Abstract
While our understanding of the molecular mechanisms underlying cancer has significantly improved, most of our knowledge focuses on protein-coding genes that make up a fraction of the genome. Recent studies have uncovered thousands of long noncoding RNAs (lncRNAs) that populate the cancer genome. A subset of these molecules shows striking cancer- and lineage-specific expression patterns, suggesting they may be potential drivers of cancer biology and have utility as clinical biomarkers. Here, we discuss emerging modalities of lncRNA biology and their interplay with cancer-associated concepts, including epigenetic regulation, DNA damage and cell cycle control, microRNA silencing, signal transduction pathways, and hormone-driven disease. Additionally, we highlight the translational impact of lncRNAs, tools for their mechanistic investigation, and directions for future lncRNA research.
The emergence of lncRNAs in cancer
Cancer is a complex disease consisting of multiple factors that lead to the development of malignant tumors [1]. While much progress has been made in identifying the major contributors to cancer progression, the clinical picture remains bleak. Current research efforts aim to better understand the interplay between cancer cells, tumor microenvironments, and defense mechanisms involved in cancer development, immune evasion, and therapeutic susceptibility [1]. However, the majority of these studies focus on protein-coding genes as the crucial components in disease progression, overlooking the vast landscape of noncoding genes.
Among these noncoding transcripts are long noncoding RNAs (lncRNAs). LncRNAs are RNA species greater than 200 base pairs in length commonly characterized by poly-adenylation, splicing of multiple exons, promoter trimethylation of histone H3 at lysine 4 (H3K4me3), and transcription by RNA polymerase II [2, 3]. LncRNA-mediated biology has been implicated in a wide variety of cellular processes, including pluripotency in mouse embryonic stem cells [4] and X chromosome inactivation [5]. While some lncRNAs, such as XIST, appear to operate exclusively in the nucleus as regulators of gene expression [5, 6], others function predominantly in the cytoplasm to regulate signal transduction and the stability of mRNAs [7–9]. Several distinct mechanisms of lncRNA activity have been described. Most prominently, lncRNAs have been shown to collaborate with protein partners to form ribonucleoprotein complexes [10] (see Glossary). For example, XIST interacts with the Polycomb repressive complex 2 (PRC2), resulting in PRC2 recruitment and subsequent trimethylation of histone H3 at lysine 27 (H3K27me3) of the inactive X chromosome [11]. Air and Kcnq1ot1 bind to G9a, a histone H3 lysine 9 methylase, to regulate gene expression [12, 13]. ANRIL associates with PRC1 to regulate the INK4a locus [14]. LincRNA-p21 and PANDA are two p53-regulated lncRNAs that interact with hnRNP-K and NF-YA to regulate transcription [15, 16]. LncRNA-LET is downregulated across several cancers and functions by binding to and degrading nuclear factor 90 (NF90) protein, which enhances hypoxia-induced cancer cell invasion [17]. Given this tendency to engage proteins, lncRNAs are surfacing as decoys, scaffolds, and guides [18].
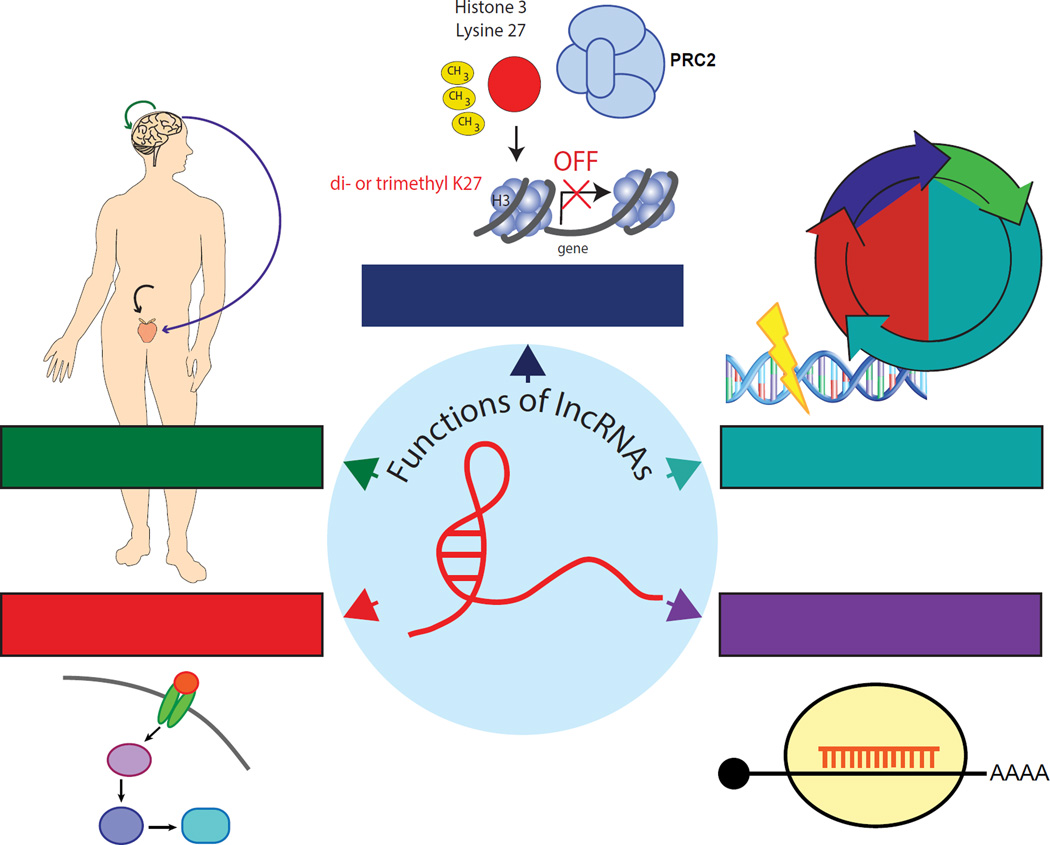
In cancer, lncRNAs are emerging as a prominent layer of previously underappreciated transcriptional regulation that function as both oncogenes and tumor suppressors [2] (Table 1). For example, overexpression of the HOTAIR lncRNA correlates with aggressive breast [19], colorectal [20], hepatocellular [21], and gastrointestinal stromal tumors [22], while lncRNA TARID prevents cancer formation through promoter demethylation at tumor suppressors [23]. In this review, we discuss emerging themes of lncRNA-mediated function within major areas of cancer progression and metastasis, focusing on advances made over the last several years (Fig. 1).
Table 1.
Examples of long noncoding RNAs in cancer
| Name | Cancer Type(s) | Tumor Suppressor/ Oncogene |
Mechanistic Theme(s) | Reference(s) |
|---|---|---|---|---|
| ANRIL (ANtisense noncoding RNA in the INK4 Locus) |
gastric | oncogene | cell cycle regulation, epigenetic complex, miRNA regulation |
14, 34, 85 |
| BANCR (BRAF Activated NonCoding RNA) |
melanoma, NSCLC | oncogene, tumor suppressor | chemokine signaling, EMT | 99–100 |
| BCAR4 (Breast Cancer Anti-estrogen Resistance-4) |
breast, multiple | oncogene | Hedgehog signaling pathway | 90, 104–105 |
| CARLo-5 (Cancer-Associated Region Long non-coding RNA-5) |
colorectal, gastric, NSCLC, prostate |
oncogene | apoptosis, cell cycle regulation, EMT | 65–67 |
| CCAT1 (Colon Cancer Associated Transcript 1) |
colorectal | oncogene | MYC | 119 |
| CCAT2 (Colon Cancer Associated Transcript 2) |
breast, colorectal, esophageal squamous cell, NSCLC |
oncogene | chromosomal instability, MYC, Wnt signaling pathway, |
92–95 |
| CTBP1-AS (C-Terminal Binding Protein 1 - AntiSense) |
prostate | oncogene | hormone-regulated | 113 |
| DRAIC (Downregulated-RNA in Androgen-Independent Cells) |
multiple, prostate | tumor suppressor | hormone-regulated | 112 |
| FAL1 (Focally Amplified LncRNA on chromosome 1) |
multiple epithelial type, ovarian | oncogene | cell cycle regulation, epigenetic complex |
31 |
| gadd7 (growth-arrested DNA damage- inducible lncRNA) |
non-specific | tumor suppressor | cell cycle regulation, DNA damage response |
68 |
| GAPLINC (Gastric Adenocarcinoma Predictive Long Intergenic NonCoding RNA) |
gastric | oncogene | ceRNA | 84 |
| GAS5 (Growth Arrest Specific 5) | non-specific, mesothelioma, prostate |
tumor suppressor | apoptosis | 71–72, 108 |
| H19 | colorectal, gastric, glioma, osteosarcoma, pancreatic |
oncogene | miRNA interaction, signal transduction | 76–80, 91 |
| HOTAIR (HOX Transcript Antisense Intergenic RNA) |
breast, colorectal, hepatocellular, GIST |
oncogene | epigenetic complex, miRNA regulation | 19–22 |
| HOTTIP (HOXA Transcript at the distal TIP) |
hepatocellular | oncogene | epigenetic complex | 53–54 |
| HULC (Hepatocellular Upregulated Long nonCoding RNA) |
hepatocellular | oncogene | ceRNA | 82, 126–127 |
| LED (LncRNA activator of Enhanced Domains) |
leukemia, non-specific | tumor suppressor | cell cycle regulation, epigenetic regulation |
63 |
| lincRNA-p21 | non-specific | tumor suppressor | cell cycle regulation, epigenetic regulation |
15, 59 |
| lncRNA-ATB (lncRNA-Activated by TGF- β) |
hepatocellular | oncogene | ceRNA, EMT, TGF-β signaling | 83 |
| lncRNA-HEIH (lncRNA-High Expression In HCC) |
hepatocellular | oncogene | cell cycle regulation, epigenetic complex |
33 |
| lncRNA-LET (Low Expression in Tumor) | colorectal, hepatocellular, lung squamous |
tumor suppressor | hypoxia, metastasis | 17 |
| lncTCF7 | hepatocellular | oncogene | epigenetic complex, Wnt signaling pathway |
42 |
| MALAT1 (Metastasis Associated Lung Adenocarcinoma Transcript-1) |
endometrioid endometrial, lung, renal cell |
oncogene | EMT, metastasis, Wnt signaling pathway |
96–98, 129–132 |
| MEG3 (Maternally Expressed 3) | colorectal, gastric, hepatocellular, meningioma, NSCLC |
tumor suppressor | DNA damage response, miRNA interaction |
60–61 |
| MIR31HG | melanoma | tumor suppressor | cell cycle regulation, OIS | 64 |
| NBAT-1 (NeuroBlastoma Associated Transcript-1) |
neuroblastoma | tumor suppressor | epigenetic complex | 32 |
| NEAT1 (Nuclear Enriched Abundant Transcript-1) |
breast, multiple solid type, prostate |
oncogene | epigenetic regulation, hormone- regulated, hypoxia |
106–107 |
| NKILA (NF-KappaB Interacting LncRNA) | breast | tumor suppressor | inflammation in tumor microenvironment, regulation of signal transduction |
122 |
| PANDA (P21 associated NcRNA DNA damage Activated) |
non-specific, leukemia | tumor suppressor | cell cycle regulation, DNA damage response |
16, 40 |
