Abstract
Rhizobium leguminosarum bv. trifolii strain CC275e is a highly effective, N2-fixing microsymbiont of white clover (Trifolium repens L.). The bacterium has been widely used in both Australia and New Zealand as a clover seed inoculant and, as such, has delivered the equivalent of millions of dollars of nitrogen into these pastoral systems. R. leguminosarum strain CC275e is a rod-shaped, motile, Gram-negative, non-spore forming bacterium. The genome was sequenced on an Illumina MiSeq instrument using a 2 × 150 bp paired end library and assembled into 29 scaffolds. The genome size is 7,077,367 nucleotides, with a GC content of 60.9 %. The final, high-quality draft genome contains 6693 protein coding genes, close to 85 % of which were assigned to COG categories. This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession JRXL00000000. The sequencing of this genome will enable identification of genetic traits associated with host compatibility and high N2 fixation characteristics in Rhizobium leguminosarum. The sequence will also be useful for development of strain-specific markers to assess factors associated with environmental fitness, competiveness for host nodule occupancy, and survival on legume seeds (New Zealand Ministry of Business, Innovation and Employment program, ‘Improving forage legume-rhizobia performance’ contract C10X1308 and DairyNZ Ltd.).
Electronic supplementary material
The online version of this article (doi:10.1186/s40793-015-0110-1) contains supplementary material, which is available to authorized users.
Keywords: Root-nodule bacteria, Microsymbiont, Nitrogen fixation, Rhizobia, Alphaproteobacteria
Introduction
White clover (Trifolium repens) is the most widely established and important legume in pastures in New Zealand [1] and globally [2]. In symbiosis with nodule-forming Rhizobium leguminosarum bacteria of the biovar trifolii (hereafter R. leguminosarumbv trifolii), clover plants fix atmospheric nitrogen into a plant-available, thus providing an economically and environmentally sustainable method of maintaining soil fertility and pasture production. Across New Zealand there are 11,400+ farms using pastures containing forage legumes (mostly white clover), covering 7.88 million hectares [3]. This constitutes about 29 % of the total land area and excludes hill country/tussock grasslands. Estimates of nitrogen input from legumes vary, however average at 185 kg N ha−1 yr−1 for pastures with a slope less than 12° [4]. Based on recent average costs of urea fertilizer (2013–14 average), the value of N2 fixation into New Zealand pastures is 1.8 billion per year; this is highly conservative as it does not encompass the value of increased forage quality, N2 fixation in extensive hill country systems, and reduced environmental costs.
R. leguminosarumbv trifolii strains vary extensively in their ability to form nodules with white clover [5], and also their effectiveness at fixing nitrogen during symbiosis [6]. As such, dedicated selection and screening programs have played a vital role in ensuring clover (and, of course, other legume species) are matched with an optimal rhizobia symbiont [7]. These are most commonly delivered into farming systems as rhizobia-inoculated seed [8].
The inoculation of white clover seed with rhizobia commenced in New Zealand in the early 20th century [8]. In addition to New Zealand produced inoculant strains, R. leguminosarumbv trifolii strain CC275e was sourced from Australia [9]. From 1974, the inoculant production in New Zealand industry was phased-out and the sole commercial strain for inoculation of white clover seed was strain CC275e, which was then replaced with R. leguminosarumbv trifolii strain TA1 (also from Australia) around 2005. Thus, R. leguminosarumbv trifolii strain CC275e was in widespread use in New Zealand for approximately three decades, and is likely to have contributed billions of dollars of nitrogen into New Zealand’s pastoral systems. On white clover, R. leguminosarumbv trifolii strain CC275e has been reported to fix more nitrogen than strain TA1 and has greater persistence in soils [9]. The decision by the inoculant industry to replace strain CC275e with strain TA1 was based on ease of production.
A number of synonyms of strain R. leguminosarumbv trifolii strain CC275e exist. In New Zealand, a culture of strain CC275e was received by the Plant Diseases Division of the Department of Scientific and Industrial Research in 1974 and a re-isolate of this culture is referred to as strain PDD2163. Furthermore, in New Zealand, strain CC275e has also been referred to as strain W16 [10], but when used commercially was most commonly known as strain NZP561 [11]. In Australia, where the bacterium originates, early work referred to it as strain W16 or Strain Hastings T71 [10]. However, strain CC275e was the designation used when the bacterium was deposited in the CSIRO (Canberra) culture collection [12], and this is the most commonly used synonym. In the American Type Culture Collection, the bacterium is referred to as ATCC 35181. For this study, an original R. leguminosarumbv trifolii strain CC275e culture was obtained from the Australian Inoculant Research Group (Gosford, NSW, Australia). These sequence data complements those of Trifolium-nodulating R. leguminosarumbv trifolii strain WSM1325 (GenBank ID 241202755), strain WSM2304 (GenBank ID 209547612), strain WSM1689 (GenBank ID 752843554), and strain TA1 (GenBank ID 653806106).
