Summary
Legionella pneumophila is a pathogenic bacterium commonly found in water. Eventually, it could be transmitted to humans via inhalation of contaminated aerosols. Iron is known as a key requirement for the growth of L. pneumophila in the environment and within its hosts. Many studies were performed to understand iron utilization by L. pneumophila but no global approaches were conducted. In this study, transcriptomic analyses were performed, comparing gene expression in L. pneumophila in standard vs. iron restricted conditions. Among the regulated genes, a newly described one, lpp_2867, was highly induced in iron restricted conditions. Mutants lacking this gene in L. pneumophila were not affected in siderophore synthesis or utilization. On the contrary, they were defective for growth on iron depleted solid media and for ferrous iron uptake. A sequence analysis predicts that Lpp_2867 is a membrane protein, suggesting that it is involved in ferrous iron transport. We thus named it IroT, for iron transporter. Infection assays showed that the mutants are highly impaired in intracellular growth within their environmental host Acanthamoeba castellanii and human macrophages. Taken together, our results show that IroT is involved, directly or indirectly, in ferrous iron transport and is a key virulence factor for L. pneumophila.
Introduction
Legionella pneumophila is a human pathogen responsible for Legionnaires’ disease, a serious form of pneumonia (Fields et al., 2002). These Gram-negative bacteria are found in freshwater environments, as well as in anthropogenic niches, like cooling towers, air conditioning systems or hot water pipes (Koide et al., 1993; Borella et al., 2004). L. pneumophila mainly multiplies within protozoa like amoebae (Rowbotham, 1980) and is also able to colonize biofilms (Declerck et al., 2007). L pneumophila infects humans via inhalation of contaminated aerosols (Steinert et al., 2002). Iron is a key nutrient for most bacteria in particular for many pathogens (Leong et al., 1974; Weinberg, 1978; Reeves et al., 1981; Ratledge and Dover, 2000; Schaible and Kaufmann, 2004). It is essential as a cofactor in various enzymatic reactions like respiration, oxidative stress response or DNA replication, but can also be a toxic element at elevated intracellular concentrations by catalyzing the Fenton reaction. Thus, to better understand L. pneumophila growth in the environment and in the host it is of primary importance to study how these bacteria are acquiring iron. It has been shown that the ability of L. pneumophila to replicate in mammalian cells or in amoebae is dependent upon iron (Cianciotto, 2007). This element is found in different chemical forms (i.e. ferric or ferrous iron) for which bacteria have developed sophisticated pathways to assimilate it (Aisen, 1976; Andrews et al., 2003).
In L. pneumophila, many genes are involved in iron acquisition pathways (for a review see (Cianciotto, 2007)). The expression of most of them is regulated by the ferric uptake regulator, Fur (Hickey and Cianciotto, 1994, 1997; Liles et al., 2000; Allard et al., 2006). Fur forms a homodimer complex with ferrous iron and acts as a transcriptional repressor by binding to specific DNA sequences (Fur boxes) in the operator region of target genes (Escolar et al., 1999). Under low iron conditions, transcriptional repression is relieved, because ferrous iron is dissociated from the Fur complex, and affinity for the Fur box is reduced.
For iron acquisition, many bacteria produce and secrete iron chelators, called siderophores that are able to bind iron with high affinity. The ferric-siderophore complex is then transported into the bacteria where the iron can be released in the cytoplasm. L. pneumophila produces a siderophore named legiobactin (Liles et al., 2000) whose expression is dependent on the lbtABC operon. The lbtA gene encodes a protein with high homology to siderophore synthetases (Allard et al., 2006). Based on this homology, lbtA has been proposed to be involved in the biosynthesis of legiobactin. The lbtB gene encodes a protein that is homologous to members of the major facilitator superfamily (MFS) of multidrug efflux pumps. Recently, it has been shown that LbtC is involved in the uptake of legiobactin and its transport across the inner membrane (Chatfield et al., 2012). Also, the lbtU gene has been described directly upstream of the lbtABC operon (Chatfield et al., 2011). Cell fractionation experiments and in silico analysis indicated that LbtU is an outer membrane protein consisting of a 16-stranded transmembrane-barrel, multiple extracellular domains, and a short periplasmic tail. LbtU is a new type of receptor that likely participates in legiobactin uptake (Chatfield et al., 2011). The Fur-regulated frgA gene encodes a protein that has sequence similarity to LbtA. FrgA is not required for production of legiobactin but may be involved in the production of another, yet-to-be-defined siderophore. FrgA mutants, unlike LbtA mutants, are defective in macrophage infection suggesting a role for the protein in intracellular iron acquisition (Hickey and Cianciotto, 1997; Liles et al., 2000; Allard et al., 2006). Another way for L. pneumophila to assimilate ferrous iron is via the FeoA–FeoB complex. This inner membrane complex participates in the ferrous iron transport and is involved in extracellular growth and intracellular infection (Robey and Cianciotto, 2002). Also, L. pneumophila secretes a pyomelanin pigment that confers ferric reductase activity and thereby helps to promote ferrous iron assimilation (Zheng et al, 2013).
In this study, we used a transcriptomic approach to investigate the effect of iron depletion on the global gene expression in L. pneumophila and further analyzed the role of a newly identified iron-regulated gene in iron acquisition and host cell infection.
Results
Iron limitation results in induction of Fur-regulated and transmissive phase genes
To identify additional iron regulated genes in L. pneumophila, we performed transcriptome analyses using whole genome microarrays carrying all genes of strain L. pneumophila Paris and Lens (Cazalet et al., 2004) and L. pneumophila strain Philadelphia (Chien et al., 2004) in low iron medium. L. pneumophila Paris was grown in BYE-iron to OD600 0.8 and then the iron chelator DFX 20 μM was added. After growth of 30 and 180 min in these iron-depleted conditions, total RNA was isolated and used for microarray analysis. Microarray results are based on three independent experiments hybridised in duplicates with dye swap. At 180 min, 113 genes were induced and 246 genes were repressed significantly (Supplementary Table S1 and S2 respectively). A selection of these genes is listed in Tables 1, 2 and 3.
Table 1.
Genes induced upon iron depletion
| Gene ID | Description | FC 30min | FC 180min |
|---|---|---|---|
| Iron related genes | |||
| lpp0651_sufA | Iron sulfur cluster assembly transcriptional regulator (SufA) | 1.94 | |
| lpp0652_sufB | Similar to ABC transporter- permease component (SuFB) | 2.05 | |
| lpp0653_sufC | Similar to ABC transporter ATP-binding protein | 2.03 | |
| lpp0654_sufD | Similar ABC transporter- permease component | 1.97 | |
| lpp0655_sufS | Similar 10 cysteine desulfurase and 10 selenocysteine lyase | 1.81 | |
| lpp0656+fifU | NifU protein family- involved in the formation or repair of [Fe-S] clusters | 1.88 | |
| lpp0657_sufT | Similar to conserved hypothetical protein | 2.10 | |
| lpp0658_poxA | Similar to putative lysyl-tRNA synthetase | 1.44 | |
| lpp1278_lbtC | Similar to drug resistance transporter- MFS superfamily | 4.28 | 3.30 |
| lpp1279_lbtB | Similar to multidrug resistance efflux pump protein | 6.98 | 4.55 |
| lpp1280_lbtA | Similar to FrgA (Iron- and Fur- regulated gene)- iron repressed gene | 10.09 | 12.69 |
| lpp1281_lbtU | Similar to protein | 1.54 | |
| lpp2710_lpp2710 | Hypothetical gene | 2.28 | 2.59 |
| lpp2711_feoB | Ferrous iron transporter B | 2.18 | 2.59 |
| lpp2712_feoA | Ferrous iron transporter A | 1.94 | |
| lpp2846_frgA | FrgA protein | 11.07 | 24.88 |
| lpp2867_lpp2867 | Putative membrane protein | 4.66 | 6.01 |
| Tramissive phase related genes | |||
| lpp0541_lpp0541 | Similar to putative sigma-54 modulation protein | 1.64 | |
| lpp0542_rpoN | RNA polymerase sigma-54 factor (sigma-L) | 1.25 | |
| lpp0602_letE | Transmission trait enhancer protein LetE | 1.57 | |
| lpp1224_flgB | Flagellar basal-body rod protein FlgB | 1.61 | |
| lpp1230_flgH | Flagellar L-ring protein precursor FlgH | 1.52 | |
| lpp1294_flaA | Flagelline | 1.32 | |
| lpp1723_fliG | Flagellar motor switch protein | 1.39 | |
| lpp1725_fliE | Flagellar hook-basal body complex protein | 1.60 | |
| lpp1726_fleR | Similar to two-component response regulator | 1.38 | |
| Miscelaneous genes | |||
| lpp0608_lpp0608 | Similar to putative outer membrane lipoproteins | 1.42 | |
| lpp0642_glnB | Nitrogen regulatory protein | 1.37 | |
| lpp0677_lpp0677 | Similar to hypothetical protein- predicted membrane protein | 1.35 | |
| lpp0699_rtxA-1 | Structural toxin protein RtxA | 1.24 | |
| lpp1117_lpp1117 | Similar to chitinase | 2.34 | |
| lpp1118_lpp1118 | Similar to B. subtilis PaiA transcriptional repressor of sporulation | 1.28 | |
| lpp1170_lpp1170 | Regulatory protein (GGDEF and EAL domains) | 1.27 | |
| lpp1209_lpp1209 | Similar to conserved hypothetical protein | 1.45 | |
| lpp1738_rir1 | Similar to ribonucleoside-diphosphate reductase- alpha subunit | 1.80 | 2.57 |
| lpp1739_rir2 | Similar to ribonucleoside-diphosphate reductase- beta subunit | 2.22 | 2.73 |
| lpp1774_lysAC | Similar to diaminopimelate decarboxylase- aspartate kinase | 1.57 | |
| lpp1883_gst | Glutathione S-transferase | 1.40 | |
| lpp2350_cecA1 | Chemiosmotic efflux system C protein A | 1.49 | |
| lpp2591_lpp2591 | lpp2591 | 1.39 | |
| lpp2594_lpp2594 | lpp2594 | 3.21 | |
| lpp2595_aroF | phospho-2-dehydro-3-deoxyheptonate aldolase | 1.43 | |
| lpp2675_lpp2675 | Weakly similar to cysteine protease | 1.36 | |
| lpp2692_enhC | Enhanced entry protein EnhC | 1.30 | |
| lpp2781_lpp2781 | Some similarity with eukaryotic proteins | 1.40 | |
| lpp2804_recR | Recombination and repair protein recR | 1.37 | |
| plpp0124 | Bifunctional protein. similar to acetyl transferase and to methyl transferase | 1.60 | |
| plpp0125 | Similar to acetyltransferase. GNAT family | 1.65 | |
| plpp0126 | Similar to conserved hypothetical protein | 1.85 | |
| plpp0127 | Similar to acetyltransferase (C-terminal part) | 2.14 | |
| plpp0128 | Unknown | 2.43 | |
| plpp0129 | Some similarity with transcriptional regulator. MerR family | 3.20 | |
Table 2.
