Abstract
Polyphenism is a form of developmental plasticity in which organisms respond to environmental cues by producing adaptive, discrete, alternative phenotypes known as morphs. The phenomenon is common and important as both a form of adaptation and a source of variation for natural selection. Understanding the evolution of polyphenism will require understanding the proximate factors that regulate alternative morph production. Renewed interest and technological advances have fueled multiple approaches to the latter, including hormone manipulation studies, targeted transcriptomic studies, and epigenetic profiling. We review these studies and suggest that integration of multilayered approaches will be necessary to understand the complex mechanisms involved in regulating alternative morphologies.
Introduction
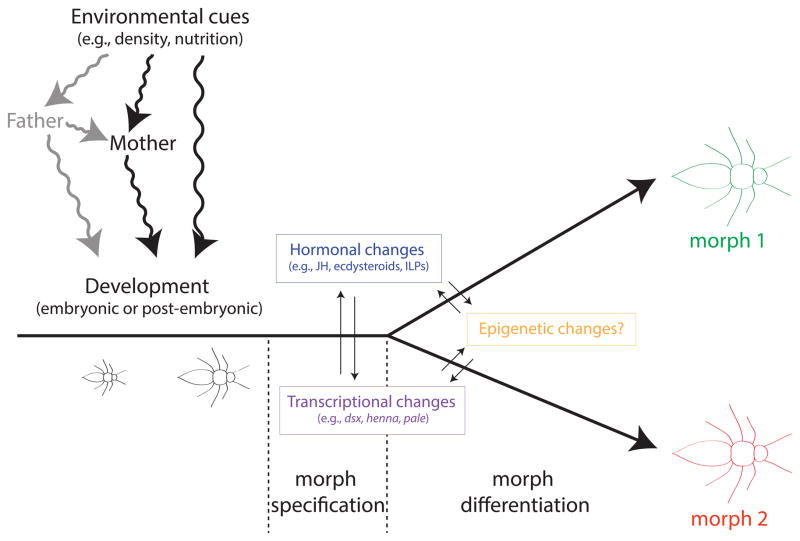
Many organisms have evolved the ability to respond to changing environmental conditions by altering development to produce adaptive phenotypes. Such developmental plasticity can be a decisive boon, allowing individuals within populations to adapt to changing environmental circumstances on short, non-evolutionary time scales. If, in addition to being adaptive, the developmental response is discrete and stable, then the phenomenon is referred to as a polyphenism, with alternative phenotypes, referred to as morphs, typically exhibiting correlated suites of traits [1–3]. The production of alternative phenotypes by the same genetic material is inherently a regulatory process: external cues are perceived by the organism and then converted into a molecular signal or signals that specify development of the appropriate morph (Figure 1). Once morph identity is specified, alternative morphs develop by proceeding down different ontogenies.
Figure 1.
The process of morph specification and differentiation during development involves a branching ontogeny influenced by hormonal, transcriptional and epigenetic changes, all of which may influence each other. Although images of post-embryonic development are shown, specification and differentiation can just as easily take place during embryonic development. These can influence development directly, or be mediated by a mother, who may receive the cue and transmit this information on to her progeny as a maternal effect. Although less supported and less investigated, paternal mediation (shown in grey) is also possible [55], either in combination with or independently of the mother.
Evolutionary studies of polyphenism typically aim to understand how polyphenic responses evolve and how such plasticity can, in turn, affect the course of evolution. Answers to both of these questions will undoubtedly be informed by a deeper understanding of the proximate mechanisms of polyphenism, in particular its underlying genetics, epigenetics, and development. Fortunately, renewed interest in phenotypic plasticity, combined with technological advances, has provided insights into this area. Here we review recent contributions to the regulation of polyphenic morph development in insects with an eye toward identifying common themes in how these alternative phenotypes are regulated.
The classic approach: hormone manipulation studies
Although technological advances promise continued progress in understanding the proximate basis of polyphenism, important insights can still be gained through the classic technique of hormone manipulation. As polyphenisms often involve manifold changes throughout the organism, it is not surprising that hormones have long been known to coordinate the development of alternative morphs. In insects, two classes of hormones, ecdysteroids and juvenile hormones (JH), underlie the developmental decisions inherent in many polyphenisms [4,5].