Organism information
Classification and features
Rhizobium leguminosarum bv. trifolii strain CC275e is a Gram-negative, motile, non-spore forming, non-encapsulated, rod shaped bacterium (Fig. 1). Colonies of R. leguminosarumbv trifolii strain CC275e form within 4 to 5 days when grown on yeast mannitol agar (YMA; [13]) at 25 °C. Colonies are white-opaque, domed and glassy in appearance, with smooth margins.
Fig. 1.
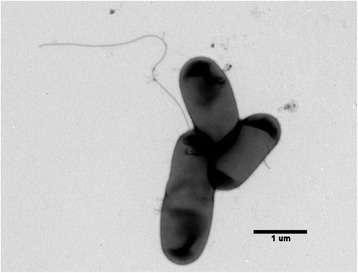
TEM micrograph of three Rhizobium leguminosarum bv. trifolii CC275e cells. The length of the bar = 1 um
Rhizobium leguminosarum and closely related species are generally regarded as non-fastidious, chemo-organotrophic bacteria [14]. Although the wider substrate requirements for strain CC275e have not been formally described, the authors support this classification based on personal experience in the handling, cultivation and fermentation of R. leguminosarumbv trifolii strain CC275e.
The R. leguminosarumbv trifolii strain CC275e genome contains three (100 % identical) copies of the 16S rRNA gene. Alignment of these nucleotide sequences against other species supports close 16S rRNA phylogeny with R. leguminosarum originating from other legume hosts (Fig. 2). The 16S rRNA gene sequence has highest similarity to other accessions of R. leguminosarum biovars trifolii (99.8 %) and phaseoli (99.6 %) (Fig. 2) - the GenBank accession numbers for these are provided in Additional file 1: Table S1. The species is placed within the order Rhizobiales of the class Alphaproteobacteria [15]. Minimum information about the Genome Sequence (MIGS) is provided in Table 1.
Fig. 2.
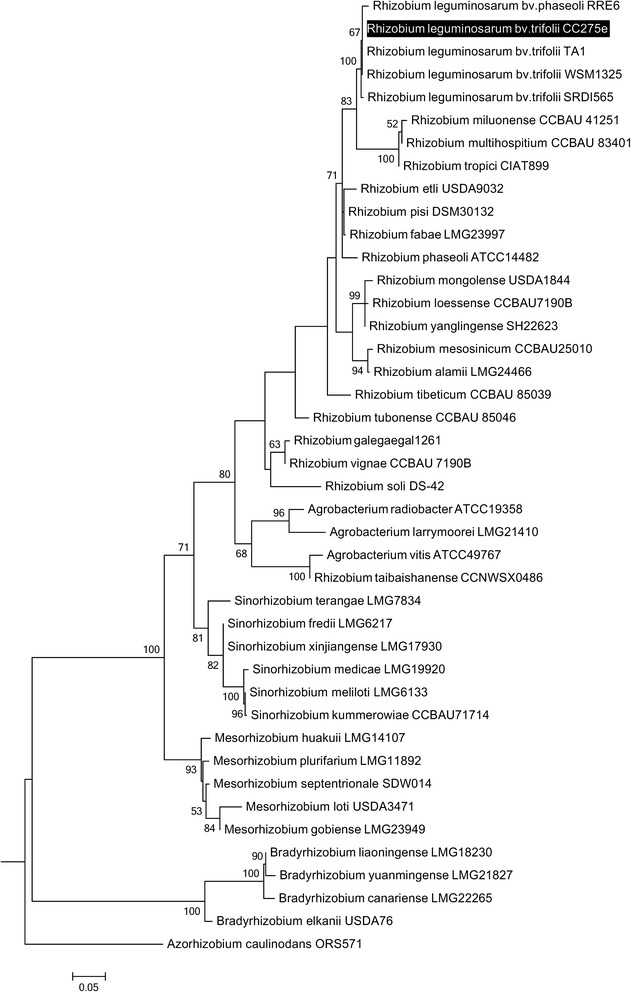
Phylogenetic tree showing relationship of R. leguminosarum bv trifolii CC275e with closely and distantly related taxa in the order Rhizobiales. The tree is based on 1498 bp length alignment of the 16S rRNA gene using MUSCLE with default parameters [31]. The tree was constructed using maximum likelihood method, with the General Time Reversible model (rate 4 classes; [32]). Nodes with bootstrap (1000 repetitions) support > 50 % are shown [33]. Accession numbers relating to the nucleotide sequences for each of the strains are listed in Additional file 1: Table S1
Table 1.
