Abstract
Physiological noise, if unaccounted for, can drastically reduce the statistical significance of detected activation in FMRI. In this paper, we systematically optimize physiological noise regressions for multi-shot 3D FMRI data. First, we investigate whether 3D FMRI data are best corrected in image space (RetroICor) or k-space (RetroKCor), in which each k-space segment can be assigned its unique physiological phase. In addition, the optimal regressor set is determined using the Bayesian Information Criterion (BIC) for a variety of 3D acquisitions corresponding to different image contrasts and k-space readouts.
Our simulations and experiments indicate that: (a) k-space corrections are more robust when performed on real/imaginary than magnitude/phase data; (b) k-space corrections do not outperform image-space corrections, despite the ability to synchronize physiological phase to acquisition time more accurately; and (c) the optimal model varied considerably between the various acquisition techniques. These results suggest the use of a tailored set of volume-wide regressors, determined by BIC or other selection criteria, that achieves optimal balance between variance reduction and potential over-fitting.
Keywords: Functional MRI, 3D EPI, GRE, SSFP, SPGR, Brainstem, Physiological noise
Introduction
Multi-slice gradient-echo echo-planar imaging (GRE-EPI) has long been the dominant acquisition method for functional MRI. The high readout efficiency combined with the relatively long effective echo time (TE) makes 2D single-shot EPI an ideal sequence for blood oxygen level dependent (BOLD) FMRI. However, the continual search for techniques that can support higher spatial resolution has spurred the exploration of multi-shot 3D acquisition strategies (Barth et al., 2010; Golay et al., 2000; Lutti et al., 2013; Miller et al., 2006; Stenger et al., 1999). When acquired with segmented readouts, 3D imaging enables high isotropic resolution images with minimal distortion. The multi-shot nature of these sequences, however, makes them more sensitive to physiological fluctuations as errors between the k-space segments result in strong temporal fluctuations in image space, particularly in the inferior regions of the brain (Tijssen et al., 2011).
Model-based physiological noise reduction methods acquire external measures of the cardiac and respiratory cycles, which are used to retrospectively remove the spurious fluctuations from the FMRI data. Assuming quasi-periodicity of the physiological cycles, simple low-order Fourier terms are fitted to the data based on the time of each image acquisition relative to the phase of the cardiac and respiratory cycles. Although initially proposed as a correction in k-space (RetroKCor;Hu et al., 1995), to date 2D EPI data are almost always corrected in image space (RetroICor) as this method showed better results for spatially localized fluctuations and avoids handling large complex k-space data sets (Glover et al., 2000). Typically, a general linear model (GLM) approach is used, in which the physiological noise measures are included as confound regressors to remove the spurious fluctuations, thereby improving statistical significance.
Over the last decade much effort has been put into optimizing RetroICor for 2D GRE-EPI: new physiological noise regressors have been identified (Birn et al., 2006a; Brooks et al., 2008; Friese et al., 2004; Shmueli et al., 2007), and optimized regressor sets have been proposed for various brain regions (Harvey et al., 2008; Hutton et al., 2011; Jones et al., 2008). Recently, RetroICor has also been used to correct 3D FMRI data (Barry et al., 2012; Jorge et al., 2013; Lutti et al., 2013). However, RetroICor inherently assumes that each image is acquired instantaneously. Although this assumption is justified for single-shot acquisitions, where the entire slice is acquired in a few tens of milli-seconds, it is not for 3D imaging methods, in which the data of a single volume are typically acquired over a period of seconds. A k-space based correction, in which each k-space segment can be assigned with its unique cardiac and respiratory phases (i.e., the relative time point in the cardiac or respiratory cycle), may be better suited to 3D FMRI than an image-based correction. Furthermore, not all regressors optimized for GRE-EPI may be as efficient for 3D FMRI data, since the BOLD-related physiological fluctuations are likely to be different due to the typically shorter repetition time (TR) and echo time (TE) as well as the segmented nature of the acquisition.
The aim of this paper is to carefully assess the existing retrospective correction methods on their performance to correct a range of different 3D FMRI acquisitions. First, the performance of RetroICor and RetroKCor on single-shot and multi-shot readouts is compared by means of in-vivo data and simulations to examine in which domain 3D FMRI data are best corrected. Second, a regressor selection method based on the Bayesian Information Criterion (BIC) (Jenkinson et al., 2009) is used to assess the efficacy of commonly used regressors and find the optimal physiological model for a range of 3D acquisitions with varying contrasts and readout types, including 3D GRE-EPI, spoiled gradient echo (SPGR), and balanced steady-state free-precession (bSSFP).