| PCAT-1 (Prostate Cancer Associated Transcript 1) |
prostate | oncogene | miRNA like function, regulation of DNA damage repair, repression of a tumor suppressor |
69–70, 86 |
| PCAT29 (Prostate Cancer Associated Transcript 29) |
prostate | tumor suppressor | hormone-regulated | 111–112 |
| PCGEM1 (prostate-specific transcript 1) | prostate | oncogene | hormone-regulated, MYC | 114–116, 118 |
| PRNCR1 (PRostate cancer associated Non Coding RNA 1) |
prostate | oncogene | hormone-regulated | 114–115 |
| PVT1 | colorectal | oncogene | MYC | 120 |
| SChLAP1 (Second Chromosome Locus Associated with Prostate 1) |
prostate | oncogene | epigenetic complex | 41, 50–52 |
| TARID (TCF21 Antisense RNA Inducing Demethylation) |
non-specific | tumor suppressor | DNA demethylation, epigenetic regulation |
23 |
| TUG1 (Taurine UpreGulated 1) | esophageal squamous cell, NSCLC |
oncogene | DNA damage response, epigenetic complex |
35 |
| XIST (X Inactive Specific Transcript) | breast, hematologic | tumor suppressor | epigenetic complex | 11,36 |
ceRNA (competing endogenous-RNA), EMT (epithelial-mesenchymal transition), GIST (gastrointestinal stromal tumor), NSCLC (non-small cell lung cancer), OIS (oncogene induced senescence)
Figure 1. LncRNAs play a crucial role within major areas of cancer progression and metastasis.
Long noncoding RNA (lncRNAs) mediate several cancer-associated processes, including epigenetic regulation, DNA damage and cell cycle control, microRNA (miRNA) silencing, signal transduction pathways, and hormone-driven disease.
Epigenetic regulation
Cancer results from an accumulation of modified genes, either by mutation or epigenetic alterations such as methylation, acetylation, and phosphorylation [24]. Growing evidence suggests key cellular genes involved in proliferation, apoptosis, and stem cell differentiation are epigenetically modified in cancer [25]. However, the mechanisms underlying precise epigenetic control are poorly understood.
An evolving model of lncRNA activity centers on their ability to bind and regulate epigenetic complexes [26]. Specifically, several lncRNAs have been shown to function by interacting with Polycomb group complexes [18]. This is especially relevant in cancer, as PRC1 and 2 are known oncogenic drivers in several types of malignancies [27–30]. For example, FAL1 (Focally Amplified LncRNA on chromosome 1), a novel oncogenic lncRNA present across several epithelial tumors, associates with BMI1, a core subunit of the PRC1 complex [31]. In ovarian cancer, FAL1 was shown to mediate cancer progression and was associated with decreased patient survival. FAL1’s interaction with BMI1 stabilizes the PRC1 complex by preventing BMI1 degradation, allowing PRC1 to occupy and repress promoters of target genes such as p21, resulting in loss of cell cycle regulation and increased tumorigenesis.
Similarly, NBAT-1, lncRNA-HEIH, HOTAIR, ANRIL, TUG1, and XIST have all been shown to interact with the enzymatic subunit of the PRC2 complex, EZH2, to modulate the repressive H3K27me3 histone mark on downstream target genes. This subsequently leads to either oncogenesis or tumor suppression in a multitude of cancer types, including neuroblastoma [32], hepatocellular [33], breast [19], gastric [34], non-small cell lung carcinoma (NSCLC) [35], and hematologic malignancies [36], respectively. In fact, up to 20% of all lncRNAs have been implicated in PRC2 binding [37], suggesting that PRC2 promiscuously binds to lncRNAs [38]. Recent studies have shown both specific and non-specific binding of PRC2 to lncRNAs, and emerging evidence suggests that these activities are not mutually exclusive [39]. However, the in vivo binding specificity of PRC2 remains to be elucidated.
One of the most studied lncRNAs, HOTAIR (HOX transcript antisense RNA), recruits the PRC2 complex to a set of genes involved in suppressing breast cancer metastasis [19]. This genome-wide retargeting of PRC2 results in repression of genes that prevent cancer progression. Additionally, HOTAIR-mediated genetic reprogramming results in gene expression signatures that resemble embryonic fibroblast gene signatures, which promotes cell migration, invasion, and metastasis. LncRNAs can also interact with Polycomb group complexes indirectly. For example, PANDA (P21 Associated ncRNA DNA Damage Activated) physically interacts with scaffold-attachment-factor-A (SAFA) to indirectly recruit both the PRC1 and PRC2 complexes to the promoters of genes involved in cellular senescence [40]. This suggests that lncRNAs can facilitate epigenetic changes through interaction with protein intermediates.
In addition to Polycomb group complexes, several lncRNAs have been linked to the SWI/SNF nucleosome-remodeling complex in cancer and other diseases [41–44]. SWI/SNF is a multi-subunit complex that uses the energy of ATP hydrolysis to redistribute and rearrange nucleosomes to influence gene expression [45, 46]. In cancer, SWI/SNF is widely considered a tumor-suppressor, as deleterious mutations are present in approximately 20% of all cancers [45, 47–49]. Indeed, SChLAP1 (Second Chromosome Locus Associated with Prostate-1), a prostate cancer-specific lncRNA that is highly expressed in 15–30% of localized and metastatic tumors [41], is significantly associated with poor clinical outcomes and lethal disease. Moreover, SChLAP1 expression enhances tumor invasion and metastasis, in part, by interacting with and abrogating genome-wide binding of the SWI/SNF complex. Subsequent studies have defined SChLAP1 as one of the best prognostic genes in prostate cancer and have also shown the clinical utility of SChLAP1 as both a tissue- and urine-based biomarker [50–52]. Comparably, lncTCF7 is highly expressed in hepatocellular carcinoma (HCC) and required for the maintenance of self-renewal capacity in liver cancer stem cells (CSC) [42]. Functionally, lncTCF7 triggers the Wnt signaling pathway by binding to and recruiting the SWI/SNF complex to the TCF7 promoter to activate gene expression. This preserves the self-renewal capabilities of liver CSCs and promotes tumor initiation in HCC. LncRNA-mediated SWI/SNF regulation has also been described in other cellular and disease processes. For example, Pol V-transcribed lncRNAs indirectly interact with the SWI/SNF complex to mediate transcriptional silencing [43]. Additionally, the cardio-protective lncRNA Mhrt directly interacts with BRG1, the catalytic subunit of SWI/SNF, to prevent cardiac hypertrophy [44]. Taken together, these studies suggest that lncRNAs play an important role in SWI/SNF regulation, and systematic efforts to characterize similar lncRNA mediators of SWI/SNF in other cancers warrants further investigation.
Additionally, HOTTIP (HOXA transcript at the distal tip) is another lncRNA upregulated in HCC [53]. HOTTIP expression is associated with clinical progression of HCC and is also an independent predictor of overall survival. Mechanistically, HOTTIP regulates the HOXA locus by interacting with the WDR5/MLL epigenetic complex to drive H3K4me3 [54]. Previous studies have identified an RNA binding pocket on WDR5 [55], suggesting that direct binding of lncRNAs to WDR5/MLL may similarly promote other cancers.
Epigenetic control by lncRNAs is not only exercised via interactions with chromatin remodelers. For example, TARID (TCF21 antisense RNA inducing demethylation) directs promoter demethylation of the tumor suppressive transcription factor TCF21 [23]. TARID is normally expressed in benign lung, oral, and ovarian epithelium but suppressed in cancer due to hypermethylation of its promoter. TARID acts as a scaffold to recruit GADD45A, a DNA demethylator, to the TCF21 promoter, resulting in demethylation of the TCF21 promoter through the base-excision repair pathway. The physical interaction between the TCF21 promoter, TARID, and GADD45A is critical for TCF21 expression and tumor suppression.