Two studies highlight the power of this technique and the insights they can provide. First, at least two species of the ant genus Pheidole possess a “supersoldier” subcaste that is affected by nutritional state and mediated by JH. In several Pheidole species that do not normally produce supersoldiers, application of a JH analog can induce supersoldiers. This result suggests that there is an ancestral developmental potential for this plastic response and that activating this latent response likely facilitated parallel evolution of the supersoldier subcaste in multiple Pheidole species [6]. Second, a hormonal approach has linked seasonal changes to color and life history adaptation in the butterfly Bicyclus anynana. In this species, wet season or dry season morphs are induced depending on the temperature that larvae and pupae experience during development [7]. A recent study has functionally confirmed what had long been suspected about this polyphenism based on titer correlations: ecdysteroids act as part of the switch that controls progression down developmental pathways leading to alternative life histories [8]. This latter study is also one of the first to examine carefully whether each member of the suite of ecologically relevant traits that characterize the alternative morphs is affected by ecdysteroids.
In general, hormonal manipulation studies are most informative when combined with attempts to correlate morph development with direct measurements of hormone titer, with differential expression of hormone regulators, and with correlated changes of morph-specific downstream genes [reviewed in 9], These two examples and a myriad of hormonal investigations in other polyphenic insects [reviewed in 5] reveal a common theme: hormonal signaling is a critical aspect of regulating polyphenisms. That said, polyphenic ontogenies involve much more than hormones and recent technological advances are revealing some of these additional regulators.
Transcriptomic approaches
The establishment of model systems for studying plasticity and advances in our ability to describe transcriptomes have fueled rapid progress toward understanding the details of alternative developmental trajectories and the nature of environmentally controlled developmental switches [sensu 10]. In a number of polyphenisms, for example, transcriptomic approaches have identified a large number of gene expression differences between morphs [e.g., 11,12–14], indicating that different genes from a shared genome contribute to each phenotype. Although most of these studies have focused on the differentiation of adult phenotypes, long after morph identity has been specified, a few have used transcriptomics to identify differences in gene expression that regulate alternative morph specification by precise temporal profiling of the transcriptome in response to inducing cues [15–18]. Even with these more targeted studies, however, it can be challenging to identify the changes in gene expression that are indeed responsible for morph specification because often there are numerous correlated changes in gene expression.
The emergence of new model insect systems has allowed us to go beyond correlation and test the potential roles played by particular factors in generating plasticity. The dung beetle, Onthophagus taurus, provides an excellent example. These beetles exhibit a male-specific horn polyphenism controlled by nutrition [20]. Small males develop virtually no horns, while large males develop large horns; females have no horns. Kijimoto et al. [21] profiled transcript levels from developing horn tissues and used the data to identify a number of candidate genes for polyphenic horn growth, including the sex determination gene doublesex (dsx). Subsequent work [18] revealed that, as in Drosophila, dsx transcripts are sex-specific, and play a role in the expression of primary and secondary sexual traits. Interestingly, male dsx is also found at higher levels in the horn primordia of large males than of small males and RNAi against male dsx results in the loss of horns in large males while leaving small males unaffected. These experiments suggest that dsx is a key regulator of both sexual dimorphism and male horn polyphenism. Presumably dsx somehow interacts with JH, which is known to mediate the nutritional control of the horn polyphenism [22,23]. But how does this interaction occur? Studies of male-specific exaggerated trait growth (either horns or mandibles) in other beetle species suggest that dsx might act to sensitize a tissue to respond or not respond to a circulating hormone such as JH or insulin-like peptides [24–28]. Whether dsx acts in this way in the horn polyphenism is a question for future research.
Transcriptional profiling of the transition between density-induced solitarious and gregarious morphs in the migratory locust provides another example. Genome-wide expression assays led to the identification of the dopamine pathway as an exciting candidate for controlling the behavioral changes that accompany phase transitions [29]. RNAi knockdown of genes in the pathway confirmed their causative role, and pharmacological manipulation showed that dopamine promotes gregariousness [29]. In addition to behavioral differences, solitarious and gregarious locusts differ in morphological, physiological, and neurochemical features [30,31]. As pointed out in recent reviews of the locust dispersal polyphenism [32,33], despite the established importance of the dopamine pathway in regulating behavioral changes, no one factor controls all of the differences between the morphs. Years of study and the application of wide variety of approaches have revealed that there must be changes in neuroendocrine factors, peptides, metabolites, transcripts, and epigenetic factors.
