukhopadhyay A. K. Tyrosine kinase-Role and significance in Cancer. Int J Med Sci 1, 101–115 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlader N. et al. SEER Cancer Statistics Review, 1975–2010. (National Cancer Institute, Bethesda, MD; 2013). [Google Scholar]
- Christensen K., Doblhammer G., Rau R. & Vaupel J. W. Ageing populations: the challenges ahead. Lancet 374, 1196–1208 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressler R. & Bahl J. J. Principles of drug therapy for the elderly patient. Mayo Clin Proc 78, 1564–1577 (2003). [DOI] [PubMed] [Google Scholar]
- Costa D. B. & Huberman M. S. Improvement of type 2 diabetes in a lung cancer patient treated with Erlotinib. Diabetes Care 29, 1711 (2006). [DOI] [PubMed] [Google Scholar]
- Williams J. A. et al. Drug-drug interactions for UDP-glucuronosyltransferase substrates: a pharmacokinetic explanation for typically observed low exposure (AUCi/AUC) ratios. Drug Metab Dispos 32, 1201–1208 (2004). [DOI] [PubMed] [Google Scholar]
- Izukawa T. et al. Quantitative analysis of UDP-glucuronosyltransferase (UGT) 1A and UGT2B expression levels in human livers. Drug Metab Dispos 37, 1759–1768 (2009). [DOI] [PubMed] [Google Scholar]
- Miners J. O., Mackenzie P. I. & Knights K. M. The prediction of drug-glucuronidation parameters in humans: UDP-glucuronosyltransferase enzyme-selective substrate and inhibitor probes for reaction phenotyping and in vitro-in vivo extrapolation of drug clearance and drug-drug interaction potential. Drug Metab Rev 42, 196–208 (2010). [DOI] [PubMed] [Google Scholar]
- Kiang T. K., Ensom M. H. & Chang T. K. UDP-glucuronosyltransferases and clinical drug-drug interactions. Pharmacol Ther 106, 97–132 (2005). [DOI] [PubMed] [Google Scholar]
- Strassburg C. P., Oldhafer K., Manns M. P. & Tukey R. H. Differential expression of the UGT1A locus in human liver, biliary, and gastric tissue: identification of UGT1A7 and UGT1A10 transcripts in extrahepatic tissue. Mol Pharmacol 52, 212–220 (1997). [DOI] [PubMed] [Google Scholar]
- Harbourt D. E. et al. Quantification of human uridine-diphosphate glucuronosyl transferase 1A isoforms in liver, intestine, and kidney using nanobore liquid chromatography-tandem mass spectrometry. Anal Chem 84, 98–105 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Ramirez J., House L. & Ratain M. J. Comparison of the drug-drug interactions potential of erlotinib and gefitinib via inhibition of UDP-glucuronosyltransferases. Drug Metab Dispos 38, 32–39 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Ramirez J., House L. & Ratain M. J. The UGT1A1*28 polymorphism correlates with erlotinib’s effect on SN-38 glucuronidation. Eur J Cancer 46, 2097–2103 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita K., Sugiyama M., Akiyama Y., Ando Y. & Sasaki Y. The small-molecule tyrosine kinase inhibitor nilotinib is a potent noncompetitive inhibitor of the SN-38 glucuronidation by human UGT1A1. Cancer Chemother Pharmacol 67, 237–241 (2011). [DOI] [PubMed] [Google Scholar]
- Li W., Xing Y. & Liu Y. Inhibition of SN-38 glucuronidation by gefitinib and its metabolite. Cancer Chemother Pharmacol 75, 1253–1260 (2015). [DOI] [PubMed] [Google Scholar]
- Midgley R. S. et al. A phase I and pharmacokinetic study of lapatinib in combination with infusional 5-fluorouracil, leucovorin and irinotecan. Ann Oncol 18, 2025–2029 (2007). [DOI] [PubMed] [Google Scholar]
- Pfizer Laboratories Div Pfizer Inc., INLYTA: Package Insert and Label Information. (2014) Available at: http://druginserts.com/lib/rx/meds/inlyta/. (Accessed: 15th October 2015). [Google Scholar]
- Udomuksorn W. et al. Influence of mutations associated with Gilbert and Crigler-Najjar type II syndromes on the glucuronidation kinetics of bilirubin and other UDP-glucuronosyltransferase 1A substrates. Pharmacogenet Genomics 17, 1017–1029 (2007). [DOI] [PubMed] [Google Scholar]
- Hanioka N. et al. Human liver UDP-glucuronosyltransferase isoforms involved in the glucuronidation of 7-ethyl-10-hydroxycamptothecin. Xenobiotica 31, 687–699 (2001). [DOI] [PubMed] [Google Scholar]
- USFDA Drug Interaction Studies—Study Design, Data Analysis, Implications for Dosing, and Labeling Recommendations. (USFDA, Silver Spring, MD 2012). [Google Scholar]
- Peer C. J. et al. Sorafenib is an inhibitor of UGT1A1 but is metabolized by UGT1A9: implications of genetic variants on pharmacokinetics and hyperbilirubinemia. Clin Cancer Res 18, 2099–2107 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchaipichat V. et al. Human udp-glucuronosyltransferases: isoform selectivity and kinetics of 4-methylumbelliferone and 1-naphthol glucuronidation, effects of organic solvents, and inhibition by diclofenac and probenecid. Drug Metab Dispos 32, 413–423 (2004). [DOI] [PubMed] [Google Scholar]
- Miners J. O., Knights K. M., Houston J. B. & Mackenzie P. I. In vitro-in vivo correlation for drugs and other compounds eliminated by glucuronidation in humans: pitfalls and promises. Biochem Pharmacol 71, 1531–1539 (2006). [DOI] [PubMed] [Google Scholar]