Insight into the biology and mechanism of lncRNAs provides a basis for the understanding of the global epigenetic modifications that occur in cancer.
DNA damage and cell cycle regulation
Proper responses to DNA damage and appropriate regulation of cell cycle checkpoints are essential for maintenance of cell integrity [56]. With alterations in more than 50% of all cancers, the p53 tumor suppressor mediates responses to DNA damage to prevent tumor-associated changes in cell metabolism, cell cycle checkpoint regulation and cell motility during cancer development [57]. While our current knowledge of these pathways is guiding targeted drug development in cancer [58], a thorough understanding of the mechanisms governing p53-related function in early tumorigenesis remains elusive.
LncRNAs have surfaced as important regulators of p53 action and cell cycle regulation in cancer. For example, lincRNA-p21 is regulated by p53 and serves as a repressor in p53-dependent transcriptional responses by physically associating with and guiding hnRNP-K to precise genomic targets [15]. Functionally, lincRNA-p21 is crucial to p53-mediated apoptosis in response to DNA damage. LincRNA-p21 recruits hnRNP-K in cis to promote p53-dependent transcription of p21, which is a well-known checkpoint regulator in the p53 pathway [59]. The absence of lincRNA-p21 compromises the G1/S checkpoint and results in increased proliferation.
Several lncRNAs are related to p53 regulation in response to cell stress, including MEG3, TUG1, PANDA, and LED. The imprinted lncRNA MEG3 (Maternally Expressed Gene 3) regulates cell proliferation and apoptosis by activating p53 in meningioma [60] and NSCLC [61]. TUG1 [35] and PANDA [16] are directly regulated by p53 binding to their promoters following DNA damage, and TUG1 and PANDA expression are reduced in primary lung and breast tumors, respectively, compared to normal tissue. Mechanistically, TUG1 recruits PRC2 to the promoter of HOXB7, reducing HOX-mediated cell proliferation; PANDA binds to and abrogates chromatin binding of NF-YA, leading to repression of apoptotic gene expression programs.
Upon cellular stress, p53 also directly regulates enhancer RNAs (eRNAs), which function by altering the expression of neighboring genes [62]. While many p53-induced eRNAs have p53-binding sites, some do not, suggesting another mediator is involved in regulating this subset of p53-responsive eRNAs. Recently, LED (LncRNA activator of Enhancer Domains) was identified as a p53-induced lncRNA that associates with and activates several of these remaining enhancers [63]. LED prominently associates with the p21 enhancer and LED knockdown significantly influences G1 checkpoint arrest and increases cell proliferation. Mechanistically, LED impacts eRNA production by epigenetically increasing the deposition of the active enhancer histone mark, H3K9ac, at specific loci. Interestingly, LED expression is downregulated by hypermethylation in 44% of cancer cell lines, suggesting LED is a p53-responsive lncRNA that regulates the p53 transcriptional response and has tumor suppressive function.
Other lncRNAs play a vital role in mediating senescence and cell cycle arrest. The lncRNA MIR31HG is upregulated during oncogene-induced senescence (OIS) and antagonizes tumor suppressive function of P16INK4A, resulting in decreased cell progression to S phase of the cell cycle [64]. MIR31HG functions by mediating Polycomb group protein-mediated repression of the INK4A locus. CARLo-5 (Cancer-Associated Region Long non-coding RNA), a lncRNA implicated in colorectal cancer [65], prostate cancer [65], gastric cancer [66], and NSCLC [67], functions by blocking cell cycle arrest at the G1 phase, resulting in uninhibited cell proliferation. LncRNA gadd7 (Growth-Arrested DNA Damage-inducible gene 7) inhibits the G1/S cell cycle transition and its expression is induced in response to DNA-damaging agents, including UV irradiation, cisplatin, and growth arrest [68]. Prostate cancer-specific lncRNA PCAT-1 (Prostate Cancer-Associated Transcript 1) is involved in the transcriptional repression of many genes related to mitosis and the cell cycle [69]. PCAT-1 expression is inversely correlated with BRCA2, and cells overexpressing PCAT-1 accumulate double-strand breaks (DSB) after treatment with DNA-damaging agents, suggesting its involvement in homologous recombination and DSB repair [70]. Downregulation of the tumor-suppressive lncRNA GAS5 (Growth Arrest-Specific 5) promotes cell proliferation, in part, by regulating cell cycle factors such as CDK6, E2F1, and p21 [71, 72].
Taken together, these mechanisms suggest that a subclass of lncRNAs are crucial gatekeepers of DNA damage repair, cell cycle progression, and apoptosis, and lncRNA dysregulation in this context contributes, in part, to cancer cell immortality.
MicroRNA silencing
MicroRNAs (miRNA) are small transcripts that have emerged as a prominent class of regulatory genes in numerous diseases, including cancer [73]. MiRNAs bind to complementary sequences on target RNAs, leading to repressed gene expression and blocked protein synthesis. Several lncRNAs mediate cancer progression by altering miRNA function. In the competing endogenous RNA (ceRNA) model, lncRNAs that harbor miRNA response elements can bind to and sequester miRNAs, preventing target transcript degradation [9, 74]. While some experimental evidence has questioned the validity of the ceRNA hypothesis [75], many lncRNAs function via miRNA pathways, both directly and indirectly.
The H19 lncRNA has been studied for decades as an important genetic factor in development and cancer [76]. Two miRNA-based mechanisms have been described regarding its function. First, H19 encodes for and produces miR-675 to promote gastric cancer [77], colorectal cancer [78], and glioma [79]. Next, H19 modulates the let-7 family of miRNAs [80], which have vital roles in development, cancer, and metabolism [81]. Specifically, H19 was found to harbor both canonical and non-canonical binding sites for let-7 and acts as a miRNA sponge to sequester and regulate the let-7 family of miRNAs.
In HCC, HULC (Highly Upregulated in Liver Cancer) and lncRNA-ATB (Activated By TGF-β) have been shown to function by miRNA-facilitated modalities. HULC, one of the most highly expressed lncRNAs in HCC, is a CREB (cAMP respone element binding protein)-regulated transcript that acts as a miRNA sponge to downregulate several miRNAs, including miR-372, leading to decreased translational repression of PRKACB and induced activation of CREB [82]. This results in an auto-regulatory loop in which HULC promotes its own expression. LncRNA-ATB enhances epithelial-mesenchymal transition(EMT), leading to cancer progression and tumor metastasis [83]. High lncRNA-ATB expression is correlated with decreased recurrence-free survival and overall survival in HCC patients. LncRNA-ATB interacts with several miR-200s, which have been previously shown to play a role in EMT suppression. Increased lncRNA-ATB expression results in decreased miR-200 level, suggesting that lncRNA-ATB functions as a microRNA sponge. Remarkably, in vivo xenograft studies showed that mutating miR-200 target sites on lncRNA-ATB decreased the abundance of circulating tumor cells in mice [83].
In gastric cancer, GAPLINC (Gastric Adenocarcinoma Predictive Long Intergenic Noncoding RNA) was identified as the most upregulated lncRNA in cancer compared to normal tissue and correlates with poor patient outcomes [84]. Mechanistically, GAPLINC regulates cell migration pathways by acting as a decoy for miR211-3p, a miRNA implicated in CD44 oncogene degradation.
Moreover, lncRNAs can alter miRNA biology indirectly. The lncRNA ANRIL (Antisense Noncoding RNA in the INK4 Locus), which is known to function in tumor development and progression [85], is highly overexpressed in gastric cancer and correlates with worse disease prognosis [34]. ANRIL binds to PRC2 and is required for PRC2-mediated silencing of miR-99a and miR-449a. Downregulation of these miRNAs releases inhibition of E2F1 and CDK6, allowing cell cycle progression and cell proliferation. Subsequently, E2F1 reactivates ANRIL, forming a positive auto-regulatory loop.
Additionally, PCAT-1, one of the most differentially expressed lncRNAs in prostate cancer compared to benign tissues [69], promotes cell proliferation, in part, by interfering with the regulation of cMyc by miR-34a [86]. Studies showed that PCAT-1 binds to MYC 3’-UTR, preventing miR-34a from engaging its target sequence. When PCAT-1 was knocked down or a PCAT-1-specific miRNA was introduced into cells, cMYC stabilization was compromised, suggesting that PCAT-1 plays a crucial post-transcriptional role in cMYC regulation.
These studies suggest that lncRNAs significantly influence miRNA biology by acting as a precursor for miRNAs, directly binding to and sequestering miRNAs, or indirectly interfering with miRNA expression and regulation. While the ceRNA hypothesis remains controversial, it is clear that miRNAs are one of several avenues by which lncRNAs mediate cancer progression and metastasis.
Signaling Pathways
The aberrant activation and propagation of cellular signals is a well-documented phenomenon in cancer. LncRNAs that function in these signaling pathways are becoming a major component of cancer mechanisms. As a key target of drug development, further investigation in this area will potentially reveal therapeutic vulnerabilities that can be targeted with novel compounds.
Cellular signaling
Several studies have highlighted the role of transforming growth factor-β (TGF-β) [87], Hedgehog [88], and Wnt [89] signaling pathways in tumor development. For example, TGF-β signaling promotes cancer cell metastasis in HCC via lncRNA-ATB. In addition to regulating miRNAs, lncRNA-ATB is induced by TGF-β and stabilizes IL-11 mRNA [83]. This allows increased IL-11 secretion and downstream IL-11/STAT3 signaling in an autocrine fashion, leading to enhanced cell colonization at distant metastatic sites.