These examples provide two lessons, one methodological and one biological. First, a transcriptomic approach can be quite useful in identifying candidate genes and pathways when none are known, if the profiling is done at developmentally appropriate stages and within relevant tissues. Second, while the “switch” metaphor is apt for some polyphenisms (e.g., male beetle horns), it fails to capture the reality of others (e.g., solitarious and gregarious locust morphs). In the case of male beetle horns, the environmental cues that trigger the development of one morph versus the other are received during a relatively short window of development and the effects are irreversible. In contrast, the transition between locust morphs is arguably more complicated; it can be intraindividual or transgenerational, partial or complete, and reversible [30]. A greater understanding of the regulation of alternative morphs in a variety of species should open our eyes to the full spectrum of ways that such developmental decisions get made and how such mechanisms can evolve.
Epigenetic approaches
One of the more exciting and recent approaches to understanding the mechanistic basis of polyphenism has been to investigate whether epigenetic modifications (e.g., acetylation or methylation of histones and methylation of CpGs in DNA) might play a role in either specification or subsequent differentiation of morphs. Such modifications are likely to correlate with any differential transcription involved in the initial morph specification after receiving the inducing environmental cue, since epigenetic changes accompany transcriptional changes [34]. The mitotic inheritance of chromatin states may also then be needed to perpetuate morph identity to the various tissues of a developing organism and allow for the differential transcription required to differentiate alternative phenotypes. A question often posed is whether epigenetic modifications play a role in polyphenism. The answer is undoubtedly “yes”, insofar as such modifications are now taken to be part and parcel of differential transcription [35], which almost certainly plays a role in morph differentiation if not specification. A more specific and perhaps more fruitful question is whether epigenetic modifications stand in the causal chain leading from environmental cue to morph specification and, if so, what types?
Of the various insect models, social insects have thus far contributed most to our knowledge of the role of epigenetic modifications in polyphenism. Inspired by the transcriptional regulatory role that DNA (mostly CpG) methylation plays in vertebrates, most work has focused on DNA methylation. Kucharski et al. [36] were the first to demonstrate a functional role for DNA methylation in morph specification, specifically diet-induced caste specification in honeybees. RNA interference of the DNA methyltransferase Dnmt3, responsible for de novo methylation of CpG cytosines, causes queens to develop from presumptive worker larvae, suggesting that de novo CpG methylation mediates between diet and caste specification. A more recent study has shown that DNA methylation may contribute to caste differentiation post-specification [37]. In the ant species Camponotus floridanus, environmental conditions cause workers to vary quantitatively in size. Alvarado et al. [37] discovered that this size variation was regulated, at least in part, by global levels of DNA methylation. Further, they implicated DNA methylation levels at the size-regulating gene Egfr and its associated expression level as a significant mediator of this process. Size variation is an important component of caste differences in ants, so this epigenetic effect could be a critical factor underlying morph differentiation.
Although in insects the functional significance of DNA methylation for gene expression is still not entirely clear, in part because methylation tends to occur within the gene body rather than at promoters [reviewed in 39], it may act by preventing or promoting the binding of regulatory proteins and by changing how tightly DNA binds to nucleosomes [46,47]. Another possible role for DNA methylation in insects has been suggested by data from social insects; namely, that it regulates alternative splicing. Several studies in honeybees and ants have noted a correlation between DNA methylation and alternative splicing at the genome-wide level [42,48,49], while a study in the jewel wasp (Nasonia vitripennis) has not [50]. Further, interfering with de novo DNA methylation alters patterns of exon skipping and intron retention in the honeybee [51]. Could DNA methylation foster plasticity by regulating the production of morph-specific transcripts? The idea is tantalizing, yet so far unsupported. It is notable, for example, that so far no study in insects has functionally tested whether methylation status has an effect on splicing for a particular transcript.
