Abstract
Gemmata massiliana is a new Planctomycetes bacterium isolated from a hospital water network in France, using a new culture medium. It is an aerobic microorganism with optimal growth at pH 8, at 30 °C and salinity ≤ 1.25 % NaCl. G. massiliana is resistant to β-lactam antibiotics, due to lack of peptidoglycan in its cell wall.G. massiliana shares a 97 % 16S rRNA gene sequence similarity with the nearest species, Gemmata obscuriglobus; and 99 % similarity with unnamed soil isolates. Its 9,249,437-bp genome consists in one chromosome and no detectable plasmid and has a 64.07 % G + C content, 32.94 % of genes encoding for hypothetical proteins. The genome contains an incomplete 19.6-kb phage sequence, 26 CRISPRs, 3 CAS and 15 clusters of secondary metabolites. G. massiliana genome increases knowledge of a poorly known world of bacteria.
Electronic supplementary material
The online version of this article (doi:10.1186/s40793-015-0103-0) contains supplementary material, which is available to authorized users.
Keywords: Gemmata massiliana, Gemmata obscuriglobus, Gemmata, Planctomycetes, Hospital water network, Culture, Genome
Introduction
Gemmata obscuriglobus, the sole cultured representative of the genus PlanctomycetesGemmata, was first isolated in a freshwater dam in Queensland, Australia [1]. Gemmata isolates were thereafter cultivated from the leakage water of a compost heap [2], an Australian soil specimen and freshwater [3] and sphagnum peat bogs [4]. Moreover, 16S rRNA gene sequences of Gemmata were detected in various environments, including a municipal wastewater treatment plant [5], rivers in Germany [6], soil [7], sphagnum peat bogs [4], clean rooms where spacecraft are assembled [8], a water specimen of the Western Pacific Ocean and sediments [9], a South African water spring [10], the gastrointestinal tract of carp [11], nonsulfur, sulfur and iron geothermal steam vents [12] and recently from human stool specimens [13].
G. obscuriglobus exhibits intriguing features, including a condensed chromatin that is surrounded by a double membrane, a rare feature among bacteria, which evokes a nucleus-like compartmentalization [14]. It has been recently debated that membrane invaginations may be the actual cause of this intracellular membranous organization [15]. Moreover, G. obscuriglobus is remarkable for its capacity to survive high doses of ionizing radiation and ultraviolet light at energy values generally depicting an ability to maintain genomic integrity [16]. It also possesses membrane coat-like proteins that are implicated in endocytosis-like processes, a feature long thought to be exclusive to eukaryotes [17]. G. obscuriglobus exhibits a large number of extracytoplasmic function sigma factors illustrating its skilled adaptation to stress and reactivity to environmental stimulus [18]. G. obscuriglobus shared many eukaryotic homologous genes including a homolog of integrin alpha-V which is implicated in signal transduction and cytoskeleton organization [19].
Herein, we describe a new isolate as a representative of a new species Gemmata massiliana, with the aim of enlarging the scope of our knowledge regarding this fascinating bacterial genus. This isolate, which has 99 % 16S rRNA gene similarity with Australian soil and freshwater strains which have been isolated but neither described nor sequenced [3], was this time isolated from a hospital water distribution system. Evidently, the choice of the culture medium had a primary effect on the growth of this bacterium [20]. It was actually elaborated using some of the filtered sample water itself as a medium basis that could simulate the natural environment and provide the microorganism with the necessary chemical components for its growth. Furthermore, antibiotics were added to the culture medium for a selective isolation of Planctomycetes which are broadly resistant to antibiotics [21]. Phenotypic and genomic features of G. massiliana sp. nov. strain CSUR P189T are presented hereafter.