In breast cancer, BCAR4 (Breast Cancer Anti-estrogen Resistance 4) was recently identified as the most upregulated lncRNA expressed in stage III breast cancer versus normal tissue, and increased expression was seen in later stage and metastatic samples, correlating with shorter survival time in breast cancer patients [90]. In vitro and in vivo experiments showed that BCAR4 increases breast cancer cell migration and invasion through interactions with two transcription factors, leading to the activation of a non-canonical Hedgehog signaling pathway. Additionally, overexpression of lncRNA H19 due to aberrant Hedgehog signaling promotes osteosarcoma development in mice [91].
The lncRNAs CCAT2 (Colon Cancer-Associated Transcript 2) and MALAT1 (Metastasis-Associated Lung Adenocarcinoma Transcript 1) drive tumor progression and metastasis in breast [92], NSCLC [93], esophageal [94], colorectal [95], renal cell [96], endometrial [97], and lung cancers [98] through general activation of the Wnt signaling pathway. Moreover, lncTCF7 (described above) recruits the SWI/SNF complex to promote Wnt signaling in HCC [42].
Additionally, some lncRNAs are involved in chemokine signaling. For example, BANCR (BRAF-regulated lncRNA 1) is upregulated in cancer tissues with the active BRAFV600E mutant and increases cell migration in melanoma through increased CXCL11 chemokine signaling [99]. BANCR also increases cell migration and invasion in NSCLC by regulating E-cadherin, N-cadherin, and Vimentin, which play key roles in EMT [100]. These studies suggest that lncRNAs may promote tumorigenesis through varying mechanisms of signal transduction
Hormonal regulation
Several cancers are driven by hormone regulation [101]. In particular, estrogen and androgen steroid hormones stimulate breast and prostate cancers [102, 103]. Given the pivotal role of these hormone receptor pathways in propelling cancer progression, it comes as no surprise that lncRNAs are also involved in their function.
Prior to being described as a mediator of non-canonical Hedgehog signaling (described above), BCAR4 was identified in a functional screen for genes involved in tamoxifen resistance [104]. Subsequent studies found that BCAR4 expression is associated with shorter metastasis free survival and overall survival in breast cancer patients, and BCAR4 mediates estrogen-independent tumor growth [105]. In prostate cancer, NEAT1 (Nuclear Enriched Abundant Transcript 1), a lncRNA necessary for nuclear paraspeckle formation [106], was identified as an estrogen receptor alpha (ER-α)-regulated lncRNA with increased expression in prostate cancers compared to normal tissues [107]. NEAT1 coordinates prostate cancer oncogenesis by interacting at promoters of prostate-cancer associated genes. Importantly, prostate cancers expressing high levels of NEAT1 are unresponsive to androgen antagonists, suggesting that NEAT1 may play a role in metastatic castrate-resistant prostate cancers (mCRPC). Additionally, lncRNA GAS5 mediates apoptosis in hormone-driven prostate and breast cancers through binding steroid receptors [108].
The androgen receptor (AR) plays a central role in establishing an oncogenic cascade that drives prostate cancer progression [109]. In fact, the mainstay of treatment for prostate cancer involves androgen deprivation therapy (ADT) [110]. PCAT29 (Prostate Cancer-Associated Transcript 29) [111] and DRAIC (Downregulated RNA In Cancer) [112] are two androgen-suppressed lncRNAs located within 20kb of each other on chromosome 15q23. Upon androgen stimulation, AR binds to the promoters of both lncRNAs to repress their transcription. Lower PCAT29 and DRAIC expression correlates with poor prognostic outcomes in prostate cancer patients. Tumors treated with ADT showed higher levels of PCAT29, and tumors that progressed after ADT had lower expression of DRAIC, suggesting that these lncRNAs may play a role in mediating mCRPC.
CTBP1-AS is another androgen-regulated lncRNA that mediates AR activity by directly inhibiting the expression of the AR co-repressor CTBP1 [113]. CTBP1-AS functions by recruiting histone deacetylases via the RNA-binding PTB-associated splicing factor (PSF) to target gene promoters. CTBP1-AS knockdown suppresses androgen-dependent cell proliferation in vitro and reduces xenograft tumor growth in vivo. Furthermore, upregulation of CTBP1-AS and downregulation of CTBP1 is detected in primary and metastatic prostate cancer samples, but not benign tissues, suggesting that this lncRNA directly contributes to prostate cancer progression.
Other lncRNAs have also been shown to directly mediate AR activity in prostate cancer. PRNCR1 and PCGEM1 are two lncRNAs that bind successively to AR to strongly enhance both ligand-dependent and ligand-independent AR-mediated gene activation and proliferation in prostate cancer cells [114]. However, the critical role of these lncRNAs in mCRPC remains questionable, as subsequent studies found extremely low levels of PRNCR1 expression in metastatic prostate tumors and lack of lncRNA binding to AR in prostate cells [115]. Nevertheless, PCGEM1 may play a role in mediating disease progression during early stages of prostate cancer [116], and further experimentation is necessary to delineate the precise role of these lncRNAs in mediating AR function.
Downstream mediators
The MYC proto-oncogene is a downstream effector of many signal transduction pathways and alterations in MYC are known to be oncogenic [117]. The human chromosomal 8q24 region includes a gene desert that contains enhancer elements that regulate MYC activity through long range chromatin looping. LncRNAs are also implicated in these processes. In prostate cancer, PCAT-1 mediates cMYC regulation [86] and PCGEM1 (Prostate Cancer Gene Expression marker 1) co-activates AR and cMYC to regulate tumor metabolism [118]. The colorectal cancer-specific lncRNA CCAT1-L mediates chromatin looping to allow the MYC promoter to interact with its enhancer elements [119]. LncRNA PVT1 is transcribed from the gene desert associated with MYC, and PVT1 expression is required for the oncogenic potential of MYC-driven human cancers [120]. Specifically, in 98% of MYC-amplified human tumors, PVT1 expression is also upregulated, and PVT1 knockdown abolishes the tumorigenicity of cancers with MYC amplification. However, PVT1 itself is not sufficient to cause tumor development without concurrent MYC upregulation, suggesting a synergistic effect exists between PVT1 and MYC in cancer development.
The transcription factor NF-κB is also highly upregulated in a variety of cancers and plays a role in tumor microenvironment inflammation, resulting in cancer development, metastasis, and invasion [121]. NKILA (NF-kB Interacting LncRNA) is upregulated by NF-κB and binds to NF-κB to form a stable complex, preventing degradation of IκB and subsequent NF-κB activation [122]. Low NKILA expression is correlated with cancer metastasis, advanced stage, higher grade, increased tumor size, and decreased patient survival, suggesting a clinically important function of NKILA in mediating inflammation-stimulated breast cancer.
Translational implications of lncRNAs
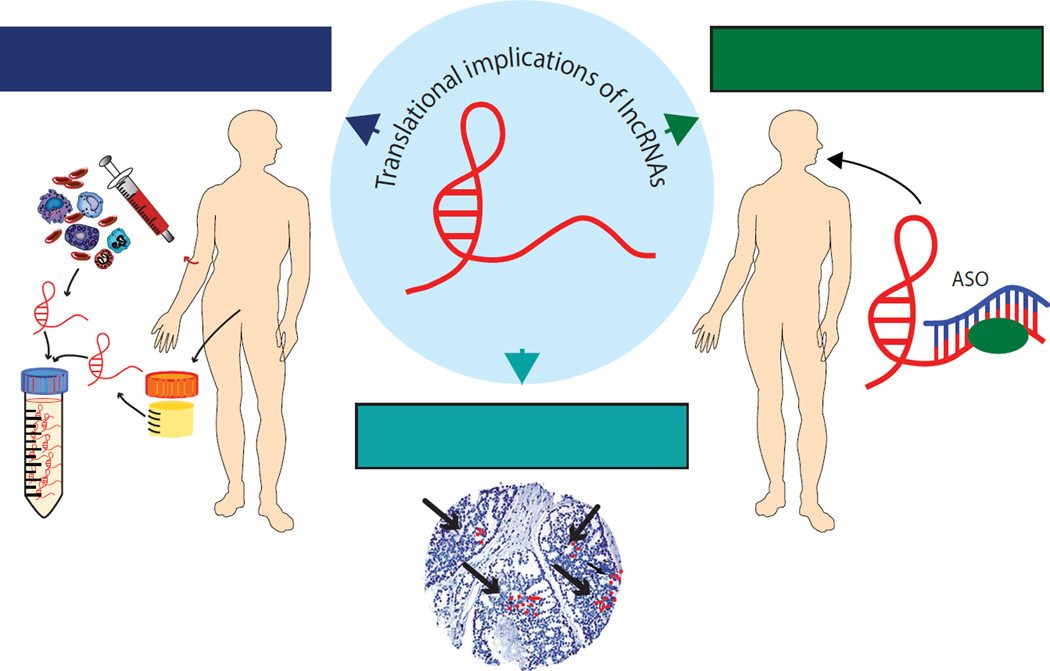
LncRNAs are beginning to show translational utility as both biomarkers and therapeutic targets (Fig. 2 and Table 2). Dozens of lncRNAs show promise as diagnostic and prognostic markers across several types of cancers [123]. In general, lncRNAs show higher tissue- and disease-specific expression compared to protein-coding genes [124]. In cancer, lncRNAs show striking cancer- and lineage-specificity, suggesting these molecules may be powerful biomarkers in the clinical setting [125]. Additionally, lncRNAs can be measured in blood, urine, and tissue, justifying the development of non-invasive tests [2]. For example, HULC is not only associated with poor prognosis in pancreatic cancer [126], but it is also highly detectable in the plasma of patients with HCC compared to healthy controls [127]. In prostate cancer, PCA3 has proven to be a powerful diagnostic urine marker [128]. Similarly, initial data show SChLAP1 can be detected in both tissue and urine of patients with more aggressive prostate cancer [50–52]. In addition to its prognostic value in colorectal carcinoma [129], renal cell carcinoma [130], and glioma [131], MALAT1 can be detected in patient serum and may serve as a diagnostic marker in prostate cancer [132]. Furthermore, AA174084 [133] and a set of oral lncRNAs [134] are found in gastric juices and saliva, and may serve as potential non-invasive biomarkers in gastric and oral squamous cell cancers, respectively.
