Intermediate clusters may cause disease in areas where dengue and malaria are present, so differential diagnosis is necessary.
Keywords: Leptospira spp., Ecuador, intermediate cluster, intermediate species, fever, carrier animals, rrs gene, urban areas, rural areas, semiurban areas, bacteria, humans, febrile, zoonoses, prevalence
Abstract
Leptospira spp., which comprise 3 clusters (pathogenic, saprophytic, and intermediate) that vary in pathogenicity, infect >1 million persons worldwide each year. The disease burden of the intermediate leptospires is unclear. To increase knowledge of this cluster, we used new molecular approaches to characterize Leptospira spp. in 464 samples from febrile patients in rural, semiurban, and urban communities in Ecuador; in 20 samples from nonfebrile persons in the rural community; and in 206 samples from animals in the semiurban community. We observed a higher percentage of leptospiral DNA–positive samples from febrile persons in rural (64%) versus urban (21%) and semiurban (25%) communities; no leptospires were detected in nonfebrile persons. The percentage of intermediate cluster strains in humans (96%) was higher than that of pathogenic cluster strains (4%); strains in animal samples belonged to intermediate (49%) and pathogenic (51%) clusters. Intermediate cluster strains may be causing a substantial amount of fever in coastal Ecuador.
Leptospirosis, caused by spirochetes of the genus Leptospira, is a neglected and potentially fatal disease that burdens impoverished communities of developing nations in tropical regions (1–4). The bacteria cause 1.7 million human cases of severe disease worldwide each year (1,2); outbreaks frequently occur during the rainy season in cities in the tropics (4–8). Domestic, peridomestic, and wild mammals harbor diverse Leptospira spp. in their kidneys, and their urine contaminates water sources and soil (6,8).
Leptospira comprises 20 species that are phylogenetically arranged in 3 clusters: pathogenic, saprophytic, and intermediate (6,9). Nine pathogenic and 5 intermediate species, comprising >200 serovars, have been characterized (6,9,10). Some reports associate intermediate cluster strains with mild (11–14) to severe (15,16) leptospirosis; however, this cluster is not well characterized (3,11,15,16). Furthermore, the current notion is that human leptospirosis is mainly caused by strains of the pathogenic cluster (2,4,6,9,10).
Many aspects of leptospirosis epidemiology remain unknown because only limited information exists regarding leptospiral population genetics and the role of environmental factors, including environmental persistence of leptospires, in disease occurrence. These deficiencies in knowledge result from the complexity of the disease (e.g., many animal reservoirs carry 1 of the 14 species of potentially infectious leptospires) and technical difficulties associated with classical diagnostics, such as cumbersome isolation of bacteria from clinical samples, complex standard serologic methods, and a lack of culture techniques to obtain isolates from environmental samples. We present a molecular approach to address some of these shortcomings.
Leptospirosis is common in tropical areas of Ecuador (17). The most severe documented outbreak occurred in 1998 in Guayaquil, where 80% of case-patients required hospitalization and 12% died (J. Leake, pers. comm., 2004). During 2010–2012 in Portoviejo, Ecuador, >2,000 serologically confirmed cases of febrile leptospirosis were reported by local health authorities (M. Morales, pers. comm., 2013). We used molecular methods to amplify and sequence the leptospiral 16S rrs gene from clinical samples from patients in 3 coastal communities in Ecuador that vary in their levels of urbanization.
Methods
Human Samples
During February 2011–December 2012, a total of 464 serum and blood spot samples were collected from acute, febrile patients attending hospitals or health posts in rural, semiurban, and urban communities in Ecuador. Samples from Esmeraldas, a rural community, were provided by Hospital de Borbón (Esmeraldas Province) and the Ecuador Ministry of Health (MoH). The hospital provided 108 serum samples from febrile patients; the samples had been tested for dengue virus (IgM ELISA; PanBio, Brisbane, Queensland, Australia) but not Leptospira spp.; 33 were positive for dengue virus. During the same time period, the Ecuador MoH collected 102 blood spot samples from febrile patients in Esmeraldas. The samples were collected onto filter paper (Whatman 903 Specimen Collection Paper; Whatman, Springfield Mill, UK), dried at room temperature, and stored at −20°C in plastic zipper bags. Twenty serum samples obtained from nonfebrile persons during March 2012 (rainy season) in the same locality were also provided. Protocols used to obtain human samples from Esmeraldas were approved by the Universidad San Francisco de Quito Bioethics Committee and the University of Michigan Institutional Review Board.
A total of 100 serum samples from febrile patients in Portoviejo, a semiurban community, were provided by the Ecuador MoH; 34 were positive for Leptospira spp. (IgM ELISA; PanBio). The other 66 samples were not tested for Leptospira spp., but they were tested for dengue virus by IgM ELISA (9 were positive). The samples had been collected during the rainy season, March–June 2012.
