Summary
Single nucleotide variants (SNVs), particularly loss-of-function mutations, are significant contributors to autism spectrum disorder (ASD) risk. Here we report the first systematic deep sequencing study of 55 postmortem ASD brains for SNVs in 78 known ASD candidate genes. Remarkably even without parental samples, we find more ASD brains with mutations that are protein-altering (26/55 cases vs 12/50 controls, p=0.015), deleterious (16/55 vs 5/50, p =0.016), or loss-of-function (6/55 vs 0/50, p =0.028) compared to controls, with recurrent deleterious mutations in ARID1B, SCN1A, SCN2A, and SETD2, suggesting these mutations contribute to ASD risk. In several cases, the identified mutations and medical records suggest syndromic ASD diagnoses. Two ASD and one Fragile X premutation case showed deleterious somatic mutations, showing that somatic mutations occur in ASD cases and supporting a model in which a combination of germline and/or somatic mutations may contribute to ASD risk on a case-by-case basis.
Introduction
Autism spectrum disorder (ASD) affects ≥1% of American children and is characterized by 1) deficits in social interaction and social communication and 2) restricted and repetitive behaviors, interests, or activities (Wingate et al., 2014). ASD is genetically heterogeneous, with contributions from heritable mutations—including monogenic conditions, common variants, and rare recessive mutations—as well as from de novo mutations (Huguet et al., 2013). Whereas genetic studies of ASD risk have analyzed DNA from blood, saliva, or cell lines (De Rubeis et al., 2014; Iossifov et al., 2014; O'Roak et al., 2011), postmortem ASD brain samples have been utilized mostly to study neuropathology, transcriptomics, and splicing (Chen et al., 2015; Irimia et al., 2014). Apart from a set of postmortem autism brain samples analyzed for copy number variants (CNVs) (Wintle et al., 2011), we know little of the underlying genetics of these postmortem autism brains. Identifying pathogenic mutations in these brains would aid genotype-phenotype correlation and interpretation of other alterations. In addition, somatic mutations—de novo mutations occurring post-fertilization—are of growing interest in neurodevelopmental disorders that have similarly high de novo mutation rates to ASD, or with similar or overlapping symptoms, and somatic mutations have been reported in case studies with ASD (Poduri et al., 2013). Targeted deep sequencing allows detection of germline and somatic mutations in developmental brain disorders, especially when brain tissue is directly analyzed (D'Gama et al., 2015; Lee et al., 2012; Poduri et al., 2012; Riviere et al., 2012) and sequence coverage is very high (Jamuar et al., 2014). We used targeted deep sequencing of brain tissue from ASD patients and neurotypical controls to investigate genetic mechanisms underlying ASD risk in these patients, including possible contributions of somatic mutations.
Results
We performed ultra-deep sequencing on DNA from postmortem cortical and/or cerebellar brain samples from 55 ASD cases, 50 neurotypical controls, and 21 cases with suspected ASD or related diagnoses (Figure 1A, Table S1). In order to achieve the high coverage necessary to sensitively detect somatic mosaic mutations, we used a targeted panel of 78 ASD candidate genes showing dominant or X-linked inheritance (see Supplemental Experimental Procedures) (Basu et al., 2009; Betancur, 2011; O'Roak et al., 2012). We achieved 547X average coverage, with 95.7% of the analyzable target regions covered by ≥100 reads and 48.2% covered by ≥1000 reads, providing sensitivity to detect somatic mutations in these genes to a level of ≈5% alternate allele frequency (AAF). We identified 408 ± 69 SNVs and indels per sample and validated all rare, high-quality, protein-altering variants (missense, nonsense, splice site, indel, and frameshift) using Sanger sequencing; any potential somatic variants were also validated with subcloning followed by Sanger sequencing of individual clones, to determine mosaicism using a technology independent of the discovery platform.
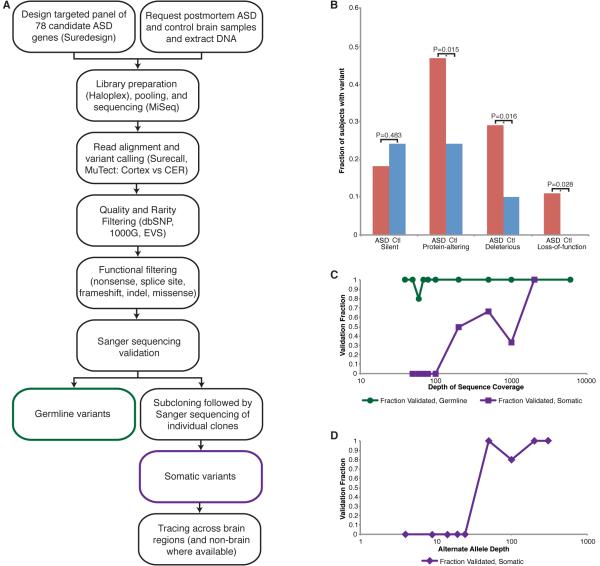
Figure 1. Targeted deep sequencing identifies germline and somatic mutations in ASD candidate genes in postmortem ASD brain.
