Summary
Noncoding expansions of a hexanucleotide repeat (GGGGCC) in the C9orf72 gene are the most common cause of familial amyotrophic lateral sclerosis and frontotemporal dementia. Here we report transgenic mice carrying a bacterial artificial chromosome (BAC) containing the full human C9orf72 gene with either a normal allele (15 repeats) or disease-associated expansion (~100-1000 repeats; C9-BACexp). C9-BACexp mice displayed pathologic features seen in C9orf72 expansion patients, including widespread RNA foci and repeat associated non-ATG (RAN) translated dipeptides, which were suppressed by antisense oligonucleotides targeting human C9orf72. Nucleolin distribution was altered supporting that either C9orf72 transcripts or RAN dipeptides promote nucleolar dysfunction. Despite early and widespread production of RNA foci and RAN dipeptides in C9-BACexp mice, behavioral abnormalities and neurodegeneration were not observed even at advanced ages, supporting the hypothesis that RNA foci and RAN dipeptides occur presymptomatically, and are not sufficient to drive neurodegeneration in mice at levels seen in patients.
Introduction
Amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD) are disorders with overlapping clinical presentation, genetics, and pathology (Chen-Plotkin et al., 2010). ALS is a form of motor neuron disease that leads to muscle wasting with spasticity from loss of upper and lower motor neurons, while FTLD patients typically develop behavioral and language disturbances due to frontal and temporal cortical degeneration. Expansions of a GGGGCC hexanucleotide repeat in the first intron/promoter of the C9orf72 gene are the most commonly identified genetic cause of both familial and sporadic ALS and FTLD (DeJesus-Hernandez et al., 2011; Renton et al., 2011). More recently, C9orf72 repeat expansions were reported in other diseases, including Alzheimer's disease, Parkinson's disease, corticobasal syndrome, Lewy body dementia, and Huntington-like syndromes (Cooper-Knock et al., 2014).
It remains uncertain whether the repeat expansion in C9orf72 causes neurodegeneration primarily through a toxic gain of function, loss of function, or both. The repeat is transcribed in both the sense and antisense directions and leads to accumulations of repeat containing RNA foci in patient tissues (Gendron et al., 2013). These RNA foci may bind RNA binding proteins and alter RNA metabolism (Donnelly et al., 2013; Lee et al., 2013; Sareen et al., 2013), and aborted transcripts containing the repeat can also disrupt nucleolar function (Haeusler et al., 2014). Additionally simple dipeptide repeats can be generated by repeat associated non-ATG dependent (“RAN”) translation of both the sense and antisense strands that have a variety of toxic effects (Ash et al., 2013; Mori et al., 2013). While it is clear that high levels of exogenous of DPR proteins (GA or GR/PR) can be toxic to cells, it remains unclear whether they can do so at the lower levels at which they are present in human tissues and CSF (Su et al., 2014).
Here we generated transgenic mice that express the C9orf72 hexanucleotide repeat in the context of the normal genetic control elements driven from a bacterial artificial chromosome (BAC) isolated from a C9orf72 expansion patient. We observed sense and antisense transcription of the repeat with development of RNA foci, as well as production of DPR protein levels similar to those seen in C9orf72 FTLD patient tissues, but without the development of neurodegeneration. These data suggest that RNA foci and RAN translation occur presymptomatically in patients, and are not sufficient to drive neurodegeneration in the absence of other stressors or overexpression.
Results
Generation of C9orf72 BAC transgenic mice
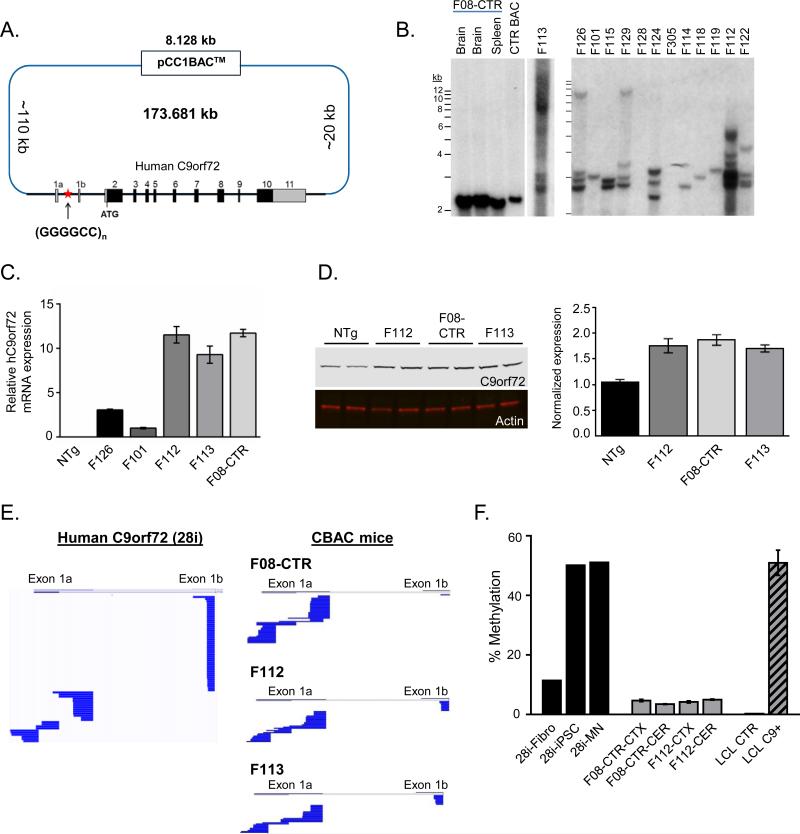