A total of 154 serum samples from febrile patients in Guayaquil, an urban community, were provided by the Ecuador MoH. Samples were collected from different medical posts and hospitals around the city during the rainy season, July–October 2011. The samples had been tested for dengue virus by IgM ELISA (all were negative); no samples were tested for Leptospira spp. All samples from Portoviejo and Guayaquil were collected by government officials and were deidentified before being sent to our laboratory.
Animal Samples
In Portoviejo, during the dry season in 2009 and the wet season in 2013, we collected urine samples from domestic animals (27 pigs, 30 dogs, and 27 cows in 2009; 30 pigs and 26 cows in 2013) and kidney samples from rats (6 in 2009 and 60 in 2013). We administered 2.5 mg/kg of furosemide (a diuretic) to animals to collect their urine during micturition or by cystocentesis. Rats were captured inside the homes of Portoviejo residents by using traps from Tomahawk Live Trap (Hazelhurst, WI, USA) or snap traps, and as needed, they were euthanized by using chloroform. Urine samples collected in 2009 from cattle, pigs, and dogs were obtained from residential areas. Cattle and pig urine samples collected in 2013 were obtained from a local slaughterhouse that processed animals from the same location.
Overview of the Molecular Analyses
Our overall analytic goal was to ensure detection of leptospires of the intermediate and pathogenic clusters. To this end, we adapted a previously used protocol to amplify the rrs genes from pathogenic and intermediate clusters. We then sequenced the rrs gene to detect leptospiral species and anomalous amplification products. A sample was considered positive when its amplicon comprised sequences for leptospira bacteria.
DNA Extraction
Frozen animal urine samples (10 mL) were thawed on ice and pelleted by centrifugation at 3,287 × g for 15 min. DNA was extracted from the pellets by using the QIAamp DNA Mini Kit (QIAGEN, Valencia, CA, USA) and stored at −80°C. Frozen serum samples were thawed on ice, and 200 μL was used for DNA extraction (QIAamp DNA Mini Kit); the DNA was stored at −80°C. Eight punches (6-mm diameter) from blood spots were placed in a 1.5-mL microcentrifuge tube and incubated in 180 μL of ATL buffer (QIAGEN) for 10 min at 85°C, and the supernatant was transferred into a new 1.5-mL microcentrifuge tube and processed for DNA extraction.
A 2-mm3 section of rat kidney was cut and washed 3 times with 1 mL of PBS. DNA was extracted by dissolving the kidney tissue in 700 µL of CTAB extraction buffer, followed by incubation (with shaking every 15 min) for 2 h at 65°C. The tubes were cooled to room temperature, and 700 µL of a chloroform–isoamyl alcohol (24:1) mixture was added to each tube. Contents were mixed and then centrifuged at 6,000 × g for 5 min, and the aqueous phase was transferred to another tube. DNA was precipitated with a 3 M sodium acetate (pH 5) solution and ethanol, and the pellet was washed with 70% ethanol, dried, and dissolved in 50 μL of Tris-EDTA buffer.
Amplification of Leptospiral rrs Gene
Leptospiral DNA from samples was detected by using 1 of the following primer sets (AB or CD), both of which amplify the same small fragment target of 16S rrs gene specific to leptospiral species: forward A 5′-GGCGGCGCGTCTITAAACATG-3′, reverse B 5′-TTCCCCCCATTGAGCAAGATT-3′, forward C 5′-CAAGTCAAGCGGAGTAGCAA-3′, reverse D 5′-CTTAACCTGCTGCCTCCCGTA-3′ (18). Amplicon sizes were 332 bp for primers AB and 290 bp for primers CD. We adapted this protocol for real-time PCR using the CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The PCR master mix included iQ SYBR Green Supermix (Bio-Rad), 0.5 μM each primer and molecular biology–grade water, and 1 μL of DNA template for a final reaction volume of 10 μL. Our amplification protocol used an initial enzyme activation step at 95°C for 3 min, followed by 45 amplification cycles (30 s at 95°C, 30 s at 62.5°C, 30 s at 72°C). To detect the presence of amplified rrs gene amplicon, we performed a melting curve analysis (65°C to 85°C, with a ramp of 0.5°C/5 s). To increase the concentration of the rrs gene amplicon, we subjected the PCR products from samples positive for rrs gene amplification to a second round of PCR amplification by using the conventional PCR protocol (18). Using the same AB or CD primer pairs and 0.5 μL of rrs gene amplicons from the first PCR amplification, we initiated the amplification protocol for the second PCR with denaturation at 94°C for 3 min, followed by 29 cycles of 94°C for 1 min, 63°C for 1.5 min, and 72°C 2 min, and then ended with a final 10-min elongation at 72°C. To rule out accidental contamination of PCR reagents, we included negative controls in all reactions. In addition, all reagents used in our analyses performed during 2012–2013 were subjected DNA amplification by using primer sets AB and CD (real-time and conventional PCR).