Surprisingly little research has investigated the role that histone modification and noncoding RNAs might play in polyphenism, making it an obvious focus of future studies. Two reports are notable exceptions. First, Simola et al. [52] found a possible role for H3K27 acetylation in caste differentiation in the carpenter ant (Camponotus floridanus) by profiling different histone modifications at the genome-wide level in different castes. Second, an excellent, thorough study by Yang et al. [53] found that a microRNA controls dopamine synthesis in the migratory locust (Locusta migratoria) by regulating two genes in the dopamine synthesis pathway (henna and pale). As noted above, dopamine levels have been shown to control the production of solitarious versus gregarious forms in this species [29]. Similar approaches promise further insights in the coming years.
Concluding remarks
Not surprisingly, the regulation of alternative developmental pathways is a complicated, multilayered process, involving both initial specification and subsequent differentiation. Changes occur in hormones, biogenic amines, transcripts, and epigenetic marks. Post-transcriptional changes may also play important roles. Because of this, approaches to dissecting the mechanism of regulation must be multilayered. To date, interactions between changes in gene expression and hormones are well established. For example, studies reviewed above suggest that morph-specific gene expression can sensitize relevant tissues to hormonal action in a morph-specific manner. Further support for this idea is provided by a recent study in the brown planthopper, which showed that two insulin receptors regulate how the wing buds respond to an insulin-like peptide secreted by the brain, producing long-winged or short-winged forms [54]. Although the role of epigenetic modifications in regulating polyphenism is still being investigated, integration is clearly on the horizon.
Proximal studies of insect polyphenism ultimately address the mechanisms that underlie a fundamental feature of life on earth: organisms can sense changes their environment and, in response, alter development to produce adaptive changes in phenotype. An understanding of these mechanisms ought to provide a sound basis for understanding how such plasticity evolves and how it may constrain or facilitate future evolution.
Highlights.
Many insects can produce adaptive, polyphenic morphs in response to environmental cues
The regulatory mechanisms underlying polyphenism are slowly being revealed
Hormonal and transcriptomic approaches have revealed specific mechanisms
Epigenetic approaches are promising, yet further study is needed
Acknowledgments
GKD is supported by award IOS-1051643 from the National Science Foundation. JAB is supported by a startup award from the University of Rochester and award R01GM116867 from the National Institute of General Medical Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jennifer A. Brisson, Email: jennifer.brisson@rochester.edu.
Gregory K. Davis, Email: gdavis@brynmawr.edu.
References and recommended reading
- 1.Michener CD. Social polymorphism in Hymenoptera. Symp R Entomol Soc Lond. 1961;1:43–56. [Google Scholar]
- 2.Mayr E. Animal Species and Evolution. Cambridge: Harvard University Press; 1963. [Google Scholar]
- 3.Moran NA. The evolutionary maintenance of alternative phenotypes. Am Nat. 1992;139:971–989. [Google Scholar]
- 4.Nijhout HF. Control mechanisms of polyphenic development. Bioscience. 1999;49:181–192. [Google Scholar]
- 5.Simpson SJ, Sword GA, Lo N. Polyphenism in insects. Curr Biol. 2011;21:R738–749. doi: 10.1016/j.cub.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 6.Rajakumar R, San Mauro D, Dijkstra MB, Huang MH, Wheeler DE, Hiou-Tim F, Khila A, Cournoyea M, Abouheif E. Ancestral Developmental Potential Facilitates Parallel Evolution in Ants. Science. 2012;335:79–82. doi: 10.1126/science.1211451. [DOI] [PubMed] [Google Scholar]
- 7.Brakefield PM, Reitsma N. Phenotypic plasticity, seasonal climate and the population biology of Bicyclus butterflies (Satyridae) in Malawi. Ecol Entomol. 1991;16:291–303. [Google Scholar]