A. Study schematic. DNA extracted from ASD and control brain samples was sequenced at high coverage using a targeted panel of candidate ASD genes. After variant calling and filtering, protein-altering variants were validated using Sanger sequencing and subcloning, and somatic variants were traced across the brain and non-brain tissues, when available. B. Fraction of cases and controls harboring synonymous, protein-altering, deleterious (subset of protein-altering), or loss-of-function (subset of deleterious) variants. p values were calculated using a two-tailed Fisher’s exact test. C. Validation of germline and somatic variants according to total depth of sequence coverage. Essentially all germline variants validated; somatic variants were more likely to validate at higher total depth (≥100X). D. Validation of somatic variants according to alternate allele depth of coverage. Somatic variants were more likely to validate at alternate allele depth ≥25X. Each data point represents the validation fraction for all variants with b. total or c. alternate allele depth from the previous data point to that data point. See also Tables S1, S3-S5.
Remarkably, despite the modest sample of ASD brains available for sequencing, and the absence of familial samples for testing segregation, damaging mutations were strikingly more common in ASD brains than control brains. Overall, we identified 34 protein-altering variants in 25 genes in ASD cases and 15 protein-altering variants in 13 genes in neurotypical controls (Table S2). Compared to controls, significantly more ASD cases harbored protein-altering variants (26/55 vs 12/50, p=0.015), deleterious variants (16/55 vs 5/50, p=0.016)—i.e., loss-of-function and missense predicted to be deleterious by at least three prediction tools—and loss-of-function variants (nonsense, splice site, and frameshift, 6/55 vs 0/50, p=0.028), whereas functionally neutral variants were equally common in cases and controls (10/55 vs 12/50, p=0.483, two-tailed Fisher’s exact tests) (Figure 1B, Table S3). The excess of protein-altering variants in ASD cases compared to controls was driven by the deleterious and particularly the loss-of-function variants, consistent with previous reports (Sanders et al., 2012). Recurrent loss-of-function and/or deleterious missense variants occurred in four genes (ARID1B, SCN1A, SCN2A, and SETD2) in ASD cases (Table 1), consistent with previous WES studies (De Rubeis et al., 2014; Iossifov et al., 2014). We also identified protein-altering variants in cases with suspected ASD or related diagnoses (Table S2). Although parental samples are not available to test for the de novo nature of these variants, the large excess of deleterious variants in ASD brains strongly suggests that most of the deleterious and loss of function variants contributed to ASD pathogenesis.
Table 1.
Deleterious Mutations in ASD and Fragile X Cases
| Case | Diagnosis | Gene | Mutation | cDNA | Protein | Type |
|---|---|---|---|---|---|---|
| 797 | Autism | ADNP | Ms | c.3281G>T | p.G1094V | Germline |
| AN16641 | Autism | ARID1B | Ms | c.4237C>T | p.P1413S | Germline |
| AN17515 | Autism | ARID1B | Ms | c.1463C>G | p.P488R | Germline |
| 5340 | Autism/Parkinson's Disease | ATP10A | Ms | c.622C>T | p.R208W | Germline |
| AN16641 | Autism | CACNA1C | Ms | c.3416G>A | p.R1139H | Germline |
| 5006 | Fragile X, premutation | CACNA1C | St | c.2T>C | (start loss) | Somatic |
| 5278 | Autism | CACNA1H | Ms | c.5909C>G | p.S1970C | Germline |
| UK28768 | Autism | CHD8 | Ms | c.2230G>A | p.V744I | Germline |
| 5297 | Autism | GRIN2A | Ms | c.2650G>A | p.D884N | Germline |
| 797 | Autism | PQBP1 | Ms | c.731C>T | p.P244L | Germline |
| AN01570 | Autism | PTEN | Ns | c.195C>A | p.Y65* | Germline |
| 5278 | Autism | SCN1A | Sp | c.602+1G>A | N/A | Somatic |
| AN16115 | Autism | SCN1A | Ms | c.4319C>T | p.A1440V | Germline |
| 4849 | Autism | SCN2A | Ms | c.2021C>A | p.T674K | Germline |
| AN04166 | Autism | SCN2A | Ms | c.5230G>A | p.G1744R | Germline |
| AN08043 | Autism | SCN2A | Sp | c.4551+1G>C | N/A | Germline |