Organism information
Classification and features
From September 2011 to August 2012, 15 points located along the water network in two hospitals in Marseille, France were sampled on a weekly basis. Water samples were collected into sterile, 500-mL containers (Dominique Dutscher, Brumath, France) containing sodium thiosulfate used to neutralize free chlorine. The water specimens were inoculated on the same day of the sampling into the Marine-like (ML) and Isosphaera-like (IL) enrichment broths incubated at 22 °C and 30 °C, in the presence of negative controls (enrichment broth without water sample) as previously described [22]. The enrichment broth consisted of the specimen water itself, passed through a 0.2-μm membrane filter (Thermo Fisher Scientific, Saint Herblain, France) complemented with a 10 % vol/vol antibiotic solution containing 40 mg/L vancomycin, 100 mg/L imipenem, 1 mg/L penicillin G and 32 mg/L amphotericin B; in addition to an enrichment solution (5 g of peptone and 1 g of yeast extract per 100 mL) for ML broth and a vitamin solution for the IL broth made of 60 μg β-aminobenzoic acid, 6 μg biotin, 3 μg vitamin B12, 600 μg nicotinamide, 300 μg thiamin, 150 mg glucose and 150 mg peptone from casein per liter of specimen filtered water (Sigma-Aldrich, Saint-Quentin Fallavier, France). A 2 mL-volume of the water sample was centrifuged at 17,000 × g using the Heraeus Pico 17 centrifuge (Thermo Fisher Scientific) for 5 min and the pellet was inoculated into 5 mL of the enrichment broth. Presence of any turbidity was monitored daily for four months. Once the turbidity was detected, 10 μL of inoculated broth were spread on solid medium that had the same composition as broth, complemented with 1.5 % agar (Sigma-Aldrich). All colonies were identified by matrix-assisted laser-desorption/ionization time-of-flight mass spectrometer (MALDI-TOF-MS) (Bruker Daltonics, Bremen, Germany) as previously described [23]. Further identification was based on 16S rRNA gene PCR amplification and sequencing [13]. Observations by electron microscopy were done as previously described [24]. Briefly, the bacteria were suspended and then washed in phosphate buffer and stained with 1 % (w/v) phosphotungstic acid. Afterwards examination was carried on using Morgagni 268D (Philips) electron microscope at an operating voltage of 60 kV. Also, a 106 bacterial suspension of G. massiliana was examined for cell size variation using BD LSRFortessa cell analyzer (Becton Dickinson, Le Pont de Claix, France) and FACSDiva software (version 6.2) as previously described [25]. Further characterization of the isolate comprised the observation of growth under anaerobic, aerobic, microaerophilic and presence of 5 % CO2 atmosphere; inoculation of Api 20E, 20NE, ZYM, and 50CH strips, (bioMérieux, La Balme les grottes, France) E-test (bioMérieux), pH, salinity and temperature tolerance. G. massiliana strain CSUR P189T sequenced in this study (Table 1) was isolated in December 2011 after 2-month incubation at 30 °C in Isosphaera-like agar preceded by 4 weeks incubation in Isosphaera-like broth. MALDI-TOF-MS yielded insignificant scores below 0.3. This isolate exhibited 97 % 16SrRNA gene nucleotide sequence (GenBank accession number JX088244) similarity with G. obscuriglobus (GenBank accession number X81957), a value lower than the threshold that was defined by Stackebrandt and Ebers to depict a new species [26]. And as stated above, G. massiliana also displayed 16S rRNA gene nucleotide sequence similarity of 99 % with unnamed isolates [3]. Those bacteria are most likely various G. massiliana strains (Fig. 1).
Table 1.
Classification and general features of Gemmata massiliana strain IIL30T
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Classification | Domain: Bacteria | TAS [41] | |
| Phylum: Planctomycetes | TAS [42] | ||
| Class: Planctomycetia | TAS [43] | ||
| Order: Planctomycetales | TAS [43] | ||
| Family: Planctomycetaceae | TAS [44] | ||
| Genus: Gemmata | TAS [1] | ||
| Species: Gemmata massiliana | IDA | ||
| Type strain: IIL30 CSUR P189T | IDA | ||
| Gram stain | Negative | IDA | |
| Cell shape | Coccus | IDA | |
| Motility | Motile | IDA | |
| Sporulation | Nonsporulating | IDA | |
| Temperature range | Mesophile | IDA | |
| Optimum temperature | 30°C | IDA | |
| pH range; Optimum | 6–8; 8 | IDA | |
| Carbon source | Unknown | ||
| GS-6 | Habitat | Hospital water | IDA |
| MIGS-6.3 | Salinity | 0 % NaCl (w/v) | IDA |
| MIGS-22 | Oxygen requirement | Aerobic | IDA |
| MIGS-15 | Biotic relationship | Free-living | IDA |
| MIGS-14 | Pathogenicity | Unknown | |
| MIGS-4 | Geographic location | France/Marseille | IDA |
| MIGS-5 | Sample collection | October 2011 | IDA |
| MIGS-4.1 | Latitude | 43.3 | IDA |
| MIGS-4.2 | Longitude | 5.4 | |
| MIGS-4.4 | Altitude | Unknown |
a Evidence codes - IDA Inferred from Direct Assay, TAS Traceable Author Statement (i.e., a direct report exists in the literature), NAS Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [45]
Fig. 1.