Theory
In principle, RetroKCor and RetroICor employ a similar method to extract physiological signals from the data. Both techniques use a linear regression with confound regressors that are generated based on external monitoring of the respiratory and cardiac cycles. Practical differences between the techniques arise due to the fact that the corrections are applied at different stages in the reconstruction pipeline.
Definition of the regressors
The confound regressors are generated based on the underlying models that are used to describe the different sources of physiological fluctuations. These models (i.e., regressors) are applicable to both RetroKCor and RetroICor. We can separate the regressors into two groups: ‘phase’ and ‘rate’ regressors.
The phase regressors are used to model signal fluctuations that vary throughout the cardiac and respiratory cycles: for example, signal from pulsatile blood flow. Regressors are typically generated both at the principal frequency of the physiological cycle and its higher harmonics. To generate the regressors a cardiac and a respiratory phase are assigned to each TR period based on the acquisition time relative to the physiological cycle. The phase regressors can be subdivided into two types: low-order Fourier terms and interaction terms. The low-order Fourier terms model the independent effects of the cardiac and respiratory fluctuation using sine and cosine values of the principal frequency and its higher harmonics (i.e., sin(φc,r),cos(phic,r),sin(2φc,r),cos(2φc,r),…, where φc,r is the cardiac or respiratory phase). The interaction terms describe the interactions between the cardiac and respiratory cycles, for example the effect of respiratory-induced pressure changes on the cardiac rhythm (Brooks et al., 2008), and are calculated by sin(φc ± φr) and cos(φc ± φr). The use of both sine and cosine terms accounts for temporal shifts.
Rate regressors are used to account for signal dependence on the rate of physiological processes. The respiratory rate has been shown to introduce low frequency variations in the BOLD signal through fluctuations in blood flow and arterial CO2 (Birn et al., 2006a; Wise et al., 2004), which can effectively be modeled with a regressor that describes the respiration volume per unit time (RVT). Recently, the heart rate regressor was also found to be an effective confound regressor (Bianciardi et al., 2009; Chang et al., 2009; Shmueli et al., 2007). Although the physiological mechanism behind this regressor is not yet well understood, it has been hypothesized that BOLD signal changes that correlate with the heart rate could be due to arousal-related neuronal activity (Chang et al., 2009).
Processing pipeline
In RetroICor the corrections are performed on the reconstructed magnitude images, at the end of the reconstruction pipeline (Fig. 1a). In 2D single-shot EPI one can reasonably make the approximation that each image is acquired instantaneously. For multi-slice acquisitions the regressors are often generated on a slice-by-slice basis to account for the small timing differences between the slices. The regressions can either be run prior to or in parallel with the task regression. For 3D acquisitions, a slice-by-slice regression is not possible as the entire volume is sampled with every RF pulse during the volume acquisition. A single physiological phase is therefore assigned to the entire volume. Best results are obtained when the phase is determined by the time halfway through the acquisition (Lutti et al., 2013). The only difference from 2D RetroICor, therefore, is that regressors are common to entire volumes, rather than specific regressors being applied to different slices.
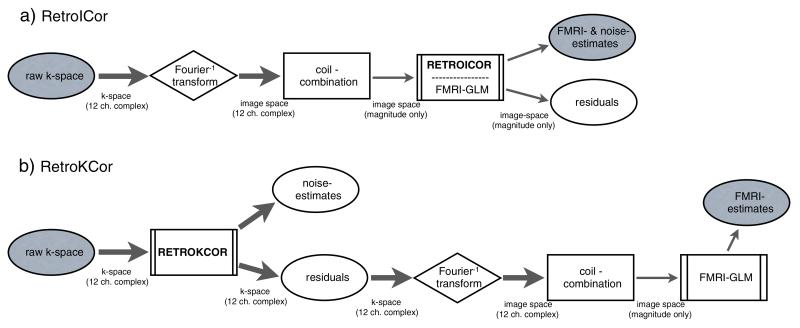
Fig. 1.
Reconstruction flowchart for RetroKCor and RetroICor. a: RetroICor corrections take place at the end of the reconstruction pipeline. The physiological noise regression can be included in the FMRI GLM analysis and is (typically) only performed on the magnitude data. b: RetroKCor corrections are performed on the complex k-space data. The residuals are the corrected data, which are further reconstructed to produce the eventual corrected images. Thick arrows represent multi-channel, complex, data, whereas the thin arrows represent combined, magnitude-only, data.