| AN09714 | Autism | SCN2A | Ns | c.4543C>T | p.R1515* | Germline |
| 1349 | Autism | SEMA5A | Ns | c.586C>T | p.R196* | Germline |
| UK20244 | Autism | SETD2 | Ms | c.4871C>G | p.S1624C | Somatic |
| AN00090 | Autism/Angelman Syndrome | SETD2 | Ms | c.1463A>G | p.Y488C | Germline |
| AN16641 | Autism | SLC6A8 | Fs | c.205_206delG | p.A69Pfs*28 | Germline |
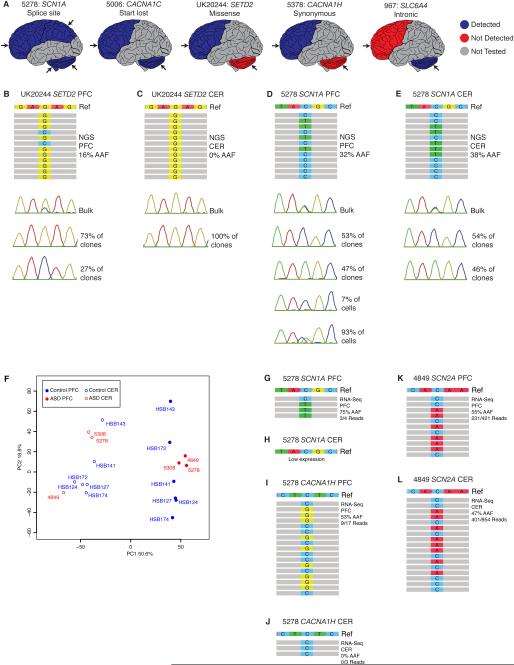
To identify de novo somatic variants, we validated all rare, high quality, protein-altering variants with AAF <40% (see Supplemental Experimental Procedures). Although NGS identified many potential somatic variants, extensive validation showed some to be false positives or germline and not bona fide somatic (Tables S4-5). Potential somatic variants with a higher total read depth (≥100, Figure 1C) and higher alternate allele read depth (≥25, Figure 1D) were more likely to validate. We identified protein-altering somatic variants in this 78-gene targeted panel in 2 ASD cases (Figure 2A and Table S6), with no protein-altering somatic variants identified in control brains even after extensive validation. Case UK20244, a 16-year-old male with ASD, harbors a somatic missense variant in SETD2, predicted to be deleterious, present in frontal cortex but undetectable in cerebellum (Figure 2B-C), which may have contributed to his ASD symptoms and death. Case 5278, a 15-year-old female with ASD, harbors a somatic splice site mutation in SCN1A, present across brain and in non-brain tissues at relatively high AAF (Figure 2D-E). We confirmed the mosaic nature of this high AAF mutation by fluorescent-activated nuclear sorting (FANS), using antisera to NeuN to separate neuronal and non-neuronal nuclei from the prefrontal cortex, followed by multiple displacement amplification of single cell genomes (Dean et al., 2001; Evrony et al., 2012). Genotyping genome-amplified DNA from single nuclei confirmed that the mutation was mosaic, with 93 ± 1.6% of cells (corrected for allelic dropout, see Experimental Procedures) harboring the mutation and the remainder not carrying the mutation (Figure 2D). This mutation has been previously reported in epileptic encephalopathies (Allen et al., 2013; Depienne et al., 2010; Fujiwara et al., 2003), and indeed Case 5278 suffered generalized epilepsy in addition to ASD. While we focused our attention on protein-altering somatic variants, we also validated a synonymous variant in CACNA1H present in frontal cortex but not detected in cerebellum of Case 5378, a 22-year-old male with Social Anxiety Disorder, and an intronic variant in SLC6A4, predicted to be benign, present in cerebellum but not detected in frontal cortex of Case 967, a 32-year-old male with suspected autism (Figure 2A and Table S6).
Figure 2. Regional distribution of somatic mutations and transcriptomic analysis of ASD brains.
A. Protein-altering somatic mutations in ASD (5278, UK20244), suspected ASD (967), Social Anxiety Disorder (5378) or Fragile X premutation (5006) brain. Blue indicates brain region tested where the mutation was detected, red indicates brain region tested where the mutation was not detected, and grey indicates regions not tested. Arrows show approximate location of the sample tested. B-E. Detection and validation of protein-altering somatic mutations in ASD brains. Each panel depicts representative NGS reads, Sanger sequencing trace from bulk brain tissue, and Sanger sequencing traces from individual clones following subcloning for Case UK20244, mutation in SETD2, B. PFC and C. CER; Case 5278, mutation in SCN1A, D. PFC and E. CER. In addition, D depicts single nuclei Sanger sequencing traces for Case 5278, mutation in SCN1A, PFC. F. PCA plot of samples analyzed for RNA-sequencing. G-L. Detection of mutations in RNA-seq data. Each panel depicts representative RNA seq reads and read counts for Case 5278, mutation in SCN1A, G. PFC and H. CER; Case 5278, mutation in CACNA1H, I.PFC and J. CER; Case 4849, mutation in SCN2A, J. PFC and L. CER. CER: cerebellum; PFC: prefrontal cortex. See also Tables S6-7.