Genes repressed upon iron depletion, related to translation.
| Gene ID | Description | FC 30 min | FC 180 min |
|---|---|---|---|
| Translation | |||
| lpp0381_secE | Preprotein translocase secE subunit | 0.77 | 0.42 |
| lpp0382_nusG | Transcription antitermination protein NusG | 0.50 | |
| lpp0383_rplK | 50S ribosomal protein L11 | 0.48 | |
| lpp0384_rplA | 50S ribosomal protein L1 | 0.41 | |
| lpp0385_rplJ | 50S ribosomal subunit protein L1 | 0.51 | |
| lpp0386_rplL | 50S ribosomal subunit protein L7/L12 | 0.44 | |
| lpp0387_rpoB | RNA polymerase B-subunit | 0.45 | |
| lpp0388_rpoC | RNA polymerase beta subunit | 0.74 | |
| lpp0389_rpsL | 30S ribosomal protein S12 | 0.69 | |
| lpp0390_rpsG | 30S ribosomal protein S7 | 0.76 | 0.55 |
| lpp0391_fusA | Translation elongation factor G | 0.70 | 0.49 |
| lpp0392_tufA2 | Translation elongation factor Tu | 0.65 | |
| lpp0393_rpsJ | 30S ribosomal subunit protein S1 | 0.47 | |
| lpp0394_rplC | 50S ribosomal subunit protein L3 | 0.79 | 0.45 |
| lpp0395_rplD | 50S ribosomal subunit protein L4 | 0.74 | 0.48 |
| lpp0396_rplW | 50S ribosomal subunit protein L23 | 0.74 | 0.47 |
| lpp0397_rplB | 50S ribosomal subunit protein L2 | 0.74 | 0.46 |
| lpp0398_rpsS | 30S ribosomal subunit protein S19 | 0.77 | 0.47 |
| lpp0399_rplV | 50S ribosomal subunit protein L22 | 0.44 | |
| lpp0400_rpsC | 30S ribosomal protein S3 | 0.76 | 0.45 |
| lpp0401_rplP | 50S ribosomal protein L16 | 0.52 | |
| lpp0402_rpmC | 50S ribosomal subunit protein L29 | 0.73 | 0.49 |
| lpp0403_rpsQ | 30S ribosomal protein S17 | 0.74 | 0.44 |
| lpp0404_rplN | 50S ribosomal protein L14 | 0.77 | 0.50 |
| lpp0405_rplX | 50S ribosomal protein L24 | 0.50 | |
| lpp0406_rplE | 50S ribosomal protein L5 | 0.77 | 0.47 |
| lpp0407_rpsN | 30S ribosomal protein S14 | 0.70 | 0.42 |
| lpp0408_rpsH | 30S ribosomal protein S8 | 0.71 | 0.45 |
| lpp0409_rplF | 50S ribosomal subunit protein L6 | 0.48 | |
| lpp0410_rplR | 50S ribosomal subunit protein L18 | 0.70 | 0.42 |
| lpp0411_rpsE | 30S ribosomal subunit protein S5 | 0.73 | 0.43 |
| lpp0412_rpmD | 50S ribosomal subunit protein L3 | 0.79 | 0.44 |
| lpp0413_rplO | 50S ribosomal subunit protein L15 | 0.78 | 0.48 |
| lpp0416_rpsM | 30S ribosomal protein S13 | 0.71 | |
| lpp0417_rpsK | 30S ribosomal protein S11 | 0.76 | |
| lpp0418_rpsD | 30S ribosomal subunit protein S4 | 0.54 | |
| lpp0419_rpoA | DNA-directed RNA polymerase alpha chain | 0.78 | 0.45 |
| lpp0420_rplQ | 50S ribosomal protein L17 | 0.43 | |
| lpp0463_rplS | 50S ribosomal protein L19 | 0.54 | |
| lpp0466_rpsP | Highly similar to 30S ribosomal protein S16 | 0.64 | |
| lpp0526_purH | Similar to purH | 0.59 | |
| lpp0527_prmA | Ribosomal protein L11 methyltransferase | 0.61 | |
| lpp1376_rpsA | 30S ribosomal protein S1 | 0.60 | |
| lpp1547_rplI | 50S ribosomal protein L9 | 0.76 | 0.49 |
| lpp1548_lpp1548 | Similar to protein | 0.70 | |
| lpp1549_rpsR | 30S ribosomal subunit protein S18 | 0.71 | |
| lpp1676_rrf | Ribosome recycling factor | 0.75 | 0.47 |
| lpp1677_pyrH | Uridylate kinase (UK) (Uridine monophosphate kinase) | 0.72 | 0.44 |
| lpp1678_tsf | Elongation factor Ts (EF-Ts) | 0.74 | 0.45 |
| lpp1679_rpsB | 30S ribosomal protein S2 | 0.48 | |
| lpp2703_rpmA | 50S ribosomal protein L27 | 0.60 | |
| lpp2704_rplU | 50S ribosomal protein L21 | 0.79 | |
| lpp2706_pth | Similar to peptidyl-tRNA hydrolase | 0.80 | 0.47 |
| lpp2819_rbfA | Ribosome-binding factor A | 0.57 | |
| lpp2820_infB | Translation initiation factor IF-2 | 0.65 | |
| lpp2821_nusA | Transcription elongation protein nusA | 0.50 |
Table 3.