Classification and general features of Rhizobium leguminosarum bv. trifolii strain CC275e according to the MIGS recommendations [34]
| MIGS ID | Property | Term | Evidence codesa |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS [35] | |
| Phylum Proteobacteria | TAS [36] | ||
| Class Alphaproteobacteria | TAS [37] | ||
| Order Rhizobiales | TAS [38] | ||
| Family Rhizobiaceae | TAS [15] | ||
| Genus Rhizobium | TAS [15] | ||
| Species Rhizobium leguminosarum | TAS [14] | ||
| Strain CC275e | TAS [12] | ||
| Gram Stain | Negative | TAS [15] | |
| Cell Shape | Rod | TAS [15] | |
| Motility | Motile | TAS [15] | |
| Sporulation | Non spore-forming | TAS [15] | |
| Temperature range | Mesophile | TAS [15] | |
| Optimum temperature | 28 °C | NAS | |
| pH range; optimum | Unknown | NAS | |
| Carbon source | Varied, chemoorganotrophic | TAS [15] | |
| MIGS-6 | Habitat | Soil, root nodule | TAS [12] |
| MIGS–6.3 | Salinity | Non–halophile | NAS |
| MIGS–22 | Oxygen requirement | Aerobic | TAS [15] |
| MIGS–15 | Biotic relationship | Free living, legume symbiotic | TAS [15] |
| MIGS–14 | Pathogenicity | Non–pathogen | TAS [15, 39] |
| MIGS–4 | Geographic location | Tasmania, Australia | TAS [12] |
| MIGS–5 | Sample collection date | 1966 | TAS [12] |
| MIGS–4.1 | Latitude | Not recorded | |
| MIGS–4.2 | Longitude | Not recorded | |
| MIGS–4.3 | Depth | Not recorded | |
| MIGS–4.4 | Altitude | Not recorded |
aEvidence codes – IDA Inferred from Direct Assay, TAS Traceable Author Statement (i.e., a direct report exists in the literature), NAS Non–traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [34]
Symbiotaxonomy
R. leguminosarumbv trifolii strain CC275e is nodule forming (Nod+) and N2 fixing (Fix+) on a range of annual and perennial clover host species. The original isolation of R. leguminosarumbv trifolii strain CC275e was from Trifolium repens L. collected from Montague, North Western Tasmania [12], and has been used commercially due to its efficacy at forming symbioses and fixation of nitrogen on white clover hosts [9]. The strain is also moderately effective (sensu Brockwell et al. [12]) on T. fragiferum L. (strawberry clover; perennial), and T. michelianum Savi, (balansa clover; annual). On T. subterraneum L. (subterranean clover; annual), T. purpureum Lois. (purple clover; annual), and T. hirtum All. (rose clover; annual), strain CC275e has been described as effective [12].
Genome sequencing information
Genome project history
R. leguminosarumbv trifolii strain CC275e was selected for sequencing based on its long history of commercial use as an inoculant for various clover (Trifolium spp.) hosts in Australia and New Zealand. In symbiosis with clover, this strain of bacteria has provided biologically-fixed nitrogen into soils for several decades, and thereby contributed to the fertility and productivity of pastoral agricultural systems in two countries. As part of a New Zealand MBIE-funded program, ‘Improving forage legume-rhizobia performance’ (C10X1308), the genomics of elite host nodulating (nod+) and N2 fixing (fix+) strains are being compared with closely related, ineffective strains. The aim is to identify markers to facilitate rhizobia selection programs, and to provide experimental tools for host colonization/competition experiments. Based on efforts in other R. leguminosarumbv trifolii strains (see accessions listed in the introduction) a sequencing strategy was developed using a predicted genome size of approximately 7 Mb. The genome sequencing and assembly was completed in 2014; summary information on the project is given in Table 2. The final R. leguminosarumbv trifolii CC275e genome assembly is a high-quality draft on 29 scaffolds, and resulted from approximately 150× sequencing coverage.