Figure 2. Translational implications of lncRNAs.
Long noncoding RNAs (lncRNAs) are emerging as both diagnostic and prognostic biomarkers that can be detected in tissue, serum, and urine. Antisense oligonucleotides (ASOs) can be used to directly target lncRNAs and are a promising therapeutic strategy in cancer.
Table 2.
Biomarker potential of long noncoding RNAs in cancer
| Name | Cancer Type(s) | Diagnostic/Prognostic | Blood/Tissue/Urine | Reference(s) |
|---|---|---|---|---|
| ANRIL (ANtisense noncoding RNA in the INK4 Locus) |
gastric | prognostic | tissue | 34 |
| BANCR (BRAF Activated NonCoding RNA) | NSCLC | prognostic | tissue | 100 |
| BCAR4 (Breast Cancer Anti-estrogen Resistance-4) | breast | prognostic | tissue | 90 |
| CARLo-5 (Cancer-Associated Region Long non- coding RNA-5) |
NSCLC | prognostic | tissue | 67 |
| CCAT2 (Colon Cancer Associated Transcript 2) | breast | prognostic | tissue | 92 |
| DRAIC (Downregulated-RNA in Androgen- Independent Cells) |
multiple, prostate | prognostic | tissue | 112 |
| FAL1 (Focally Amplified LncRNA on chromosome 1) | ovarian | prognostic | tissue | 31 |
| GAPLINC (Gastric Adenocarcinoma Predictive Long Intergenic NonCoding RNA) |
gastric | diagnostic, prognostic | tissue | 84 |
| GAS5 (Growth Arrest Specific 5) | gastric | prognostic | tissue | 72 |
| HOTAIR (HOX Transcript Antisense Intergenic RNA) | breast, colorectal, GIST, hepatocellular |
prognostic | tissue | 19–22 |
| HOTTIP (HOXA Transcript at the distal TIP) | hepatocellular | prognostic | tissue | 53 |
| HULC (Hepatocellular Upregulated Long nonCoding RNA) |
hepatocellular, pancreatic | diagnostic, prognostic | blood, tissue | 126–127 |
| lncRNA-ATB (lncRNA-Activated by TGF-β) | hepatocellular | prognostic | tissue | 83 |
| lncRNA-HEIH (lncRNA-High Expression In HCC) | hepatocellular | prognostic | tissue | 33 |
| MALAT1 (Metastasis Associated Lung Adenocarcinoma Transcript-1) |
colorectal, glioma, prostate, renal cell |
diagnostic, prognostic | blood, tissue | 96, 129–132 |
| MEG3 (Maternally Expressed 3) | NSCLC | prognostic | tissue | 61 |
| NBAT-1 (NeuroBlastoma Associated Transcript-1) | neuroblastoma | prognostic | tissue | 32 |
| NEAT1 (Nuclear Enriched Abundant Transcript-1) | prostate | prognostic | tissue | 107 |
| NKILA (NF-KappaB Interacting LncRNA) | breast | prognostic | tissue | 122 |
| PCA3 (Prostate Cancer Antigen 3) | prostate | diagnostic | urine | 128 |
| PCAT29 (Prostate Cancer Associated Transcript 29) | prostate | prognostic | tissue | 111 |
| SChLAP1 (Second Chromosome Locus Associated with the Prostate 1) |
prostate | prognostic | tissue, urine | 41, 50–52 |
| TUG1 (Taurine UpreGulated 1) | NSCLC | prognostic | tissue | 35 |
GIST (gastrointestinal stromal tumor), NSCLC (non-small cell lung cancer)
Direct targeting of lncRNAs may be a viable therapeutic strategy in cancer. Antisense technology has gained considerable traction over the past few years as several antisense oligonucleotides (ASOs) have been introduced into clinical trials and some have been FDA-approved for clinical use [135–138]. ASOs function by basepair hybridizing to target RNAs, resulting in transcript-specific RNAse H-mediated catalytic degradation [139]. ASOs are a particularly attractive therapeutic modality for several reasons, including predictable human pharmacokinetics, prolonged tissue elimination half-lives, enhanced specificity compared to small molecule inhibitors, and lack of cytochrome P450 enzyme metabolism [91, 139–141]. These characteristics are thought to make ASOs safer for patients and also more suitable for combination therapies with other drugs. Given the important role of lncRNAs across several cancer pathways, ASO-mediated therapies are likely to surface as a promising class of new cancer drugs over the next few years.
Concluding Remarks
While we attempted to classify lncRNAs by their predominant mechanistic modality, most transcripts could fit into multiple categories, suggesting that lncRNAs may form important regulatory networks that can coordinate numerous aspects of cancer progression simultaneously. Although our understanding of lncRNA-mediated cancer biology has increased significantly in the last several years, we believe this is only the tip of the iceberg (Outstanding Questions Box). A continued understanding of the role of lncRNAs in cancer will be enhanced by new tools that uncover novel lncRNAs, better annotate known lncRNAs, as well as assess lncRNA localization, structure and function (Text Box 1).
Outstanding Questions Box.
- How do lncRNA variants contribute to cancer progression?
-
○Differential expression patterns in cancer have guided lncRNA discovery and investigation thus far. LncRNA transcript variants such as mutations, amplifications, deletions, and fusions remain unexplored.
-
○
- How do lncRNAs coordinate various molecular networks to drive cancer?
-
○The majority of lncRNA studies to date have focused on a single mechanism of action. However, several transcripts have numerous functions, suggesting that lncRNAs may form important regulatory networks that can coordinate many aspects of cancer progression simultaneously.
-
○
- Do a subset of lncRNAs have protein-coding potential?
-
○Some lncRNAs have been identified as TUCPs (Transcripts of Unknown Coding Potential). Previously undiscovered small peptides produced from these lncRNAs may have significant roles in cancer biology.
-
○
- How can lncRNAs be used to guide precision medicine approaches in cancer?
-
○An emerging area of cancer therapeutics utilizes genomic signatures to guide treatment choices for patients. Current efforts employ aberrations in protein-coding genes; incorporating lncRNAs into these analyses may improve therapeutic response and patient outcomes.
-
○
- How can lncRNAs be utilized in the clinical setting?
-
○LncRNAs have been identified as powerful diagnostic and prognostic biomarkers. Targeting lncRNAs directly with antisense oligonucleotides may also be a promising therapeutic strategy.
-
○
Text Box 1. Tools for lncRNA investigation.
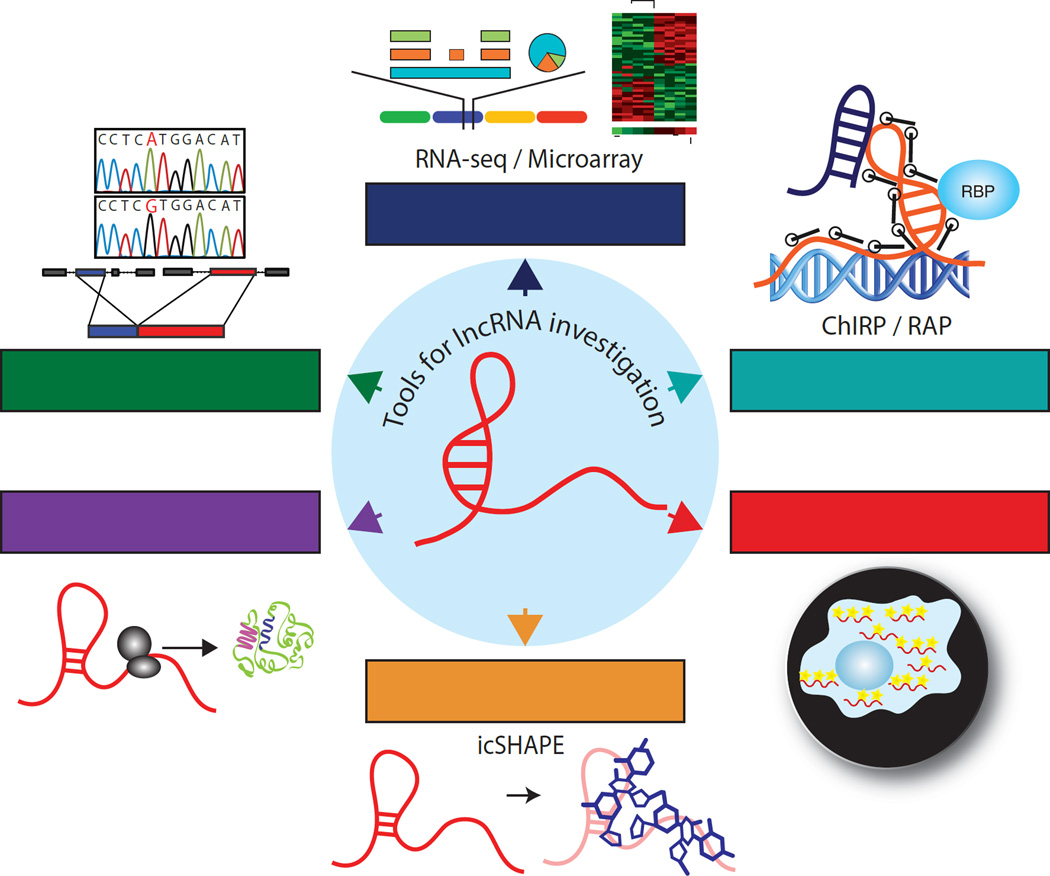
Several techniques and tools have been developed to discover and study lncRNAs (Fig. 3). RNA-seq has emerged as the most powerful method of lncRNA discovery. Recently, a large-scale lncRNA annotation effort identified nearly 60,000 lncRNAs across the cancer genome, suggesting that a large portion of the human transcriptome remains unexplored [125]. The MiTranscriptome portal (www.mitranscriptome.org) was developed using RNA-seq data from over 6,500 samples comprised of benign and malignant tissues as well as cell lines, making it the most comprehensive lncRNA annotation to date. Furthermore, a Sample Set Enrichment Analysis (SSEA) revealed approximately 8,000 previously unknown lncRNAs showing tissue- and/or cancer-specificity, providing the scientific community with a vast database of potentially critical molecules for future study. While this new resource will provide a foundation for lncRNA genomics and cancer disease mechanisms, it is limited to poly-adenylated transcripts. Future sequencing and annotation efforts will need to focus on also identifying non-poly-adenylated lncRNAs.