Sequence Analysis of Leptospiral rrs Gene
Concentrated rrs gene amplicons of 277 samples were sequenced at Functional Biosciences (Madison, WI, USA) by using primers AB or CD. To analyze DNA sequences, we used MEGA 5.08 (http://www.megasoftware.net) and BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). For sequencing, we selected 19 amplicons from samples collected in 2009 from different animals in Portoviejo. Overall, 11 Leptospira spp. sequences were submitted to GenBank under accession nos. JN377490 and JN377491 (L. inadai from animals in Portoviejo, 2009); JN377492 (L. borgpetersenii from an animal in Portoviejo, 2009); KF303505 (L. wolffii from a human in Esmeraldas, 2012); KF285460 (L. wolffii from a human in Portoviejo, 2012); KF285460 (L. wolffii from a human in Guayaquil, 2011); KM259910 (L. wolffii from an animal in Portoviejo, 2013); KF303504 (L. noguchii from a human in Esmeraldas, 2012); KF303503 (L. borgpetersenii from a human in Guayaquil, 2011); KJ573104 (L. noguchii from an animal in Portoviejo, 2013); and KJ573105 (L. borgpetersenii from an animal in Portoviejo, 2013).
Design of Intermediate Leptospira spp.–Specific Assay
We designed a Leptospira spp. assay to target only intermediate Leptospira spp. We used Primer3 (19) to design a reverse primer (R Inter: 5′-TCTTTACCTATCARATCYTGTGATCCA-3′) to be used with A or C forward primers; amplicon sizes were 160 bp for A and 143 bp for C. The specificity of this assay was validated with 17 leptospiral DNA samples from reference strains of pathogenic, saprophytic, and intermediate leptospiral species obtained from the Royal Tropical Institute (Table 1). We tested for the intermediate leptospiral genotype in 75 human serum samples from Portoviejo that were real-time PCR positive for Leptospira spp. The real-time PCR amplicons were subjected to a second PCR amplification using R-Inter reverse primer. The total reaction volume of the intermediate-specific real-time PCR assay, using GoTaq Flexi Polymerase (Promega, Madison, WI, USA), was 20 μL; 0.5 μL of the real-time PCR amplicon was used as template for the conventional PCR.
Table 1. PCR results by using primer Inter- R combined with primers A and C*.
| Leptospira spp. | Strain | PCR result | |
|---|---|---|---|
| Intermediate | |||
| L. broomii | 5399 | + | |
| L. fainei | BUT 6 | + | |
| L.inadai | 10 | + | |
| L. wolffii | Korat-H2T | + | |
|
L. licerasiae
|
VAR010 |
+ |
|
| Pathogenic | |||
| L. interrogans | Pomona | − | |
| L. kirschneri | Kambale | − | |
| L. borgpetersenii | MUS 127 | − | |
| L. noguchii | M7 | − | |
| L. alexanderi | A85 | − | |
| L. santarosai | CZ 390 | − | |
|
L. weilii
|
Sarmin |
− |
|
| Saprophytic | |||
| L. vanthielii | WazHolland | − | |
| L. biflexa | Patoc I | − | |
| L.meyeri | ICF | − | |
| L. wolbachii | CDC | − | |
| L. kmetyi | Bejo-Iso9T | − | |
*Only intermediate Leptospira species were amplified by using Inter-R specific primer. +, positive; –, negative.
Results
Human Samples
Leptospiral DNA was detected in 73 (68%) of 108 serum samples and 59 (57%) of 102 blood spots from febrile patients in the rural study site (Esmeraldas) (Table 2; Figure). All Leptospira spp.–positive amplicons from blood spots and 70 (96%) of the 73 Leptospira spp.–positive amplicons from serum samples showed 100% DNA sequence identity with L. wolffii (intermediate cluster). The remaining 3 positive amplicons from serum samples showed 99% identity with L. noguchii (pathogenic cluster). Of the 108 serum samples, 31 (29%) were positive for Leptospira spp. (PCR) and dengue virus (IgM ELISA), 4 (3.7%) were positive for dengue virus only (IgM ELISA), and 42 (39%) were positive for Leptospira spp. only (PCR). DNA sequences of 6 (4%) of 135 amplicons showed anomalous amplification products. All serum samples from nonfebrile patients were either PCR negative for Leptospira rrs gene (n = 15) or determined to be negative for Leptospira spp. because of anomalous amplification products (n = 5).