Genes repressed upon iron depletion, related to various functions
| Gene ID | Description | FC 30 min | FC 180 min |
|---|---|---|---|
| Lipids | |||
| lpp0528_accC | Biotin carboxylase (A subunit of acetyl-CoA carboxylase) | 0.54 | |
| lpp0529_accB | acetyl-CoA carboxylase biotin carboxyl carrier protein | 0.60 | |
| lpp0572_fabZ | (3R)-hydroxymyristoyl-[acyl carrier protein]dehydratase | 0.77 | |
| lpp1347_plsX | Fatty acid/phospholipid synthesis protein | 0.61 | |
| lpp1348_fabH | 3-oxoacy1-[acyl-carrier-protein] synthase III | 0.58 | |
| lpp1349_fabD | Malonyl CoA-acyl carrier protein transacylase | 0.58 | |
| lpp1350_fabG | 3-oxoacyl-[acyl-carrier protein] reductase | 0.59 | |
| lpp1351_acp | Acyl carrier protein (ACP) | 0.66 | |
| lpp1352_fabF2 | 3-oxoacyl-[acyl-carrier-protein] synthase II (Beta-ketoacyl-ACP synthase II) | 0.57 | |
| Nucleotide synthesis | |||
| lpp0004_gyrB | DNA gyrase- subunit B (type II topoisomerase) | 0.75 | |
| lpp0113_polA | DNA polymerase I | 0.70 | |
| lpp0320_rhlE | Similar to ATP-dependent RNA helicase RhlE | 0.74 | 0.72 |
| lpp3002_rho | Transcription termination factor Rho | 0.48 | |
| Membrane bioenergetics | |||
| lpp0920_ccmC | Heme exporter protein CcmC | 0.75 | |
| lpp0923_ccmF | Cytochrome C-type biogenesis protein CcmF | 0.73 | |
| lpp0924_ccmG | Cytochrome C biogenesis protein | 0.73 | |
| lpp2824_nuoM | NADH-quinone oxidoreductase chain M | 0.69 | |
| lpp2825_nuoL | NADH-quinone oxidoreductase chain L | 0.60 | |
| lpp2827_nuoJ | NADH-quinone oxidoreductase chain J | 0.78 | 0.49 |
| lpp2829_nuoH | NADH-quinone oxidoreductase chain H | 0.65 | |
| lpp2832_nuoE | NADH dehydrogenase I chain E | 0.72 | |
| lpp2833_nuoD | NADH dehydrogenase I chain D | 0.72 | |
| lpp2834_nuoC | NADH dehydrogenase I chain C | 0.75 | |
| lpp2961_coxA | Cytochrome c oxidase- subunit I | 0.73 | |
| lpp2962_coxB | Cytochrome c oxidase- subunit II | 0.70 | |
| lpp3052_atpC | Highly similar to H+transporting ATP synthase epsilon chain | 0.66 | |
| lpp3053_atpD | Highly similar to H+transporting ATP synthase beta chain | 0.75 | 0.45 |
| lpp3054_atpG | Highly similar to H+transporting ATP synthase chain gamma | 0.70 | 0.36 |
| lpp3055_atpA | Highly similar to H+transporting ATP synthase chain alpha | 0.76 | 0.44 |
| lpp3056_atpH | Highly similar to H+transporting ATP synthase chain delta | 0.78 | 0.41 |
| lpp3057_atpF | Highly similar to H+transporting ATP synthase chain b | 0.73 | |
| lpp3058_atpE | Highly similar to H+transporting ATP synthase chain c | 0.59 | |
| Phase modification | |||
| lpp0845_csrA | Global regulator CsrA | 0.78 | |
| lpp2757_sspA | Similar to stringent starvation protein A | 0.74 | |
| Carbohydrate metabolism | |||
| lpp1388_deoC | Similar to 2-deoxyribose-5-phosphate aldolase | 0.74 | |
| lpp1389_xapA | Similar to xanthosine phosphorylase | 0.77 | 0.68 |
| lpp1390_lpp1390 | Similar to cytidine/deoxycytidine deaminase Cdd | 0.69 | 0.62 |
| lpp1460_aceF | Pyruvate dehydrogenase (dihydrolipoyltransacetylase component) E2p | 0.71 | 0.63 |
| lpp1461_aceE | Pyruvate dehydrogenase (decarboxylase component) E1p | 0.72 | 0.55 |
| lpp1462_lpp1462 | Cystein rich protein | 0.77 | |
| Miscallenous | |||
| lpp1500_lpp1500 | Similar to conserved hypothetical protein | 0.79 | |
| lpp1502_lpp1502 | Hypothetical protein | 0.77 | |
| lpp1504_lpp1504 | Similar to conserved hypothetical protein | 0.61 | 0.44 |
| lpp1505_ndk | Similar to nucleoside diphosphate kinase | 0.71 | 0.56 |
Among the induced genes, those known to be regulated by iron were found the most highly induced ones, already after 30 min of growth (Table 1). First, frgA (lpp_2846) corresponded to the most highly induced gene in our conditions. FrgA is similar to LbtA and might be involved in siderophore synthesis (Hickey and Cianciotto, 1997; Allard et al., 2006). Second, lbtABC (lpp_1278-1280) were also highly induced. They are involved in the legiobactin siderophore synthesis and transport (Allard et al., 2006; Chatfield et al., 2012). Third were feoA and feoB (Robey and Cianciotto, 2002; Chatfield and Cianciotto, 2007) that form an operon. FeoB is a ferrous iron transporter, important for extracellular and intracellular growth. Fourth, lbtU (lpp_1281) encodes a legiobactin transporter allowing L. pneumophila to uptake ferric iron (Chatfield et al., 2011). Fifth, the entire operon lpp_0651-0658 was induced, which encodes proteins similar to SufABCDST and NifU. The Suf proteins are involved in iron-sulfur cluster biogenesis (Roche et al., 2013). These clusters are found in the so called iron-sulfur proteins that are involved in various pathways such as electron transfer, redox catalysis and regulation of gene expression (Py and Barras, 2010; Roche et al., 2013).
Many genes defined as transmissive phase genes (Bruggemann et al., 2006) were also induced in our transcriptome results (Table 1). The transmissive phase of L. pneumophila is induced due to nutritional deprivation, like it is the case at the end of the replication cycle within the host. At this stage, bacteria become flagellated, more cytotoxic and infectious (Byrne and Swanson, 1998; Molofsky and Swanson, 2004). Our data show that LetE and sigma 54 were induced already in exponential growth phase as well as many genes involved in flagella synthesis, suggesting that iron limitation triggers phase transition in L. pneumophila. Also, enhC and rtxA expressions were significantly up-regulated. These genes have been described to be involved in host entry (Cirillo et al., 2000). Finally, rir1 and rir2, encoding the ribonucleoside diphosphate reductase, were induced even after 30 min. These enzymes are usually iron-dependent and are involved in the reductive synthesis of deoxyribonucleotides from their corresponding ribonucleotides. Recently, similar enzymes have been shown to be also induced response to iron limitation in E. coli, these enzymes are Mn-dependent (Andrews, 2011).
Iron limitation results in repression of translation and metabolic pathways
Among the repressed genes, many are involved in protein biosynthesis (Table 2). Most ribosomal proteins were repressed, even after 30 min. This suggests that iron limitation leads to a quasi shutdown of the translation machinery. Also, many genes involved in membrane bioenergetics (respiratory chain and ATP synthase) and in metabolism of carbon, lipids and nucleotides were repressed (Table 3). Taken together, the genes identified as repressed indicate that there is an arrest of the major metabolic activity in the bacteria in response to iron limitation.
lpp_2867, a newly identified iron regulated gene
lpp_2867, which was not described to be regulated by iron in the literature, was actually one of the highest induced genes (fold change 6.01 at 180 min) as seen by microarray analyses, and was already induced even after 30 min (Table 1). As regulation of iron metabolism is mainly controlled by the Fur regulator (Hickey and Cianciotto, 1994), we searched for Fur-boxes as described for E. coli, in the genome of L. pneumophila. Sequences highly similar to putative Fur boxes were found upstream of genes already known to be iron regulated (feoA, frgA, lbtA, lbtU) and, importantly, upstream of lpp_2867 (Fig. 1). Furthermore, when comparing the location of the Fur-boxes and the transcriptional start sites (TSS) defined for these genes by TSS mapping using directional RNAseq (Sahr et al., 2012) these correlated perfectly. In order to confirm the regulation of these genes upon iron limitation, qRT-PCR experiments were performed confirming the results obtained from our transcriptome analyses (Table S3 Supporting information).
Figure 1. Potential lpp_2867 Fur box as compared to the consensus Fur box.
Alignment of the putative Fur-boxes of all genes found to be iron-regulated are given. Conserved residues are coloured in the same way.