Table 2.
Genome sequencing project information for Rhizobium leguminosarum bv. trifolii strain CC275e
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | High-quality draft |
| MIGS-28 | Libraries Used | Illumina TruSeq™ DNA Sample Preparation Kit V2, 2 × 150 bp paired end library |
| MIGS-29 | Sequencing platform | Illumina MiSeq™ |
| MIGS-31.2 | Fold coverage | 3.75 million reads, ≈150 × genome coverage |
| MIGS-30 | Assemblers | A5, SSPACE, Velvet Optimiser |
| MIGS-32 | Gene calling method | Glimmer 3 |
| Locus Tag | ||
| Genbank ID | JRXL00000000 | |
| Genbank Date of Release | 27st October, 2014 | |
| GOLD ID | Gp0113226 | |
| BIOPROJECT | 259682 | |
| MIGS-13 | Source Material Identifier | ATCC 35181 |
| Project relevance | Symbiotic N2 fixation, agriculture |
Growth conditions and genomic DNA preparation
A loop of a single colony of R. leguminosarumbv trifolii CC275e was inoculated into YM broth [13] and grown to mid-log phase via incubation at 28 °C at 200 rpm for 12 h. DNA was extracted from the cell culture using a Gentra Puregene Cell kit (Qiagen). Spectrophotometry was used to quantify the DNA and ensure quality was sufficient for sequencing analysis (Nanodrop Thermo Scientific).
Genome sequencing and assembly
Genome sequencing was conducted through NZGL (contract NZGL00940) at Massey University (MGS). Sequencing was performed on an Illumina MiSeqTM instrument (details in Table 2), using 2 × 150 bp paired-end (PE) library with an average insert size of 420 bp. The sequencing run generated 3,751,285 reads totaling 1088 Mb of data.
Reads were assembled using the Java Assembling and Scaffolding Tool (JAST; [16]). Quality control of the sequence reads was conducted in Flexbar [17], and initial de novo assembly in A5 [18]; this resulted in 52 contigs. Bowtie2 [19] and Velvet [20] were further used to optimize the assembly, using the genome of the closely strain R. leguminosarum strain WSM1325 (Fig. 2) as a reference (NCBI accession 241202755). SSPACE [21] was used to assemble the 35 contigs into 29 scaffolds (Table 3). Summary details of the sequencing process are given in Table 2.
Table 3.
Genome statistics for Rhizobium leguminosarum bv. trifolii strain CC275e
| Attribute | Value | % of total |
|---|---|---|
| Genome size (bp) | 7,077,367 | 100.00 |
| DNA coding (bp) | 6,201,447 | 87.62 |
| DNA G + C (bp) | 4,306,744 | 60.90 |
| DNA scaffolds | 29 | |
| Total genes | 6747 | 100.00 |
| Protein coding genes | 6693 | 99.00 |
| RNA genes | 54 | 0.80 |
| Pseudo genes | not determined | not determined |
| Genes in internal clusters | not determined | not determined |
| Genes with function prediction | 5018 | 74.37 |
| Genes assigned to COGs | 5722 | 84.80 |
| Genes with Pfam domains | 5682 | 84.22 |
| Genes with signal peptides | 531 | 7.87 |
| Genes with transmembrane helices | 1584 | 23.48 |
| CRISPR repeats | 0 |
Genome annotation
Annotation was added by the NCBI Prokaryotic Genome Annotation Pipeline (http://www.ncbi.nlm.nih.gov/genome/annotation_prok/). Clusters of orthologous groups of proteins (COGs) were predicted using COGnitor [22], and the presence of signal peptides was detected using SignalP [23]. Pfam domains were predicted using HMMER [24] against the Pfam-A database [25]. Transmembrane predictions and CRISPR repeats were found in Genious [26] using the Transmembrane prediction (http://www.geneious.com/plugins/transmembrane-prediction-plugin) and CRT plugins [27] respectively.