We generated a BAC library from skin fibroblasts of a C9-ALS patient that had ~800 repeats by southern blot (Sareen et al., 2013). BAC clone 239 was ~174 kb, and contained the complete C9orf72 gene with the expansion (Figure 1A). Southern blot confirmed that the GGGGCC expansion was present in the BAC, but also highly unstable in bacteria (Figure S1A). To generate control mice with a normal (2-30) repeat length, we injected a subclone derived from BAC 239 that had stably contracted to 15 repeats in bacteria, generating line F08-CTR (Figure S1A, Figure 1B). To generate mice with expansions in the disease range (C9-BACexp), we injected a subclone which had a major repeat size of ~800 (Figure S1A). The resulting founders showed a variety of C9orf72 repeat sizes in the genome on southern blot, ranging from ~100 to 1000 (Figure 1B). The banding pattern remained stable between littermates (Figure S1B), and line F112 showed little change across four generations, indicating that the multiple repeat sizes represented BACs that were inserted into the genome as a single transgene array (Figure S1C). No major shifts in repeat sizes were seen across tissues or brain regions in individual mice, indicating that the repeats showed relatively little somatic instability (Figure S1C). Quantitative RT-PCR and western blot confirmed that the human C9orf72 transcript and protein were expressed (Figure 1C,D), and C9orf72 protein levels were modestly increased (~2-fold). For further analysis we chose two C9-BACexp lines (F112 and F113) as they showed a range of expansion sizes all in the disease range (~100 to 1000), and a similar level of transcript and protein expression to the F08-CTR line. The C9orf72 hexanucleotide repeat expansion occurs between two alternatively used upstream exons (1a and 1b), and is only transcribed in exon 1a containing isoforms. We therefore performed 5’ rapid amplification of cDNA ends (5’RACE) on spinal cord tissue, and confirmed that the human BAC expressed predominantly 1a (repeat containing) transcripts, similar to iPSC-derived motor neurons from the patient from which BAC 239 was derived (Figure 1E). Given that repeat expansions can induce methylation and silencing of C9orf72 transcription in humans, we performed a methylation sensitive restriction enzyme-qPCR to examine transgene methylation levels (Figure 1F) and observed that methylation was very low in C9-BAC transgenic mouse lines. Of note in the lines with multiple insertions, the degree of expression from each of the different transgenes cannot be ascertained.
Figure 1. Generation of C9orf72 BAC transgenic mice.
(A) Schematic diagram of C9-BAC construct. The pCC1 BAC backbone (8.128 kb) was used to isolate a region contained the entire human C9orf72 gene (~36 kb) from a patient with C9-ALS, including ~110 kb upstream and ~20 kb downstream. >30 different C9-BAC founder lines were generated with either the contracted control BAC (15 repeats) or expansions of various sizes.
(B) Southern blot analysis of the repeat length in C9-BAC founder lines. A contracted subclone was used to generate the control (F08-CTR) line with 15 GGGGCC repeats. Lines F08-CTR, F112 and F113 showed comparable expression, and lines F112 and F113 had a variety of repeat sizes ranging from ~100-1000.
(C) Relative expression of human C9orf72 mRNA in cortex from C9-BAC transgenic lines measured by qPCR.
(D) Western blot analysis of C9orf72 protein (48kD) expressed in C9-BAC mouse cortex (left). Anti-C9orf72 (top), and anti-Actin (bottom). Western blot quantitation (right) normalized to nontransgenic (NTg) control.
(E) Sequence alignments of exon 1a and exon 1b utilization from 5’RACE analysis of C9orf72 transcripts. iPSC-derived motor neurons are shown on the left (from BAC donor patient) to demonstrate exon 1a and 1b utilization in human cells.
(F) C9orf72 promoter methylation assay as assessed by MSRE-qPCR. Fibroblasts (Fibro), induced pluripotent stem cells (iPSC) and iPSC-derived motor neurons (MN) are shown for BAC donor patient (~800 repeats), which revealed variable methylation from ~10% in fibroblasts to ~50% in iPSC and MNs. Methylation levels were low (~5%) in C9-BAC transgenic lines. Lymphoblastoid cell lines (LCL) from a non-expanded patient (CTR) and C9orf72 expansion carrier (C9+) are shown for comparison. *All data are shown as mean ± SEM. See also Figure S1.
C9-BACexp transgenic mice develop RNA foci and DPR proteins similar to human C9orf72 expansion carriers
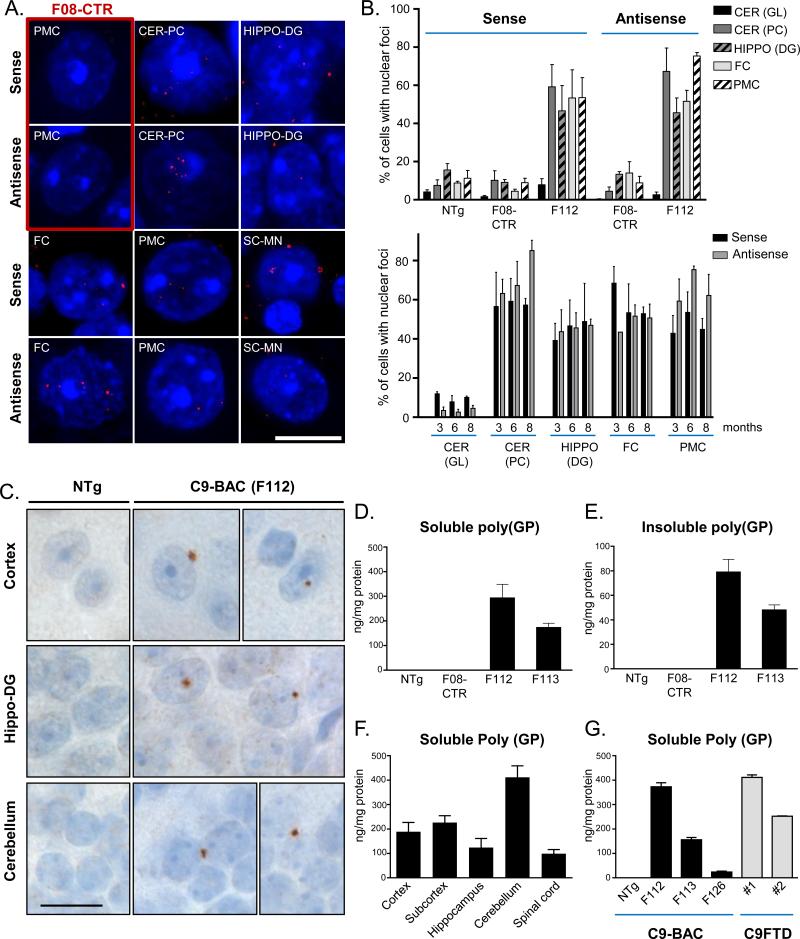
To examine the core pathologic features seen in C9orf72 expansion patients, we surveyed brain regions in C9-BACexp transgenic mice for sense and antisense RNA foci. We observed that C9-BACexp lines F112 and F113 showed both sense and antisense RNA foci throughout the nervous system (Figure 2A). By contrast, RNA foci were not observed in the F08-CTR line carrying 15 repeats (Figure 2A). In most brain regions, ~40-80% of cells exhibited sense and antisense foci starting at 3 months of age, and they did not change in distribution or frequency over time (Figure 2B).