Gemmata massiliana phylogenetic position amongst other Planctomycetes species. The following Phylogenetic tree shows G. massiliana strain CSUR P189T position relatively to Gemmata obscuriglobus, to undescribed Gemmata species and to other Planctomycetes. Sequences were aligned using CLUSTALW [39], and phylogenetic inferences obtained using the neighbor-joining method within the MEGA software [40] only bootstrap values ≥95 % are indicated at nodes. T. maritima (M21774) was used as an outgroup. The scale bar represents a 5 % nucleotide sequence divergence
In all culture-based observations, the negative controls remained sterile. G. massiliana grew at 25 °C, 30 °C and 37 °C; no growth was observed at 4 °C or at 45 °C, and growth was optimal at 30 °C. The diameter of the colonies varied between 0.1 mm and 1 mm on Isosphaera-like agar. Growth was observed in all tested atmospheres for the exception of the anaerobic atmosphere, optimal growth occurred in aerobic conditions. Tolerable salinity varied between 0 and 1.25 % with an optimal growth in the absence of salt; pH tolerance varied from pH 6 to pH 8, with an optimal growth at pH 8. Cells grown on agar are motile and Gram-negative (Fig. 2). Negative staining showed two populations of cells, including a small-cell-variant with a 1.1-μm diameter and a large-cell variant with a 2.1-μm diameter (Fig. 3). This feature is not an electron microscopy artifact since FACS scan further disclosed two populations within the G. massiliana cultured on Caulobacter agar for 7 days (Fig. 4). The isolate tested negative for catalase and oxydase and positive for esculinase, alkaline phosphatase, naphtol-AS-BI-phosphohydrolase, valine, trypsin, acid phosphatase and leucine arylamidase. It was resistant to β-lactam antibiotics at concentrations of 32 mg/L for penicillin G and imipenem, 256 mg/L for vancomycin and 66 mg/L for amphotericin B. G. massiliana exhibited intermediate susceptibility to chloramphenicol (MIC, 6 mg/L), colistin (MIC, 6 mg/L) and is susceptible to tetracycline (MIC, 0.016 mg/L), ciprofloxacin (MIC, 0.016 mg/L) cotrimoxazole (MIC, 0.016 mg/L), gentamicin (MIC, 0.016 mg/L), minocycline (MIC, 0.016 mg/L), erythromycin (MIC, 0.016 mg/L) and rifampicine (MIC, 0.016 mg/L). Matrix-assisted laser-desorption/ionization time-of-flight mass spectrometer (MALDI-TOF-MS) yielded a unique peptidic profile (Fig. 5). The isolate has been deposited in two public collections as G. massiliana strain IIL30T, in the Collection de Souches de l’Unité des Rickettsies (Marseille, France; CSUR P189T) and Deutsche Sammlung von Mikrorganismen and Zellkuturen (Braunshwing, Germany; DSM 26013T).
Fig. 2.
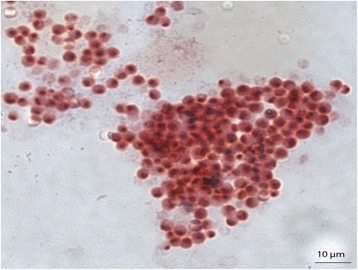
Gram staining of G. massiliana strain CSUR P189T. The bar scale reprensents 10 μm
Fig. 3.
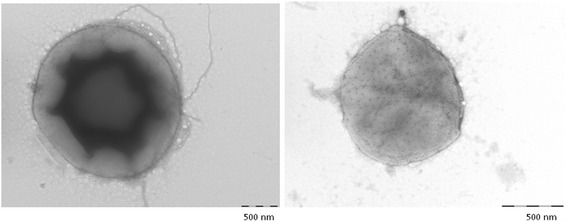
G. massiliana transmission electron microscopy. We observed two size-different populations of G. massiliana strain CSUR P189T, using a Morgagni 268D (Philips) at an operating voltage of 60kV. The scale bar represents 500 nm
Fig. 4.

G. massiliana two cell variants as demonstrated by flow cytometry. Representative histogram of the two cell variants in a G. massiliana sample shown in purple, was generated by a BD LSRFortessa (Le Pont de Claix, France) and FACSDiva software (version 6.2)
Fig. 5.
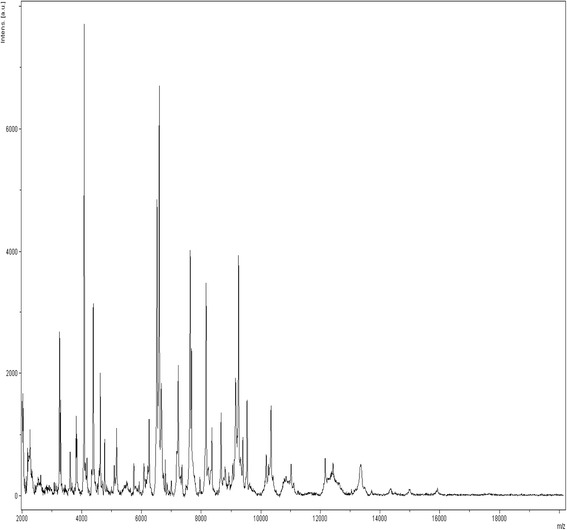
Reference mass spectrum of G. Massiliana strain CSUR P189T. This reference spectrum was generated from 10 spectra corresponding to 10 deposited colonies of G. massiliana
Genome sequencing information
Genome project history
The organism was selected for sequencing on the basis of its phylogenetic position and 16S rRNA gene similarity to G. obscuriglobus, the sole named species in this genus, and its isolation was done in the context of a study on the detection of Planctomycetes bacteria in the hospital water network. The bioproject Genbank accession number is PRJEB621 and consists of 417 contigs and 22 scaffolds. Table 2 shows the project information and its association with MIGS version 2.0 compliance [27].