Genome properties
The genome of R. leguminosarumbv trifolii strain CC275e is estimated to be 7,077,367 nucleotides in size (Table 3). The GC content is 60.9 % which is similar to closely related strains such as R. leguminosarumbv trifolii strain TA1 (60.74 %; [28]). The final draft consists of 29 scaffolds, the largest of which is 1,609,666 bp and the smallest 1167 bp. In total, 6747 genes were identified, 99 % of these were protein coding and the rest rRNA genes (Table 3). The majority of protein coding genes (84.22 %) have functionality predicted against COG categories; these are listed in Table 4. The remainder are listed as hypothetical.
Table 4.
Number of protein coding genes of Rhizobium leguminosarum bv. trifolii strain CC275e associated with the general COG functional categories
| Code | Value | % of total | COG category |
|---|---|---|---|
| J | 189 | 2.69 | Translation |
| A | 0 | 0.00 | RNA processing and modification |
| K | 624 | 8.88 | Transcription |
| L | 186 | 2.65 | Replication |
| B | 2 | 0.03 | Chromatin structure and dynamics |
| D | 38 | 0.54 | Cell cycle control |
| Y | 0 | 0.00 | Nuclear structure |
| V | 64 | 0.91 | Defense mechanisms |
| T | 361 | 5.14 | Signal transduction mechanisms |
| M | 297 | 4.23 | Cell wall/membrane/ biogenesis |
| N | 96 | 1.37 | Cell motility |
| Z | 0 | 0.00 | Cytoskeleton |
| W | 0 | 0.00 | Extracellular structures |
| U | 74 | 1.05 | Intracellular trafficking |
| O | 185 | 2.63 | Posttranslational modification |
| C | 295 | 4.20 | Energy production and conversion |
| G | 646 | 9.19 | Carbohydrate transport and metabolism |
| E | 672 | 9.56 | Amino acid transport and metabolism |
| F | 108 | 1.54 | Nucleotide transport and metabolism |
| H | 151 | 2.15 | Coenzyme transport and metabolism |
| I | 238 | 3.39 | Lipid transport and metabolism |
| P | 234 | 3.33 | Inorganic ion transport and metabolism |
| Q | 95 | 1.35 | Secondary metabolites biosynthesis |
| R | 623 | 8.87 | General function prediction only |
| S | 544 | 7.74 | Function unknown |
| - | 1305 | 18.57 | Not in COGs |
Analysis of the genome by Eckhart gel electrophoresis [29] (Fig. 3) revealed the presence of six mega-plasmids. Mega-plasmids are typical of the ‘ancillary genome’ present in many R. leguminosarum strains [30] and commonly host many of the recognition factors associated with host compatibility, and nitrogen fixation. Based on the known mega-plasmid profile of R. leguminosarumbv trifolii strain WSM1325 (Fig. 3), the mega-plasmids in R. leguminosarumbv trifolii strain CC275e are approximately >1000, 500, 280, 280, 150, and 140 kb in size. As yet it is unknown to which scaffolds these mega-plasmids are associated.
Fig. 3.
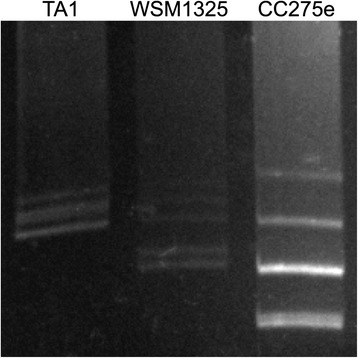
Eckhardt gel electropherogram showing ‘mega-plasmid’ profiles of R. leguminosarum bv trifolii strain CC275e against strains TA1 and WSM1325. The bright central band for strain CC275e represents co-migration of two similarly sized plasmids. Also, note double band at bottom of strain CC275e lane profile. The size of plasmids in reference strain WSM1325 are 294, 350, 516, and 829, 661, 516, 350, and 294 kb
Conclusions
Rhizobiumleguminosarium bv. trifolii bacteria are an important resource for agricultural production [1, 2, 4]. In symbiosis with a suitable legume host (legume root nodules), atmospheric nitrogen fixed by these bacteria provides a source of plant nutrition that increases the farming system fertility in an economically and environmentally sustainable manner. Strains of R. leguminosarumbv trifolii vary in host-compatibility between legume species [5], and their nitrogen fixation efficacy when in symbiosis [6]. Understanding the genetic factors controlling these, and other phenotypes such as saprophytic survival, and desiccation tolerance, will enable increased utilization of R. leguminosarumbv trifolii for farming systems. The strain R. leguminosarumbv trifolii strain CC275e has been commercially used as an inoculant for white-clover for several decades [9]. The genome sequencing of this ‘highly efficacious’ bacterium, allows for the identification of genetic factors associated with desirable phenotypes (see previous). This will be achieved by comparison of the R. leguminosarumbv trifolii strain CC275e with closely related stains (e.g. based on 16S rRNA similarity) that differ in one or more phenotypes.
