Summary
One of the clearest functions of the gut microbiota in humans is resistance to colonization by enteric bacterial pathogens. Reconstitution of the microbiota offers an exciting therapeutic approach, but great challenges must be overcome.
We are all covered by and filled with bacteria. The first decade of sequence-based exploration of the human microbiome has established the importance of this dense and diverse ecosystem that we all carry. Entire fields of biology, such as physiology, immunology and behavior are in the process of realigning to account for the effects of the microbiota within their existing intellectual frameworks. In enteric infectious diseases, where the microbiota was generally believed to serve as a physical barrier to pathogens, our understanding of microbiota-mediated resistance has evolved considerably. It is now clear that the microbiota are active participants in preventing, and sometimes in driving disease, depending on the state of the system. Work in the field has been driven by the idea that if beneficial missing microbes were added back to the intestine, it may force out the offending microbes, rebalance the system, and prevent disease. Targeted approaches to rehabilitating the intestinal ecosystem, with fully defined indications, therapeutics, and diagnostics, may still be years away, but the remarkable success of early trials treating recurrent Clostridium difficile infection by reconstitution of the gut microbiota is cause for measured but realistic hope. Here we will survey the known functions of the gut microbiota in defense against enteric infectious diseases with a focus on C. difficile and we will address some of the major challenges facing those who hope to target the gut microbiota for therapy.
The Gut Microbiota
Germ-free laboratory animals can survive and reproduce in a sterile environment, however, the realities of life on a microbe-dominated planet led to the co-evolution of animals with bacteria (McFall-Ngai et al., 2013). Ever since the evolution of the coelomate digestive tract, microbial populations have co evolved within the intestines of animal hosts to form a complex ecosystem: many bacterial taxa and their phages intermingle with a wide array of viruses, and a smaller number of fungi and archaea. The presence and abundance of protists and nematodes among the microbiota are likely to vary between populations, in part based on access to health care and local sanitation standards.
Although microbiologists have been isolating and studying gut bacteria since the beginnings of the discipline (Rajilić-Stojanović and Vos, 2014), it is fair to say that pathogenic bacteria have received greater attention and are better understood thanbacterial species that comprise the microbiota (Box 1). In the 1950s, antibiotics coupled with gnotobiotic mouse rearing techniques led to the loss-of-function studies that established the functional importance of the microbiota in resistance to infections. During this period it became clear that susceptibility to infection varied between hosts and that antibiotics can sensitize hosts to infection. In the 1970s investigators determined the density of Enterococci and Enterobacteriaceae following various antibiotic treatments and demonstrated the importance of the commensals in controlling human infections (Littman and Pamer, 2011; Smith et al., 2007).
Box 1 - Microbes, hosts and damage.
Terminology becomes complicated when incorporating the microbiota into microbial pathogenesis, which has focused narrowly on interactions between exogenous pathogens and hosts. The damage-response framework proposed by Pirofsky and Casadevall, which considers only microbes, hosts, and damage, is a method of fully integrating the microbiota into infectious disease (Casadevall and Pirofski, 2003, 2015). The source of damage can be microbes or host. Rather than defining individual microbes as pathogens, mutualists, or commensals, the damage response framework focuses on the outcome of interactions between microbes and host. The state of the host becomes a function of all interactions with its microbes, resulting in net benefit, damage or indifference. This allows the incorporation of host inflammatory feed-forward loops that cause damage and exacerbate some acute infections (Stecher et al., 2007). The damage response framework also pairs nicely with emerging concepts from immunology such as disease tolerance, which is defined as host responses to infection that prevent damage, while not directly lowering levels of the offending microbe (Medzhitov et al., 2012). Approaches that focus on outcomes rather than strict classification of microbes will be especially important in determining the roles of microbiota members in chronic diseases such as inflammatory bowel disease. The neutral term microbiont, defined as any microbe living inside a host, has been proposed as an alternative to traditional host-centric classifications of microbes (Miles et al., 2015).
Prior to the wide application of second-generation sequencing technology to analysis of the microbiota, David Relman’s group performed a seminal study characterizing the colonic microbiota of three healthy individuals by cloning and sequencing 13,355 bacterial 16S rRNA gene sequences. They found that many of the bacteria had never been cultivated, that most of the sequences derived from species that had never been associated with human disease, and that each individual harbored a distinct colonic microbiota (Eckburg et al., 2005). In the last 10 years high throughput sequencing has prompted the adaptation of tools from microbial ecology to profile the entire gut microbiota population. This work has determined that the gut microbiota comprises 100s of strains, the majority of which come from just 2 bacterial phyla: the Bacteroidetes and Firmicutes. Differences in microbiota composition have been noted between body sites and individuals, and between cultures around the world. Knowledge of baseline compositions of the microbiota has linked changes in composition to phenotypes in laboratory models and pathology in humans. Infectious diseases, particularly hospital-acquired infections that follow antibiotic treatment, represent one of the clearest linkages between changes in the microbiota and health.