Interestingly, a BLAST analysis shows that Lpp_2867 is conserved (identity >96%) in all L. pneumophila genomes sequenced to date. This protein was also found in all Legionella sp. genomes although less conserved (identity >56%) but not in other bacteria present in public databases. This suggests that Lpp_2867 is a protein specific for the genus Legionella. The ortholog of lpp_2867 in L. pneumophila 130b (98% identity), which is the best characterized strain for iron metabolism, is designated lpw_30711. The genomic organization is conserved in both strains (Paris and 130b) and they are monocistronic. Lpp_2867/Lpw_30711 is a protein of 660 amino acids. The BLAST analysis predicted the presence of a conserved domain of the DUF3816 family (Pfam database). This family of proteins includes membrane transporters, suggesting that Lpp_2867/ Lpw_30711 might be a transporter.
lpw_30711 and lpp_2867 mutant strains are impaired in growth on iron-depleted solid medium
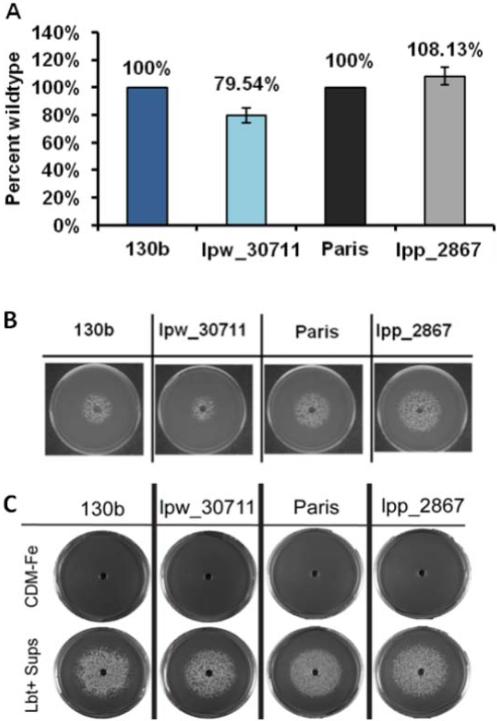
In order to understand the role of lpw_30711 and lpp_2867, mutants of strains 130b and Paris were made by allelic exchange. A gentamycin cassette was inserted in these two genes, leading to disruption and deletion from positions 538 to 1326 of the genes. To determine the capability of the mutants to grow on iron-restricted media, L. pneumophila Paris and 130b were cultured on BCYE plates lacking iron supplementation with or without DFX (ferric iron chelator). The wild type strains and their mutants grew similarly on standard BCYE agar, which is routinely supplemented with 0.25 g of ferric pyrophosphate per liter (Fig. 2A). The mutants also grew similarly to wild type when cultured on BCYE agar that lacks the iron supplementation, suggesting that yeast extract contains traces of iron (Fig. 2B). In contrast, an feoB mutant, used as a control, was impaired for growth on this medium. Together, these data indicate that the mutants do not have a generalized growth defect. However, the two mutants were impaired for growth on BCYE plates lacking iron supplementation and containing DFX (Fig. 2C), indicating that the lpp_2867 / lpw_30711 gene is required for optimal growth on iron-depleted conditions in both strains. Besides, complemented strains were tested in the same conditions and there was no evidence of complementation (data not shown). It could be that plasmid copy was toxic and actually reduced growth in these conditions. In summary, Lpw_30711 and Lpp_2867 are not absolutely required for extracellular growth in bacteriological media but, as iron becomes restricted in agar media, the proteins are needed for optimal extracellular replication.
Figure 2. Growth of the lpw_30711 and lpp_2867 mutants is impaired on low iron BCYE plates.
Growth of L. pneumophila strains on BCYE plates with various amount of iron (A) on standard BCYE plates, (B) on BCYE without iron supplementation, and (C) on BCYE without iron supplementation and with 14 μM DFX. Dilutions of wild type 130b and Paris strains, lpw_30771 and lpp_2867 mutants, or feoB mutant were spotted on each medium and incubated for growth. To avoid interference between different strains, a single column of each strain was spotted on one plate. The results are representative of three independent experiments.
Mutants are not defective for siderophore production or utilization
As a first step towards understanding the impaired growth of the lpw_30711 and lpp_2867 mutants on low-iron media, we tested the strains for production of siderophore (Fig. 3). For this purpose, the different strains were grown in deferrated CDM at 37 °C leading to the production of legiobactin, and then at 24 h post-inoculation the cell-free supernatants were filter-sterilized. Siderophore activity in the supernatants was confirmed using the CAS (Chrome Azurol S) assay, with DFX serving as the standard (Allard et al., 2006). The CAS assay did not show significant differences for the siderophore production between the wild type and the mutant strains (Fig. 3A). Subsequently, the supernatants of cultures where L. pneumophila WT and mutants had been grown were assessed for legiobactin bioactivity by examining their ability to rescue the growth of an L. pneumophila ferrous transport (feoB) mutant on BCYE plates without iron supplementation. The mutant supernatants had bioactivity comparable to wild type (Fig. 3B). In order to analyse the ability of the L. pneumophila strains to use legiobactin, each strain was spread on BCYE plates without iron supplementation but containing 10 μM DFX. A well was made at the centre of each plate and 75 μl of legiobactin containing supernatant was added to each well. Negative-control wells contained equal volumes of deferrated CDM. The CAS-positive supernatant facilitated the growth of the strains (Fig. 3C) in iron limited condition. Taken together, our data suggest that mutants are not impaired for siderophore production or utilization.
Fig 3. lpw_30711 and lpp_2867 mutants are not impaired in siderophore production or utilization.
(A) CAS assay comparing wild type and mutant supernatants for levels of siderophore activity. Data represent the mean +/- SD of duplicate cultures. Siderophore activity of the mutants was not statistically different from that of its parental strains after three independent experiments. (B) Siderophore production: the bioassay was performed on both wild type strains and their respective mutants. Supernatants of each strain were tested for siderophore biological activity by examining their ability to promote the growth of the NU269 feoB mutant on non-iron supplemented BCYE agar. The results are representative of three independent experiments. (C) Siderophore utilization: wild type 130b and Paris strains and the corresponding lpw_30771 and lpp_2867 mutants were spread on BCYE agar without iron supplementation and containing 10 μM DFX. A well was made at the center of each plate and 75 μl of deferrated CDM (upper row) or legiobactin-containing supernatant (lower row) were added to each well. Growth was recorded after 6 days of incubation at 37°C. The results are representative of three independent experiments.
lpw_30711 and lpp_2867 are required for optimal acquisition of ferrous iron
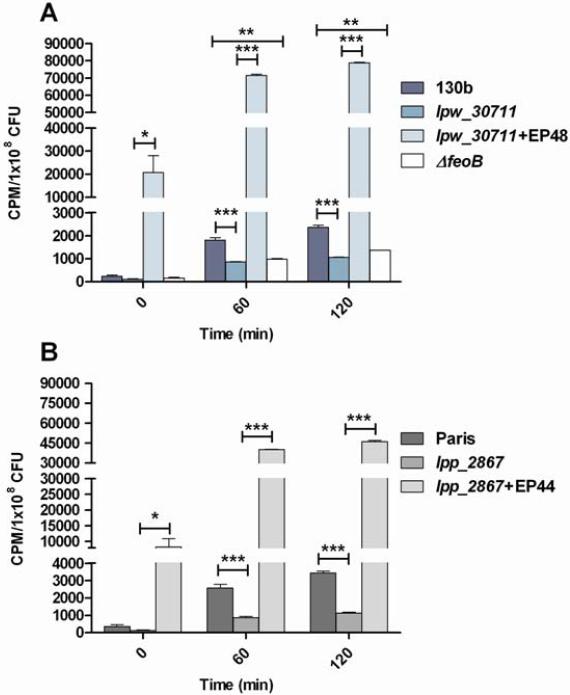
As a next step towards investigating the role of lpw_30711 / lpp_2867 in iron metabolism, we compared the wild type,mutant strains and complemented strains in ferrous iron uptake assays. Both the lpw_30711 and lpp_2867 mutants were impaired for ferrous iron uptake, but not as much as the feoB mutant (Fig. 4A-B). The lpw_30711 / lpp_2867 mutants incorporated radioactive ferrous iron at a level that was significantly below that of the wild-type after both 60 min and 120 min (Fig. 4). Also, the addition of lpw_30711 or lpp_2867 in trans (plasmid) allowed to complement the transport defect. In the complemented strains the transport of ferrous iron was largely higher than in wild type strains suggesting that the number of gene copy influenced iron transport. These data indicate that lpw_30711 / lpp_2867, directly or indirectly, promotes acquisition of ferrous iron.
Figure 4. The lpw_30711 and lpp_2867 mutants are defective for ferrous iron uptake.
Strains 130b (A) and Paris (B), their respective mutant strains lpw_30711 and lpp_2867 and the complemented strains with plasmids EP 48 or EP 44 were grown in deferrated CDM, and resuspended in buffer mixed with 55FeCl3. The feoB mutant was also used as a control. After 60 and 120 min of incubation, the levels of intracellular radiolabelled Fe2+ were determined. Data represent the mean +/− SEM from triplicate, and the results are representative of three independent experiments. (*** p < 0.001 (t-test)).