Table 2. Leptospira spp.–positive samples from febrile patients in 3 communities along the coast of Ecuador, 2011–2012*.
| Location, year | No. samples analyzed |
Leptospira spp.–positive samples |
No. (%) spurious PCR products† | ||||
|---|---|---|---|---|---|---|---|
| Pathogenic cluster |
Intermediate cluster |
||||||
| No. (%) | Species | No. (%) | Species | ||||
| Esmeraldas, 2011–2012‡ | 108§ | 3 (2.7) | L. noguchii | 73 (68) | L. wolffii | 6 (4) | |
|
|
102¶ |
0 |
– |
|
59 (58) |
L. wolffii
|
0 |
| Portoviejo, 2012# | 100 | 0 | – | 24 (24) | L. wolffii | 0 | |
|
|
|
|
|
|
1 (1) |
L. inadai
|
15 (32) |
| Guayaquil 2011** | 154 | 3 (1.9) | L. borgpetersenii | 28 (18) | L. wolffii | 9 (21) | |
| 1 (0.6) | L. kirschneri/L. interrogans†† | – | – | 0 | |||
*The 3 communities were in rural (Esmeraldas), semiurban (Portoviejo), and urban (Guayaquil) locations. Leptospiral DNA in patient samples was detected by PCR. Molecular methods were used to amplify and sequence the leptospiral rrs gene from DNA. –, not applicable/no value. †The spurious products represent serum samples that produced amplicons of the correct size but with DNA sequences different from Leptospira (for the pathogenic and intermediate cluster). ‡Of samples from Esmeraldas, 27% were positive for dengue virus (IgM ELISA) and Leptospira sp. (PCR). §Serum samples. ¶Blood spot samples. #Sixty-six samples were tested for dengue virus by IgM ELISA (57 negative, 9 positive) but were not tested for Leptospira sp.; 34 samples were IgM ELISA–positive for Leptospira sp. but were not tested for dengue virus. **Samples tested negative for dengue virus by IgM ELISA. ††The amplicon showed the same degree of identity to both species.
Figure.
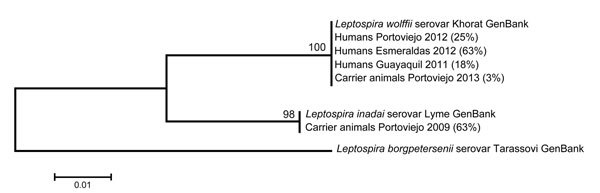
Maximum-likelihood tree for DNA sequences of the Leptospira spp. rrs gene recovered from serum samples from febrile humans and from urine and kidney samples from animal carriers in Ecuador. Esmeraldas, Portoviejo, and Guayaquil are 3 rural, semiurban, and urban communities, respectively, along the coast of Ecuador. Pathogenic L. borgpetersenii was used as an outgroup. Numbers in parentheses indicate the percentage of samples per community that contained DNA signatures highly similar to GenBank reference strains L. wolffii (NR_044042), L. inadai (accession no. JQ988844.1), and L. borgpetersenii (accession no. JQ988861.1). Scale bar indicates the degree of nucleotide substitutions.
Leptospiral DNA was detected in 25 (25%) of 100 serum samples from the semiurban study site (Portoviejo): 7 of these were also IgM positive for Leptospira spp. (dengue IgM unknown), 16 were IgM ELISA negative for dengue virus (Leptospira spp. IgM unknown), and 2 were IgM ELISA positive for dengue virus (Leptospira IgM unknown). Twenty-four Leptospira spp.–positive amplicons showed 100% DNA sequence identity to L. wolffii, 1 amplicon showed 98% identity to L. inadai, and 15 amplicons (of the expected size) were anomalous amplification products.
Leptospiral DNA was detected in 32 (21%) of 154 serum samples from the urban study site (Guayaquil) (Table 2). As with samples from Portoviejo and Esmeraldas, most samples from Guayaquil had amplicon sequences that shared 100% identity with L. wolffii (intermediate cluster). Only 3 (2%) samples shared amplicon sequence identity with pathogenic Leptospira spp. and 2 with L. borgpetersenii (99% identity); 1 could not be differentiated as L. kirschneri or L. interrogans (both 99% identity) (Table 2). Of 43 amplicons displaying the expected size, 9 were anomalous amplification products.
Animal Samples
Of the 90 animal samples collected from Portoviejo during the 2009 dry season, 65 (72%) were PCR positive for Leptospira spp.: 21 (70%) of 30 samples from dogs, 18 (67%) of 27 from pigs, 20 (74%) of 27 from cattle, and all 6 rat kidney samples. However, we sequenced only 19 amplicons from these samples (3 from dogs, 3 from pigs, 7 from cattle, and 6 from rats). BLAST analysis of amplicon sequences from these 19 samples showed that 14 (74%) had 100% sequence identity to L. inadai (intermediate cluster), whereas amplicons from 5 animals (3 cows, 1 pig, and 1 rat) had 100% identity to L. borgpetersenii (pathogenic cluster) (Figure; Table 3). During 2009–2013, the dominant species of leptospires shifted from L. inadai (intermediate cluster) to L. borgpetersenii (pathogenic cluster) (Table 3). In addition, among intermediate types, we observed a population shift from L. inadai to L. wolffii (Table 3; Figure); the identity for L. wolffii sequences was 99%.