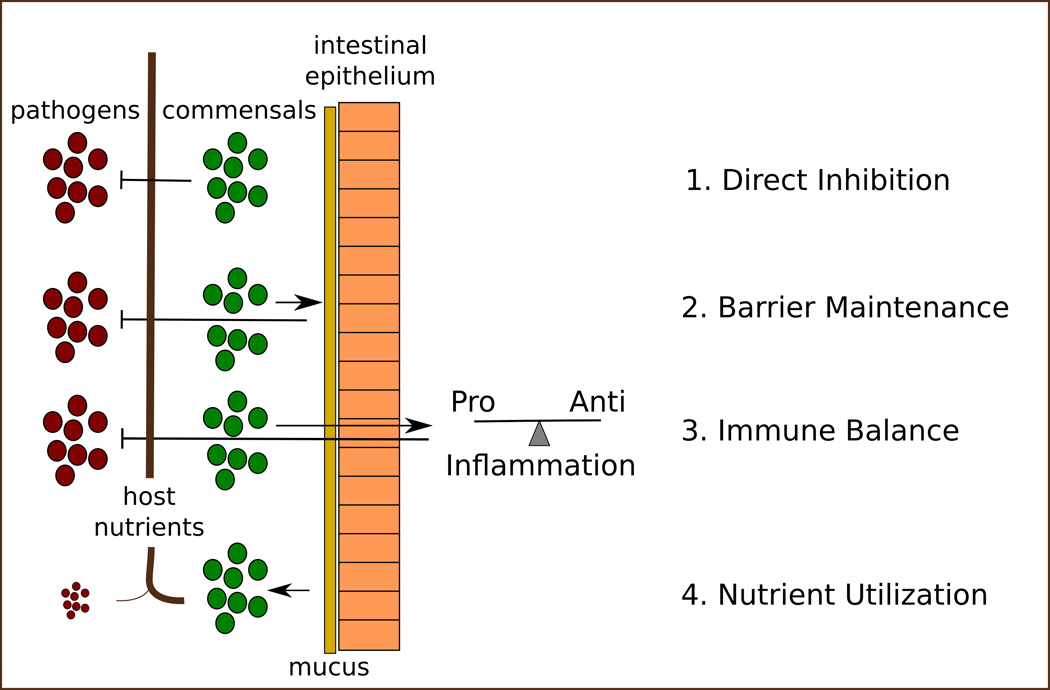
Here we will review four major functions of the microbiota that are relevant to bacterial infectious disease: direct inhibition, barrier maintenance, immune modulation and metabolism (Figure 1). These functions all contribute to colonization resistance, defined as the native ability of a host to suppress invasion by exogenous microorganisms, including potential pathogens (Stecher et al., 2013.
Figure 1. Colonization resistance.
The gut microbiome contributes to colonization resistance through at least 4 inter-related functions. 1) Direct inhibition of neighboring bacteria via the production of toxic compounds. 2) Maintenance of the mucus barrier and underlying intestinal epithelium. 3) Regulation of the immune reponse. 4) Efficient utilization of ingested and endogenous host nutrients.
Direct inhibition
Bacteria produce myriad bioactive small molecules and have long been the primary source of antibiotic candidates for the pharmaceutical industry (Milshteyn et al., 2014). One example are antimicrobial peptides, the bacteriocins, which selectively kill and inhibit the growth of competing bacteria (Kommineni et al., 2015). A computational prediction of biosythetic gene clusters in the human microbiota identified over 3,000 candidate clusters, including many newly annotated antimicrobial peptides, suggesting that these molecules may play a major role in shaping the population structure of the microbiota (Donia et al., 2014).
Bacteria also produce growth inhibitory molecules in secondary metabolism through modification of non-toxic host molecules, which has the potential for wide systemic effects (Box 2). Bile acids are one such example. They are produced in the liver and are secreted into the intestine in milligram quantities where they are modified by the gut microbiota into dozens of different secondary bile acids, each with its own unique spectrum of chemical and biological activities (Devlin and Fischbach, 2015; Ridlon et al., 2006). Secondary bile acid production is drastically reduced by antibiotic treatment, after which mice and humans are susceptible to C. difficile infection (Buffie et al., 2015; Theriot et al., 2014; Weingarden et al., 2014). Reconstitution of antibiotic-treated mice with C. scindens, a bacterium capable of producing secondary bile acids, was sufficient to restore secondary bile acid levels and reduce C. difficile burden. Furthermore, levels of C. scindens were negatively correlated with C. difficile infection in antibiotic-treated hematopoietic stem cell transplant patients, suggesting that a similar mechanism may contribute to C. difficile resistance in humans (Buffie et al., 2015).