Review of available medical records and OMIM identified consistent genotype-phenotype correlations in many cases with deleterious and loss-of-function variants (Table 2). Case 797, a 9-year-old male with autism, harbors missense mutations in ADNP and PQBP1 predicted to be deleterious. Mutations (mostly loss-of-function) in both genes are associated with intellectual disability (ID) syndromes, and the same PQBP1 missense mutation we identified was recently reported as potentially causative in two brothers with ID (Redin et al., 2014). While medical records did not indicate non-nervous system symptoms, which are commonly seen in the associated syndromes, Case 797 had language and developmental delay, attention deficit disorder, and mild hypotonia, suggesting the missense mutations may contribute to a milder form of the associated syndromes. Case AN01570, an 18-year-old female with autism, harbors a nonsense mutation in PTEN previously reported in Cowden syndrome (Tan et al., 2011). She had her first seizure at age 8 and was diagnosed with epilepsy, and her autopsy records noted her large brain size (2100 grams at 18 years old; head circumference measurement was not available), suggestive of autism with macrocephaly associated with PTEN mutations. Case AN16641, a 9-year-old male with autism, harbors a frameshift mutation in SLC6A8 and missense mutations in ARID1B and CACNA1C predicted to be deleterious. He has a family history of seizures and speech delay, and developed febrile seizures at almost 3 years of age. He was noted to be hyperactive and was diagnosed with language delay, ID, and a multiplex developmental disorder. His symptoms suggest a diagnosis of cerebral creatine deficiency syndrome 1, associated with mutations in SLC6A8, and potential contribution from ARID1B, associated with ID.
Table 2.
Syndromic ASD cases suggested by genotype-phenotype correlation
| Case | Gender | Age | Cause of Death |
Mutation | Associated Syndrome | Clinical Symptoms |
|---|---|---|---|---|---|---|
| 797 | M | 9 | Drowning |
ADNP Ms PQBP1 Ms |
HVDAS RENS1, ID (Redin et al. 2014) |
Language and developmental delay, ADD, mild hypotonia |
| AN01570 | F | 18 | SUDEP | PTEN LOF | Cowden syndrome (Tan et al. 2011), autism with macrocephaly |
Large brain size noted at autopsy, epilepsy |
| AN16641 | M | 9 | SUDEP |
ARID1B Ms CACNA1C Ms SLC6A8 LOF |
MRD12 (Timothy syndrome) CCDS1 |
FH seizures and speech delay, febrile seizures from 3 years, language delay, hyperactivity, ID, multiplex developmental disorder |
| 5278 | F | 15 | Drowning, seizure disorder |
CACNA1H Ms SCN1A LOF |
Epilepsy susceptibility Dravet syndrome, GEFSP2, FEB3A (Fujiwara et al. 2003, Depienne et al. 2010, Allen et al. 2013) |
Progressed from febrile seizures in infancy to non-febrile and generalized tonic-clonic seizures |
| AN16115 | F | 11 | Drowning, seizure disorder |
SCN1A Ms | Dravet syndrome (Gaily et al. 2013), GEFSP2, FEB3A |
FH seizures and neuropsychiatric disease, progressed from febrile seizures in infancy to febrile, tonic-clonic, and potentially myoclonic seizures, mild ID, hyperlexia |
| AN09714 | M | 60 | Pancreatic cancer |
SCN2A LOF (SGSM3 Ms predicted to be benign) |
EIEE11, BFIS3 | Seizures from 3 years old, likely episode of status epilepticus, delayed motor milestones, ID |
Three cases harbor mutations in ion channel genes associated with seizure disorders, and presented with symptoms suggestive of epilepsy. Case 5278, a 15-year old female with autism, harbors a missense mutation in CACNA1H predicted to be deleterious and a somatic splicing mutation in SCN1A, previously reported in epileptic encephalopathies, as mentioned above (Allen et al., 2013; Depienne et al., 2010; Fujiwara et al., 2003). After febrile seizures in early infancy, she developed nonfebrile and generalized tonic-clonic seizures, suggestive of generalized epilepsy with febrile seizures plus type 2 (GEFSP2). Case AN16115, an 11-year old female with autism, harbors a missense mutation in SCN1A predicted to be deleterious; a mutation to a different amino acid at the same position was previously reported in Dravet syndrome (Gaily et al., 2013). She had a family history of seizures and neuropsychiatric disease, and was diagnosed with mild ID and hyperlexia. She had her first febrile seizure at 4.5 months old, and developed febrile, tonic-clonic, and potentially myoclonic seizures, consistent with a SCN1A mutation. Case AN09714, a 60-year-old male with autism and a nonsense mutation in SCN2A, showed delayed motor milestones and ID. His first seizure was at age 3, and at 15 he was hospitalized with pneumonia and experienced seizures every 5 minutes for a 12-hour period, suggestive of an SCN2A-associated syndrome like epileptic encephalopathy, early infantile, 11 (EIEE11). These three cases highlight the complex relationship between ASD and seizures (Jeste and Geschwind, 2014). Based on available medical records, almost half of the ASD cases had seizures while there was no evidence of seizures in controls.