Growth of the lpw_30711 and lpp_2867 mutants is impaired in macrophages and A. castellanii
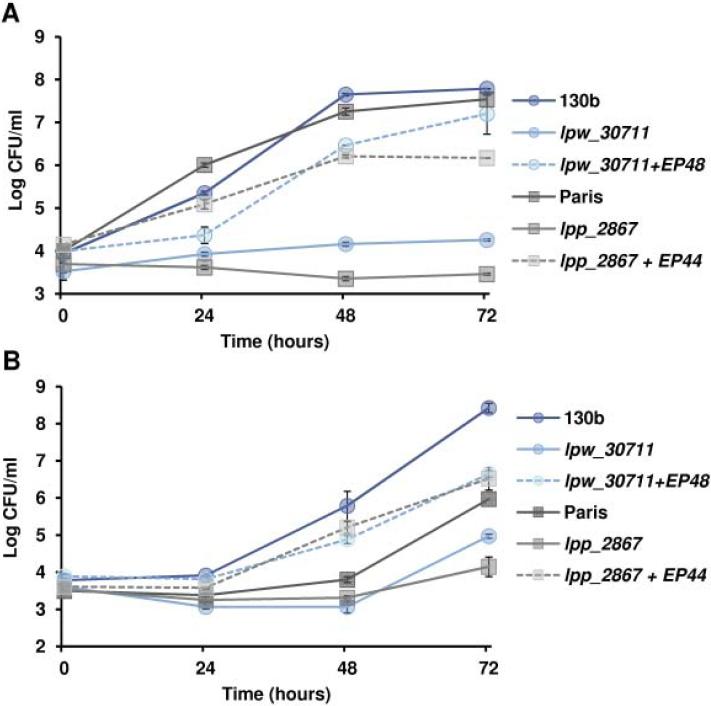
Because iron acquisition and iron-related genes are important for intracellular infection, we examined the relative ability of the lpw_30711 mutant and lpp_2867 mutant to infect macrophages or A. castellanii. Both hosts were infected with L. pneumophila and the growth was followed for 72 hours by CFU count (Fig. 5). The two mutants clearly showed a defect in intra-macrophage growth (Fig. 5A). Indeed, they hardly grew even after 72 hours. The lpw_30711 mutant exhibited a 10-fold reduced growth compared to the 130b strain at 24 hours, increasing to 104-fold at 48 and 72 hours. Results were similar for the Paris strain and its mutant. Both of the mutants also displayed a significant growth defect compared to wild type in A. castellanii, although the difference between the mutant and the parental strains was less pronounced than in macrophages (Fig. 5B).. Finally, complementation restored partly the infection ability in both macrophages and A. castellanii. Taken together, these data indicate that lpw_30711 / lpp_2867 are essential to infect both macrophages and amoebae.
Figure 5. The lpw_30711 and lpp_2867 mutants are defective for intracellular growth.
(A) Intra-macrophage (U937 cells) growth or (B) intra-amoebal (A. castellanii) growth of the wild-type strains 130b and Paris, their respective mutant strains lpw_30711 and lpp_2867 or the complemented strains with plasmids EP 48 or EP 44. Intracellular growth was monitored by CFU determination. Data represent the mean CFU +/− SEM from triplicate wells. The results are representative of three independent experiments (p < 0.05 at both the 24- 48- and 72-hour time points in (A) and 48- and 72-hour time points in (B) (t-test)).
Discussion
Iron is an essential nutrient for bacteria and for L. pneumophila in particular. This bacterium needs iron for extracellular as well intracellular growth. Many studies have described iron metabolism in L. pneumophila but no global approaches have been performed yet. Thus, here we undertook transcriptome analyses on the whole genome level to extend our knowledge of the iron response and the number of iron-regulated genes in L. pneumophila. Our whole genome expression analyses under iron limiting conditions indeed identified the genes, which had been previously described as iron regulated and we also identified, a new iron regulated gene, lpp_2867, for which we show that it plays an important role in growth and virulence of L. pneumophila.
A general analysis of the transcriptional response of L. pneumophila to iron limitation revealed that lack of iron leads to a shutdown of the translation machinery and to the induction of many transmissive phase genes as defined previously (Bruggemann et al., 2006). These results suggest that iron limitation triggers the stringent response, similar to that triggered in response to amino acid or fatty acid limitation (Dalebroux et al., 2009; Edwards et al., 2009). A similar effect of iron limitation on stringent response has been described for E. coli (Magnusson et al., 2005; Vinella et al., 2005). The stringent response is regulated in bacteria by RelA and SpoT, which produce under different stress conditions the alarmone (p)ppGpp. SpoT is the enzyme implicated in response to iron (Vinella et al., 2005; Dalebroux and Swanson, 2012). In L. pneumophila, (p)ppGpp activates the LetA-LetS two component system and the alternative sigma factor RpoS, both involved in the switch to transmissive phase. Indeed, LetA induces expression of two small RNAs (RsmY and RsmZ) that inhibit the activity of the global repressor CsrA (Sahr et al., 2009). It leads to relieve expression of transmission traits. In agreement with this hypothesis, CsrA was repressed and LetE induced in our study. Also, flagella expression, a well defined transmission trait, was induced.
Iron metabolism is regulated by the global repressor Fur, which binds promoters, at Fur boxes, in presence of iron (Escolar et al., 1999). In case of iron limitation this repression is relieved. Our study identified already known Fur-regulated genes and a new Fur regulated candidate, lpp_2867. This gene is highly induced under iron-deficient conditions and furthermore, exhibits a potential Fur box sequence upstream its transcriptional start site.
In addition to its iron-regulation, several lines of evidence indicate that lpw_30711 / lpp_2867 is involved in ferrous iron acquisition. First, mutants lacking lpw_30711 or lpp_2867 were defective for growth on solid media depleted for iron by the addition of DFX, a ferric iron chelator. The lpw_30711 and lpp_2867 mutants grew similarly to the wild type bacteria in standard BYE broth and BCYE agar, indicating that this mutant phenotype was not the result of a generalized growth defect. Second, iron uptake assays showed that the lpw_30711 and lpp_2867 mutants have decreased ferrous iron accumulation, although their decrease in iron assimilation was less pronounced compared to a feoB mutant. The fact that the two mutant phenotypes were observed for two independently derived mutants confirms that the observed phenotypes are due to inactivation of the lpw_30711 / lpp_2867 gene rather than spontaneous second-site mutation(s). That the lpw_30711 / lpp_2867 gene is monocistronic in strain 130b and strain Paris further indicates that the mutant phenotypes are due to the specific loss of lpw_30711 / lpp_2867 rather than any polar effect of the mutation. To confirm this complementation was performed and the complemented strains had a restored phenotype. Thus, the lpw_30711 / lpp_2867 gene is required, directly or indirectly, for optimal extracellular growth on low-iron agar media and ferrous iron uptake. The fact that DFX-treated broth cultures of the lpp_2867 mutant attained wild-type levels of growth by stationary phase indicates the ability of other iron acquisition systems to compensate, in some cases, for the defect in Fe2+ uptake. CAS assay revealed that lpw_30711 / lpp_2867 is not involved in siderophore production. This result was confirmed by the bioassay, in which the CAS-positive supernatant from the mutants were able to rescue the growth of the feoB mutant. Thus, Lpw_30711 / Lpp_2867 plays a conditional role in extracellular growth under iron depleted conditions and appears to be specifically involved in ferrous rather than ferric iron assimilation. Based upon bioinformatic analyses of the Lpw_30711 / Lpp_2867 protein and the iron-uptake defect, we would hypothesize that under extracellular growth conditions Lpw_30711 / Lpp_2867 promotes iron assimilation by being (part of) a membrane transporter of ferrous iron or by facilitating the formation of such a membrane transporter.
Our in vitro infection data demonstrate the importance of the lpw_30711 and lpp_2867 genes in intracellular replication. The lpw_30711 and lpp_2867 mutants exhibited a 104-fold decreased intracellular growth in human U937 cell macrophages. A defect was also noted in A. castellanii co-culture conditions, but less important than in macrophages. The fact that two independent mutants showed this intracellular growth defect coupled with the monocistronic nature of lpw_30711 / lpp_2867 indicate that Lpw_30711 / Lpp_2867 is required, directly or indirectly, for optimal intracellular infection by L. pneumophila. In addition, the complemented strains had a restored phenotype. With respect to its role in infection, the simplest hypothesis would be that Lpp_2867 / Lpw_30711 promote the membrane transport of ferrous iron that is needed for intracellular growth. However, the orthologous protein (MavN) in the L. pneumophila Philadelphia strain has been implicated as a possible substrate for the Dot/Icm type IV secretion system (Huang et al., 2011), suggesting that Lpw_30711 / Lpp_2867 might be secreted under some circumstances. Summarizing the results of our mutant analysis, the lpw_30711 / lpp_2867 gene is required, directly or indirectly, for optimal i) extracellular growth in low-iron agar media, ii) ferrous iron uptake, and iii) intracellular infection of macrophages and amoebae.