Box 2: The microbiota as an endocrine organ.
Bile acid metabolism by the microbiota has the potential to affect systemic physiology. Bile acids function as signaling molecules through multiple nuclear hormone receptors and G-protein coupled receptors expressed in a variety of tissue types (Zhou and Hylemon, 2014) and cells of the immune system (Brestoff and Artis, 2013). Secondary bile acids tend to outcompete primary forms for binding to identified receptors. Therefore, the balance of primary to secondary bile acids has the potential to regulate signaling through these pathways (Ridlon and Bajaj, 2015). In addition, C. scindens may convert glucocorticoids (corticosteroids) to androgens, thus this one bacterium may affect the balance of multiple classes of exogenous and endogenous steroid hormones (Ridlon et al., 2013). Microbial metabolites produced by spore-forming Clostridia, including bile acids, influence the levels of serotonin produced by enterochromaffin cells, suggesting that system-wide modulation of physiology and behavior may be mediated by the gut microbiota (Hsiao et al., 2013; Yano et al., 2015). Going forward it will be important for the developers of orally dosed drug treatments to consider the effects of the microbiota on the bioavailability of drugs. The variability of the microbiota in the population may underlie some of the heterogeneity of treatment outcomes observed in clinical trials (Mani et al., 2014).
Barrier Maintenance
Gut bacteria facilitate their own sequestration within the intestine by regulating the strength of the intestinal barrier. The microbiota are restricted to the intestinal lumen by a mucus layer that overlays the intestinal epithelium and separates the microbiota from the patrolling immune cells of the intestinal lamina propria. Treatment of mice with antibiotics reduces the thickness of the mucus layer and results in increased contact between the underlying intestinal epithelium and gut bacteria (Wlodarska et al., 2011). Exposure to bacterial products is sufficient to stimulate mucus synthesis in germ free mice (Johansson et al., 2015; Petersson et al., 2011) and alterations to host diet were sufficient to reduce the thickness of the mucus layer, further emphasizing the importance of microbial metabolism to barrier maintenance (Earle et al., 2015). Maintenance of the mucus barrier may be relevant clinically, as defects in mucus permeability have been observed in mouse mutants susceptible to colitis (Johansson et al., 2014). The microbiota directly affects the health of underlying intestinal epithelial cells by producing short chain fatty acids (SCFAs), which are a primary nutrient for the colonic epithelium. Decreases in SCFAs may result in degradation of the epithelial barrier. In the absence of a strong epithelial barrier Fusobacterium necrophorum and Bacteroides fragilis, common species among the microbiota, cause abscesses and tissue infections.
Immune Maturation & Inflammation
One of the first demonstrations that individual bacteria of the microbiota contribute to immune development came from studies with Bacteroides fragilis which demonstrated that monocolonization of germ-free mice promoted development of CD4 T lymphocytes (Mazmanian et al., 2005). Subsequent studies have demonstrated that murine intestinal bacterial species promoted the differentiation of CD4 T cells into Th17 cells (Ivanov et al., 2009). Th17 cells can contribute to colonization resistance against pathogens, but also contribute to the development of autoimmune pathology. Bacteria belonging to the cluster XIVa Clostridium group are associated with the development of anti-inflammatory T regulatory cells (Atarashi et al., 2013) as are bacterial consortia consisting of Bacteroides species (Faith et al., 2014). Short chain fatty acids derived from gut microbiota metabolism maintains the balance of inflammatory and anti-inflammatory T cell subsets (Arpaia et al., 2013; Furusawa et al., 2013; Smith et al., 2013). Thus, the balance of the bacterial population structure in the gut and the resulting metabolome have the potential to drive pro-inflammatory or anti-inflammatory responses to immune stimuli.
The microbiota is also a source of ligands for innate immune signaling. Treatment with antibiotics increases susceptibility of mice to dextran sodium sulfate (DSS)-mediated colitis, a phenotype that can be rescued by the administration of Toll-like receptor ligands. This suggests that pattern recognition receptor signaling originating from the microbiota is necessary for protection against epithelial damage (Rakoff-Nahoum et al., 2004). Intestinal microbes stimulate innate immune receptors to promote expression of bactericidal C-type lectins by intestinal epithelial cells that act directly to suppress bacterial growth (Vaishnava et al., 2011). Stimulation by intestinal bacteria can also enhance systemic antiviral responses (Abt et al., 2012; Ganal et al., 2012), while intestinal viruses can enhance antibacterial immunity (Kernbauer et al., 2014).