Table 3. Species and cluster of leptospiral DNA sequences recovered from animals in 2009 and 2013, Portoviejo, Ecuador*.
| Location, year, animal, no analyzed samples |
Leptospira spp.–positive samples |
No. (%) spurious PCR products† | ||||
|---|---|---|---|---|---|---|
| Pathogenic cluster |
Intermediate cluster |
|||||
| No. (%) | Species | No. (%) | Species | |||
| Portoviejo, 2009‡ | ||||||
| Cattle, n = 7 | 3 (43) | L. borgpetersenii | 4 (57) | L. inadai | 0 | |
| Rats, n = 6 | 1 (17) | L. borgpetersenii | 5 (83) | L. inadai | 0 | |
| Dogs, n = 3 | 0 | – | 3 (100) | L. inadai | 0 | |
| Pigs, n = 3 |
1 (33) |
L. borgpetersenii
|
|
2 (67) |
L. inadai
|
0 |
| Portoviejo, 2013§ | ||||||
| Cattle, n = 26 | 5 (19) | L. borgpetersenii | 1 (4) | L.wolffii | 3 (27) | |
| 1 (4) | L. kirschneri | – | – | |||
| Rats, n = 60 | 3 (5) | L. borgpeterseni | 1 (1.7) | L.wolffii | 3 (21) | |
| 2 (3.3) | L. kirschner¶ | – | – | |||
| Pigs, n = 30 |
2 (6.7) |
L. borgpetersenii
|
|
1 (3.3) |
L.wolffii
|
5 (50) |
| *–, not applicable/no value. †Percentage of amplicons (obtained from samples of each animal species) which showed expected size but the DNA sequences were different from Leptospira spp. ‡Dry season. §Rainy season. ¶One amplicon also showed the same degree of identity to L. interrogans. | ||||||
Verification of the Intermediate Leptospira spp.–Specific Assay
The R-Inter primers amplified only intermediate Leptospira sequences when tested against 17 leptospiral DNA from reference strains (Table 1). Of the 75 human serum samples with a supportive real-time PCR melting curve, 12 were positive for leptospiral sequence when primer pair AB was used, but 23 were positive when primer R-Inter was used. Of the 12 PCR reaction products that were positive by primer pair AB, 10 were also positive when using primer pairs A/R-Inter or C/R-Inter, and 2 DNA samples positive for leptospiral sequence using primers AB were negative when using R-Inter primer. The amplified sequences showed 100% identity to L. wolffii. We did not run this test with L. inadai–positive samples collected from animals in Portoviejo in 2009 because the samples were unavailable for this analysis. Nevertheless, in silico testing showed that the nucleotide sequence of R-Inter primer was identical to L. inadai sequences.
Discussion
Our findings show that leptospiral DNA was present in various proportions in febrile patients living in 3 communities in Ecuador; the DNA was present in 63% of samples from persons at a rural site and in 25% and 21% of samples from persons at semiurban and urban sites, respectively. The use of leptospiral rrs DNA amplification and subsequent sequencing enabled us to detect leptospiral DNA (pathogenic and intermediate clusters) and rule out false-positive reactions. Of note, 96% of leptospiral DNA from human serum showed identity with intermediate rather than pathogenic clade strains. This finding is in contrast with the current notion that human leptospirosis is mainly caused by pathogenic cluster strains (2,4,6,9,10). One reason our findings contrast with those of prior studies is that we sequenced the amplified rrs gene to identify false-positive reactions and to identify intermediate cluster Leptospira spp.
Although our study lacked serologic data to determine acute leptospirosis (seroconversion using paired serum samples), the presence of leptospiral DNA in febrile persons combined with no evidence of dengue infection (a major cause of fever in coastal Ecuador) makes it plausible that the fever was caused by leptospirosis. The finding that none of the serum samples from asymptomatic persons contained leptospiral DNA also supports this finding.
The presence of intermediate leptospiral DNA and the absence of more serious symptoms of leptospirosis (jaundice, hemorrhages, renal failure) in our study are consistent with reports of mild disease linked to intermediate Leptospira spp., such as L. licerasiae in Peru (11), L. wolffii in Thailand (14), and L. inadai (12). Severe leptospirosis symptoms have been associated only with the intermediate cluster species L. broomii (15).