- Mano Y., Usui T. & Kamimura H. In vitro inhibitory effects of non-steroidal anti-inflammatory drugs on 4-methylumbelliferone glucuronidation in recombinant human UDP-glucuronosyltransferase 1A9–potent inhibition by niflumic acid. Biopharm Drug Dispos 27, 1–6 (2006). [DOI] [PubMed] [Google Scholar]
- Cao Y. F. et al. Drug-Drug Interactions Potential of Icariin and Its Intestinal Metabolites via Inhibition of Intestinal UDP-Glucuronosyltransferases. Evid Based Complement Alternat Med 2012, 395912 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel K. E. & Wilkinson G. R. In vitro and in vivo drug interactions involving human CYP3A. Annu Rev Pharmacol Toxicol 38, 389–430 (1998). [DOI] [PubMed] [Google Scholar]
- Radominska-Pandya A., Czernik P. J., Little J. M., Battaglia E. & Mackenzie P. I. Structural and functional studies of UDP-glucuronosyltransferases. Drug Metab Rev 31, 817–899 (1999). [DOI] [PubMed] [Google Scholar]
- Turgeon D., Carrier J. S., Chouinard S. & Belanger A. Glucuronidation activity of the UGT2B17 enzyme toward xenobiotics. Drug Metab Dispos 31, 670–676 (2003). [DOI] [PubMed] [Google Scholar]
- Balliet R. M. et al. Characterization of UGTs active against SAHA and association between SAHA glucuronidation activity phenotype with UGT genotype. Cancer Res 69, 2981–2989 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Chen G., Dellinger R. W., Sharma A. K. & Lazarus P. Characterization of 17-dihydroexemestane glucuronidation: potential role of the UGT2B17 deletion in exemestane pharmacogenetics. Pharmacogenet Genomics 20, 575–585 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter J. K., Crawford J. M. & Owens I. S. Cloning of two human liver bilirubin UDP-glucuronosyltransferase cDNAs with expression in COS-1 cells. J Biol Chem 266, 1043–1047 (1991). [PubMed] [Google Scholar]
- Strassburg C. P., Manns M. P. & Tukey R. H. Expression of the UDP-glucuronosyltransferase 1A locus in human colon. Identification and characterization of the novel extrahepatic UGT1A8. J Biol Chem 273, 8719–8726 (1998). [DOI] [PubMed] [Google Scholar]
- Strassburg C. P., Nguyen N., Manns M. P. & Tukey R. H. Polymorphic expression of the UDP-glucuronosyltransferase UGT1A gene locus in human gastric epithelium. Mol Pharmacol 54, 647–654 (1998). [PubMed] [Google Scholar]
- Uchaipichat V., Mackenzie P. I., Elliot D. J. & Miners J. O. Selectivity of substrate (trifluoperazine) and inhibitor (amitriptyline, androsterone, canrenoic acid, hecogenin, phenylbutazone, quinidine, quinine, and sulfinpyrazone) “probes” for human udp-glucuronosyltransferases. Drug Metab Dispos 34, 449–456 (2006). [DOI] [PubMed] [Google Scholar]
- Ritter J. K. Intestinal UGTs as potential modifiers of pharmacokinetics and biological responses to drugs and xenobiotics. Expert Opin Drug Metab Toxicol 3, 93–107 (2007). [DOI] [PubMed] [Google Scholar]
- Gagez A. L., Rouguieg-Malki K., Sauvage F. L., Marquet P. & Picard N. Simultaneous evaluation of six human glucuronidation activities in liver microsomes using liquid chromatography-tandem mass spectrometry. Anal Biochem 427, 52–59 (2012). [DOI] [PubMed] [Google Scholar]
- Ito K., Brown H. S. & Houston J. B. Database analyses for the prediction of in vivo drug-drug interactions from in vitro data. Br J Clin Pharmacol 57, 473–486 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunton L. L., Lazo J. S. & Parker K. L. Goodman & Gilman’s The Pharmacological Basis of Therapeutics (McGraw-Hill, New York; 2005). [Google Scholar]
- Peng B., Lloyd P. & Schran H. Clinical pharmacokinetics of imatinib. Clin Pharmacokinet 44, 879–894 (2005). [DOI] [PubMed] [Google Scholar]
- Rugo H. S. et al. Phase I trial of the oral antiangiogenesis agent AG-013736 in patients with advanced solid tumors: pharmacokinetic and clinical results. J Clin Oncol 23, 5474–5483 (2005). [DOI] [PubMed] [Google Scholar]
- GlaxoSmithKline LLC., TYKERB: Package Insert and Label Information. (2015) Available at: http://druginserts.com/lib/rx/meds/tykerb-1/. (Accessed: 15th October 2015). [Google Scholar]
- Smith D. A. et al. Effects of ketoconazole and carbamazepine on lapatinib pharmacokinetics in healthy subjects. Br J Clin Pharmacol 67, 421–426 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- EMA Assessment report of Caprelsa/vandetanib. (European Medicines Agency, London, UK; 2011). [Google Scholar]
- Martin P. et al. Pharmacokinetics of vandetanib: three phase I studies in healthy subjects. Clin Ther 34, 221–237 (2012). [DOI] [PubMed] [Google Scholar]
- Garrett M. et al. Population pharmacokinetic analysis of axitinib in healthy volunteers. Br J Clin Pharmacol 77, 480–492 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Erp N. P. et al. Influence of CYP3A4 inhibition on the steady-state pharmacokinetics of imatinib. Clin Cancer Res 13, 7394–7400 (2007). [DOI] [PubMed] [Google Scholar]