Figure 2. C9-BAC transgenic mice develop RNA foci and RAN peptides similar to human C9orf72 expansion carriers.
(A) Fluorescence in situ hybridization (FISH) of sense and antisense RNA foci in F08-CTR (red box) and F112 C9-BAC mice. CER-GL: cerebellar granular layer; CER-PC: cerebellar purkinje cells, hippo-DG: hippocampus dentate gyrus; FC: frontal cortex; PMC: primary motor cortex; SC-MN: spinal cord motor neuron. Scale bar = 10μm.
(B) Quantification of sense and antisense foci in nontransgenic (NTg), F08-CTR and F112 C9-BACexp transgenic mice across brain regions. Lower graph shows the percent of cells with foci across different brain regions at 3, 6, and 8 months of age.
(C) Immunostaining of poly(GP) in 20 month old NTg or F112 C9-BACexp mice showing inclusions in brain regions including cortex, hippocampus dentate gyrus (Hippo-DG), and cerebellar granular layer.
(D) Soluble poly(GP) is present in the cortex of 6 month-old C9-BACexp transgenic mice (F112 and F113), but not in NTg mice, nor F08-CTR mice, as assessed using a poly(GP) immunoassay. (n=2 for F112, F113, NTG, and F08-CTR)
(E) Insoluble poly(GP) levels are lower than soluble poly(GP) levels (C) in the cortex of C9-BACexp transgenic mice, as determined by immunoassay of the insoluble fraction. (n=2 for F112, F113, NTG, and F08-CTR)
(F) Immunoassay of Poly(GP) levels in different brain regions. Cerebellum showed the highest levels, while levels were lowest in the spinal cord. (n=3)
(G) Comparison of poly(GP) levels in the cortex from C9-BAC mouse lines and C9orf72-positive FTLD cases. Lines F112 and F113 produce poly(GP) at similar levels to those detected in two different human C9orf72+ FTD frontal cortex samples (FTD #1 and FTD #2). *All data are shown as mean ± SEM. See also Figure S2.
Immunostaining showed that poly(GP) inclusions were rare in brains of young mice (6 months), but were more frequent in the brains of older mice (20 months) suggesting they accumulate with age (Figure 2C). To quantitatively assess DPR production, we utilized an immunoassay to detect poly(GP), a DPR protein produced from both sense and antisense strands of the repeat, and abundantly expressed in C9-FTD brains. We found that C9-BACexp mice showed abundant poly(GP) DPR proteins in both the soluble and insoluble fractions from as early as 6 months of age, which were not detected in nontransgenic mice, or in the F08-CTR line (Figure 2D,E). Poly(GP) DPR proteins were most abundant in cerebellar tissue, and least abundant in spinal cord tissue, mirroring the distribution of DPR pathology observed in C9orf72 expansion carriers (Davidson et al., 2014) (Figure 2F). When run in parallel in the same immunoassay, cortical tissue from C9-BACexp mice and C9-FTLD patients showed quantitatively similar levels of soluble poly(GP) DPR proteins (Figure 2G). These data indicate that C9-BACexp mice develop sense and antisense RNA foci, and widespread DPR production in a distribution similar to that seen in human C9orf72 expansion carriers.