Fragile X accounts for ~2% of ASD cases (Huguet et al., 2013), and the Fragile X mutation is subject to somatic expansion (Lokanga et al., 2013), so we also tested postmortem brain samples from ASD and 10 Fragile X cases for expansion of the CGG repeat in the 5’ UTR of FMR1. We did not detect unsuspected expansion in any ASD cases, but did find mosaic expansion in three Fragile X mutation and premutation cases (Figure S1). Case 1421, a 69-year-old male with Fragile X, harbored a mosaic full mutation in prefrontal cortex. Case 5006, an 85-year-old male diagnosed with a premutation, harbored a mosaic full mutation in both prefrontal cortex and cerebellum. We also detected a somatic null mutation in CACNA1C in Case 5006 that disrupts the start methionine, present in both prefrontal cortex and cerebellum (Table S6), suggesting that this mutation might contribute to pathogenesis. Case 4664, a 71-year-old male diagnosed with a premutation and an unspecified neurological disorder, harbored a mosaic full mutation in prefrontal cortex not detected in cerebellum. These data show that Fragile X premutations and mutations can show somatic instability in the brain, with potentially distinct effects in different brain regions.
To test if mutations were detectable in RNA and hence affect the transcriptome, we performed RNA-sequencing (RNA-seq) on total RNA with acceptable RNA integrity (RIN≥5) extracted from prefrontal cortex and cerebellar samples of ASD cases with protein-altering somatic mutations and/or sodium channel mutations. The ASD cases were compared to matched (age and sex) neurotypical controls (Table S1). After quality control, batch correction, and quantile normalization (see Supplemental Experimental Procedures), the expression values for protein coding genes were calculated. Consistent with previous findings (Kang et al., 2011), principal component analysis (PCA) confirmed that both ASD and control PFC and CER samples have distinctive transcriptional profiles. PCA also revealed that the ASD PFC and CER samples clustered based on their regional identity and not the disease state (Figure 2F), indicating that changes in the transcriptome associated with ASD are smaller than global transcriptional differences between the two analyzed regions.
RNA-seq data analysis confirmed the presence of several germline and somatic mutations in the ASD cases (Figure 2G-L). The germline CACNA1H and somatic SCN1A mutations were detected in case 5278, and the germline SCN2A mutation was detected in case 4849. Low gene expression, low read depth in RNAseq, and/or potential destabilization of RNA by the mutation might contribute to instances where mutant alleles were not detected (e.g., Figure 2H). To further elucidate transcriptomic differences, we identified genes that exhibited the highest fold differences in expression between ASD and control samples (see Supplemental Experimental Procedures). As each case sample had distinct mutations and there were no biological replicates, each case sample was compared with two control samples and only genes that had an absolute fold change of 2 and were changed in the same direction between case and both control samples were considered (Table S7). GO enrichment analysis identified neuron and synapse-related annotation clusters (data not shown), but larger sample sizes would be required to fully characterize global effects of these mutations on transcriptome.
Discussion
Our analysis provides a unique resource of genetically characterized, publically available human postmortem ASD brains for functional studies, and supports a heterogeneous model of ASD risk, where germline and/or de novo somatic mutations contribute on a case-by-case basis. In many cases, we identify a single variant predicted to be deleterious or loss-of function, including four independent variants in SCN2A, currently one of the strongest ASD candidate genes (Hoischen et al., 2014). Similarly to previous studies (O'Roak et al., 2011), we also identify several ASD cases with multiple germline variants predicted to be deleterious. As discussed above, case AN16641 harbors a frameshift mutation in SLC6A8 and missense variants in ARID1B and CACNA1C predicted to be deleterious, and Case 797 harbors missense variants in ADNP and PQBP1 predicted to be deleterious. In addition, Case AN00090 harbors a germline missense variant in SETD2 predicted to be deleterious and a previously identified 15q deletion consistent with her diagnosis of Angelman syndrome (Wintle et al., 2011). Finally, we identify a case with a somatic mutation and a case with a germline and somatic variant: Case UK20244 harbors a somatic missense variant in SETD2 predicted to be deleterious and Case 5278 harbors a somatic splice site mutation in SCN1A and a germline missense variant in CACNA1H predicted to be deleterious. These cases underscore the complex genetic architecture of ASD, and the need for detailed phenotypic information to aid interpretation of identified genetic variants.