Febrile symptoms could be caused by many infectious agents (17), as evidenced by our finding that in Guayaquil and Portoviejo, 80% and 57% of the febrile population, respectively, did not show evidence of leptospirosis or dengue infection. In addition, environmental factors in these communities may facilitate exposure of inhabitants to multiple infectious agents; thus, febrile symptoms may be due to co-infections. We found evidence of concurrent dengue virus (IgM ELISA) and Leptospira spp. (PCR) infection in 27% of serum samples from Esmeraldas and 22% from Portoviejo. However, concomitant positive diagnostic outcomes for leptospirosis and dengue might be due to persistent presence of antibodies. Detection of IgM antibodies to dengue virus starts 4–5 days after the onset of symptoms and extends for up to 5 months after infection (20), whereas PCR for Leptospira spp. on blood samples mainly detects acute infection (21) and, as reported by others (22,23), is unsuitable for detecting asymptomatic renal colonization. Thus, while co-infection cannot be ruled out, it is conceivable that most, if not all, of these co-infected febrile patients had acute leptospirosis.
We also found high carriage rates of intermediate leptospires (L. inadai and L. wolffii) among domestic and peridomestic animals in Portoviejo in 2009 and 2013 (Figure; Table 3). This finding concurs with those in published reports showing intermediate leptospires carried by domestic and peridomestic animals (11,13,24). The meaning of the relative proportion of intermediate cluster strains observed in this animal study must be considered with caution as we cannot exclude the possibility of selection bias, given the fact that animals were not randomly sampled.
We present molecular evidence of the presence of a similar intermediate Leptospira spp. (L. wolffii) among animal populations and humans in the same locality (Portoviejo). However, because the sampling was conducted at different times, we were unable to directly link Leptospira spp. carriage among animals and humans. This linkage is further complicated by a difference in prevalence rates of L. wolffii DNA among humans in 2012 and animals in 2013 (24% and 2.6%, respectively) (Figure). The difference in distribution of Leptospira spp. in humans and animals may be caused by human lifestyle, which can reduce direct or indirect exposure to the animals, or by different environmental survival capacities of pathogenic and intermediate Leptospira spp. (25). Nevertheless, we showed presence of the same intermediate DNA species of Leptospira in humans and animals, which is consistent with findings in other studies that suggest a link between human disease caused by intermediate leptospiral species (L. licerasiae) from rats and water sources in Peru (3,11).
The presence of leptospiral species in animals appears to be temporally dynamic. In Portoviejo, we observed that the dominant leptospiral species shifted from L. inadai in 2009 to L. borgpetersenii and L. wolffii in 2013 (Table 3). Temporal changes in leptospiral sequence types have been previously reported (26). It is possible that environmental conditions (e.g., humidity, intensity of rainy season, abundance of some animal species, chemical changes in natural water sources) may favor colonization of reservoir animals with a given type of leptospires. These temporal dynamics may explain the apparent sporadic nature of leptospirosis outbreaks; the circulation of pathogenic Leptospira spp. may cause typical and easily recognizable disease, whereas the circulation of intermediate species may cause a generally milder disease with a broad spectrum of symptoms, which makes the disease prone to misdiagnosis. These results highlight the need to conduct longitudinal surveys of leptospiral populations.
Differences in sanitary infrastructure may explain the higher prevalence of leptospiral infection in the rural community as compared with the more urban communities (Table 2). Communities in Esmeraldas tend to rely more on rivers for fresh water and transportation, increasing the probability for leptospiral exposure. Because both urban and rural communities co-exist with animals that carry leptospires, the difference in prevalence we observed likely reflects the efficiency of leptospiral dispersal by water. Unfortunately, we were unable to obtain information about water exposure or occupation of the febrile patients.
Our study also lacked clinical data for febrile patients, which prevented the investigation of presumptive leptospirosis–dengue co-infections or of the difference between infections with pathogenic or intermediate leptospiral species. Although these limitations prevent us from drawing stronger conclusions, our study clearly showed compelling evidence of the abundant presence of intermediate Leptospira spp. in humans and animals. This finding warrants further investigation of the effect of these species on the disease burden observed in veterinary and human public health.
Other limitations in our study were the low number of DNA sequences obtained in 2009 from animals in Portoviejo and the lack of leptospiral isolates belonging to the intermediate cluster. A year later, we attempted without success to amplify leptospiral sequences from positive samples. Also, despite many attempts, we failed to isolate intermediate Leptospira spp. from febrile humans and domestic animals, although we isolated L. santarosai (pathogenic cluster) from a dog urine sample collected in Portoviejo in 2009 (data not shown). It is possible that intermediate species circulate at lower numbers than pathogenic counterparts or that some of these species may be more fastidious than pathogenic species.