Utilization of nutrients
Host nutrition is the primary source of energy for the gut microbiota and changes in host dietary patterns can result in rapid changes in the population structure of the microbiota (Carmody et al., 2015; David et al., 2014). As nutrition shapes population structure and alters the gut metabolome, it may shift the microbiota to a state that is sensitive to infection (Ferreyra et al., 2014a). The ability to successfully colonize a mouse with a given bacterial strain has been correlated with abundance of close relatives in the target host (Stecher et al., 2010). These data suggest that high levels of a particular taxon may shift the gut metabolome to a state that is permissive to growth of closely related strains due to similar metabolic requirements. The microbiota can also produce nutrients that directly feed pathogens. Bacteroides thetaiotaomicron can cleave sialic acid moieties from mucin glycan strands and produce high levels of succinate in mono-colonized gnotobiotic mice. The production of both of these metabolites by B. thetaiotaomicron increased the burden of Clostridium difficile in co-colonized mice (Ferreyra et al., 2014b; Ng et al., 2013). Such syntrophic interactions may be widespread between pathogens and the microbiota and represent potential targets for treatment (Box 3).
Box 3: Infectious Disease Ecology.
Our deeper understanding of the microbiome’s impact on the host forces us to reconsider infectious diseases from the perspective of interactions between species within a complex ecosystem. Infectious diseases are likely to occur by two major mechanisms: acquisition of an exogenous bacterium, or expansion of an endogenous strain that causes harm at high levels (so-called pathobionts). C. difficile and other pathogens are carried asymptomatically in the population and the question of how often cases are caused by de novo infection versus expansion remains a challenging exercise in epidemiology. Acquisition of a pathogen endemic to a hospital is conceptually similar to an invasive species. A foreign, but well-adapted bacterium encounters a favorable environment (in this case the GI tract of a patient made sensitive through antibiotics), and expands to fill the niche. The process of pathobiont expansion is similar to the loss of a predator, which allows expansion of former prey species. Mathematical models inspired by ecology and environmental engineering are being developed with increasing sophistication (Bucci and Xavier, 2014; Manor et al., 2014) and such models will be critical in guiding investigators in designing experiments in the laboratory and in testing hypotheses generated from model organisms on human sample sets (Buffie et al., 2015; Stein et al., 2013).
Enteric Infectious Diseases in the Post-Microbiome Era
The impact of the full range of medical treatments on the microbiota remains largely unexplored. Prophylactic antibiotic administration often begins at birth, and the average child has many courses of antibiotics during development. Profiling of the microbiota in response to a single course of the commonly used antibiotic ciprofloxacin revealed that some changes to the microbiota were reversible within weeks, while others were permanent (Dethlefsen and Relman, 2011). Memory effects based on previous history have also been observed due to diet (Carmody et al., 2015), shifting of feeding time, (Zarrinpar et al., 2014) and sleep-wake cycles (Thaiss et al., 2014). The effects of diurnal rhythms on microbiota composition and immunity are likely to be significant, especially in light of recent reports of diurnal fluctuations in immune cell subsets and expression of immune signaling receptors in intestinal epithelial cells (Mukherji et al., 2013; Nguyen et al., 2013; Yu et al., 2013). Models derived from data at single time-points may be missing key biology. All of these variables may contribute to the development of infectious disease through their effects on the microbiota and colonization resistance.
Treating acute infections
C. difficile is the most prominent microbiont exploiting antibiotic-mediated injury to the microbiota, with greater than 250,000 cases annually. Cases of C. difficile are almost always associated with prior antibiotic-treatment, thus it seemed clear from the beginning that susceptibility to this infection resulted from the loss of protective microbiota (Britton and Young, 2014). The importance of the microbiota is reflected in the ability of 16S sequencing data alone to distinguish between C. difficle-positive diarrhea, C. difficile-negative diarrhea, and controls (Schubert et al., 2014). If there were any doubts about the ability of microbiota reconstitution to cure recurrent C. difficile infection, they were put to rest with a randomized, controlled clinical trial of fecal microbiota transplantation (FMT) demonstrating over 90% effectiveness compared to 30% for conventional antibiotic treatment (van Nood et al., 2013). A recent meta-analysis suggests a cumulative cure rate of 89% in immunocompromized patients (Kelly et al., 2014). Mechanistic correlation between mouse and man came from an FMT study in humans that examined levels of bile acids during treatment, which found that levels of growth-inhibitory and spore germination-inhibitory secondary bile acids were inversely correlated with sensitivity to C. difficile (Weingarden et al., 2014).
These data suggest that FMT represents a viable treatment option for patients with recurrent C. difficile. A fecal sample, banked before hospitalization for surgery or other planned interventions that involve antibiotics, may provide a method of preventing hospital-acquired infections, while ensuring replenishment of the patient’s own personalized microbiota. Such an approach would require an infrastructure of service labs to become economically viable. The relative effectiveness of donor stool transplants (heterologous) versus self stool transplants (autologous) has not been tested and it is possible that undesirable phenotypes may be transferred from donors to recipients (Alang and Kelly, 2015). The stability of the transferred microbiota over time is unclear. Follow up studies are beginning to appear that suggest post-FMT variance in population structure of recipients is not significantly elevated over normal baseline variance in the donor (Fuentes et al., 2014; Weingarden et al., 2015).