Acknowledgements
This work was funded through the New Zealand MBIE and DairyNZ funded programme “Improving forage legume-rhizobia performance” (C10X1308). Clément Delestre acknowledges the AgResearch bioinformatics team for internship funding. Sequencing was performed by MGS, and was coordinated by Lorraine Berry under NZGL contract NZGL00940 within the public-good funding stream. Sample QC, library QC, and library preparation was performed by Xiaoxiao Lin, sequencing by Richard Fong, and data QC by Mauro Truglio. Prof. Michael Hynes (University of Calgary) provided useful knowledge on R. leguminosarum mega-plasmids. TEM was conducted by Dr Duane Harland and James Vernon (AgResearch).
Abbreviations
- CSIRO
Commonwealth scientific and industrial research organisation
- Fix+
Nitrogen fixation positive
- NZGL
New Zealand genomics Ltd
- Nod+
Nodulation positive
- MGS
Massey genome service
- MBIE
Ministry of business, innovation and employment
- R. leguminosarum bv trifolii
Rhizobium leguminosarum symbiovar trifolii
- TEM
Transmission electron microscopy
- YM
Yeast mannitol
Additional file
List of strain names and associated NCBI GenBank accession numbers for bacterial isolates in Fig. 2. (XLSX 13 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SW, HR, CR, MO, BB, RB, and AG conceived of this study, participated in design, and helped draft the manuscript. CD and AL conducted genome assembly and associated bioinformatic analysis. SY, CB, and EG coordinated and conducted all microbiology, cell handling for TEM, DNA extraction and purification, and Eckhardt gel electrophoresis. All authors read and approved the final manuscript.
References
- 1.Caradus JR, Woodfield DR, Stewart AV. Overview and vision for white clover. Grassl Res Pract Ser. 1996;6:1–6. [Google Scholar]
- 2.Annicchiarico P, Barrett B, Brummer EC, Julier B, Marshall AH. Achievements and challenges in improving temperate perennial forage legumes. Crit Rev Plant Sci. 2015;34:327–80. doi: 10.1080/07352689.2014.898462. [DOI] [Google Scholar]
- 3.Statistics New Zealand . Agricultural areas in hectares, by usage and region, at 30 June 2012. 2012. [Google Scholar]
- 4.Ledgard SF, Sprosen MS, Penno JW, Rajendram GS. Nitrogen fixation by white clover in pastures grazed by dairy cows. Plant Soil. 2001;299:177–87. doi: 10.1023/A:1004833804002. [DOI] [Google Scholar]
- 5.Howieson JG, Yates RJ, O’Hara GW, Ryder M, Real D. The interactions of Rhizobium leguminosarum biovar trifolii in nodulation of annual and perennial Trifolium spp. from diverse centres of origin. Aust J Exp Agric. 2005;45:199–207. doi: 10.1071/EA03167. [DOI] [Google Scholar]
- 6.Rhys GJ, Bonish PM. Effectiveness of Rhizobium trifolii populations associated with Trifolium species in Taranaki. New Zeal J Exp Agr. 1981;9:327–35. [Google Scholar]
- 7.Bullard GK, Roughley RJ, Pulsford DJ. The legume inoculant industry and inoculant quality control in Australia: 1953–2003. Aust J Exp Agric. 2005;45:127–40. doi: 10.1071/EA03159. [DOI] [Google Scholar]
- 8.Cunningham GH. Certification of legume seed inoculants. N Z J Agric. 1957;94:578. [Google Scholar]
- 9.Lowther WL, Kerr GA. White clover seed inoculation and coating in New Zealand. Proc N Z Grassl Assoc. 2011;73:93–102. [Google Scholar]
- 10.Brockwell J, Gibson AH. Root nodule bacteria for some cultivated species of Trifolium. J Aust Inst Agric Sci. 1968;34:224–7. [Google Scholar]
- 11.McIntyre HJ, Davies H, Hore TA, Miller SH, Dufour JP, Ronson CW. Trehalose biosynthesis in Rhizobium leguminosarum bv. trifolii and its role in desiccation tolerance. Appl Environ Microbiol. 2007;73:3984–92. doi: 10.1128/AEM.00412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brockwell J, McIlroy R, Hebb DM. The Australian collection of Rhizobium strains for temperate legumes. Catalogue 1998. Canberra: CSIRO Publishing; 1998. [Google Scholar]
- 13.Vincent JM. The cultivation, isolation and maintenance of rhizobia. In: Vincent JM, editor. A manual for the practical study of root-nodule bacteria. Oxford: Blackwell Scientific; 1970. pp. 1–13. [Google Scholar]