Novel methods to isolate RNA in vivo have led to the discovery of chromatin, RNA, and protein interacting partners of lncRNA transcripts. RNA pulldown methods such as chromatin isolation by RNA purification (ChIRP) [154] and RNA antisense purification (RAP) [155] have aided in the discovery of lncRNA function. These methods utilize antisense complementary oligonucleotides to isolate a target RNA and its associated molecules. Downstream sequencing and mass-spectrometry analysis can then be used to identify novel interactors in an unbiased manner.
Another useful tool to delineate lncRNA function is direct visualization. Single-molecule RNA-FISH (fluorescence in situ hybridization) is a powerful method to localize and visualize lncRNAs expression patterns in cells and tissues [156, 157]. Multiplexing RNA-FISH with protein immunofluorescence can also be used to identify and confirm RNA-protein interactions.
One of the greatest biological challenges has been the structural analysis of RNA molecules in vivo. A new approach, termed icSHAPE (in vivo click selective 2’-hydroxyl acylation and profiling experiment) enables RNA structure analysis in vivo at nucleotide resolution for all four bases and can identify RNA strandedness [158]. Perhaps the most relevant aspect of this technique to lncRNAs is the ability to differentiate structural changes in RNA at protein-binding sites.
Traditionally, computational methods are used to determine whether RNA is coding or noncoding. A variety of tools analyze sequence features such as open reading frame (ORF) length and the presence of a protein domain within a transcript. A subset of lncRNAs has been classified as TUCPs (Transcript of Unknown Coding Potential) [125, 143]. This is especially relevant as several examples of novel small peptides produced from putative lncRNAs have been described [159]. Improved bioinformatics tools and experimental methods, such as ribosomal profiling [160], should be employed to thoroughly assess the protein-coding capacity of a transcript.
While new biological and computational techniques have greatly accelerated our ability to investigate RNAs in cancer research, most lncRNA discovery and annotation efforts in cancer have been severely limited, with poor overlap between different catalogues [142], avoidance of monoexonic transcripts and complex regions of the genome [143], poor bioinformatics tools for ab initio assembly of novel transcripts [144], and small cohorts from which to reconstruct the cancer transcriptome [69]. Moreover, several studies continue to rely on microarray-based platforms for the identification of disease-associated lncRNAs [90, 145]; however, their use in discovering new lncRNAs is limited because gene expression probes are designed against previously annotated transcripts. Therefore, RNA-seq remains the most powerful tool to discover new lncRNAs in an unbiased fashion [146, 147].
Additionally, almost all lncRNA studies to date have focused on the aberrant expression patterns of novel transcripts in cancer. While this is an essential first step to identify important lncRNAs in cancer, future analyses will need to include transcript variants that populate and drive cancer. Protein alterations such as point mutations, deletions and amplifications, and gene fusions have emerged as key regulators in cancer [148, 149]. As our understanding of cancer-associated lncRNAs expands, similar variants will also need to be explored in noncoding transcripts. Furthermore, studying isoform-specific functions may reveal new insights on lncRNA gene function [10, 125]. Uncovering the precise function of lncRNAs in cell and animal models also cannot be overlooked. While methods to knockdown (ASOs and Locked Nucleic Acids (LNAs) [150]) and knockout (CRISPR) [151, 152] lncRNA genes are improving, caution should be taken when employing these tools to explore lncRNA function [153].
Less than a decade ago, lncRNAs were mostly ignored, often considered “junk” DNA and attributed to leaky transcription. Now, functional, mechanistic, and translational insights have revealed the crucial role of lncRNAs in cell biology and disease pathogenesis. Importantly, lncRNAs are emerging as critical players in cancer progression and metastasis. Given the tissue- and disease-specific nature of these transcripts, their abundance throughout the genome, and the relatively recent discovery of the majority of these transcripts, it is likely that lncRNAs hold the answer to questions in cancer that have eluded us for years.
Figure 3. Tools for lncRNA investigation.
Emerging areas for investigation in the long noncoding RNA (lncRNA) field include improved discovery methods, unbiased interactome analysis, transcript visualization and localization, RNA structure determination, discovery of small peptides produced from short open reading frames (sORF), and identification and comprehension of lncRNA variants.
Trends Box.
Thousands of long noncoding RNAs (lncRNAs) populate the cancer genome and show cancer-specific expression patterns
LncRNAs drive cancer biology and mediate several cancer-associated concepts, including epigenetic regulation, DNA damage and cell cycle control, miRNA silencing, signal transduction pathways, and hormone-driven disease
New tools are emerging as powerful methods for lncRNA discovery and mechanistic investigation, including RNA-seq, ChIRP, RAP, RNA-FISH and icSHAPE
The MiTranscriptome compendium provides a comprehensive annotation of over 8,000 previously undiscovered cancer-associated lncRNAs that may be critical molecules for future study
Uncovering lncRNA function may reveal new translational opportunities for biomarker development and therapeutic targeting
Acknowledgements
We thank Robin Kunkel for assistance with figure preparation and Karen Giles for critically reading the manuscript and for the submission of documents. This work was supported in part by US National Institutes of Health Prostate Specialized Program of Research Excellence grant P50 CA69568, Early Detection Research Network grant UO1 CA111275, US National Institutes of Health grant R01 CA132874 (A.M.C.), and US Department of Defense grant PC100171 (A.M.C.). A.M.C. is supported by the Prostate Cancer Foundation and the Howard Hughes Medical Institute. A.M.C. is an American Cancer Society Research Professor and a Taubman Scholar of the University of Michigan. A.S. is supported by a Prostate Cancer Foundation Young Investigator Award and by a National Institutes of Health Ruth L. Kirschstein National Research Service Award F30 CA180376. A.S. is a Fellow of the University of Michigan Medical Scientist Training Program. U.S. is supported by the Howard Hughes Medical Institute Medical Research Fellows Program. A.M.C. serves on the scientific advisory board of Paradigm. He is a co-inventor on patents filed by the University of Michigan covering the diagnostic and therapeutic field of use for T2-ERG in prostate cancer which has been licensed to Hologic and the diagnostic and therapeutic field of use for SChLAP1 which has been co-licensed to Genome Dx. Paradigm, Hologic, and Genome Dx did not play roles in the design and conduct of this study, in the collection, analysis, or interpretation of the data, or in the preparation, review, or approval of the article.