- 8*.Oostra V, Mateus ARA, van der Burg KRL, Piessens T, van Eijk M, Brakefield PM, Beldade P, Zwaan BJ. Ecdysteroid Hormones Link the Juvenile Environment to Alternative Adult Life Histories in a Seasonal Insect. American Naturalist. 2014;184:E79–E92. doi: 10.1086/677260. First study to functionally link ecdysteroids to the seasonal color polyphenism in Bicyclus, a polyphenism well-studied from an ecological perspective. The authors also performed careful analysis of the full array of traits affected by the polyphenism to determine which characteristics were directly changed by hormone manipulation. [DOI] [PubMed] [Google Scholar]
- 9.Zera AJ. Endocrine analysis in evolutionary-developmental studies of insect polymorphism: hormone manipulation versus direct measurement of hormonal regulators. Evol Dev. 2007;9:499–513. doi: 10.1111/j.1525-142X.2007.00181.x. [DOI] [PubMed] [Google Scholar]
- 10.Stearns SC. The evolutionary significance of phenotypic plasticity. BioScience. 1989;39:436–445. [Google Scholar]
- 11.Wang X, Fang X, Yang P, Jiang X, Jiang F, Zhao D, Li B, Cui F, Wei J, Ma C, et al. The locust genome provides insight into swarm formation and long-distance flight. Nature Communications. 2014;5:1–9. doi: 10.1038/ncomms3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallot A, Shigenobu S, Hashiyama T, Jaubert-Possamai S, Tagu D. Sexual and asexual oogenesis require the expression of unique and shared sets of genes in the insect Acyrthosiphon pisum. Bmc Genomics. 2012;13 doi: 10.1186/1471-2164-13-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daniels EV, Murad R, Mortazavi A, Reed RD. Extensive transcriptional response associated with seasonal plasticity of butterfly wing patterns. Molecular Ecology. 2014;23:6123–6134. doi: 10.1111/mec.12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nanoth Vellichirammal N, Zera AJ, Schilder RJ, Wehrkamp C, Riethoven JJ, Brisson JA. De novo transcriptome assembly from fat body and flight muscles transcripts to identify morph-specific gene expression profiles in Gryllus firmus. PLoS One. 2014;9:e82129. doi: 10.1371/journal.pone.0082129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, Oakley TH, Tokishita S, Aerts A, Arnold GJ, Basu MK, et al. The ecoresponsive genome of Daphnia pulex. Science. 2011;331:555–561. doi: 10.1126/science.1197761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishikawa A, Ishikawa Y, Okada Y, Miyazaki S, Miyakawa H, Koshikawa S, Brisson JA, Miura T. Screening of upregulated genes induced by high density in the vetch aphid Megoura crassicauda. J Exp Zool A. 2012;317:194–203. doi: 10.1002/jez.1713. [DOI] [PubMed] [Google Scholar]
- 17.Le Trionnaire G, Jaubert-Possamai S, Bonhomme J, Gauthier JP, Guernec G, Le Cam A, Legeai F, Monfort J, Tagu D. Transcriptomic profiling of the reproductive mode switch in the pea aphid in response to natural autumnal photoperiod. J Insect Physiol. 2012;58:1517–1524. doi: 10.1016/j.jinsphys.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 18.Kijimoto T, Moczek AP, Andrews J. Diversification of doublesex function underlies morph-, sex-, and species-specific development of beetle horns. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:20526–20531. doi: 10.1073/pnas.1118589109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitman D, Ananthakrishnan TN, editors. Phenotypic Plasticity of Insects: Mechanisms and Consequences. Enfield: Science Publishers; 2009. [Google Scholar]
- 20.Moczek AP, Emlen DJ. Proximate determination of male horn dimorphism in the beetle Onthophagus taurus (Coleoptera: Scarabaeidae) Journal of Evolutionary Biology. 1999;12:27–37. [Google Scholar]
- 21.Kijimoto T, Costello J, Tang Z, Moczek AP, Andrews J. EST and microarray analysis of horn development in Onthophagus beetles. BMC Genomics. 2009;10:504. doi: 10.1186/1471-2164-10-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emlen DJ, Nijhout HF. Hormonal control of male horn length dimorphism in Onthophagus taurus (Coleoptera: Scarabaeidae): a second critical period of sensitivity to juvenile hormone. J Insect Phys. 2001;47:1045–1054. doi: 10.1016/s0022-1910(01)00084-1. [DOI] [PubMed] [Google Scholar]