Given the heterogeneous nature of fecal samples, resistance from clinicians treating chronically ill patients, and regulatory uncertainty, work has focused on defining the microbes responsible for colonization resistance versus C. difficile. Multiple labs have identified commensal microbes that confer resistance to C. difficile in germ free (Reeves et al., 2012) and antibiotic treated mice (Buffie et al., 2015; Lawley et al., 2012). Two of these studies identified strains from Clostridial Cluster XIVa as sufficient to reduce C. difficile burden. C. scindens, assigned to cluster XIVa, has the potential to directly inhibit C. difficile growth through production of growth-inhibitory and spore germination-inhibitory secondary bile acids. Thus, C. scindens could form the base of a consortium designed to inhibit C. difficile. A mix of 17 Clostridial strains, 12 from cluster XIVa including an isolate of C. scindens, induced the development of anti-inflammatory T regulatory (Treg) cells in mice and reduced colitis (Atarashi et al., 2013), as have combinations of Bacteroides (Faith et al., 2014). Inclusion of such strains could potentially synergize with C. scindens to reduce morbidity by suppressing the damage-producing inflammation caused by C. difficile. In order to return an acutely infected patient to a state of health, it will be necessary for tailored treatments to to both drive out the offending microbe and to fully restore colonization resistance to a broad spectrum of damage-causing microbionts. While it is likely that single strains will be identified that can eliminate microbes of interest, if a patient has lost a significant portion of baseline microbiota diversity, it is unlikely that single strains will be sufficient to fully restore baseline levels of colonization resistance.
Treating chronic dysbiosis
Patients undergoing allogeneic hematopoietic stem cell transplantation, which includes a regimen of antibiotic prophylaxis, often lose microbiota diversity and become dominated by antibiotic-resistant organisms such as Streptococcus viridans, Enterococcus faecium and bacteria belonging to the Enterobacteriaceae family. And while patients can appear healthy with a low-diversity microbiota, the loss of microbiota diversity at the time of stem cell engraftment is associated with markedly increased mortality over two years following transplantation (Taur et al., 2012, 2014; Ubeda et al., 2010). In these cases, eliminating the dominant strains and restoring microbiota diversity would be desirable. In mouse models of E. faecium domination, FMT is is effective in restoring diversity and eliminating E. faceium (Ubeda et al., 2013).
In order to eliminate unwanted strains in an otherwise healthy patient it will be necessary for the donor microbiota to overcome the colonization resistance of the host and to expel the offending strain. This may not be possible in the absence of a treatment to lower colonization resistance. Colonization of germ free mice with B. fragilis renders mice highly resistant to subsequent challenge with the same B. fragilis strain, however, mice mono-colonized with B. fragilis were not resistant to colonization by other closely related Bacteroides species. Furthermore, in these experiments B. fragilis was neither expelled, nor even reduced in CFU burden (Lee et al., 2013). These data suggest the existence of multiple distinct niches for closely related bacteria within the intestine and illustrate the challenges faced by prospective microbiome engineers in even simple distillations of the problem. Further investigations into the basic mechanisms of colonization and colonization resistance will be crucial to translating these ideas to the clinic.
Prospects
A simple internet search returns links to do-it-yourself FMT advocacy web sites, instructional videos and unsettling images of home blenders filled with brown liquid. This may have contributed to a recent decision by the Food and Drug Administration to regulate feces as a drug and to require an Investigational New Drug application for all applications of FMT, except in the case of severe recurrent C. difficile. FMT outside of a healthcare setting is not safe and should not be attempted. This statement may seem at odds with the tremendous success of FMT trials in treating C. difficile; however, the donors in these trials were carefully selected and screened in a process that eliminated over 90% of candidates and samples were processed according to Current Good Manufacturing Practices to minimize the potential of contamination (Petrof and Khoruts, 2014). A significant risk exists for transfer of pathogenic microbes and viruses, particularly in immune-compromised patients.