Intermediate leptospires are rarely detected in humans, probably because many PCR protocols amplify genes that are present only in pathogenic species (21). Genetic characterization of Leptospira spp. makes it possible to understand disease transmission patterns and to obtain new insights by reinterpreting serologic and clinical epidemiologic data within a genetic context. Correct identification of the etiologic agent is critical for disease management in regions where dengue, malaria, leptospirosis, and, more recently, chikungunya are present (27,28). Our finding of a high number of false-positive reactions reveals the risks of using the 16S PCR (without amplicon sequencing) for diagnosis of leptospirosis.
Acknowledgments
We thank Ministerio de Salud Pública–Ecuador, Celia Riera, Erika Keim, Daisy Parrales, María Inés Baquero, Nadia López, Iván Haro, and Soraya Reyes for their contributions to this investigation; William Cevallos for providing samples from nonfebrile patients in Esmeraldas; and Josefina Coloma for her contribution to the dengue virus detection in Esmeraldas.
This work was funded by the National Institute of Allergy and Infectious Disease, National Institutes of Health (grants AI050038 and AI101913); Organización Panamericana de la Salud cede Ecuador; Universidad San Francisco de Quito Ecuador; and Center for Microbial Genetics and Genomics, Northern Arizona University.
Biography
Mr. Chiriboga has an engineering degree in biotechnology processes and is currently working as a laboratory technician at the Microbiology Institute, Universidad San Francisco de Quito. He is interested in the development of new molecular techniques to be applied in the diagnosis of infectious diseases.
Footnotes
Suggested citation for this article: Chiriboga J, Barragan V, Arroyo G, Sosa A, Birdsell DN, España K, et al. High prevalence of intermediate Leptospira spp. DNA in febrile humans from urban and rural Ecuador. Emerg Infect Dis. 2015 Dec [date cited]. http://dx.doi.org/10.3201/eid2112.140659
References
- 1.Grace D, Mutua F, Ochungo P, Kruska R, Jones K, Brierley L, et al. Mapping of poverty and likely zoonoses hotspots. Zoonoses project 4; report to Department for International Development. July 2012. [cited 2013 May 16]. http://www.bbsrc.ac.uk/documents/dfid-zoonoses-report-4/
- 2.Hartskeerl RA. Leptospirosis: current status and future trends. Indian J Med Microbiol. 2006;24:309. 10.4103/0255-0857.29404 [DOI] [PubMed] [Google Scholar]
- 3.Ganoza CA, Matthias MA, Collins-Richards D, Brouwer KC, Cunningham CB, Segura ER, et al. Determining risk for severe leptospirosis by molecular analysis of environmental surface waters for pathogenic Leptospira. PLoS Med. 2006;3:e308. 10.1371/journal.pmed.0030308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maciel EA, de Carvalho AL, Nascimento SF, de Matos RB, Gouveia EL, Reis MG, et al. Household transmission of Leptospira infection in urban slum communities. PLoS Negl Trop Dis. 2008;2:e154. 10.1371/journal.pntd.0000154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mota CS, Fernandez J, Guzman G, Peña C, Gomez V, Coradin H, et al. Outbreak of leptospirosis in children after tropical storm in the Dominican Republic. Int J Infect Dis. 2008;12(Suppl 1):e334–5. 10.1016/j.ijid.2008.05.894 [DOI] [Google Scholar]
- 6.Ko AI, Goarant C, Picardeau M. Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat Rev Microbiol. 2009;7:736–47. 10.1038/nrmicro2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oliveira DS, Guimarães MJ, Portugal JL, Medeiros Z. The socio-demographic, environmental and reservoir factors associated with leptospirosis in an urban area of north-eastern Brazil. Ann Trop Med Parasitol. 2009;103:149–57. 10.1179/136485909X398221 [DOI] [PubMed] [Google Scholar]
- 8.Trevejo RT, Rigau-Pérez JG, Ashford DA, McClure EM, Jarquín-González C, Amador JJ, et al. Epidemic leptospirosis associated with pulmonary hemorrhage–Nicaragua, 1995. J Infect Dis. 1998;178:1457–63. 10.1086/314424 [DOI] [PubMed] [Google Scholar]