- 23.Emlen DJ, Nijhout HF. Hormonal control of male horn length dimorphism in the dung betle Onthophagus taurus (Coleoptera: Scarabaeidae) J Insect Phys. 1999;45:45–53. doi: 10.1016/s0022-1910(98)00096-1. [DOI] [PubMed] [Google Scholar]
- 24.Emlen DJ, Warren IA, Johns A, Dworkin I, Corley Lavine L. A mechanism of extreme growth and reliable signaling in sexually selected ornaments and weapons. Science. 2012;337:860–864. doi: 10.1126/science.1224286. [DOI] [PubMed] [Google Scholar]
- 25.Gotoh H, Miyakawa H, Ishikawa A, Ishikawa Y, Sugime Y, Emlen DJ, Lavine LC, Miura T. Developmental link between sex and nutrition; doublesex regulates sex-specific mandible growth via juvenile hormone signaling in stag beetles. PLoS Genet. 2014;10:e1004098. doi: 10.1371/journal.pgen.1004098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gotoh H, Cornette R, Koshikawa S, Okada Y, Lavine LC, Emlen DJ, Miura T. Juvenile hormone regulates extreme mandible growth in male stag beetles. PLoS One. 2011;6:e21139. doi: 10.1371/journal.pone.0021139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito Y, Harigai A, Nakata M, Hosoya T, Araya K, Oba Y, Ito A, Ohde T, Yaginuma T, Niimi T. The role of doublesex in the evolution of exaggerated horns in the Japanese rhinoceros beetle. Embo Reports. 2013;14:561–567. doi: 10.1038/embor.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lavine L, Gotoh H, Brent CS, Dworkin I, Emlen DJ. Exaggerated trait growth in insects. Annual Review of Entomology. 2015;60:453–472. doi: 10.1146/annurev-ento-010814-021045. [DOI] [PubMed] [Google Scholar]
- 29.Ma Z, Guo W, Guo X, Wang X, Kang L. Modulation of behavioral phase changes of the migratory locust by the catecholamine metabolic pathway. Proc Nat Acad Sci. 2011;108:3882–3887. doi: 10.1073/pnas.1015098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pener M, Simpson S. Locust phase polyphenism: an update. Anv Insect Phys. 2009;23:1–79. [Google Scholar]
- 31.Pener MP, Yerushalmi Y. The physiology of locust phase polymorphism: an update. J Insect Physiol. 1998;44:365–377. doi: 10.1016/s0022-1910(97)00169-8. [DOI] [PubMed] [Google Scholar]
- 32.Ernst UR, Van Hiel MB, Depuydt G, Boerjan B, De Loof A, Schoofs L. Epigenetics and locust life phase transitions. J Exp Biol. 2015;218:88–99. doi: 10.1242/jeb.107078. [DOI] [PubMed] [Google Scholar]
- 33*.Wang X, Kang L. Molecular mechanisms of phase change in locusts. Annu Rev Entomol. 2014;59:225–244. doi: 10.1146/annurev-ento-011613-162019. Comprehensive review of the extensive amount of outstanding work that has been done to elucidate the many factors underlying phase change in locusts. [DOI] [PubMed] [Google Scholar]
- 34.Bonasio R, Tu S, Reinberg D. Molecular Signals of Epigenetic States. Science (New York, NY) 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 36.Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319:1827–1830. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- 37**.Alvarado S, Rajakumar R, Abouheif E, Szyf M. Epigenetic variation in the Egfr gene generates quantitative variation in a complex trait in ants. Nat Commun. 2015;6 doi: 10.1038/ncomms7513. Exciting study linking DNA methylation levels both globally and at the Egfr gene to quantitative differences in environmentally-cued size variation in a species of ant. [DOI] [PubMed] [Google Scholar]
- 38.Glastad KM, Hunt BG, Yi SV, Goodisman MA. DNA methylation in insects: on the brink of the epigenomic era. Insect Mol Biol. 2011;20:553–565. doi: 10.1111/j.1365-2583.2011.01092.x. [DOI] [PubMed] [Google Scholar]