Engineering the microbiome for health is clearly desirable. Association studies have identified microbial candidates underlying a diverse array of acute and chronic diseases. A major challenge going forward will be to determine efficient, minimally invasive methods of colonizing patients, coupled with the elimination of unwanted resident microbes. Methods of colonization may arise from studies aimed at understanding the phenomena that contribute to the stability of the microbiota (Lahti et al., 2014) and synthetic biology may uncover genetic modules that increase the efficiency of colonization (Yaung et al., 2015). Future laboratory investigations should include studies not only of colonization dynamics in germ-free and antibiotic treated mice, but of naïve specific pathogen free mice as well (Kommineni et al., 2015). Most useful would be the discovery of ‘gain-of-function’ variants of intestinal bacteria that have the ability to outcompete and displace closely related strains in antibiotic naïve humans. This will be a great challenge. Over 70 years of antibiotic chemistry have not produced a small molecule capable of selectively eliminating bacteria at the strain level; however, one of the world’s densest microbial ecosystems is within us and may contain fantastic new molecules and genetic modules that have evolved for just this purpose. Designer microbial consortia have the potential to reduce the risks involved in FMT, but the challenges involved in their development are as great as with any other drug.
References
- Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, Paley MA, Antenus M, Williams KL, Erikson J, et al. Commensal Bacteria Calibrate the Activation Threshold of Innate Antiviral Immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alang N, Kelly CR. Weight gain after fecal microbiota transplantation. Open Forum Infect. Dis. 2015;2:ofv004. doi: 10.1093/ofid/ofv004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, et al. Metabolites produced by commensal bacteria promote peripheral regulatory Tcell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat. Immunol. 2013;14:676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton RA, Young VB. Role of the Intestinal Microbiota in Resistance to Colonization by Clostridium difficile. Gastroenterology. 2014 doi: 10.1053/j.gastro.2014.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci V, Xavier JB. Towards Predictive Models of the Human Gut Microbiome. J. Mol. Biol. 2014;426:3907–3916. doi: 10.1016/j.jmb.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody RN, Gerber GK, Luevano JM, Jr, Gatti DM, Somes L, Svenson KL, Turnbaugh PJ. Diet Dominates Host Genotype in Shaping the Murine Gut Microbiota. Cell Host Microbe. 2015;17:72–84. doi: 10.1016/j.chom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A, Pirofski L. The damage-response framework of microbial pathogenesis. Nat. Rev. Microbiol. 2003;1:17–24. doi: 10.1038/nrmicro732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A, Pirofski L. What Is a Host? Incorporating the Microbiota into the Damage- Response Framework. Infect. Immun. 2015;83:2–7. doi: 10.1128/IAI.02627-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. U.S.A. 2011;108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin AS, Fischbach MA. A biosynthetic pathway for a prominent class of microbiotaderived bile acids. Nat. Chem. Biol. 2015;11:685–690. doi: 10.1038/nchembio.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donia MS, Cimermancic P, Schulze CJ, Wieland Brown LC, Martin J, Mitreva M, Clardy J, Linington RG, Fischbach MA. A Systematic Analysis of Biosynthetic Gene Clusters in the Human Microbiome Reveals a Common Family of Antibiotics. Cell. 2014;158:1402–1414. doi: 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earle KA, Billings G, Sigal M, Lichtman JS, Hansson GC, Elias JE, Amieva MR, Huang KC, Sonnenburg JL. Quantitative Imaging of Gut Microbiota Spatial Organization. Cell Host Microbe. 2015;18:478–488. doi: 10.1016/j.chom.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the Human Intestinal Microbial Flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI. Identifying Gut Microbe–Host Phenotype Relationships Using Combinatorial Communities in Gnotobiotic Mice. Sci. Transl. Med. 2014;6:220ra11, ra220ra11. doi: 10.1126/scitranslmed.3008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreyra JA, Ng KM, Sonnenburg JL. The Enteric Two-Step: nutritional strategies of bacterial pathogens within the gut. Cell. Microbiol. 2014a;16:993–1003. doi: 10.1111/cmi.12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreyra JA, Wu KJ, Hryckowian AJ, Bouley DM, Weimer BC, Sonnenburg JL. Gut Microbiota-Produced Succinate Promotes C. difficile Infection after Antibiotic Treatment or Motility Disturbance. Cell Host Microbe. 2014b;16:770–777. doi: 10.1016/j.chom.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes S, van Nood E, Tims S, Heikamp-de Jong I, Braak CJ, ter Keller JJ, Zoetendal EG, de Vos WM. Reset of a critically disturbed microbial ecosystem: faecal transplant in recurrent Clostridium difficile infection. ISME J. 2014;8:1621–1633. doi: 10.1038/ismej.