Table 2.
Project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Draft |
| MIGS-28 | Libraries used | 454 paired end 3-kb and shotgun XL+ libraries |
| MIGS-29 | Sequencing platforms | 454 Roche Titanium |
| MIGS-31.2 | Fold coverage | 22.4× |
| MIGS-30 | Assemblers | Newbler version 2.8 |
| MIGS-32 | Gene calling method | CLC genomics workbench 6.0.1 |
| Genbank ID | CBXA000000000.1 | |
| Genbank Date of Release | January 20,2014 | |
| GOLD ID | Gp0033443 | |
| BIOPROJECT | PRJEB621 | |
| MIGS-13 | Source Material Identifier | DSM 26013 |
| Project relevance | Medical, hospital water network |
Growth conditions and genomic DNA preparation
G. massiliana strain IIL30 was grown aerobically on Caulobacter agar (peptone enzymatic hydrolysate type I: from meat; 2 g/L, yeast extract; 1 g/L, MgSO4; 0.2 g/L, agar; 15 g/L) at 30 °C. A 300 μL-bacterial suspension was diluted in 1 mL TE buffer for lysis treatment: a lysozyme incubation of 30 min at 37 °C followed by an overnight Proteinase K incubation at 37 °C. The DNA was purified by three phenol-chloroform extractions and ethanolic precipitation at -20 °C overnight. After centrifugation, the DNA was resuspended in 144 μL TE buffer. The concentration was measured by the Quant-it Picogreen kit (Invitrogen, Saint Aubin, France) on the Genios_Tecan fluorometer at 67.4 ng/μl.
Genome sequencing and assembly
A shotgun XL+ and 3-kb paired-end libraries were pyrosequenced on the 454_Roche_Titanium. This project was loaded twice on a 1/4 region for the paired end application on PTP Picotiterplate and once ¼ region for the shotgun XL+ strategy. The XL+ shotgun library was constructed with 1μg of DNA as described by the manufacturer (Roche, Meylan, France) with the GS Rapid library XL+ Prep kit. The fragmentation was performed on a Covaris device (KBioScience-LGC Genomics, Queens Road, Teddington, Middlesex, TW11 0LY, UK) through microTUBE and the size was read at 1.58 Kb on the Agilent 2100 BioAnalyzer on a DNA labchip High Sensitivity, as expected in the range of 1.5 Kb. The concentration of the shotgun library was measured on the fluorometer TBS and determined at 2.02E + 09 molecules/μL. The paired-end library was constructed from 5 μg of DNA. It was mechanically fragmented on Covaris device through miniTUBE-Red 3 Kb. The DNA fragmentation was visualized using the Agilent 2100 BioAnalyzer on a DNA labchip 7500 with an optimal size of 3 kb. The library was constructed according to the 454_Titanium paired end protocol and manufacturer (Roche). Circularization and nebulization were performed and generated a pattern with an optimal at 440 bp, respectively. After PCR amplification through 17 cycles followed by double size selection, the single stranded paired end library was quantified on the RNA pico 6000 labchip on the BioAnalyzer at 386 pg/μL. The library concentration equivalence was calculated at 1.61E + 09 molecules/μL. The library was stocked at -20 °C until used.
The XL+ shotgun library was clonal amplified with 0.5, 1 and 2 cpb in 2 emPCR reactions per conditions and the 3 kb paired end library was amplified with 0.5 and 2 cpb in 2 emPCR reactions per conditions and 4 reactions in 1cpb with the GS Titanium SV emPCR Kit (Lib-L) v2 (Roche). The yields of the emPCR were 5, 6.9 and 10 % respectively for the shotgun XL+, and 5.62, 10.27 and 14.92 % for the clonal amplification of the 3kb paired end libraries according to the quality expected by the range of 5 to 20 % from the Roche procedure. 790,000 beads of each library were loaded on a ¼ region from the GS Titanium PicoTiterPlate PTP Kit with the GS Titanium Sequencing Kit XLR70.
The runs were performed overnight and then analyzed on the cluster through the gsRunBrowser and gsAssembler_Roche. The global 566 858 passed filter sequences generated 201.6 Mb with a length average of 409 bp. These sequences were assembled on the gsAssembler from Roche with 90 % identity and 40 bp as overlap. It lead to 22 scaffolds, 417 large contigs (>1,500 bp) and 475 all contigs (>0.5 kb) and generated a genome size of 9.249 Mb which corresponds to a coverage of 22.4x equivalent genome.