Glossary
- Ribonucleoprotein complex (RNP)
a cellular complex containing RNAs and proteins
- Polycomb Repressive Complex 2 (PRC2)
a multi-protein complex containing the core components of SUZ12, EED, RbAp48 and EZH2. PRC2 primarily functions as a histone methyltransferase, adding a trimethyl group to histone H3 on lysine 27 (H3K27me3) to produce transcriptionally silent chromatin
- Epigenetics
the heritable variations in gene expression that result due to differences in how DNA is read rather than in the DNA sequence itself. Epigenetic alterations include DNA methylation and histone modifications
- Chromatin remodeler
a protein complex that physically changes DNA architecture to allow or restrict access of regulatory proteins and transcription machinery to DNA. The function of chromatin remodelers is often carried out by epigenetic modifications using the energy of ATP hydrolysis
- Enhancer RNA (eRNA)
a class of non-coding RNAs that plays a role in enhancer DNA-mediated transcriptional regulation. eRNAs have been shown to recruit RNA polymerase II to these DNA regions to assist in the initiation of gene transcription
- Oncogene induced senescence (OIS)
a sustained induction of the Rb and p53 tumor suppressive pathways in response to an activating mutation in an oncogene or loss of tumor suppressive activity within a cell. The mechanism by which OIS occurs has not been fully elucidated, however it is known to protect against the progression to cancer in response to oncogenic stress
- Epithelial-mesenchymal transition (EMT)
the cellular mechanism by which epithelial cells gain migratory and invasive properties to form mesenchymal cells. This phenomenon is often seen when cancer cells gain the ability to invade and metastasize to distant organs. EMT can also occur in normal biological processes, such as wound healing
- Transforming growth factor-beta signaling pathway (TGF- β)
a cellular signaling pathway involved in numerous physiological processes, including cell differentiation, cell growth, and apoptosis. Ligand binding initiates a cascade of signaling through serine/threonine receptor kinase activity
- Hedgehog signaling pathway
a cellular signaling pathway that is required for the regulation of embryonic cell development. Aberrations in Hedgehog signaling may result in developmental or growth defects
- Wnt signaling pathway
a cellular signaling pathway initiated by binding of Wnt ligand to a Frizzled family receptor, resulting in transmission of this signal to Dishevelled within the cell. This leads to downstream regulation of genes involved in embryonic development, cell differentiation, cell migration, and cell proliferation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer discovery. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guttman M, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011 doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JT. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23:1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poliseno L, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salmena L, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao J, et al. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagano T, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 13.Pandey RR, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Molecular cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 14.Yap KL, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Molecular cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huarte M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hung T, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nature genetics. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang F, et al. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Molecular cell. 2013;49:1083–1096. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 18.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Molecular cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kogo R, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 21.Yang Z, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18:1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 22.Niinuma T, et al. Upregulation of miR-196a and HOTAIR Drive Malignant Character in Gastrointestinal Stromal Tumors. Cancer Res. 2012;72:1126–1136. doi: 10.1158/0008-5472.CAN-11-1803. [DOI] [PubMed] [Google Scholar]
- 23.Arab K, et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Molecular cell. 2014;55:604–614. doi: 10.1016/j.molcel.2014.06.031. [DOI] [PubMed] [Google Scholar]
- 24.Sharma S, et al. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Esteller M. Epigenetics in cancer. The New England journal of medicine. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 26.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 27.Kleer CG, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varambally S, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 29.Schwartz YB, Pirrotta V. A new world of Polycombs: unexpected partnerships and emerging functions. Nature reviews. Genetics. 2013;14:853–864. doi: 10.1038/nrg3603. [DOI] [PubMed] [Google Scholar]
- 30.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu X, et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer cell. 2014;26:344–357. doi: 10.1016/j.ccr.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pandey GK, et al. The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer cell. 2014;26:722–737. doi: 10.1016/j.ccell.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 33.Yang F, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 34.Zhang EB, et al. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5:2276–2292. doi: 10.18632/oncotarget.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang EB, et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell death & disease. 2014;5:e1243. doi: 10.1038/cddis.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yildirim E, et al. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152:727–742. doi: 10.1016/j.cell.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davidovich C, et al. Promiscuous RNA binding by Polycomb repressive complex 2. Nature structural & molecular biology. 2013;20:1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davidovich C, et al. Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA. Molecular cell. 2015;57:552–558. doi: 10.1016/j.molcel.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puvvula PK, et al. Long noncoding RNA PANDA and scaffold-attachment-factor SAFA control senescence entry and exit. Nature communications. 2014;5:5323. doi: 10.1038/ncomms6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prensner JR, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nature genetics. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, et al. The Long Noncoding RNA lncTCF7 Promotes Self-Renewal of Human Liver Cancer Stem Cells through Activation of Wnt Signaling. Cell stem cell. 2015;16:413–425. doi: 10.1016/j.stem.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 43.Zhu Y, et al. A SWI/SNF chromatin-remodeling complex acts in noncoding RNA-mediated transcriptional silencing. Molecular cell. 2013;49:298–309. doi: 10.1016/j.molcel.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han P, et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514:102–106. doi: 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nature reviews. Cancer. 2011;11:481–492. doi: 10.1038/nrc3068. [DOI] [PubMed] [Google Scholar]
- 46.Lu P, Roberts CW. The SWI/SNF tumor suppressor complex: Regulation of promoter nucleosomes and beyond. Nucleus. 2013;4:374–378. doi: 10.4161/nucl.26654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reisman D, et al. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 48.Shain AH, Pollack JR. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PloS one. 2013;8:e55119. doi: 10.1371/journal.pone.0055119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kadoch C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nature genetics. 2013;45:592–601. doi: 10.1038/ng.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prensner JR, et al. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol. 2014;15:1469–1480. doi: 10.1016/S1470-2045(14)71113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mehra R, et al. A novel RNA in situ hybridization assay for the long noncoding RNA SChLAP1 predicts poor clinical outcome after radical prostatectomy in clinically localized prostate cancer. Neoplasia. 2014;16:1121–1127. doi: 10.1016/j.neo.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bottcher R, et al. Novel long non-coding RNAs are specific diagnostic and prognostic markers for prostate cancer. Oncotarget. 2015 doi: 10.18632/oncotarget.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quagliata L, et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology. 2014;59:911–923. doi: 10.1002/hep.26740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang YW, et al. Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. eLife. 2014;3:e02046. doi: 10.7554/eLife.02046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 57.Muller PA, Vousden KH. p53 mutations in cancer. Nature cell biology. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 58.Vazquez A, et al. The genetics of the p53 pathway, apoptosis and cancer therapy. Nature reviews. Drug discovery. 2008;7:979–987. doi: 10.1038/nrd2656. [DOI] [PubMed] [Google Scholar]
- 59.Dimitrova N, et al. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Molecular cell. 2014;54:777–790. doi: 10.1016/j.molcel.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang X, et al. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer research. 2010;70:2350–2358. doi: 10.1158/0008-5472.CAN-09-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu KH, et al. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC cancer. 2013;13:461. doi: 10.1186/1471-2407-13-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Younger ST, et al. Integrative genomic analysis reveals widespread enhancer regulation by p53 in response to DNA damage. Nucleic acids research. 2015 doi: 10.1093/nar/gkv284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leveille N, et al. Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nature communications. 2015;6:6520. doi: 10.1038/ncomms7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Montes M, et al. The lncRNA MIR31HG regulates p16(INK4A) expression to modulate senescence. Nature communications. 2015;6:6967. doi: 10.1038/ncomms7967. [DOI] [PubMed] [Google Scholar]
- 65.Kim T, et al. Long-range interaction and correlation between MYC enhancer and oncogenic long noncoding RNA CARLo-5. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:4173–4178. doi: 10.1073/pnas.1400350111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y, et al. Enhanced expression of long noncoding RNA CARLo-5 is associated with the development of gastric cancer. International journal of clinical and experimental pathology. 2014;7:8471–8479. [PMC free article] [PubMed] [Google Scholar]
- 67.Luo J, et al. Long non-coding RNA CARLo-5 is a negative prognostic factor and exhibits tumor pro-oncogenic activity in non-small cell lung cancer. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:11541–11549. doi: 10.1007/s13277-014-2442-7. [DOI] [PubMed] [Google Scholar]
- 68.Liu X, et al. Long non-coding RNA gadd7 interacts with TDP-43 and regulates Cdk6 mRNA decay. The EMBO journal. 2012;31:4415–4427. doi: 10.1038/emboj.2012.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Prensner JR, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nature biotechnology. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Prensner JR, et al. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer research. 2014;74:1651–1660. doi: 10.1158/0008-5472.CAN-13-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu Z, et al. Downregulation of GAS5 promotes bladder cancer cell proliferation, partly by regulating CDK6. PloS one. 2013;8:e73991. doi: 10.1371/journal.pone.0073991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun M, et al. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC cancer. 2014;14:319. doi: 10.1186/1471-2407-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO molecular medicine. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tay Y, et al. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Denzler R, et al. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Molecular cell. 2014;54:766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cui H, et al. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer research. 2002;62:6442–6446. [PubMed] [Google Scholar]
- 77.Zhuang M, et al. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochemical and biophysical research communications. 2014;448:315–322. doi: 10.1016/j.bbrc.2013.12.126. [DOI] [PubMed] [Google Scholar]
- 78.Tsang WP, et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31:350–358. doi: 10.1093/carcin/bgp181. [DOI] [PubMed] [Google Scholar]