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, Lienenklaus S, Weiss S, Staeheli P, Aichele P, et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37:171–186. doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MEV, Gustafsson JK, Holmén-Larsson J, Jabbar KS, Xia L, Xu H, Ghishan FK, Carvalho FA, Gewirtz AT, Sjövall H, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63:281–291. doi: 10.1136/gutjnl-2012-303207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson MEV, Jakobsson HE, Holmén-Larsson J, Schütte A, Ermund A, Rodríguez-Piñeiro AM, Arike L, Wising C, Svensson F, Bäckhed F, et al. Normalization of Host Intestinal Mucus Layers Requires Long-Term Microbial Colonization. Cell Host Microbe. 2015;18:582–592. doi: 10.1016/j.chom.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly CR, Ihunnah C, Fischer M, Khoruts A, Surawicz C, Afzali A, Aroniadis O, Barto A, Borody T, Giovanelli A, et al. Fecal Microbiota Transplant for Treatment of Clostridium difficile Infection in Immunocompromised Patients. Am. J. Gastroenterol. 2014;109:1065–1071. doi: 10.1038/ajg.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernbauer E, Ding Y, Cadwell K. An enteric virus can replace the beneficial function of commensal bacteria. Nature. 2014;516:94–98. doi: 10.1038/nature13960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kommineni S, Bretl DJ, Lam V, Chakraborty R, Hayward M, Simpson P, Cao Y, Bousounis P, Kristich CJ, Salzman NH. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature. 2015;526:719–722. doi: 10.1038/nature15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahti L, Salojärvi J, Salonen A, Scheffer M, de Vos WM. Tipping elements in the human intestinal ecosystem. Nat. Commun. 2014;5 doi: 10.1038/ncomms5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C, Goulding D, Rad R, Schreiber F, Brandt C, et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 2012;8:e1002995. doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littman DR, Pamer EG. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe. 2011;10:311–323. doi: 10.1016/j.chom.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani S, Boelsterli UA, Redinbo MR. Understanding and modulating mammalianmicrobial communication for improved human health. Annu. Rev. Pharmacol. Toxicol. 2014;54:559–580. doi: 10.1146/annurev-pharmtox-011613-140007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor O, Levy R, Borenstein E. Mapping the Inner Workings of the Microbiome: Genomic- and Metagenomic-Based Study of Metabolism and Metabolic Interactions in the Human Microbiome. Cell Metab. 2014;20:742–752. doi: 10.1016/j.cmet.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. U.S.A. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Schneider DS, Soares MP. Disease tolerance as a defense strategy. Science NY. 2012;335:936–941. doi: 10.1126/science.1214935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles J, Jonathan H, Handelsman J. Allies and Adversaries: Roles of the Microbiome in Infectious Disease. Microbe. 2015;10:370–374. [Google Scholar]
- Milshteyn A, Schneider JS, Brady SF. Mining the Metabiome: Identifying Novel Natural Products from Microbial Communities. Chem. Biol. 2014;21:1211–1223. doi: 10.1016/j.chembiol.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in Intestinal Epithelium Is Orchestrated by the Circadian Clock and Microbiota Cues Transduced by TLRs. Cell. 2013;153:812–827. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen KD, Fentress SJ, Qiu Y, Yun K, Cox JS, Chawla A. Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes. Sci. N. Y. NY. 2013;341:1483–1488. doi: 10.1126/science.1240636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JFWM, Tijssen JGP, et al. Duodenal Infusion of Donor Feces for Recurrent Clostridium difficile. N. Engl. J. Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- Petersson J, Schreiber O, Hansson GC, Gendler SJ, Velcich A, Lundberg JO, Roos S, Holm L, Phillipson M. Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am. J. Physiol. - Gastrointest. Liver Physiol. 2011;300:G327–G333. doi: 10.1152/ajpgi.00422.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrof EO, Khoruts A. From Stool Transplants to Next-Generation Microbiota Therapeutics. Gastroenterology. 2014;146:1573–1582. doi: 10.1053/j.gastro.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajilić-Stojanović M, Vos WMde. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol. Rev. 2014;38:996–1047. doi: 10.1111/1574-6976.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of Commensal Microflora by Toll-Like Receptors Is Required for Intestinal Homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Reeves AE, Koenigsknecht MJ, Bergin IL, Young VB. Suppression of Clostridium difficile in the Gastrointestinal Tracts of Germfree Mice Inoculated with a Murine Isolate from the Family Lachnospiraceae. Infect. Immun. 2012;80:3786–3794. doi: 10.1128/IAI.00647-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon JM, Bajaj JS. The human gut sterolbiome: bile acid-microbiome endocrine aspects and therapeutics. Acta Pharm. Sin. B. 2015;5:99–105. doi: 10.1016/j.apsb.