Despite targeting only the best-validated ASD genes, we identified two ASD and one Fragile X premutation case with protein-altering somatic mutations, providing evidence that somatic mutations can occur in ASD, though the overall contribution of somatic mutations to ASD risk cannot yet be defined. We observed a somatic mutation that likely occurred early in embryonic development, present across the brain and non-brain tissues (SCN1A), as well as somatic mutations that likely occurred later, after separation of the three germ layers and even after separation of the forebrain, midbrain, and hindbrain. Hence, somatic mutations that are undetectable in current clinical assays using blood-derived DNA could be important mediators of disease expression or penetrance and modify germline variants. The abnormal “patches” observed in the prefrontal and temporal cortex of some autism cases (Casanova et al., 2013; Stoner et al., 2014) may represent visible consequences of somatic mutations, and it will be important to understand how the cell type and brain region in which a somatic mutation occurs modifies its effect on phenotype. With our small sample size and focused gene list, we did not detect a significant burden of rare protein-altering somatic mutations in ASD cases compared to controls (p=0.5, two-tailed Fisher’s exact test). Somatic mutations may occur in genes not targeted by our panel, since we only studied genes implicated in ASD via germline mutations. Other more severe mutations, such as chromosome trisomies or activating AKT3 mutations, can be tolerated in the mosaic state while lethal in the germline (Poduri et al., 2012; Poduri et al., 2013). Larger sample sizes examined with high-coverage (≥200X) sequencing will be needed to further assess the full contribution of somatic mutations to ASD and other neuropsychiatric diseases, like schizophrenia, where germline risk mutations have been slower to emerge.
Experimental Procedures
Human tissue and DNA samples
Research performed on de-identified postmortem human specimens was approved by the institutional review boards of Boston Children’s Hospital and Yale School of Medicine. Case details are available in Table S1.
DNA extraction and targeted sequencing
Brain DNA was extracted using the QIAamp DNA Mini kit (Qiagen) and libraries prepared using a custom Haloplex Target Enrichment Kit (Agilent) according to the manufacturer’s protocols. We performed sequencing on MiSeq sequencers (Illumina). Further details on DNA sequencing and analysis are available in the Supplemental Experimental Procedures. Targeted sequencing data is deposited in NDAR Collection 2029.
Variant validation
Rare and protein-altering (nonsynonymous, nonsense, splice-site, frameshift, and insertion-deletion) variants in the target genes were validated using Sanger sequencing. For potential somatic variants, the original DNA was amplified using polymerase chain reaction (PCR), subcloned into a TOPO TA vector (Invitrogen), and transformed into TOP10 chemically competent Escherichia coli cells (Invitrogen); multiple clones were isolated and sequenced to quantify the degree of mosaicism independently of the NGS platform.
Fragile X analysis
CGG repeat assay was performed using the AmplideX® FMR1 PCR Reagents (Asuragen) and analyzed using GeneMapper® Software (Life Technologies) according to the manufacturer’s protocol.
Single nuclei analysis
Isolation of single neuronal and non-neuronal nuclei using fluorescence-activated nuclear sorting (FANS) with antisera to NeuN, and single cell, whole genome-amplification using multiple displacement amplification (MDA) (Dean et al., 2001) were performed as described previously (Evrony et al., 2012). Single nuclei from the prefrontal cortex of Case 5278 were isolated, amplified, and sequenced for the splicing mutation in SCN1A using PCR (GoTaq Hot Start DNA Polymerase, Promega). Sequencing analysis to calculate the number of cells with the mutation, taking into account allelic dropout, was performed as described previously (Evrony et al., 2012).