- 39.Lyko F, Maleszka R. Insects as innovative models for functional studies of DNA methylation. Trends Genet. 2011;27:127–131. doi: 10.1016/j.tig.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 40.Glastad KM, Hunt BG, Goodisman MAD. Evolutionary insights into DNA methylation in insects. Current Opinion in Insect Science. 2014;1:25–30. doi: 10.1016/j.cois.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 41.Elango N, Hunt BG, Goodisman MAD, Yi SV. DNA methylation is widespread and associated with differential gene expression in castes of the honeybee, Apis mellifera. Proc Nat Acad Sci. 2009;106:11206–11211. doi: 10.1073/pnas.0900301106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lyko F, Foret S, Kucharski R, Wolf S, Falckenhayn C, Maleszka R. The Honey Bee Epigenomes: Differential Methylation of Brain DNA in Queens and Workers. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walsh TK, Brisson JA, Robertson HM, Gordon K, Jaubert-Possamai S, Tagu D, Edwards OR. A functional DNA methylation system in the pea aphid Acyrthosiphon pisum. Insect Mol Biol. 2010;2010:215–228. doi: 10.1111/j.1365-2583.2009.00974.x. [DOI] [PubMed] [Google Scholar]
- 44.Falckenhayn C, Boerjan B, Raddatz G, Frohme M, Schoofs L, Lyko F. Characterization of genome methylation patterns in the desert locust Schistocerca gregaria. Journal of Experimental Biology. 2013;216:1423–1429. doi: 10.1242/jeb.080754. [DOI] [PubMed] [Google Scholar]
- 45.Hunt B, Brisson JA, Yi S, Goodisman M. Functional conservation fo DNA methylation in the pea aphid and the honeybee. Genome Biol Evol. 2010;2:719–728. doi: 10.1093/gbe/evq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 47.Collings CK, Waddell PJ, Anderson JN. Effects of DNA methylation on nucleosome stability. Nucleic Acids Res. 2013;41:2918–2931. doi: 10.1093/nar/gks893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bonasio R, Li Q, Lian J, Mutti NS, Jin L, Zhao H, Zhang P, Wen P, Xiang H, Ding Y, et al. Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator. Curr Biol. 2012;22:1755–1764. doi: 10.1016/j.cub.2012.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Flores K, Wolschin F, Corneveaux JJ, Allen AN, Huentelman MJ, Amdam GV. Genome-wide association between DNA methylation and alternative splicing in an invertebrate. BMC Genomics. 2012;13:480. doi: 10.1186/1471-2164-13-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, Wheeler D, Avery A, Rago A, Choi J-H, Colbourne JK, Clark AG, Werren JH. Function and evolution of DNA methylation in Nasonia vitripennis. PLoS Genet. 2013;9:e1003872. doi: 10.1371/journal.pgen.1003872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li-Byarlay H, Li Y, Stroud H, Feng S, Newman TC, Kaneda M, Hou KK, Worley KC, Elsik CG, Wickline SA, et al. RNA interference knockdown of DNA methyltransferase 3 affects gene alternative splicing in the honey bee. Proceedings of the National Academy of Sciences. 2013;110:12750–12755. doi: 10.1073/pnas.1310735110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52*.Simola DF, Ye C, Mutti NS, Dolezal K, Bonasio R, Liebig J, Reinberg D, Berger SL. A chromatin link to caste identity in the carpenter ant Camponotus floridanus. Genome Research. 2013;23:486–496. doi: 10.1101/gr.148361.112. First study to profile histone modifications at the genome-wide level in a polyphenic species. The authors found that castes differed in H3K27 acetylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53**.Yang M, Wei Y, Jiang F, Wang Y, Guo X, He J, Kang L. MicroRNA-133 inhibits behavioral aggregation by controlling dopamine synthesis in locusts. PLoS Genet. 2014;10:e1004206. doi: 10.1371/journal.pgen.1004206. A meticulous investigation of how a microRNA contributes significantly to the locust dispersal polyphenism by controlling gene expression activity in the dopamine synthesis pathway. This is the first study to implicate a microRNA in polyphenism regulation in insects. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54*.Xu H-J, Xue J, Lu B, Zhang X-C, Zhuo J-C, He S-F, Ma X-F, Jiang Y-Q, Fan H-W, Xu J-Y, et al. Two insulin receptors determine alternative wing morphs in planthoppers. Nature. 2015;519:464. doi: 10.1038/nature14286. Study that revealed a role for insulin in regulating the wing polyphenism (short versus long wings) of planthoppers. The authors found that two different insulin receptors have different effects on the phenotypes. [DOI] [PubMed] [Google Scholar]
- 55.Chen B, Li S, Ren Q, Tong X, Zhang X, Kang L. Paternal epigenetic effects of population density on locust phase-related characteristics associated with heat-shock protein expression. Molecular Ecology. 2015;24:851–862. doi: 10.1111/mec.13072. [DOI] [PubMed] [Google Scholar]