- 9.Evangelista KV, Coburn J. Leptospira as an emerging pathogen: a review of its biology, pathogenesis and host immune responses. Future Microbiol. 2010;5:1413–25. 10.2217/fmb.10.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levett PN. Leptospirosis. Clin Microbiol Rev. 2001;14:296–326. 10.1128/CMR.14.2.296-326.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matthias MA, Ricaldi JN, Cespedes M, Diaz MM, Galloway RL, Saito M, et al. Human leptospirosis caused by a new, antigenically unique Leptospira associated with a Rattus species reservoir in the Peruvian Amazon. PLoS Negl Trop Dis. 2008;2:e213. 10.1371/journal.pntd.0000213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmid GP, Steere AC, Kornblatt AN, Kaufmann AF, Moss CW, Johnson RC, et al. Newly recognized Leptospira species (“Leptospira inadai” serovar lyme) isolated from human skin. J Clin Microbiol. 1986;24:484–6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zakeri S, Khorami N, Ganji ZF, Sepahian N, Malmasi AA, Gouya MM, et al. Leptospira wolffii, a potential new pathogenic Leptospira species detected in human, sheep and dog. Infect Genet Evol. 2010;10:273–7. 10.1016/j.meegid.2010.01.001 [DOI] [PubMed] [Google Scholar]
- 14.Slack AT, Kalambaheti T, Symonds ML, Dohnt MF, Galloway RL, Steigerwalt AG, et al. Leptospira wolffii sp. nov., isolated from a human with suspected leptospirosis in Thailand. Int J Syst Evol Microbiol. 2008;58:2305–8. 10.1099/ijs.0.64947-0 [DOI] [PubMed] [Google Scholar]
- 15.Levett PN, Morey RE, Galloway RL, Steigerwalt AG. Leptospira broomii sp. nov., isolated from humans with leptospirosis. Int J Syst Evol Microbiol. 2006;56:671–3. 10.1099/ijs.0.63783-0 [DOI] [PubMed] [Google Scholar]
- 16.Arzouni JP, Parola P, La Scola B, Postic D, Brouqui P, Raoult D. Human infection caused by Leptospira fainei. Emerg Infect Dis. 2002;8:865–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manock SR, Jacobsen KH, de Bravo NB, Russell KL, Negrete M, Olson JG, et al. Etiology of acute undifferentiated febrile illness in the Amazon Basin of Ecuador. Am J Trop Med Hyg. 2009;81:146–51 . [PubMed] [Google Scholar]
- 18.Mérien F, Amouriaux P, Perolat P, Baranton G, Saint Girons I. Polymerase chain reaction for detection of Leptospira spp. in clinical samples. J Clin Microbiol. 1992;30:2219–24 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–86 . [DOI] [PubMed] [Google Scholar]
- 20.Prince HE, Matud JL. Estimation of dengue virus IgM persistence using regression analysis. Clin Vaccine Immunol. 2011;18:2183–5. 10.1128/CVI.05425-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Picardeau M. Diagnosis and epidemiology of leptospirosis. Med Mal Infect. 2013;43:1–9 . 10.1016/j.medmal.2012.11.005 [DOI] [PubMed] [Google Scholar]
- 22.Ganoza CA, Matthias MA, Saito M, Cespedes M, Gotuzzo E, Vinetz JM. Asymptomatic renal colonization of humans in the Peruvian Amazon by Leptospira. PLoS Negl Trop Dis. 2010;4:e612. 10.1371/journal.pntd.0000612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bal AE, Gravekamp C, Hartskeerl RA, De Meza-Brewster J, Korver H, Terpstra WJ. Detection of leptospires in urine by PCR for early diagnosis of leptospirosis. J Clin Microbiol. 1994;32:1894–8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perolat P, Chappel RJ, Adler B, Baranton G, Bulach DM, Billinghurst ML, et al. Leptospira fainei sp. nov., isolated from pigs in Australia. Int J Syst Bacteriol. 1998;48:851–8. 10.1099/00207713-48-3-851 [DOI] [PubMed] [Google Scholar]
- 25.Bulach DM, Zuerner RL, Wilson P, Seemann T, McGrath A, Cullen PA, et al. Genome reduction in Leptospira borgpetersenii reflects limited transmission potential. Proc Natl Acad Sci U S A. 2006;103:14560–5. 10.1073/pnas.0603979103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thaipadungpanit J, Wuthiekanun V, Chierakul W, Smythe LD, Petkanchanapong W, Limpaiboon R, et al. A dominant clone of Leptospira interrogans associated with an outbreak of human leptospirosis in Thailand. PLoS Negl Trop Dis. 2007;1:e56. 10.1371/journal.pntd.0000056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ellis T, Imrie A, Katz AR, Effler PV. Underrecognition of leptospirosis during a dengue fever outbreak in Hawaii, 2001–2002. Vector Borne Zoonotic Dis. 2008;8:541–7. 10.1089/vbz.2007.0241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crump JA, Morrissey AB, Nicholson WL, Massung RF, Stoddard RA, Galloway RL, et al. Etiology of severe non-malaria febrile illness in Northern Tanzania: a prospective cohort study. PLoS Negl Trop Dis. 2013;7:e2324. 10.1371/journal.pntd.0002324 [DOI] [PMC free article] [PubMed] [Google Scholar]