RNA extraction and sequencing
Total RNA was extracted using mirVana kit (Ambion) with some modifications to the manufacturer’s protocol and we prepared libraries using TruSeq Stranded Total RNA with Ribo-Zero Gold Kit (Illumina) according to the manufacturer’s protocol. We performed sequencing on HiSeq 2000 sequencer (Illumina). Further details on RNA sequencing and analysis are available in the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Aldo Rozzo, R. Sean Hill, and Jennifer N. Partlow for logistical assistance, Timothy W. Yu and Annapurna Poduri for help with panel design, Madelen Diaz for help with DNA extractions, Jian Wang and Yiping Shen for MiSeq sequencing support, the Dana Farber Cancer Institute Flow Cytometry Core, the Biopolymers Facility at Harvard Medical School, Boston Children’s Hospital IDDRC (P30 HD18655), Stela Filipovic-Sadic of Asuragen, Inc. for help with Fragile X analysis, and Robert Johnson, Jane Pickett, and Carolyn Sloan for help obtaining samples. Postmortem human tissue was obtained from the NIH NeuroBioBank at the University of Maryland, Baltimore, MD, the Oxford Brain Bank, Oxford, UK (supported by the Medical Research Council, Brains for Dementia Research and the NIHR Oxford Biomedical Research Center), and Autism BrainNet (sponsored by the Simons Foundation and Autism Speaks; the Autism Tissue Program was the predecessor to Autism BrainNet; via the Harvard Brain Tissue Resource Center, Belmont, MA and The Centre for Applied Genomics Genome Resource Facility, The Hospital for Sick Children, Toronto, ON) and tissue collection in the Sestan lab. A.M.D. was supported by the NIGMS (T32GM007753) and NRSA (5T32 GM007226-39) and is a Stuart H.Q. & Victoria Quan Fellow at Harvard Medical School. R.E.R was supported by the NIGMS (T32GM007753). S.P., M.L. and N.S. were supported by the National Institute of Mental Health (P50MH106934, UO1MH103339, and U01MH106874). C.A.W. was supported by the National Institute of Mental Health (R01MH083565 and U01MH106883), the Simons Foundation (178093), and the Manton Center for Orphan Disease Research. C.A.W. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
A.M.D., S.S.J., and C.A.W. designed DNA experiments. A.M.D, S.S.J., and A-T.N.L. obtained tissue samples and extracted DNA, prepared sequencing libraries, analyzed sequencing data, and performed validation. A.M.D and R.E.R. isolated single nuclei and performed MDA and genotyping. S.P., M.L., and N.S. designed RNA experiments, and S.P. and M.L. extracted RNA, prepared sequencing libraries, and analyzed sequencing data. A.M.D. and C.A.W. wrote the manuscript, and all coauthors edited the manuscript.
References
- Allen AS, Berkovic SF, Cossette P, Delanty N, Dlugos D, Eichler EE, Epstein MP, Glauser T, Goldstein DB, Han Y, et al. De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu SN, Kollu R, Banerjee-Basu S. AutDB: a gene reference resource for autism research. Nucleic acids research. 2009;37:D832–836. doi: 10.1093/nar/gkn835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur C. Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain research. 2011;1380:42–77. doi: 10.1016/j.brainres.2010.11.078. [DOI] [PubMed] [Google Scholar]
- Casanova MF, El-Baz AS, Kamat SS, Dombroski BA, Khalifa F, Elnakib A, Soliman A, Allison-McNutt A, Switala AE. Focal cortical dysplasias in autism spectrum disorders. Acta neuropathologica communications. 2013;1:67. doi: 10.1186/2051-5960-1-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JA, Penagarikano O, Belgard TG, Swarup V, Geschwind DH. The emerging picture of autism spectrum disorder: genetics and pathology. Annual review of pathology. 2015;10:111–144. doi: 10.1146/annurev-pathol-012414-040405. [DOI] [PubMed] [Google Scholar]
- D'Gama AM, Geng Y, Couto JA, Martin B, Boyle EA, LaCoursiere CM, Hossain A, Hatem NE, Barry B, Kwiatkowski DJ, et al. mTOR pathway mutations cause hemimegalencephaly and focal cortical dysplasia. Annals of neurology. 2015;77:720–725. doi: 10.1002/ana.24357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean FB, Nelson JR, Giesler TL, Lasken RS. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome research. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depienne C, Trouillard O, Gourfinkel-An I, Saint-Martin C, Bouteiller D, Graber D, Barthez-Carpentier MA, Gautier A, Villeneuve N, Dravet C, et al. Mechanisms for variable expressivity of inherited SCN1A mutations causing Dravet syndrome. Journal of medical genetics. 2010;47:404–410. doi: 10.1136/jmg.2009.074328. [DOI] [PubMed] [Google Scholar]
- Evrony GD, Cai X, Lee E, Hills LB, Elhosary PC, Lehmann HS, Parker JJ, Atabay KD, Gilmore EC, Poduri A, et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Sugawara T, Mazaki-Miyazaki E, Takahashi Y, Fukushima K, Watanabe M, Hara K, Morikawa T, Yagi K, Yamakawa K, Inoue Y. Mutations of sodium channel alpha subunit type 1 (SCN1A) in intractable childhood epilepsies with frequent generalized tonic-clonic seizures. Brain : a journal of neurology. 2003;126:531–546. doi: 10.1093/brain/awg053. [DOI] [PubMed] [Google Scholar]