In conclusion, Lpp_2867 in strain Paris and its homolog Lpw_30711 in strain 130b are controlled by iron concentration in a Fur-dependent manner and promote iron assimilation by aiding in ferrous iron transport. Importantly, Lpp_2867 / Lpw_30711 are involved in L. pneumophila virulence, and it is proposed that the proteins might be involved in scavenging ferrous iron from host cells. Therefore, we suggest naming these proteins IroT/MavN for iron transporter.
Materials and Methods
Bacterial strains and growth conditions
L. pneumophila strain Paris CIP 107629T and strain 130b ATCC BAA-74 (also known as Wadsworth or AA100) and the mutants were grown in filter-sterilized BYE (10 g/l ACES, 10 g/l yeast extract, 1 g/l alpha-ketogluturate, pH 6.9) supplemented with L-cysteine at 0.4 g/l and iron pyrophosphate at 0.25 g/l (Sigma, St Louis, MO) or on solid medium BCYE, which is obtained by adding charcoal (1.5 g/l) and agar (15 g/l) to non-filtered BYE and autoclaved 15 min at 121 °C. The feoB mutant was constructed by insertion of a kanamycin resistance cassette via allelic exchange as described previously (Robey 2002). E. coli DH5α was used as the host for recombinant plasmids and was cultured on Luria Bertani broth (10 g/l tryptone, 5 g/l yeast extract, 10 g/l NaCl) or agar (15 g/l agar). When appropriate, media were supplemented with the following antibiotics at final concentrations suitable for L. pneumophila: gentamicin 2.5 μg/ml, chloramphenicol 5 μg/ml, kanamycin 25 μg/ml and for E. coli: chloramphenicol 30 μg/ml.
To compare the ability to grow on iron-restricted solid media, L. pneumophila were cultured on BCYE plates lacking iron supplementation with or without iron chelator DFX (Deferroxamine mesylate) as described previously (Chatfield et al., 2011). BCYE lacking the iron supplement contains approximately 14 μM iron, as determined by the ferrozine assay (Riemer et al., 2004). Bacteria were precultured for 3 days on standard BCYE agar and suspended in filtered sterilized base buffer (10.466 g/l MOPS, 0.212 g/l KH2PO4, 2.926 g/l NaCl, pH6.5) to 1×109 CFU per ml, and then 10 μl aliquots taken from 10-fold serial dilutions in PBS were spotted on the BCYE plates, BCYE-Fe plates, or BCYE-Fe plates with 10, 12, 14, or 16 μM DFX. Growth was recorded after 5 days of incubation at 37 °C.
Total RNA isolation
For the transcriptomic analyses, L. pneumophila Paris was grown in BYE without iron, under shaking (170 rpm), at 37 °C. When cells reached late exponential phase (OD600 0.8), 20 μM of DFX were added. After 30 min, 60 min and 180 min, 10 ml of suspension were collected and cells were pelleted and resuspended in 400 μl of resuspension buffer (12.5 mM Tris, 5 mM EDTA and 10% glucose). Then, 500 μl of acid phenol (pH 4.6) and 0.4 g of glass beads (0.2–0.3 mm diameter; Sigma) were added. The cells were sheared mechanically using a Fastprep apparatus (Thermo scientific). After centrifugation at 13,000 g for 5 min, the supernatant was transferred to a fresh tube, and 1 ml of Trizol reagent (Invitrogen) was added. The sample was incubated for 5 min at room temperature. Total RNA was extracted twice with chloroform.
Microarray hybridization and data analysis
RNA was prepared in triplicates (three independent cultures) and each RNA sample was hybridized twice to the microarrays (dye swap). RNA was reverse-transcribed and labelled with Cy5 or Cy3. The design of microarrays containing gene-specific 70 mer oligonucleotides based on all predicted genes of the genome of L. pneumophila strain Paris (CR628336) and its plasmids (CR628338) was previously described (Bruggemann et al., 2006). Hybridization was performed following the manufacturers’ recommendations (Corning) using 250 pmol of Cy3- and Cy5-labelled cDNA. Slides were scanned on a GenePix 4000A scanner (Axon Instruments). Laser power and/or PMT were adjusted to balance the two channels and the resulting files were analysed using Genepix Pro 4.0 software. Spots were excluded from analysis in case of high local background fluorescence, slide abnormalities or weak intensity.
Quantitative RT PCR
Quantitative RT PCR was performed on a LightCycler (Roche), using the LightCycler FastStart RNA MasterPLUS Sybr Green I kit (Roche), according to the manufacturer's instructions. The 16S rRNA gene was used as a reference gene to normalize gene expression. The level of the gene expression was assessed by determining the crossing point (Cp), cycle at which the amplification curve crossed the detection threshold. According to the study of Livak (Livak and Schmittgen, 2001), the relative changes in gene expression were determined by calculating 2−ΔΔCp, with ΔCp = Cp target gene - Cp reference gene (16S) and ΔΔCp = ΔCp sample 1 – ΔCp sample 2.
lpw_30711 and lpp_2867 mutant constructions
L. pneumophila is naturally competent thus transformation and subsequent homologous recombination of a DNA construct can be used for mutant construction (Lomma et al., 2010). To prepare these naturally competent cells, bacteria were grown on standard BCYE agar for three days at 37 °C, a colony was inoculated in BYE to obtain an OD600 0.01, and incubated at 37 °C, under agitation (170 rpm), up to the late exponential phase. For transformation, the antibiotic cassette flanked by 500 bp upstream and downstream the gene to mutate, was constructed by 3-step PCR using the one Taq Hot Start DNA polymerase (New England Biolabs Inc). The first step was the amplification of the upstream and the downstream extremities of the gene lpp2867. Primers used to perform the amplification of the upstream extremity were lpp2867-F1 (5’ GTTGGAGCAAGCCTGATAC 3’) and lpp2867-Gent-R1 (5’ CTGGGTTCGTGCCTTCATCAAAGGCCGGCAACTAAACTAAA 3’). The 3’ extremity of the reverse primer has complementarity with the upstream region of the gentamicin cassette. Primers used to amplify the downstream extremity of the gene lpp_2867 are lpp2867-Gen-F2 (5’ CCTAACAATTCGTTCAAGCCGTGACCTACAATGCCGTTACCG 3’) and lpp2867-R2 (5’ CAACGAGTGCGGAAAGAATC 3’). The 5’ extremity of the forward primer has complementarity with the 3’ extremity of the gentamicin cassette (Rolando et al., 2013). To amplify the gentamicin cassette, the two primers K7-Gent-F1 (5’ TTTAGTTTAGTTGCCGGCCTTTGATGAAGGCACGAACCCAG 3’) and K7-Gent-R1 (5’ GGTAACGGCATTGTAGGTCACGGCTTGAACGAATTGTTAGG 3’) were used. Then the three PCR fragments (10 nM for each fragment) were mixed, followed by a PCR amplification using the flanking primers (lpp2867-F1 and lpp2867-R2). The amplicon at the correct size was gel purified and 10 μg of linear DNA containing the recombinant allele carrying the antibiotic cassette (1885 pb) were added to the competent cells. After 24 hours at 30 °C, without shaking, potential mutants were selected on BCYE agar containing gentamicin (2.5 μg/ml). To confirm homologous recombination and correct mutant construction, PCR amplification was performed with primers lpp2867-F3 (5’ TCCAAATACGCCAGGGAAC 3’) and lpp2867-R3 (5’ TGGCAGAACAATCCCAGAG 3’).
Complementation of the lpw_30711 and lpp_2867 mutants
In order to complement mutant strains, a 2.2 kb fragment containing either the lpp_2867 or lpw_30711 gene with their promoter was amplified with primers lpp2867-pro-comp-XbaI-F (5’ TCTAGAGATACTACCTGATGAAACGAAT 3’) and lpp2867-comp-XbaI-R (5’ TCTAGAGTAAGGAGTATCATTAACTGAAC 3’). The amplicons were cloned into pGemTeasy (PROMEGA; Madison, WI) to yield pEP12 and pEP19, encoding lpp_2867 and lpw_30711, respectively. These fragments, lpp_2867 and lpw_30711 genes, were transferred on XbaI fragments into pMMB2002, chloramphenicol resistant, to yield pEP44 and pEP48 plasmids.