Mice carrying C9orf72 hexanucleotide expansions do not show evidence of neurodegeneration or functional deficits in the nervous system
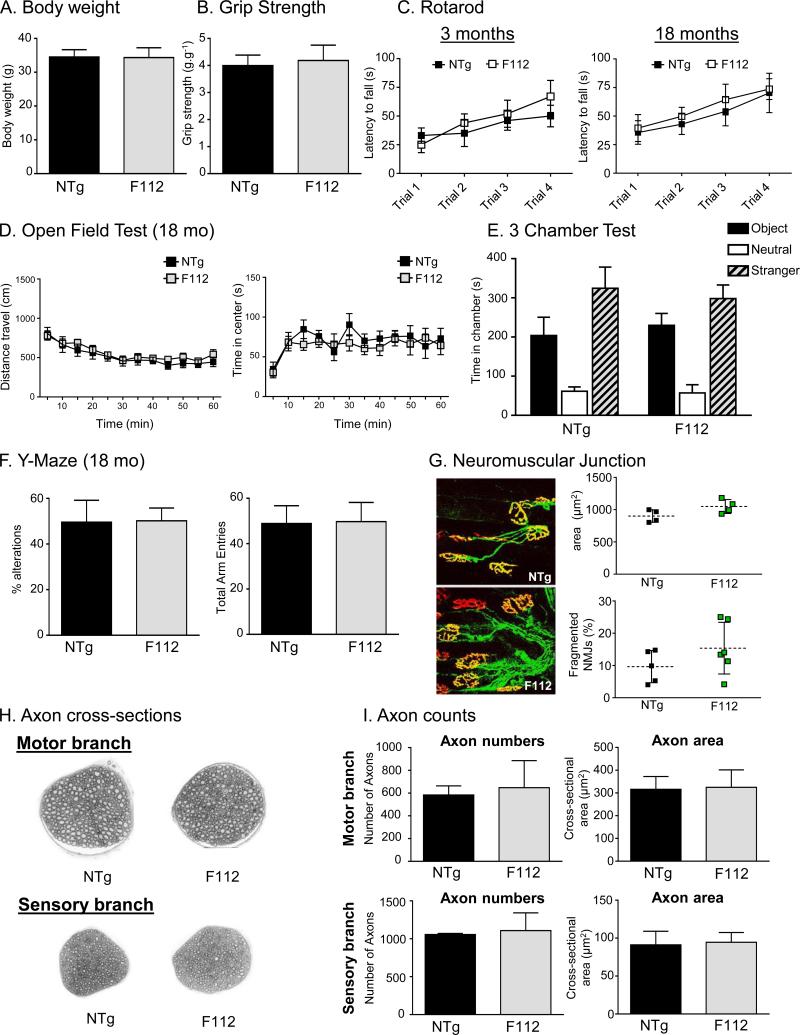
Given the widespread RNA foci and DPR protein levels observed in C9-BACexp mice, we examined line F112 for functional defects in the nervous system. No abnormalities were seen in body weight, grip strength, rotarod, or open field testing analysis at either young (3 months) or advanced ages (18 months) (Figure 3A-D). Given that rodent models of FTLD can display selective abnormalities in sociability and social novelty, we performed the 3-chamber test (Roberson, 2012) but found no difference between nontransgenic and F112 C9-BAC mice (Figure 3E). Finally we performed the Y-maze test to assess memory and novelty seeking behavior, but again we observed no difference between C9-BACexp and nontransgenic control mice (Figure 3F).
Figure 3. C9-BAC transgenic mice do not show evidence of neurodegeneration or functional deficits in the nervous system.
(A) Body weight and (B) grip strength normalized to body weight at 18 months of age. For this and all behavioral studies below, male mice (NTg, n=8; F112, n=10) were used unless otherwise indicated.
(C) Rotarod testing of cohorts of nontransgenic (NTg) and C9-BACexp mice (F112). No differences in sensorimotor coordination, or motor learning across trials, were observed in either young (3 months; NTg, n=8; F112, n=8) or aged (18 months; NTg, n=8; F112, n=10) animals.
(D) Open field test at 18 months comparing total activity (left) and time spent in the center (right) for NTg mice and C9-BACexp (F112) mice.
(E) 3 chamber test for sociability and social novelty at 16 months. F112 mice prefered spending time with the stranger and the novel object more than in the neutral zone, no different from NTg mice.
(F) Y-Maze assessment of memory and novelty seeking at 18 months was not different between C9-BACexp mice and NTg controls.
(G) Neuromuscular junction staining (bungarotoxin = red, synaptophysin/neurofilament = green) of the tibialis anterior muscle in NTg and C9-BACexp (F112) mice at 20 months showed no differences in occupancy, fragmentation or area (n=6).
(H) Femoral motor and sensory nerve plastic sections from NTg, F08-CTR, and C9-BACexp (F112) mice.
(I) Axon counts of femoral motor and sensory nerves (n=6) in C9-BAC (F112) and control mice (NTg, F08-CTR) at 20 months of age. See also Figure S2.
To interrogate the mice for evidence of “subclinical” levels of frontal cortex or motor system damage, we examined them histologically. No difference in neuromuscular junction innervation or femoral axon counts were observed in C9-BACexp mice compared to controls (Figure 3G-I). Additionally we performed immunohistochemistry on the brains and spinal cords of C9-BACexp mice, but found no evidence of protein aggregation (p62, TDP-43, ubiquitin), gliosis, inflammation, synapse or neuron loss (GFAP, IBA1, synaptophysin, and NeuN stain) (Figure S2).