- 79.Shi Y, et al. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PloS one. 2014;9:e86295. doi: 10.1371/journal.pone.0086295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kallen AN, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Molecular cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Roush S, Slack FJ. The let-7 family of microRNAs. Trends in cell biology. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 82.Wang J, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic acids research. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yuan JH, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 84.Hu Y, et al. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer research. 2014;74:6890–6902. doi: 10.1158/0008-5472.CAN-14-0686. [DOI] [PubMed] [Google Scholar]
- 85.Pasmant E, et al. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer research. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 86.Prensner JR, et al. The long non-coding RNA PCAT-1 promotes prostate cancer cell proliferation through cMyc. Neoplasia. 2014;16:900–908. doi: 10.1016/j.neo.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Massague J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Taipale J, Beachy PA. The Hedgehog and Wnt signalling pathways in cancer. Nature. 2001;411:349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- 89.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 90.Xing Z, et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. 2014;159:1110–1125. doi: 10.1016/j.cell.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chan LH, et al. Hedgehog signaling induces osteosarcoma development through Yap1 and H19 overexpression. Oncogene. 2014;33:4857–4866. doi: 10.1038/onc.2013.433. [DOI] [PubMed] [Google Scholar]
- 92.Redis RS, et al. CCAT2, a novel long non-coding RNA in breast cancer: expression study and clinical correlations. Oncotarget. 2013;4:1748–1762. doi: 10.18632/oncotarget.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Qiu M, et al. CCAT2 is a lung adenocarcinoma-specific long non-coding RNA and promotes invasion of non-small cell lung cancer. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:5375–5380. doi: 10.1007/s13277-014-1700-z. [DOI] [PubMed] [Google Scholar]
- 94.Wang J, et al. Long noncoding RNA CCAT2 correlates with smoking in esophageal squamous cell carcinoma. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015 doi: 10.1007/s13277-015-3220-x. [DOI] [PubMed] [Google Scholar]
- 95.Ling H, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome research. 2013;23:1446–1461. doi: 10.1101/gr.152942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hirata H, et al. Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205. Cancer research. 2015;75:1322–1331. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhao Y, et al. A novel wnt regulatory axis in endometrioid endometrial cancer. Cancer research. 2014;74:5103–5117. doi: 10.1158/0008-5472.CAN-14-0427. [DOI] [PubMed] [Google Scholar]
- 98.Gutschner T, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer research. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Flockhart RJ, et al. BRAFV600E remodels the melanocyte transcriptome and induces BANCR to regulate melanoma cell migration. Genome research. 2012;22:1006–1014. doi: 10.1101/gr.140061.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sun M, et al. Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Molecular cancer. 2014;13:68. doi: 10.1186/1476-4598-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocrine reviews. 2004;25:276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- 102.Clemons M, Goss P. Estrogen and the risk of breast cancer. The New England journal of medicine. 2001;344:276–285. doi: 10.1056/NEJM200101253440407. [DOI] [PubMed] [Google Scholar]
- 103.Scher HI, Sawyers CL. Biology of progressive, castration-resistant prostate cancer: directed therapies targeting the androgen-receptor signaling axis. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2005;23:8253–8261. doi: 10.1200/JCO.2005.03.4777. [DOI] [PubMed] [Google Scholar]
- 104.Meijer D, et al. Functional screen for genes responsible for tamoxifen resistance in human breast cancer cells. Molecular cancer research : MCR. 2006;4:379–386. doi: 10.1158/1541-7786.MCR-05-0156. [DOI] [PubMed] [Google Scholar]
- 105.Godinho M, et al. Characterization of BCAR4, a novel oncogene causing endocrine resistance in human breast cancer cells. Journal of cellular physiology. 2011;226:1741–1749. doi: 10.1002/jcp.22503. [DOI] [PubMed] [Google Scholar]
- 106.Clemson CM, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Molecular cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chakravarty D, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nature communications. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hudson WH, et al. Conserved sequence-specific lincRNA-steroid receptor interactions drive transcriptional repression and direct cell fate. Nature communications. 2014;5:5395. doi: 10.1038/ncomms6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Visakorpi T, et al. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nature genetics. 1995;9:401–406. doi: 10.1038/ng0495-401. [DOI] [PubMed] [Google Scholar]
- 110.Sharifi N, et al. Androgen deprivation therapy for prostate cancer. Jama. 2005;294:238–244. doi: 10.1001/jama.294.2.238. [DOI] [PubMed] [Google Scholar]
- 111.Malik R, et al. The lncRNA PCAT29 inhibits oncogenic phenotypes in prostate cancer. Molecular cancer research : MCR. 2014;12:1081–1087. doi: 10.1158/1541-7786.MCR-14-0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sakurai K, et al. The lncRNA DRAIC/PCAT29 Locus Constitutes a Tumor-Suppressive Nexus. Molecular cancer research : MCR. 2015 doi: 10.1158/1541-7786.MCR-15-0016-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Takayama K, et al. Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. The EMBO journal. 2013;32:1665–1680. doi: 10.1038/emboj.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang L, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Prensner JR, et al. The IncRNAs PCGEM1 and PRNCR1 are not implicated in castration resistant prostate cancer. Oncotarget. 2014;5:1434–1438. doi: 10.18632/oncotarget.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Parolia A, et al. The long non-coding RNA PCGEM1 is regulated by androgen receptor activity in vivo. Molecular cancer. 2015;14:46. doi: 10.1186/s12943-015-0314-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hung CL, et al. A long noncoding RNA connects c-Myc to tumor metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:18697–18702. doi: 10.1073/pnas.1415669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xiang JF, et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell research. 2014;24:513–531. doi: 10.1038/cr.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tseng YY, et al. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512:82–86. doi: 10.1038/nature13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dolcet X, et al. NF-kB in development and progression of human cancer. Virchows Archiv : an international journal of pathology. 2005;446:475–482. doi: 10.1007/s00428-005-1264-9. [DOI] [PubMed] [Google Scholar]
- 122.Liu B, et al. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer cell. 2015;27:370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 123.Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2013;26:155–165. doi: 10.1038/modpathol.2012.160. [DOI] [PubMed] [Google Scholar]
- 124.Esteller M. Non-coding RNAs in human disease. Nature reviews. Genetics. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 125.Iyer MK, et al. The landscape of long noncoding RNAs in the human transcriptome. Nature genetics. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Peng W, et al. Long noncoding RNA HULC is a novel biomarker of poor prognosis in patients with pancreatic cancer. Medical oncology. 2014;31:346. doi: 10.1007/s12032-014-0346-4. [DOI] [PubMed] [Google Scholar]
- 127.Xie H, et al. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. BioMed research international. 2013;2013:136106. doi: 10.1155/2013/136106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hessels D, et al. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. European urology. 2003;44:8–15. doi: 10.1016/s0302-2838(03)00201-x. discussion 15–16. [DOI] [PubMed] [Google Scholar]
- 129.Zheng HT, et al. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. International journal of clinical and experimental pathology. 2014;7:3174–3181. [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang HM, et al. Upregulation of long non-coding RNA MALAT1 correlates with tumor progression and poor prognosis in clear cell renal cell carcinoma. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36:2947–2955. doi: 10.1007/s13277-014-2925-6. [DOI] [PubMed] [Google Scholar]
- 131.Ma KX, et al. Long noncoding RNA MALAT1 associates with the malignant status and poor prognosis in glioma. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36:3355–3359. doi: 10.1007/s13277-014-2969-7. [DOI] [PubMed] [Google Scholar]
- 132.Ren S, et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. European journal of cancer. 2013;49:2949–2959. doi: 10.1016/j.ejca.2013.04.026. [DOI] [PubMed] [Google Scholar]
- 133.Shao Y, et al. Gastric juice long noncoding RNA used as a tumor marker for screening gastric cancer. Cancer. 2014;120:3320–3328. doi: 10.1002/cncr.28882. [DOI] [PubMed] [Google Scholar]
- 134.Tang H, et al. Salivary lncRNA as a potential marker for oral squamous cell carcinoma diagnosis. Molecular medicine reports. 2013;7:761–766. doi: 10.3892/mmr.2012.1254. [DOI] [PubMed] [Google Scholar]
- 135.Meng L, et al. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 2015;518:409–412. doi: 10.1038/nature13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Buller HR, et al. Factor XI antisense oligonucleotide for prevention of venous thrombosis. The New England journal of medicine. 2015;372:232–240. doi: 10.1056/NEJMoa1405760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Noveck R, et al. Effects of an antisense oligonucleotide inhibitor of C-reactive protein synthesis on the endotoxin challenge response in healthy human male volunteers. Journal of the American Heart Association. 2014;3 doi: 10.1161/JAHA.114.001084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gaudet D, et al. Targeting APOC3 in the familial chylomicronemia syndrome. The New England journal of medicine. 2014;371:2200–2206. doi: 10.1056/NEJMoa1400284. [DOI] [PubMed] [Google Scholar]
- 139.Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annual review of pharmacology and toxicology. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 140.Fey RA, et al. Local and systemic tolerability of a 2'O-methoxyethyl antisense oligonucleotide targeting interleukin-4 receptor-alpha delivery by inhalation in mouse and monkey. Inhalation toxicology. 2014;26:452–463. doi: 10.3109/08958378.2014.907587. [DOI] [PubMed] [Google Scholar]
- 141.Geary RS, et al. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Advanced drug delivery reviews. 2015 doi: 10.1016/j.addr.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 142.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & development. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Steijger T, et al. Assessment of transcript reconstruction methods for RNA-seq. Nature methods. 2013;10:1177–1184. doi: 10.1038/nmeth.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Du Z, et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nature structural & molecular biology. 2013;20:908–913. doi: 10.1038/nsmb.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature biotechnology. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Leary RJ, et al. Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16224–16229. doi: 10.1073/pnas.0808041105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Grasso CS, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wahlestedt C, et al. Potent and nontoxic antisense oligonucleotides containing locked nucleic acids. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5633–5638. doi: 10.1073/pnas.97.10.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gaj T, et al. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends in biotechnology. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hsu PD, et al. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bassett AR, et al. Considerations when investigating lncRNA function in vivo. eLife. 2014;3:e03058. doi: 10.7554/eLife.03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chu C, et al. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Molecular cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Engreitz JM, et al. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 2014;159:188–199. doi: 10.1016/j.cell.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Batish M, et al. Single molecule imaging of RNA in situ. Methods in molecular biology. 2011;714:3–13. doi: 10.1007/978-1-61779-005-8_1. [DOI] [PubMed] [Google Scholar]
- 157.Femino AM, et al. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- 158.Spitale RC, et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519:486–490. doi: 10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Andrews SJ, Rothnagel JA. Emerging evidence for functional peptides encoded by short open reading frames. Nature reviews. Genetics. 2014;15:193–204. doi: 10.1038/nrg3520. [DOI] [PubMed] [Google Scholar]
- 160.Ingolia NT, et al. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell reports. 2014;8:1365–1379. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]