Competent L. pneumophila cells were prepared as follows: L. pneumophila were grown on standard BCYE for three days and inoculated in 50 ml of BYE at 108 cells/ml, under shacking (170 rpm), over night, at 37 °C. When cells were in stationary phase, cells suspensions were centrifuged ( 5,000 g for 15 min at 4 °C). The supernatant w solution. Finally, cells were concentrated at 1011 cells/ml in glycerol solution. For transformation, cells were transferred into a cold electroporation cuvette, and 1 μg of purified plasmid was added. Electroporation was performed using a Biorad Micropulseur apparatus (2.5 kV). After the pulse, cold BYE was added and cuvettes were incubated at 37 °C, without shaking. After two or three hours, 100 μl of suspension were spread on BCYE plates containing 5 μg/ml of chloramphenicol. Transformants were confirmed by the presence of plasmid DNA by electrophoresis on 0.8 % agarose gel.
Chrome Azurol S (CAS) assay to analyze siderophore production
In order to assess siderophore production, L. pneumophila strains were grown in deferrated CDM (Reeves et al., 1983), at 37 °C, under shaking (225 rpm) for 18 – 24 h. Supernatants were collected after centrifugation (5,000 g for 10 min) and filtered (0.2 μm). Siderophore activities were quantified using the CAS assay, with DFX serving as standard (Liles et al., 2000; Allard et al., 2006; Allard et al., 2009). Supernatants were tested for siderophore activity by examining their ability to promote the growth of the NU269 feoB mutant on non-iron supplemented BCYE agar (Allard et al., 2009). The gene feoB encode an intra-membrane Fe2+ permease and thus the mutant NU269 is defective for uptake of Fe2+ but not Fe3+ (Robey and Cianciotto, 2002). The mutant's growth deficit can be reversed by the addition of Fe3+ salt or supernatant containing siderophore.
Siderophore utilization
In order to determine the ability to utilize legiobactin, bacteria (130b, lpw_30711, Paris and lpp_2867) were pre-cultured on BCYE plates for 3 days, suspended in base buffer, and 1×104 CFU were spread on each BCYE plate (data not shown) or BCYE plate without iron supplementation but containing 10μM DFX. A well was made at the center of each plate, and 75 μl of supernatants containing legiobactin obtained from wild-type cultures grown in deferrated CDM, were deposited into the wells. Negative-control wells contained equal volumes of deferrated CDM. The plates were cultured for 6 days at 37 °C (Chatfield et al., 2012).
Iron uptake
Following the method used by Zheng et al., bacterial strains were previously grown in non-iron supplemented BYE broth until OD660 1.0 (Zheng et al., 2013). Bacteria were centrifuged at 5,000 × g, washed, and resuspended in deferrated CDM medium to an OD660 of 0.3. After 13 hours of incubation, at 37 °C, with shaking (225 rpm), the bacterial cultures were centrifuged and washed three times in base buffer (50 mM MOPS, 2 mM monobasic potassium phosphate, 50 mM sodium chloride, pH 6.5). The final bacterial pellets were resuspended in base buffer to an OD660 of 1.0, and 55FeCl3 (PerkinElmer, Boston, MA) in 10 mM HCl was added to a final concentration of 1 μCi/ml (37 kBq/ml). Vitamin C was added to a final concentration 1 mM to reduce ferric iron to ferrous iron, and allow the measure of ferrous iron uptake. After 0, 60, 120 min of incubation at room temperature, 1 ml of the suspension (n=3) was filtered through a 0.45 μm-pore-size nitrocellulose membrane (Millipore, Billerica, MA) and washed with 5 ml of 0.5% thioglycolic acid solution. The number of counts per minute (cpm) of radioactivity associated with the bacteria was measured with a Beckman LS6500 scintillation counter, and the mean of the counts per minute over a 5 min period was recorded. The experiment was done three times and similar results were obtained.
Infection assays
Assessment of the ability of the different L. pneumophila wild type and mutant strains to establish an intracellular infection was performed in both human U937 macrophages (ATCC CRL-1593.2) and A. castellanii (ATCC 30234). Growth kinetics of L. pneumophila in U937 macrophages were recorded as described previously (Viswanathan et al., 2000; Robey and Cianciotto, 2002). Briefly, U937 macrophages were cultivated in RPMI 1640 medium with L-glutamine (Cellgro) supplemented with 10% fetal bovine serum (Atlanta Biologicals) in a 5% CO2 incubator, at 37 °C. 106 adherent U937 cells were infected with bacteria at a multiplicity of infection (MOI) of 0.5. The bacterial inoculums had been grown for three days on BCYE agar. After 2 hours, required to allow the bacterial internalization, extracellular bacteria were removed by repeated washing, and then infected monolayers were incubated at 37 °C in a 5% CO2 incubator. At 24, 48 and 72 hours post-inoculation, intracellular bacteria were released by lysis of the monolayers with 10 μl of 10% saponin (Sigma). For estimation of viable cell counts, serial 10-fold dilutions from triplicate wells for each strain were plated on standard BCYE agar.
To perform co-culture with A. castellanii, amoebae were cultured in buffer made of 2% protease bacto peptone, 0.1% yeast extract, 4 mM MgSO4, 0.5 M CaCl2, 0.1% sodium citrate dihydrate, 0.05 mM Fe(NH4)2(SO4)2. 6 H2O, 2.5 mM Na2HPO4 dibasic, 2.5 mM KH2PO4 monobasic, pH 6.5, supplemented with 0.1 M of glucose. To harvest the amoebae, cultures were centrifuged and pellets were resuspended in buffer without glucose, to a concentration of 1 × 105 cells/ml. 1 ml was placed in each well of a 24-wells culture dish (NUNC), and was incubated at 35 °C. The amoebae were allowed to adhere to the wells before the addition of the bacteria. Next, 1 ml of 104 CFU/ml bacteria was added in each well. And for estimation of viable cell counts, serial 10-fold dilutions from triplicate wells for each strain were plated on standard BCYE agar.
Supplementary Material
Acknowledgments
Work in the Buchrieser lab was supported by the Institut Carnot-Pasteur MI, the Fondation pour la Recherche Médicale (FRM), the French Region Ile de France (DIM Malinf) and grant ANR-10-LABX-62-IBEID. Work in the Cianciotto lab was funded by NIH grant AI034937. We thank Jessica Tyson for her help during experiments of amoeba infection. Emilie Portier is supported by the Région Poitou-Charentes.