C9-BACexp mice have evidence of nucleolar stress without downstream sequelae
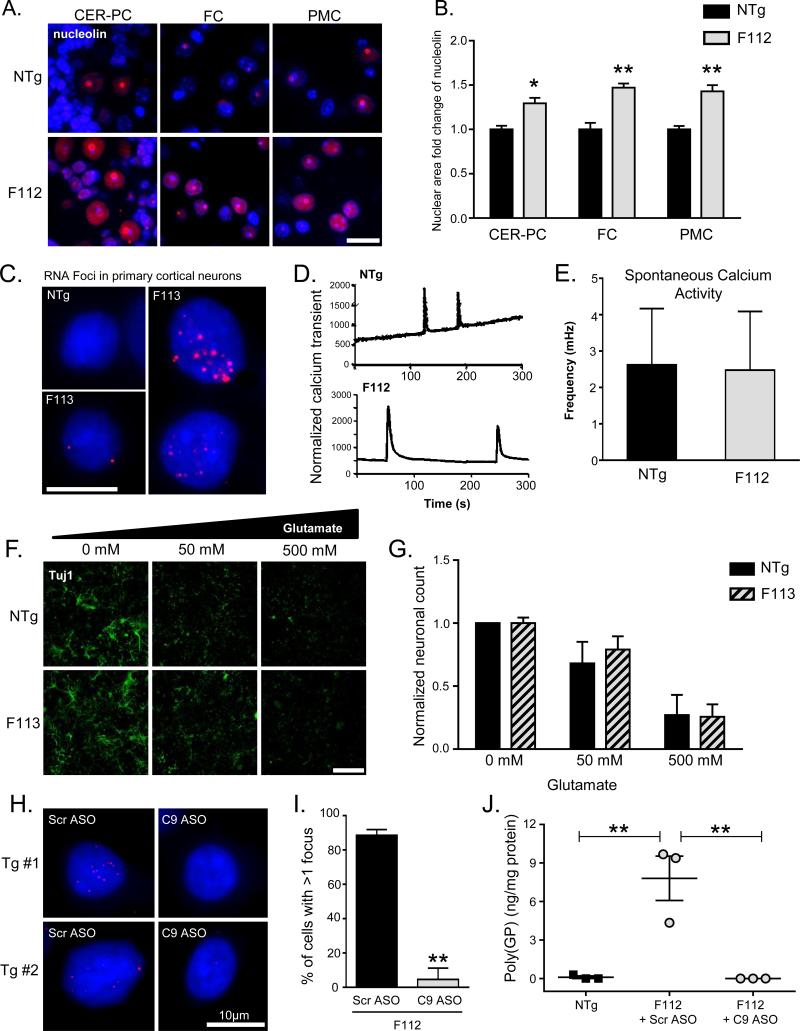
We first investigated colocalization of sense and antisense RNA foci with the RNA binding proteins Pur α, hnRNPA3, hnRNPA2/B1, and hnRNP-H, which were previously reported to bind to GGGGCC-repeat containing RNA foci in culture models and in patient tissues (Lee et al., 2013; Xu et al., 2013). Surprisingly, none of these RNA binding proteins showed sequestration, or even consistent colocalization with sense or antisense RNA foci in C9-BACexp transgenic mice (Figure S3). In addition to altering RNA binding protein function, the C9orf72 expansion was shown to disrupt nucleolar integrity by either binding the G-quadraduplex structure of aborted transcripts to nucleolin (Haeusler et al., 2014), or from arginine containing dipeptide repeat proteins which localize to the nucleolus and disrupt its function (Kwon et al., 2014; Wen et al., 2014). Interestingly we observed that C9-BACexp mouse neurons showed dispersion of nucleolin from the nucleolus, similar to what has been observed in cultured cells and nervous tissue from C9orf72 expansion patients (Figure 4A,B). While the partial mislocalization of nucleolin supports some degree of nucleolar stress, we did not observe any downstream sequelae such as altered RAN GTPase splicing or ribosomal RNA biogenesis (Figure S3). To probe further for alterations in gene expression, we analyzed transcriptomes by RNA-seq of spinal cord and cortex from 6 month old C9-BAC mice (F08-CTR, F112 C9-BACexp and nontransgenic mice), an age at which RNA foci and DPR levels are highly elevated. Gene set enrichment analysis (GSEA) identified immunomodulatory and extracellular matrix pathways as significantly enriched (FDR<0.05) among genes downregulated in the cortex of C9-BACexp compared to nontransgenic mice (Figure S4). These pathways were not enriched among genes downregulated in cortex from the F08-CTR line compared to nontransgenic mice, indicating a specific effect in the C9-BACexp mice carrying the hexanucleotide expansion. Furthermore, in comparing altered pathways with those observed in recently published datasets using human C9orf72 patient cortex or iPSC derived neurons (Donnelly et al., 2013), we found that the dendritic cell pathway regulating TH1 and TH2 cell development was downregulated in both the C9-BACexp mice and human iPSC derived neurons compared to controls (Figure S4). Together, these data suggest that the RNA foci, DPR proteins and nucleolar stress present in C9-BACexp mice alter RNA expression in specific pathways promoting cellular dysfunction that are shared with the human iPSC derived neurons, but are not sufficient to drive neurodegeneration.
Figure 4. C9-BACexp mice show evidence of nucleolar stress, and decreased RNA foci and DPR proteins after antisense oligonucleotide treatment.
(A) Immunofluorescence staining of nucleolin (red) costained with DAPI (blue) in nontransgenic (NTg) and C9-BACexp (F112) mice in the cerebellar purkinje cells (CER-PC), frontal cortex (FC), and primary motor cortex (PMC). C9-BACexp mice showed partial displacement of nucleolin from the nucleolus. (scale bar = 20 μm)
(B) Quantitation of nucleolin staining distribution revealed a shift in the ratio of the amount of staining in the nucleus vs. the nucleolus (n=4 animals; two sided t-test, NTg vs. F112; *p<0.05; **p<0.01).
(C) RNA FISH of sense foci in primary cortical cultures from neonatal NTg and F113 C9-BACexp mice. RNA foci were observed in 73% (±9%) of neurons and astrocytes.
(D) Representative calcium recording from culture cortical neurons (NTg, top and F112, bottom).
(E) Frequency of calcium transients observed quantitated using Fluo-4 live imaging (n=27, NTg and n=68, F112).
(F) Glutamate toxicity assay in primary cortical neuron cultures showing Tuj1 staining after 24 hours of glutamate treatment at the indicated dose. (scale bar = 400 μm)
(G) The number of neurons (TuJ1 positive) were divided by total number of cells (DAPI, not shown) and then normalized to the controls (no treatment) for both F113 and NTg.
(H) RNA FISH of sense foci in primary cortical cultures treated with either control antisense oligonucleotides (Scr ASO) or an ASO targeting exon 2 of the human C9orf72 transcript (C9 ASO).
(I) Quantification of RNA foci reduction after C9 ASO treatment (**p=0.004, two sided t-test).
(J) Immunoassay showing reduction of poly(GP) DPR proteins in cortical cultures from C9-BACexp (F112 line) mice treated with C9 ASO compared to Scr ASO (**p=0.004,one way ANOVA). See also Figure S3.