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon JM, Kang D-J, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- Ridlon JM, Ikegawa S, Alves JMP, Zhou B, Kobayashi A, Iida T, Mitamura K, Tanabe G, Serrano M, De Guzman A, et al. Clostridium scindens: a human gut microbe with a high potential to convert glucocorticoids into androgens. J. Lipid Res. 2013;54:2437–2449. doi: 10.1194/jlr.M038869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert AM, Rogers MAM, Ring C, Mogle J, Petrosino JP, Young VB, Aronoff DM, Schloss PD. Microbiome Data Distinguish Patients with Clostridium difficile Infection and Non-C. difficile-Associated Diarrhea from Healthy Controls. mBio. 2014;5:e01021–e01014. doi: 10.1128/mBio.01021-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 2007;19:59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Chaffron S, Käppeli R, Hapfelmeier S, Freedrich S, Weber TC, Kirundi J, Suar M, McCoy KD, von Mering C, et al. Like Will to Like: Abundances of Closely Related Species Can Predict Susceptibility to Intestinal Colonization by Pathogenic and Commensal Bacteria. PLoS Pathog. 2010;6:e1000711. doi: 10.1371/journal.ppat.1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Berry D, Loy A. Colonization resistance and microbial ecophysiology: using gnotobiotic mouse models and single-cell technology to explore the intestinal jungle. FEMS Microbiol. Rev. 2013 doi: 10.1111/1574-6976.12024. [DOI] [PubMed] [Google Scholar]
- Stein RR, Bucci V, Toussaint NC, Buffie CG, Rätsch G, Pamer EG, Sander C, Xavier JB. Ecological modeling from time-series inference: insight into dynamics and stability of intestinal microbiota. PLoS Comput. Biol. 2013;9:e1003388. doi: 10.1371/journal.pcbi.1003388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taur Y, Xavier JB, Lipuma L, Ubeda C, Goldberg J, Gobourne A, Lee YJ, Dubin KA, Socci ND, Viale A, et al. Intestinal Domination and the Risk of Bacteremia in Patients Undergoing Allogeneic Hematopoietic Stem Cell Transplantation. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2012 doi: 10.1093/cid/cis580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taur Y, Jenq RR, Perales M-A, Littmann ER, Morjaria S, Ling L, No D, Gobourne A, Viale A, Dahi PB, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. 2014;124:1174–1182. doi: 10.1182/blood-2014-02-554725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, Abramson L, Katz MN, Korem T, Zmora N, et al. Transkingdom Control of Microbiota Diurnal Oscillations Promotes Metabolic Homeostasis. Cell. 2014;159:514–529. doi: 10.1016/j.cell.2014.09.048. [DOI] [PubMed] [Google Scholar]
- Theriot CM, Koenigsknecht MJ, Carlson PE, Hatton GE, Nelson AM, Li B, Huffnagle GB, Li JZ, B YV. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 2014;5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda C, Taur Y, Jenq RR, Equinda MJ, Son T, Samstein M, Viale A, Socci ND, van den Brink MRM, Kamboj M, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J. Clin. Invest. 2010;120:4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda C, Bucci V, Caballero S, Djukovic A, Toussaint NC, Equinda M, Lipuma L, Ling L, Gobourne A, No D, et al. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect. Immun. 2013;81:965–973. doi: 10.1128/IAI.01197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O, Ley R, Wakeland EK, Hooper LV. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarden A, González A, Vázquez-Baeza Y, Weiss S, Humphry G, Berg-Lyons D, Knights D, Unno T, Bobr A, Kang J, et al. Dynamic changes in short- and long-term bacterial composition following fecal microbiota transplantation for recurrent Clostridium difficile infection. Microbiome. 2015;3:10. doi: 10.1186/s40168-015-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarden AR, Chen C, Bobr A, Yao D, Lu Y, Nelson VM, Sadowsky MJ, Khoruts A. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am. J. Physiol. - Gastrointest. Liver Physiol. 2014;306:G310–G319. doi: 10.1152/ajpgi.00282.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M, Willing B, Keeney KM, Menendez A, Bergstrom KS, Gill N, Russell SL, Vallance BA, Finlay BB. Antibiotic Treatment Alters the Colonic Mucus Layer and Predisposes the Host to Exacerbated Citrobacter rodentium-Induced Colitis. Infect. Immun. 2011;79:1536–1545. doi: 10.1128/IAI.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, Nagler CR, Ismagilov RF, Mazmanian SK, Hsiao EY. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaung SJ, Deng L, Li N, Braff JL, Church GM, Bry L, Wang HH, Gerber GK. Improving microbial fitness in the mammalian gut by in vivo temporal functional metagenomics. Mol. Syst. Biol. 2015;11:788. doi: 10.15252/msb.20145866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Rollins D, Ruhn KA, Stubblefield JJ, Green CB, Kashiwada M, Rothman PB, Takahashi JS, Hooper LV. TH17 Cell Differentiation Is Regulated by the Circadian Clock. Science. 2013;342:727–730. doi: 10.1126/science.1243884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinpar A, Chaix A, Yooseph S, Panda S. Diet and Feeding Pattern Affect the Diurnal Dynamics of the Gut Microbiome. Cell Metab. 2014;20:1006–1017. doi: 10.1016/j.cmet.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Hylemon PB. Bile acids are nutrient signaling hormones. Steroids. 2014;86:62–68. doi: 10.1016/j.steroids.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]