- Gaily E, Anttonen AK, Valanne L, Liukkonen E, Traskelin AL, Polvi A, Lommi M, Muona M, Eriksson K, Lehesjoki AE. Dravet syndrome: new potential genetic modifiers, imaging abnormalities, and ictal findings. Epilepsia. 2013;54:1577–1585. doi: 10.1111/epi.12256. [DOI] [PubMed] [Google Scholar]
- Hoischen A, Krumm N, Eichler EE. Prioritization of neurodevelopmental disease genes by discovery of new mutations. Nature neuroscience. 2014;17:764–772. doi: 10.1038/nn.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huguet G, Ey E, Bourgeron T. The genetic landscapes of autism spectrum disorders. Annual review of genomics and human genetics. 2013;14:191–213. doi: 10.1146/annurev-genom-091212-153431. [DOI] [PubMed] [Google Scholar]
- Iossifov I, O'Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Weatheritt RJ, Ellis JD, Parikshak NN, Gonatopoulos-Pournatzis T, Babor M, Quesnel-Vallieres M, Tapial J, Raj B, O'Hanlon D, et al. A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell. 2014;159:1511–1523. doi: 10.1016/j.cell.2014.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamuar SS, Lam AT, Kircher M, D'Gama AM, Wang J, Barry BJ, Zhang X, Hill RS, Partlow JN, Rozzo A, et al. Somatic mutations in cerebral cortical malformations. The New England journal of medicine. 2014;371:733–743. doi: 10.1056/NEJMoa1314432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeste SS, Geschwind DH. Disentangling the heterogeneity of autism spectrum disorder through genetic findings. Nature reviews Neurology. 2014;10:74–81. doi: 10.1038/nrneurol.2013.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Huynh M, Silhavy JL, Kim S, Dixon-Salazar T, Heiberg A, Scott E, Bafna V, Hill KJ, Collazo A, et al. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nature genetics. 2012;44:941–945. doi: 10.1038/ng.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokanga RA, Entezam A, Kumari D, Yudkin D, Qin M, Smith CB, Usdin K. Somatic expansion in mouse and human carriers of fragile X premutation alleles. Human mutation. 2013;34:157–166. doi: 10.1002/humu.22177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, Karakoc E, Mackenzie AP, Ng SB, Baker C, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nature genetics. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, Carvill G, Kumar A, Lee C, Ankenman K, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poduri A, Evrony GD, Cai X, Elhosary PC, Beroukhim R, Lehtinen MK, Hills LB, Heinzen EL, Hill A, Hill RS, et al. Somatic activation of AKT3 causes hemispheric developmental brain malformations. Neuron. 2012;74:41–48. doi: 10.1016/j.neuron.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poduri A, Evrony GD, Cai X, Walsh CA. Somatic mutation, genomic variation, and neurological disease. Science. 2013;341:1237758. doi: 10.1126/science.1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redin C, Gerard B, Lauer J, Herenger Y, Muller J, Quartier A, Masurel-Paulet A, Willems M, Lesca G, El-Chehadeh S, et al. Efficient strategy for the molecular diagnosis of intellectual disability using targeted high-throughput sequencing. Journal of medical genetics. 2014;51:724–736. doi: 10.1136/jmedgenet-2014-102554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviere JB, Mirzaa GM, O'Roak BJ, Beddaoui M, Alcantara D, Conway RL, St-Onge J, Schwartzentruber JA, Gripp KW, Nikkel SM, et al. De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nature genetics. 2012;44:934–940. doi: 10.1038/ng.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoner R, Chow ML, Boyle MP, Sunkin SM, Mouton PR, Roy S, Wynshaw-Boris A, Colamarino SA, Lein ES, Courchesne E. Patches of disorganization in the neocortex of children with autism. The New England journal of medicine. 2014;370:1209–1219. doi: 10.1056/NEJMoa1307491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MH, Mester J, Peterson C, Yang Y, Chen JL, Rybicki LA, Milas K, Pederson H, Remzi B, Orloff MS, Eng C. A clinical scoring system for selection of patients for PTEN mutation testing is proposed on the basis of a prospective study of 3042 probands. American journal of human genetics. 2011;88:42–56. doi: 10.1016/j.ajhg.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingate M, Kirby RS, Pettygrove S, Cunniff C, Schulz E, Ghosh T, Robinson C, Lee LC, Landa R, Constantino J, et al. Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years - Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2010. 2014 Mmwr Surveill Summ 63. [PubMed] [Google Scholar]
- Wintle RF, Lionel AC, Hu P, Ginsberg SD, Pinto D, Thiruvahindrapduram B, Wei J, Marshall CR, Pickett J, Cook EH, Scherer SW. A genotype resource for postmortem brain samples from the Autism Tissue Program. Autism research : official journal of the International Society for Autism Research. 2011;4:89–97. doi: 10.1002/aur.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.