Genome annotation
Open Reading Frames were predicted using CLC Genomics Workbench software package 6.0.1 (CLC, Denmark). From the 417 contigs, any ORF spanning a sequencing gap region was eliminated. As for the Clusters of Orthologous Groups, rpsblast was done by blasting all predicted proteins against the National Center for Biotechnology Information (NCBI) COG database with an e-value of 10-3. The search for tRNA genes, ribosomal RNAs, proteins and genes predictions was completed by XEGEN. Phage detection was realized using the PHAST software [28], anti-smash 2 [29] for secondary metabolite detection, Resfinder tool [30] for antibiotic resistance genes, CRISPR Finder for clustered regularly interspaced short palindromic repeats [31] and GGDC web server [32] for in silico determining of DNA-DNA hybridization (DDH) values. Cell-division and cytoskeleton-related proteins were searched by running a blastp against a database described in [33], complemented by the FtsZl1 sequences [34]. We also targeted the peptidoglycan synthesis genes as described precedently [35]. A G. massiliana suspension, with an optical density of 1,1 at 260 nm when diluted 20 times, was required to conduct the pulsed-field gel electrophoresis and the southern blotting as previously described [36]. Migration parameters where fixed as following; initial time: 5 s, final time: 20 s, run time: 20 h, voltage: 5 V/cm, angle: 120°.
Genome properties
The genome is 9,249,437-bp long with 64.07 % GC content (Fig. 6). It is composed of 417 contigs (22 scaffolds). Of the 8,065 predicted genes, 7,985 were protein-coding genes, and 80 were RNAs (2 genes are 5S rRNA, 1 gene is 16S rRNA, 1 gene is 23S rRNA, 76 genes are tRNA genes). A total of 3,890 genes (48.72 %) were assigned a putative function. 1,097 genes were identified as ORFans (13.74 %). The remaining genes were annotated as hypothetical proteins (2,630 genes = > 32.94 %). The distribution of genes into COGs functional categories is presented in Table 3. The properties and the statistics of the genome are summarized in Tables 3 and 4.
Fig. 6.

Graphical circular map of the chromosome. From outside to the center: Genes on the forward strand colored by COG categories (only genes assigned to COG), genes on the reverse strand colored by COG categories (only gene assigned to COG), RNA genes (tRNAs green, rRNAs red), GC content and GC skew
Table 3.
Genome statistics
| Attribute | Value | % of total |
|---|---|---|
| Genome size (bp) | 9,249,437 | 100 |
| DNA coding (bp) | 7,995,313 | 86.44 |
| DNA G + C (bp) | 5,925,688 | 64.07 |
| DNA scaffolds | 22 | 100 |
| Total genes | 8,065 | 100 |
| Protein-coding genes | 7,985 | 99.01 |
| RNA genes | 80 | 0.99 |
| Pseudo genes | NA | - |
| Genes in internal clusters | 3497 | 43.79 |
| Genes with function prediction | 3,237 | 40.53 |
| Genes assigned to COGs | 2,443 | 30.59 |
| Genes with Pfam domains | 6,088 | 76.24 |
| Genes with signal peptides | 1,979 | 24.78 |
| Genes with transmembrane helices | 1,515 | 18.97 |
| CRISPR repeats | 26 | 100 |
The total is based on either the size of the genome in base pairs or the total number of protein coding genes in the annotated genome
Table 4.
Number of genes associated with general COG functional categories
| Code | Value | %age | Description |
|---|---|---|---|
| J | 168 | 2.10 | Translation, ribosomal structure and biogenesis |
| A | 1 | 0.01 | RNA processing and modification |
| K | 360 | 4.51 | Transcription |
| L | 404 | 5.06 | Replication, recombination and repair |
| B | 2 | 0.3 | Chromatin structure and dynamics |
| D | 28 | 0.35 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0 | Nuclear structure |
| V | 98 | 1.23 | Defense mechanisms |
| T | 419 | 5.25 | Signal transduction mechanisms |
| M | 260 | 3.26 | Cell wall/membrane biogenesis |
| N | 94 | 1.18 | Cell motility |
| Z | 0 | 0 | Cytoskeleton |
| W | 1 | 0.01 | Extracellular structures |
| U | 99 | 1.24 | Intracellular trafficking and secretion |
| O | 170 | 2.13 | Posttranslational modification, protein turnover, chaperones |
| C | 240 | 3.01 | Energy production and conversion |
| G | 240 | 3.01 | Carbohydrate transport and metabolism |
| E | 277 | 3.47 | Amino acid transport and metabolism |
| F | 63 | 0.79 | Nucleotide transport and metabolism |
| H | 130 | 1.63 | Coenzyme transport and metabolism |
| I | 125 | 1.57 | Lipid transport and metabolism |
| P | 188 | 2.35 | Inorganic ion transport and metabolism |
| Q | 143 | 1.79 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 766 | 9.59 | General function prediction only |
| S | 368 | 4.61 | Function unknown |
| - | 4136 | 51.80 | Not in COGs |
The total is based on the total number of protein coding genes in the annotated geno
Insights from the genome sequence
An incomplete 19.6-Kb phage sequence was detected, which lacks the attachment sites. A total of 21 questionable CRISPRs and 5 confirmed ones were found in the genome.At least 3 CAS proteins (CRISPR associated proteins) were also detected. No antibiotic resistance genes could be spotted using the resfinder tool, but the 15 clusters of secondary metabolites consisting of 5 terpene clusters, 5 bacteriocin clusters, 3 type four polyketide synthase(T4PKS) clusters, 1 T4PKS-T1PKS cluster, and 2 T3PKS clusters, put G. massiliana in the frontline of the Planctomycetes phylum, followed by Schlesneria paludicola with 13 clusters. As for the search of cell division- and cytoskeleton-related planctomycetal proteins, 10 were identified: FtsK, Noc, divK, divJ, FtsZl1, MraW, ClpX, CLpP, EnvA and FtsE. A plasmid replication protein with 44 % similarity to Haliscomenobacter hydrossis plasmid encoded RepA protein was predicted in the genome. Pulsed-field gel electrophoresis yielded slightly distinct bands so we decided to run a southern blot using genomic DNA and a DiG labeled DNA probe to try to confirm the presence of detectable plasmid, which was not the case. DDH values for 10 Planctomycetes genomes are presented in Additional file 1. DDH value between G. massiliana and G. obscuriglobus was 22.0 %.