Footnotes
Conflict of interest
None to declare
References
- Aisen P. The binding and release of iron by transferrin. Birth Defects Orig Artic Ser. 1976;12:81–95. [PubMed] [Google Scholar]
- Allard KA, Viswanathan VK, Cianciotto NP. lbtA and lbtB are required for production of the Legionella pneumophila siderophore legiobactin. J Bacteriol. 2006;188:1351–1363. doi: 10.1128/JB.188.4.1351-1363.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard KA, Dao J, Sanjeevaiah P, McCoy-Simandle K, Chatfield CH, Crumrine DS, et al. Purification of Legiobactin and importance of this siderophore in lung infection by Legionella pneumophila. Infect Immun. 2009;77:2887–2895. doi: 10.1128/IAI.00087-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews SC. Making DNA without iron - induction of a manganese-dependent ribonucleotide reductase in response to iron starvation. Mol Microbiol. 2011;80:286–289. doi: 10.1111/j.1365-2958.2011.07594.x. [DOI] [PubMed] [Google Scholar]
- Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- Borella P, Montagna MT, Romano-Spica V, Stampi S, Stancanelli G, Triassi M, et al. Legionella infection risk from domestic hot water. Emerg Infect Dis. 2004;10:457–464. doi: 10.3201/eid1003.020707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggemann H, Hagman A, Jules M, Sismeiro O, Dillies MA, Gouyette C, et al. Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pneumophila. Cell Microbiol. 2006;8:1228–1240. doi: 10.1111/j.1462-5822.2006.00703.x. [DOI] [PubMed] [Google Scholar]
- Byrne B, Swanson MS. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect Immun. 1998;66:3029–3034. doi: 10.1128/iai.66.7.3029-3034.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalet C, Rusniok C, Bruggemann H, Zidane N, Magnier A, Ma L, et al. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet. 2004;36:1165–1173. doi: 10.1038/ng1447. [DOI] [PubMed] [Google Scholar]
- Chatfield CH, Cianciotto NP. The secreted pyomelanin pigment of Legionella pneumophila confers ferric reductase activity. Infect Immun. 2007;75:4062–4070. doi: 10.1128/IAI.00489-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatfield CH, Mulhern BJ, Burnside DM, Cianciotto NP. Legionella pneumophila LbtU acts as a novel, TonB-independent receptor for the legiobactin siderophore. J Bacteriol. 2011;193:1563–1575. doi: 10.1128/JB.01111-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatfield CH, Mulhern BJ, Viswanathan VK, Cianciotto NP. The major facilitator superfamily-type protein LbtC promotes the utilization of the legiobactin siderophore by Legionella pneumophila. Microbiology. 2012;158:721–735. doi: 10.1099/mic.0.055533-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien M, Morozova I, Shi S, Sheng H, Chen J, Gomez SM, et al. The genomic sequence of the accidental pathogen Legionella pneumophila. Science. 2004;305:1966–1968. doi: 10.1126/science.1099776. [DOI] [PubMed] [Google Scholar]
- Cianciotto NP. Iron acquisition by Legionella pneumophila. Biometals. 2007;20:323–331. doi: 10.1007/s10534-006-9057-4. [DOI] [PubMed] [Google Scholar]
- Cirillo SL, Lum J, Cirillo JD. Identification of novel loci involved in entry by Legionella pneumophila. Microbiology. 2000;146(Pt 6):1345–1359. doi: 10.1099/00221287-146-6-1345. [DOI] [PubMed] [Google Scholar]
- Dalebroux ZD, Swanson MS. ppGpp: magic beyond RNA polymerase. Nat Rev Microbiol. 2012;10:203–212. doi: 10.1038/nrmicro2720. [DOI] [PubMed] [Google Scholar]
- Dalebroux ZD, Edwards RL, Swanson MS. SpoT governs Legionella pneumophila differentiation in host macrophages. Mol Microbiol. 2009;71:640–658. doi: 10.1111/j.1365-2958.2008.06555.x. [DOI] [PubMed] [Google Scholar]
- Declerck P, Behets J, van Hoef V, Ollevier F. Replication of Legionella pneumophila in floating biofilms. Curr Microbiol. 2007;55:435–440. doi: 10.1007/s00284-007-9006-7. [DOI] [PubMed] [Google Scholar]
- Edwards RL, Dalebroux ZD, Swanson MS. Legionella pneumophila couples fatty acid flux to microbial differentiation and virulence. Mol Microbiol. 2009;71:1190–1204. doi: 10.1111/j.1365-2958.2008.06593.x. [DOI] [PubMed] [Google Scholar]
- Escolar L, Perez-Martin J, de Lorenzo V. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol. 1999;181:6223–6229. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields BS, Benson RF, Besser RE. Legionella and Legionnaires' disease: 25 years of investigation. Clin Microbiol Rev. 2002;15:506–526. doi: 10.1128/CMR.15.3.506-526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey EK, Cianciotto NP. Cloning and sequencing of the Legionella pneumophila fur gene. Gene. 1994;143:117–121. doi: 10.1016/0378-1119(94)90615-7. [DOI] [PubMed] [Google Scholar]
- Hickey EK, Cianciotto NP. An iron- and fur-repressed Legionella pneumophila gene that promotes intracellular infection and encodes a protein with similarity to the Escherichia coli aerobactin synthetases. Infect Immun. 1997;65:133–143. doi: 10.1128/iai.65.1.133-143.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Boyd D, Amyot WM, Hempstead AD, Luo ZQ, O'Connor TJ, et al. The E Block motif is associated with Legionella pneumophila translocated substrates. Cell Microbiol. 2011;13:227–245. doi: 10.1111/j.1462-5822.2010.01531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide M, Saito A, Kusano N, Higa F. Detection of Legionella spp. in cooling tower water by the polymerase chain reaction method. Appl Environ Microbiol. 1993;59:1943–1946. doi: 10.1128/aem.59.6.1943-1946.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong J, Neilands JB, Raymond KN. Coordination isomers of biological iron transport compounds. III. (1) Transport of lambda-cis-chromic desferriferrichrome by Ustilago sphaerogena. Biochem Biophys Res Commun. 1974;60:1066–1071. doi: 10.1016/0006-291x(74)90421-5. [DOI] [PubMed] [Google Scholar]
- Liles MR, Scheel TA, Cianciotto NP. Discovery of a nonclassical siderophore, legiobactin, produced by strains of Legionella pneumophila. J Bacteriol. 2000;182:749–757. doi: 10.1128/jb.182.3.749-757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lomma M, Dervins-Ravault D, Rolando M, Nora T, Newton HJ, Sansom FM, et al. The Legionella pneumophila F-box protein Lpp2082 (AnkB) modulates ubiquitination of the host protein parvin B and promotes intracellular replication. Cell Microbiol. 2010;12:1272–1291. doi: 10.1111/j.1462-5822.2010.01467.x. [DOI] [PubMed] [Google Scholar]
- Magnusson LU, Farewell A, Nystrom T. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 2005;13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Molofsky AB, Swanson MS. Differentiate to thrive: lessons from the Legionella pneumophila life cycle. Mol Microbiol. 2004;53:29–40. doi: 10.1111/j.1365-2958.2004.04129.x. [DOI] [PubMed] [Google Scholar]
- Py B, Barras F. Building Fe-S proteins: bacterial strategies. Nat Rev Microbiol. 2010;8:436–446. doi: 10.1038/nrmicro2356. [DOI] [PubMed] [Google Scholar]
- Ratledge C, Dover LG. Iron metabolism in pathogenic bacteria. Annu Rev Microbiol. 2000;54:881–941. doi: 10.1146/annurev.micro.54.1.881. [DOI] [PubMed] [Google Scholar]
- Reeves MW, Pine L, Neilands JB, Balows A. Absence of siderophore activity in Legionella species grown in iron-deficient media. J Bacteriol. 1983;154:324–329. doi: 10.1128/jb.154.1.324-329.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves MW, Pine L, Hutner SH, George JR, Harrell WK. Metal requirements of Legionella pneumophila. J Clin Microbiol. 1981;13:688–695. doi: 10.1128/jcm.13.4.688-695.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemer J, Hoepken HH, Czerwinska H, Robinson SR, Dringen R. Colorimetric ferrozine-based assay for the quantitation of iron in cultured cells. Anal Biochem. 2004;331:370–375. doi: 10.1016/j.ab.2004.03.049. [DOI] [PubMed] [Google Scholar]
- Robey M, Cianciotto NP. Legionella pneumophila feoAB promotes ferrous iron uptake and intracellular infection. Infect Immun. 2002;70:5659–5669. doi: 10.1128/IAI.70.10.5659-5669.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche B, Aussel L, Ezraty B, Mandin P, Py B, Barras F. Iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity. Biochim Biophys Acta. 2013;1827:455–469. doi: 10.1016/j.bbabio.2012.12.010. [DOI] [PubMed] [Google Scholar]
- Rolando M, Sanulli S, Rusniok C, Gomez-Valero L, Bertholet C, Sahr T, et al. Legionella pneumophila effector RomA uniquely modifies host chromatin to repress gene expression and promote intracellular bacterial replication. Cell Host Microbe. 2013;13:395–405. doi: 10.1016/j.chom.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Rowbotham TJ. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J Clin Pathol. 1980;33:1179–1183. doi: 10.1136/jcp.33.12.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahr T, Rusniok C, Dervins-Ravault D, Sismeiro O, Coppee JY, Buchrieser C. Deep sequencing defines the transcriptional map of L. pneumophila and identifies growth phase-dependent regulated ncRNAs implicated in virulence. RNA Biol. 2012;9:503–519. doi: 10.4161/rna.20270. [DOI] [PubMed] [Google Scholar]
- Sahr T, Bruggemann H, Jules M, Lomma M, Albert-Weissenberger C, Cazalet C, Buchrieser C. Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol Microbiol. 2009;72:741–762. doi: 10.1111/j.1365-2958.2009.06677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaible UE, Kaufmann SH. Iron and microbial infection. Nat Rev Microbiol. 2004;2:946–953. doi: 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- Steinert M, Hentschel U, Hacker J. Legionella pneumophila: an aquatic microbe goes astray. FEMS Microbiol Rev. 2002;26:149–162. doi: 10.1111/j.1574-6976.2002.tb00607.x. [DOI] [PubMed] [Google Scholar]
- Vinella D, Albrecht C, Cashel M, D'Ari R. Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol Microbiol. 2005;56:958–970. doi: 10.1111/j.1365-2958.2005.04601.x. [DOI] [PubMed] [Google Scholar]
- Viswanathan VK, Edelstein PH, Pope CD, Cianciotto NP. The Legionella pneumophila iraAB locus is required for iron assimilation, intracellular infection, and virulence. Infect Immun. 2000;68:1069–1079. doi: 10.1128/iai.68.3.1069-1079.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg ED. Iron and infection. Microbiol Rev. 1978;42:45–66. doi: 10.1128/mr.42.1.45-66.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Chatfield CH, Liles MR, Cianciotto NP. Secreted pyomelanin of Legionella pneumophila promotes bacterial iron uptake and growth under iron-limiting conditions. Infect Immun. 2013;81:4182–4191. doi: 10.1128/IAI.00858-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.