Suppression of RNA foci and DPR proteins with antisense oligonucleotides in primary cortical cultures from C9-BACexp mice
Finally to examine phenotypes in C9-BACexp mice at the cellular level, we generated primary cortical cultures from C9-BACexp mice and compared them to nontransgenic controls. FISH revealed RNA foci in 73% (SD ±9%) of cultured neurons and astrocytes from C9-BACexp mice (Figure 4C), which is higher than the 20-35% reported in human iPSC derived neurons from C9orf72 patients (Donnelly et al., 2013; Sareen et al., 2013). No difference in the frequency of calcium transients (Figure 4D,E), or sensitivity to glutamate toxicity (Figure 4F,G), was observed in cortical neurons cultured from C9-BACexp neurons. However, antisense oligonucleotides (ASOs) targeting exon 2 of human C9orf72 were able to markedly suppress both RNA foci and poly(GP) DPR proteins in primary cortical cultures (Figure 4H-J). These data indicate that cortical cells from C9-BACexp mice recapitulate key features of C9orf72 expansion pathology in vitro, and demonstrate their utility for investigating therapeutic approaches aimed at diminishing RNA foci and DPR pathology produced at physiologic levels in mice.
Discussion
Here we report that C9-BACexp mice exhibited the cardinal pathologic features seen in patients, including sense and antisense RNA foci, and soluble DPR proteins at levels similar to those seen in brains of FTLD patients. Despite this fact, C9-BACexp mice did not develop measurable neurodegeneration during their lifespan. This is consistent with animal models of many neurodegenerative diseases, which typically mimic only certain aspects of the human pathophysiology, and frequently lack overt neurodegeneration (Rockenstein et al., 2007). Additionally, many mouse models require high level overexpression driven by a transgenic promoter to produce pathology, which opens the possibility of emergent toxicities that do not relate to the human disease being modeled. While this manuscript was being prepared, mice expressing high levels of a synthetic C9orf72 repeat under control of the chicken β-actin promoter using adeno-associated virus were reported that make RNA foci, RAN peptides and develop neurodegeneration (Chew et al., 2015). Importantly, C9-BACexp mice show similar amounts of sense and antisense RNA foci and DPR peptides to human disease tissues, without requiring high levels of overexpression obtained using an exogenous promoter. This suggests that additional factors will be needed to drive neurodegeneration from expression of the C9orf72 repeat at normal physiologic levels in mice.
Studies have supported a variety of potential gain of function toxicities resulting from the C9orf72 hexanucleotide expansion expression. These include RNA toxicity from sequestration of RNA-binding proteins by RNA foci (Donnelly et al., 2013; Lee et al., 2013; Sareen et al., 2013); ER stress or Unc119 sequestration from poly(GA) DPR aggregates (May et al., 2014; Zhang et al., 2014); and nucleolar stress from either G-quadraduplex containing RNA transcripts (Haeusler et al., 2014) or arginine (PR/GR) containing DPR proteins (Kwon et al., 2014; Mizielinska et al., 2014; Tao et al., 2015; Wen et al., 2014). While sense and antisense RNA foci were widespread throughout the nervous system, C9-BACexp mice surprisingly did not show binding or sequestration of RNA-binding proteins previously reported to interact with foci. The variability in sequestered RNA-binding proteins from the different published reports suggests that none of the RNA-binding proteins reported thus far is likely a pivotal player in C9orf72 pathogenesis. By contrast we did observe altered localization of nucleolin suggestive of mild nucleolar stress, but despite this we did not observe any downstream sequelae such as altered ribosomal RNA biogenesis, or altered splicing events. This difference may be due to the severe toxicity seen in the acute overexpression culture models, or because the reported sequelae of altered nucleolin localization occur relatively late in the cascade of neurodegeneration caused by the C9orf72 expansion.
It is notable that macroscopic p62-positive DPR protein aggregates were only observed in the brain tissues of aged mice, while the poly(GP) immunoassay showed that soluble DPR protein levels in younger mice, suggesting that altered protein homeostasis with age leads to accumulation of insoluble DPR aggregates. Regardless there remains controversy as to whether the macroscopic DPR aggregates play a key role in C9orf72 repeat pathogenesis, since their distribution has little relationship to the clinical phenotype in ALS and FTLD patients, with the highest degree of pathology typically in the cerebellum, and relatively little in the spinal cord (Davidson et al., 2014; Mann, 2014).
While the C9-BACexp mice focus on modeling gain of function manifestations in C9orf72 expansion carriers, partial loss of C9orf72 function due to epigenetic silencing remains a potential contributor in disease pathogenesis. Decreased expression of C9orf72 transcripts has been reported (DeJesus-Hernandez et al., 2011; Gijselinck et al., 2012), as have epigenetic modifications of the repeat-containing allele which may be either detrimental or protective (Belzil et al., 2013; Russ et al., 2015; Xi et al., 2013). The C9orf72 protein has been reported to regulate endosomal trafficking in cultured cells (Farg et al., 2014), and loss of the C9orf72 orthologue in zebrafish or worms led to motor deficits (Ciura et al., 2013; Therrien et al., 2013). While the C9-BACexp mice cannot resolve whether C9orf72 expansion pathogenesis involves a component of loss of function, they do argue that RNA foci and RAN dipeptides alone are not sufficient to drive neurodegeneration. This is in keeping with several observations from human C9orf72 patient genetics and pathology. First, the C9orf72 expansion is not completely penetrant, appearing in both normal elderly individuals and up to 0.6% of normal control populations of European or North American origin (Galimberti et al., 2014; Harms et al., 2013). This supports the hypothesis that additional environmental or genetic factors are required for neurodegeneration to manifest as a consequence of the C9orf72 expansion mutation. Second, accumulation of DPR pathology clearly predates and does not correspond regionally to neurodegeneration in humans, which instead correlates with TDP-43 pathology (Baborie et al., 2014; Proudfoot et al., 2014). Given that the C9-BACexp mice display these early features of RNA foci and DPR proteins without neurodegeneration or TDP-43 pathology, we propose that they are recapitulating the pre-symptomatic phase of disease seen in humans, which requires additional insults (genetic, environmental, and/or aging) to fully manifest and lead to neurodegeneration. This makes them extremely useful for studying early stages of disease pathogenesis, and to identify additional stressors which promote neurodegeneration in C9orf72 expansion carriers. Finally they will be an invaluable tool for investigating therapeutic strategies which alter RNA foci and DPR pathology, as we found that ASOs targeting the human C9orf72 transcript were able to significantly diminish the RNA foci burden and poly(GP) levels in cultured cortical cells.