While mining the genome for peptidoglycan synthesizing genes, only GT28 and GH73 genes were found. This is below the three-gene set previously shown to be associated with peptidoglycan synthesis [35], a minimal set of 3 genes is required for peptidoglycan metabolism. This observation agrees with the data available on Planctomycetes that lack peptidoglycan in their cell wall [37].
G. massiliana had a slightly larger genome than G. obscuriglobus (9.249 Mb vs 9.16 Mb), a lower G + C content (64 % vs 67.2 %), it codes for a higher number of genes (8,065 vs 7,645), had six cell-shape and division proteins in common with G. obscuriglobus and the other Planctomycetes previously studied [30] and four detected for the first time in this phylum. G. massiliana encodes 15 secondary metabolite gene clusters versus 12 in G. obscuriglobus and 26 CRISPRs versus 24 in G. obscuriglobus. It, also, showed a different antibiotic resistance profile [21].
Conclusions
These results show that G. massiliana is a member of the genus Gemmata, exhibiting few features in common with the other characterized member of this genus, G. obscuriglobus. This unequivocally proves that G. massiliana is a new species of the genus Gemmata. Interestingly, G. massiliana exhibited two variants characterized by electron microscopy and FACS scan analysis a feature which has not been previously reported in other Planctomycetes. Nevertheless, we did observe this feature in another water-borne, not related β-Proteobacteria, Minibacterium massiliensis [38]. DDH in silico analysis revealed that differentisolates of a same Planctomycetes species exhibited a 66.7 % value, whereas isolates belonging to the same genus exhibited DDH values variation from 21.5 % DDH to 24.6 % DDH. Additionally, isolates affiliated to different genera exhibited DDH values variation from 17.5 % to 24 %, with the exception of Rhodopirellula baltica. The value of in silico hybridization of G. massiliana with G. obscuriglobus was much lower than the 70 % DDH limit for delineating same species. These data confirm that G. massiliana should be considered as a new Gemmata species.
Description of Gemmata massiliana sp. nov.
Gemmata massiliana (ma.si.lia.na L. fem. adj. of Massilia, taken from the old Greek and Roman name for Marseille where Gemmata massiliana was first isolated.) Pink colonies grown on IL agar varied in diameter between 0.1 mm and 1 mm. Optimal growth was observed at 30°C. Cells are gram negative cocci, motile, aerobic, with two cell populations of 1.1-μm and 2.1-μm diameter. Optimal growth occurs in absence of salt. The isolate tested negative for catalase and oxydase and positive for esculinase, alkaline phosphatase, naphtol-AS-BI-phosphohydrolase, valine, trypsin, acid phosphatase and leucine arylamidase. It was resistant to penicillin G, imipenem and amphotericin B. G. massiliana exhibited intermediate susceptibility to chloramphenicol, colistin, and is susceptible to tetracycline, ciprofloxacin, cotrimoxazole, gentamicin, minocycline, erythromycin and rifampicine.
The genome size is 9.249 Mb with a 64.07 % G + C content and 8065 predicted genes. Genome analysis identified an incomplete phage sequence, 26 CRISPRs and 3 CAS proteins, 15 clusters of secondary metabolites and 10 cell division- and cytoskeleton-related planctomycetal proteins: FtsK, Noc, divK, divJ, FtsZl1, MraW, ClpX, CLpP, EnvA and FtsE. Nonetheless no antibiotic resistance genes have been detected. The 16S rRNA gene and genome sequences have been deposited in GenBank under accession numbers JX088244 and CBXA010000001-CBXA010000171, respectively. The type strain (CSUR P189T, DSMZ 26013T) was isolated from a hospital water network.
Acknowledgements
The authors thank the Xegen Company for automating the genomic annotation process. This study was funded by the Méditerranée-Infection Foundation.