- 14.Ramírez-Bahena MH, García-Fraile P, Peix A, Valverde A, Rivas R, Igual JM, et al. Revision of the taxonomic status of the species Rhizobium leguminosarum (Frank 1879) Frank 1889AL, Rhizobium phaseoli Dangeard 1926AL and Rhizobium trifolii Dangeard 1926AL. R. trifolii is a later synonym of R. leguminosarum. Reclassification of the strain R. leguminosarum DSM 30132 (=NCIMB 11478) as Rhizobium pisi sp. nov. Int J Syst Evol Microbiol. 2008;58:2484–90. doi: 10.1099/ijs.0.65621-0. [DOI] [PubMed] [Google Scholar]
- 15.Kuykendall LD, Young JM, Martínez-Romero E, Kerr A, Sawada H. Genus I. Rhizobium. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. ‘Bergey’s manual of systematic bacteriology. 2. New York: Springer; 2005. [Google Scholar]
- 16.Delestre C. JAST: Java assembling and scaffolding tool. 2014. [Google Scholar]
- 17.Dodt M, Roehr JT, Ahmed R, Dieterich C. Flexbar − flexible barcode and adapter processing for next-generation sequencing platforms. MDPI Biol. 2012;1:895–905. doi: 10.3390/biology1030895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tritt A, Eisen JA, Facciotti MT, Darling AE. An integrated pipeline for de novo assembly of microbial genomes. PLoS One. 2012;7:e42304. doi: 10.1371/journal.pone.0042304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zerbino DR. Using the Velvet de novo Assembler for Short-Read Sequencing Technologies. Curr Protoc Bioinformatics. 2010;31:11.5.1–11.5.12. doi: 10.1002/0471250953.bi1105s31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–9. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 22.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–7. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 23.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–6. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 24.Eddy SR. Accelerated Profile HMM Searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. The Pfam protein families database: Nucleic Acids Research. Database Issue. 2014;42:D222–30. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bland C, Ramsey TL, Sabree F, Lowe M, Brown K, Kyrpides NC, et al. CRISPR Recognition Tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics. 2007;18:209. doi: 10.1186/1471-2105-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reeve W, Tian R, De Meyer S, Melino V, Terpolilli J, Ardley J, et al. Genome sequence of the clover-nodulating Rhizobium leguminosarum bv. trifolii strain TA1. Stand Genomic Sci. 2013;9:243–53. doi: 10.4056/sigs.4488254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hynes MF, McGregor NF. Two plasmids other than the nodulation plasmid are necessary for formation of nitrogen-fixing nodules by Rhizobium leguminosarum. Mol Microbiol. 1990;4:567–74. doi: 10.1111/j.1365-2958.1990.tb00625.x. [DOI] [PubMed] [Google Scholar]
- 30.Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–9. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tavaré S. Some Probabilistic and Statistical Problems in the Analysis of DNA Sequences. Lect Math Life Sci. 1986;17:57–86. [Google Scholar]
- 33.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–91. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 34.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, et al. Towards a richer description of our complete collection of genomes and metagenomes “Minimum Information about a Genome Sequence ” (MIGS) specification. Nat Biotechnol. 2008;26:541–7. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87:4576–9. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garrity GM, Bell JA, Lilburn T. Phylum XIV. Proteobacteria phyl. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. ‘Bergey’s manual of systematic bacteriology. 2. New York): Springer; 2005. [Google Scholar]
- 37.Garrity GM, Bell JA, Lilburn T. Class I. Alphaproteobacteria class. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. ‘Bergey’s manual of systematic bacteriology. 2. New York): Springer; 2005. [Google Scholar]
- 38.Kuykendall LD. Order VI. Rhizobiales ord. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. ‘Bergey’s manual of systematic bacteriology. 2. New York): Springer; 2005. [Google Scholar]
- 39.Biological Agents: Technical rules for biological agents. TRBA: 466