Experimental Procedures
All studies were conducted in accordance with the protocols described by the National Institutes of Health's Guide for the Care and Use of Animals and approved by The Jackson Laboratory and Cedars-Sinai Institutional Animal Care and Use Committees. Mice are available from The Jackson Laboratory with the following catalog numbers: 23099: C57BL/6J-Tg(C9orf72_3)112Lutzy/J; 23100: C57BL/6J-Tg(C9orf72_3)113Lutzy/J; 23088: C57BL/6J-Tg(C9orf72_2)8Lutzy/J.
Supplementary Material
Acknowledgements
We would like to thank Uthra Rajamani and Dhruv Sareen for assistance with the ImageXpress, David Rushton and Virginia Mattis for assistance with the primary cortical cultures, Vince Funari for assistance with RNA-sequencing, and Frank Bennett and Frank Rigo of Isis Pharmaceuticals for supplying antisense oligonucleotides. This work was supported by National Institutes of Health (NIH) Grants NS055980 and NS069669 (R.H.B), AG039510 (E.B.L.), AG000255 (E.Y.L.), NS089979 (T.F.G.), NS084528 (L.P.), NS063964 (L.P.); NS077402 (L.P.); NS084974 (L.P.), ES20395 (L.P.), Department of Defense ALSRP AL130125 (L.P.), Mayo Clinic Foundation (L.P.), Mayo Clinic Center for Individualized Medicine (L.P.), ALS Association (L.P., T.F.G.), Robert Packard Center for ALS Research at Johns Hopkins (L.P.), Target ALS (L.P., R.H.B.), the Robert and Louise Schwab Family (R.H.B.) and the Cedars-Sinai ALS Research Fund (R.H.B.). The project was supported by the National Center for Advancing Translational Sciences, Grant UL1TR000124.
Footnotes
Author Contributions: J.G.O. coordinated the project, was involved performance and analysis of all experiments. L.B. established and screened C9-BAC founder lines via Southern Blotting, coordinated behavioral testing, performed axon and neuromuscular junction analysis. T.G. examined RAN peptide expression. J.G.O., L.B., T.G., A.K.M.G.M. screened C9-BAC founder lines for repeat expansion and expression. A.K.M.G.M. performed RNA FISH and immunohistochemistry experiments. A.K.M.G.M., SC, J.G.O., L.B., and A.A. performed tissue collections and analysis. E.B.L. and E.L. carried out methylation assays. M.B.H. and J.C. performed Southern blots and repeat primed PCR. S.B. embedded and processed tissue for analysis. J.Z. counted RNA foci. S.G. performed RACE analysis. K.J.K. and K.W. sequenced the repeat region in the C9-BAC control line. R.H.B. and R.H. analyzed RNA Seq data. C.L. and R.H.B. planned, designed, and interpreted the experiments. J.G.O and R.H.B. wrote the manuscript.
References
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, 3rd, Rademakers R, et al. Unconventional Translation of C9ORF72 GGGGCC Expansion Generates Insoluble Polypeptides Specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baborie A, Griffiths TD, Jaros E, Perry R, McKeith IG, Burn DJ, Masuda-Suzukake M, Hasegawa M, Rollinson S, Pickering-Brown S, et al. Accumulation of dipeptide repeat proteins predates that of TDP-43 in Frontotemporal Lobar Degeneration associated with hexanucleotide repeat expansions in C9ORF72 gene. Neuropathol Appl Neurobiol. 2014 doi: 10.1111/nan.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzil VV, Bauer PO, Prudencio M, Gendron TF, Stetler CT, Yan IK, Pregent L, Daughrity L, Baker MC, Rademakers R, et al. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol. 2013;126:895–905. doi: 10.1007/s00401-013-1199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen-Plotkin AS, Lee VM, Trojanowski JQ. TAR DNA-binding protein 43 in neurodegenerative disease. Nat Rev Neurol. 2010;6:211–220. doi: 10.1038/nrneurol.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew J, Gendron TF, Prudencio M, Sasaguri H, Zhang YJ, Castanedes-Casey M, Lee CW, Jansen-West K, Kurti A, Murray ME, et al. Neurodegeneration. C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science. 2015;348:1151–1154. doi: 10.1126/science.aaa9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciura S, Lattante S, Le Ber I, Latouche M, Tostivint H, Brice A, Kabashi E. Loss of function of C9orf72 causes motor deficits in a zebrafish model of Amyotrophic Lateral Sclerosis. Ann Neurol. 2013 doi: 10.1002/ana.23946. [DOI] [PubMed] [Google Scholar]
- Cooper-Knock J, Shaw PJ, Kirby J. The widening spectrum of C9ORF72-related disease; genotype/phenotype correlations and potential modifiers of clinical phenotype. Acta Neuropathol. 2014;127:333–345. doi: 10.1007/s00401-014-1251-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson YS, Barker H, Robinson AC, Thompson JC, Harris J, Troakes C, Smith B, Al-Saraj S, Shaw C, Rollinson S, et al. Brain distribution of dipeptide repeat proteins in frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta neuropathologica communications. 2014;2:70. doi: 10.1186/2051-5960-2-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farg MA, Sundaramoorthy V, Sultana JM, Yang S, Atkinson RA, Levina V, Halloran MA, Gleeson PA, Blair IP, Soo KY, et al. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum Mol Genet. 2014;23:3579–3595. doi: 10.1093/hmg/ddu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimberti D, Arosio B, Fenoglio C, Serpente M, Cioffi SM, Bonsi R, Rossi P, Abbate C, Mari D, Scarpini E. Incomplete penetrance of the C9ORF72 hexanucleotide repeat expansions: frequency in a cohort of geriatric non-demented subjects. J Alzheimers Dis. 2014;39:19–22. doi: 10.3233/JAD-131172. [DOI] [PubMed] [Google Scholar]
- Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PE, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, Chew J, et al. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013;126:829–844. doi: 10.1007/s00401-013-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijselinck I, Van Langenhove T, van der Zee J, Sleegers K, Philtjens S, Kleinberger G, Janssens J, Bettens K, Van Cauwenberghe C, Pereson S, et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol. 2012;11:54–65. doi: 10.1016/S1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms MB, Cady J, Zaidman C, Cooper P, Bali T, Allred P, Cruchaga C, Baughn M, Libby RT, Pestronk A, et al. Lack of C9ORF72 coding mutations supports a gain of function for repeat expansions in amyotrophic lateral sclerosis. Neurobiol Aging. 2013;34:2234, e2213–2239. doi: 10.1016/j.neurobiolaging.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, Kim J, Yun J, Xie Y, McKnight SL. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014;345:1139–1145. doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YB, Chen HJ, Peres JN, Gomez-Deza J, Attig J, Stalekar M, Troakes C, Nishimura AL, Scotter EL, Vance C, et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell reports. 2013;5:1178–1186. doi: 10.1016/j.celrep.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann DM. Dipeptide repeat protein toxicity in frontotemporal lobar degeneration and in motor neurone disease associated with expansions in C9ORF72-a cautionary note. Neurobiol Aging. 2014 doi: 10.1016/j.neurobiolaging.2014.10.011. [DOI] [PubMed] [Google Scholar]
- May S, Hornburg D, Schludi MH, Arzberger T, Rentzsch K, Schwenk BM, Grasser FA, Mori K, Kremmer E, Banzhaf-Strathmann J, et al. C9orf72 FTLD/ALS-associated Gly-Ala dipeptide repeat proteins cause neuronal toxicity and Unc119 sequestration. Acta Neuropathol. 2014;128:485–503. doi: 10.1007/s00401-014-1329-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizielinska S, Gronke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IO, Pietrzyk J, et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014;345:1192–1194. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, et al. The C9orf72 GGGGCC Repeat Is Translated into Aggregating Dipeptide-Repeat Proteins in FTLD/ALS. Science. 2013 doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- Proudfoot M, Gutowski NJ, Edbauer D, Hilton DA, Stephens M, Rankin J, Mackenzie IR. Early dipeptide repeat pathology in a frontotemporal dementia kindred with C9ORF72 mutation and intellectual disability. Acta Neuropathol. 2014;127:451–458. doi: 10.1007/s00401-014-1245-7. [DOI] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson ED. Mouse models of frontotemporal dementia. Ann Neurol. 2012;72:837–849. doi: 10.1002/ana.23722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockenstein E, Crews L, Masliah E. Transgenic animal models of neurodegenerative diseases and their application to treatment development. Advanced drug delivery reviews. 2007;59:1093–1102. doi: 10.1016/j.addr.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Russ J, Liu EY, Wu K, Neal D, Suh E, Irwin DJ, McMillan CT, Harms MB, Cairns NJ, Wood EM, et al. Hypermethylation of repeat expanded C9orf72 is a clinical and molecular disease modifier. Acta Neuropathol. 2015;129:39–52. doi: 10.1007/s00401-014-1365-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sareen D, O'Rourke JG, Meera P, Muhammad AK, Grant S, Simpkinson M, Bell S, Carmona S, Ornelas L, Sahabian A, et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Science translational medicine. 2013;5:208ra149. doi: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Zhang Y, Gendron TF, Bauer PO, Chew J, Yang WY, Fostvedt E, Jansen-West K, Belzil VV, Desaro P, et al. Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron. 2014;83:1043–1050. doi: 10.1016/j.neuron.2014.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z, Wang H, Xia Q, Li K, Li K, Jiang X, Xu G, Wang G, Ying Z. Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity. Hum Mol Genet. 2015 doi: 10.1093/hmg/ddv005. [DOI] [PubMed] [Google Scholar]
- Therrien M, Rouleau GA, Dion PA, Parker JA. Deletion of C9ORF72 results in motor neuron degeneration and stress sensitivity in C. elegans. PLoS ONE. 2013;8:e83450. doi: 10.1371/journal.pone.0083450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Tan W, Westergard T, Krishnamurthy K, Markandaiah SS, Shi Y, Lin S, Shneider NA, Monaghan J, Pandey UB, et al. Antisense Proline-Arginine RAN Dipeptides Linked to C9ORF72-ALS/FTD Form Toxic Nuclear Aggregates that Initiate In Vitro and In Vivo Neuronal Death. Neuron. 2014;84:1213–1225. doi: 10.1016/j.neuron.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, Zinman L, Moreno D, Schymick J, Liang Y, Sato C, Zheng Y, Ghani M, Dib S, Keith J, et al. Hypermethylation of the CpG Island Near the GC Repeat in ALS with a C9orf72 Expansion. Am J Hum Genet. 2013 doi: 10.1016/j.ajhg.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Poidevin M, Li X, Li Y, Shu L, Nelson DL, Li H, Hales CM, Gearing M, Wingo TS, et al. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc Natl Acad Sci U S A. 2013;110:7778–7783. doi: 10.1073/pnas.1219643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Jansen-West K, Xu YF, Gendron TF, Bieniek KF, Lin WL, Sasaguri H, Caulfield T, Hubbard J, Daughrity L, et al. Aggregation-prone c9FTD/ALS poly(GA) RAN-translated proteins cause neurotoxicity by inducing ER stress. Acta Neuropathol. 2014;128:505–524. doi: 10.1007/s00401-014-1336-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.