Abbreviations
- ML
Marine-like
- IL
Isosphaera-like
- DDH
DNA-DNA hybridization
Additional files
DDH values for 10 Planctomycetes genomes. Values are expressed in percentage. (XLS 26 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
RA isolated and performed the phenotypic characterization of the new species, did some of the genomic analyses and drafted the manuscript. CC helped with the phenotypic characterization and genomic analyses. MG performed some of the genomic analysis. CR performed the genomic sequencing and helped to draft the manuscript. SA did the pulsed-field gel electrophoresis and the southern blotting and helped to draft the manuscript. DR contributed the necessary reagents and materials for this study. MD conceived and developed the study and helped to draft the manuscript. All authors read and approved the final manuscript.
References
- 1.Franzmann PD, Skerman VBD. Gemmata obscuriglobus, a new genus and species of the budding bacteria. Antonie Van Leeuwenhoek. 1984;50:261–268. doi: 10.1007/BF02342136. [DOI] [PubMed] [Google Scholar]
- 2.Ward N, Rainey FA, Stackebrandt E, Schlesner H. Unraveling the extent of diversity within the order Planctomycetales. Appl Environ Microbiol. 1995;61:2270–2275. doi: 10.1128/aem.61.6.2270-2275.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang J, Jenkins C, Webb RI, Fuerst JA. Isolation of Gemmata-like and Isosphaera-like planctomycete bacteria from soil and freshwater. Appl Environ Microbiol. 2002;68:417–422. doi: 10.1128/AEM.68.1.417-422.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kulichevskaya IS, Pankratov TA, Dedysh SN. Detection of representatives of the Planctomycetes in Sphagnum peat bogs by molecular and cultivation approaches. Mikrobiologiia. 2006;75:329–335. [PubMed] [Google Scholar]
- 5.Chouari R, Le Paslier D, Daegelen P, Ginestet P, Weissenbach J, Sghir A. Molecular evidence for novel planctomycete diversity in a municipal wastewater treatment plant. Appl Environ Microbiol. 2003;69:7354–7363. doi: 10.1128/AEM.69.12.7354-7363.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brümmer IHM, Felske ADM, Wagner-Döbler I. Diversity and seasonal changes of uncultured Planctomycetales in river biofilms. Appl Environ Microbiol. 2004;70:5094–5101. doi: 10.1128/AEM.70.9.5094-5101.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buckley DH, Huangyutitham V, Nelson TA, Rumberger A, Thies JE. Diversity of Planctomycetes in soil in relation to soil history and environmental heterogeneity. Appl Environ Microbiol. 2006;72:4522–4531. doi: 10.1128/AEM.00149-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moissl C, Osman S, La Duc MT, Dekas A, Brodie E, DeSantis T, et al. Molecular bacterial community analysis of clean rooms where spacecraft are assembled. FEMS Microbiol Ecol. 2007;61:509–521. doi: 10.1111/j.1574-6941.2007.00360.x. [DOI] [PubMed] [Google Scholar]
- 9.Shu Q, Jiao N. Different Planctomycetes diversity patterns in latitudinal surface seawater of the open sea and in sediment. J Microbiol. 2008;46:154–159. doi: 10.1007/s12275-008-0002-9. [DOI] [PubMed] [Google Scholar]
- 10.Tekere M, Lotter A, Olivier J, Jonker N, Venter SN. Metagenomic analysis of bacterial diversity of Siloam hot water spring, Limpopo. South Africa J Biotechnol. 2011;10:18005–18012. [Google Scholar]
- 11.Van Kessel MA, Dutilh BE, Neveling K, Kwint MP, Veltman JA, Flik G, et al. Pyrosequencing of 16S rRNA gene amplicons to study the microbiota in the gastrointestinal tract of carp (Cyprinus carpio L.) AMB Express. 2011;1:1–9. doi: 10.1186/2191-0855-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benson CA, Bizzoco RW, Lipson DA, Kelley ST. Microbial diversity in nonsulfur, sulfur and iron geothermal steam vents. FEMS Microbiol Ecol. 2011;76:74–88. doi: 10.1111/j.1574-6941.2011.01047.x. [DOI] [PubMed] [Google Scholar]
- 13.Cayrou C, Sambe B, Armougom F, Raoult D, Drancourt M. Molecular diversity of the Planctomycetes in the human gut microbiota in France and Senegal. APMIS. 2013;121:1082–1090. doi: 10.1111/apm.12087. [DOI] [PubMed] [Google Scholar]
- 14.Fuerst J, Sagulenko E. Beyond the bacterium: planctomycetes challenge our concepts of microbial structure and function. Nat Rev Microbiol. 2011;9:403–13. doi: 10.1038/nrmicro2578. [DOI] [PubMed] [Google Scholar]
- 15.Santarella-Mellwig R, Pruggnaller S, Roos N, Mattaj IW, Devos DP. Three-Dimensional Reconstruction of Bacteria with a complex endomembrane system. PLoS Biol. 2013;11 doi: 10.1371/journal.pbio.1001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lieber A, Leis A, Kushmaro A, Minsky A, Medalia O. Chromatin Organization and Radio Resistance in the Bacterium Gemmata obscuriglobus. J Bacteriol. 2009;191:1439–1445. doi: 10.1128/JB.01513-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lonhienne TG, Sagulenko E, Webb RI, Lee KC, Franke J, Devos DP, et al. Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus. Proc Natl Acad Sci. 2010;107:12883–12888. doi: 10.1073/pnas.1001085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeske O, Jogler M, Petersen J, Sikorski J, Jogler C. From genome mining to phenotypic microarrays: Planctomycetes as source for novel bioactive molecules. Antonie Van Leeuwenhoek. 2013;1–17. [DOI] [PubMed]
- 19.Jenkins C, Kedar V, Fuerst JA. Gene discovery within the planctomycete division of the domain Bacteria using sequence tags from genomic DNA libraries. Genome Biol. 2002;3:1–11. doi: 10.1186/gb-2002-3-6-research0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lage OM, Bondoso J. Bringing Planctomycetes into pure culture. Front Microbiol. 2012;3:405. doi: 10.3389/fmicb.2012.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cayrou C, Raoult D, Drancourt M. Broad-spectrum antibiotic resistance of Planctomycetes organisms determined by Etest. J Antimicrob Chemother. 2010;65:2119–2122. doi: 10.1093/jac/dkq290. [DOI] [PubMed] [Google Scholar]
- 22.Cayrou C, Terra A, Drancourt M. Genotyping of Rhodopirellula baltica organisms using multispacer sequence typing. Mar Ecol. 2013;34:251–254. doi: 10.1111/maec.12015. [DOI] [Google Scholar]
- 23.Cayrou C, Raoult D, Drancourt M. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for the identification of environmental organisms: the Planctomycetes paradigm. Environ Microbiol Rep. 2010;2:752–760. doi: 10.1111/j.1758-2229.2010.00176.x. [DOI] [PubMed] [Google Scholar]
- 24.Roux V, Million M, Robert C, Magne A, Raoult D. Non-contiguous finished genome sequence and description of Oceanobacillus massiliensis sp. nov. Stand Genomic Sci. 2013;9:370. doi: 10.4056/sigs.4267953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hugon P, Lagier JC, Robert C, Lepolard C, Papazian L, Musso D, et al. Molecular studies neglect apparently gram-negative populations in the human gut microbiota. J Clin Microbiol. 2013;51:3286–3293. doi: 10.1128/JCM.00473-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stackebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–155. [Google Scholar]
- 27.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26:541–7. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic Acids Res. 2011;39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, et al. AntiSMASH 2.0 a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41:W204–W212. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35:W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Auch AF, Klenk HP, Göker M. Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand Genomic Sci. 2010;2:142–8. doi: 10.4056/sigs.541628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jogler C, Waldmann J, Huang X, Jogler M, Glöckner FO, Mascher T, et al. Identification of proteins likely to be involved in morphogenesis, cell division, and signal transduction in planctomycetes by comparative genomics. J Bacteriol. 2012;194:6419–6430. doi: 10.1128/JB.01325-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Makarova KS, Koonin EV. Two new families of the FtsZ-tubulin protein superfamily implicated in membrane remodeling in diverse bacteria and archaea. Biol Direct. 2010;5:33. doi: 10.1186/1745-6150-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cayrou C, Henrissat B, Gouret P, Pontarotti P, Drancourt M. Peptidoglycan: a post-genomic analysis. BMC Microbiol. 2012;12:294. doi: 10.1186/1471-2180-12-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, et al. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- 37.König E, Schlesner H, Hirsch P. Cell wall studies on budding bacteria of the Planctomyces/Pasteuria group and on a Prosthecomicrobium sp. Arch Microbiol. 1984;138:200–205. doi: 10.1007/BF00402120. [DOI] [Google Scholar]
- 38.Audic S, Robert C, Campagna B, Parinelle H, Claverie JM, Raoult D, et al. Genome analysis of Minibacterium massiliensis highlights the convergent evolution of water-living bacteria. PLoS Genet. 2007;3 doi: 10.1371/journal.pgen.0030138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. ClustalW and ClustalX version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 40.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archae, Bacteria, and Eukarya. Proc Natl Acad Sci U S A. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garrity GM, Holt JG. The Road Map to the Manual. Bergey‘s Manual of Systematic Bacteriology. Second. New York: Springer; 2001. pp. 119–166. [Google Scholar]
- 43.Ward V, Naomi L. Phylum XXV. Planctomycetes Garrity and Holt 2001, 137 emend. Ward. Bergey’s Manual of Systematic Bacteriology. New York: Springer; 2010. pp. 879–925. [Google Scholar]
- 44.Schlesner H, Stackebrandt E. Assignment of the genera Planctomyces and Pirella to a new family Planctomycetaceae fam. nov. and description of the order Planctomycetales ord. nov. Syst Appl Microbiol. 1986;8:174–176. doi: 10.1016/S0723-2020(86)80072-8. [DOI] [Google Scholar]
- 45.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]