In RetroKCor, corrections are performed in k-space. In the case of multi-shot 3D acquisitions, each k-space segment, or “shot”, can be reasonably considered to have been acquired instantaneously, even though the entire k-space volume is acquired over several seconds. The corrections are therefore performed on a segment-by-segment basis by assigning a unique cardiac phase and respiratory phase to each segment. The regressors are thus implemented as 4D point-wise regressors (with a unique time-series for each k-space segment).
As shown in Fig. 1b the regressions are performed on complex k-space data. In addition, individual channels in multi-channel coils must be analyzed separately to account for the (complex) coil profiles. Given that both magnitude/phase (or real/imaginary) data are analyzed, each regressor requires a total number of Nc × 2 regressions (where Nc is the number of channels). Unlike RetroICor, where one multiple regression analysis can be used to fit both task and physiological confound regressors, FMRI analysis with RetroKCor is performed in two stages. In the first stage, a regression is performed with only the physiological regressors. The output of this stage is the residuals, that is, the k-space signal with physiological effects removed. The corrected k-space data are then Fourier transformed into image space (each channel separately, followed by a sum of squares coil combination). In the second stage, the magnitude images are then subjected to conventional FMRI analysis.
Methods
This paper is structured around three experiments: 1) correction of single-shot 2D data, in order to obtain insight in the differences between image based and k-space based regressions, 2) correction of 3D simulated data, in order to investigate the effect of regressor resolution, and 3) correction of 3D in-vivo data to validate the optimizations and find the optimal physiological model for a range of 3D FMRI acquisitions.
Optimization of the regression on 2D data
RetroICor and RetroKCor were tested on the Shepp-Logan digital phantom in Matlab (MathWorks, Natick, MA). The phantom simulated a time-series of single-slice images (matrix size = 64 × 64, 300 time points) to which thermal and physiological noise was added. The artificially generated time-series were then corrected in image space or k-space and assessed on residual variance.
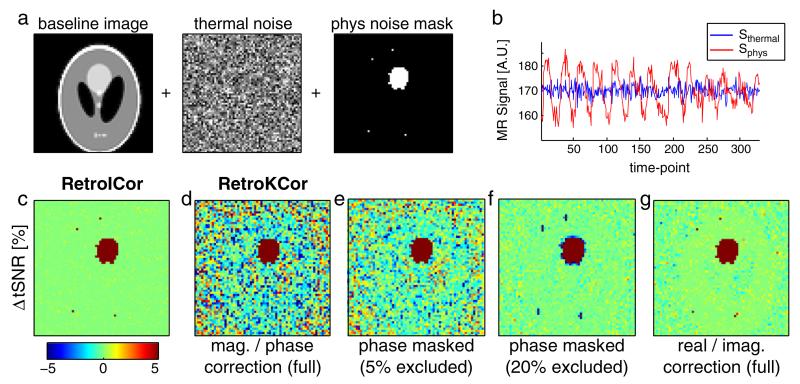
First, thermal and physiological noise were added to the time-series data in image space. Different levels of Gaussian random noise were added to the real and imaginary channels of the data to generate images with signal-to-noise (SNR) values of 10, 25, 50, and 100. A respiratory trace measured in one of the in-vivo scans (see below) was used to simulate the physiological signal fluctuations by adding the trace to each data set in predefined locations. The added signal was scaled such that the physiological contrast-to-noise ratios (pCNR) were 1, 2.5, 5, and 10 for the data sets with SNR values of 10, 25, 50 and 100, respectively. We were particularly interested to test whether the size of the physiological noise regions in image space impacted the ability to regress this noise out in k-space, since the signal power of effects that are highly-localized in image space is spread throughout k-space. The mask, within which physiological noise was added, therefore consisted of one large contiguous region and a few isolated voxels (Fig. 2a).
Fig. 2.
Simulation experiments. a: the simulation pipeline showing the addition of thermal noise (globally) and physiological fluctuations (within the mask shown on the right). b: example time courses of the simulated noise (SNR = 100, pCNR = 10). c–g: the difference in tSNR after correction with RetroICor (panel c) compared to various optimizations of RetroKCor (panel d–g). Red and blue denote increase and decrease in tSNR after noise correction, respectively.
Second, the RetroICor and RetroKCor corrections were performed with the regressor being the same respiratory trace as was used to simulate the signal fluctuations (i.e., simulating a perfect physiological noise model). The RetroICor regressions were performed on the magnitude image time-series. To test RetroKCor, the simulated image-space data were first Fourier transformed into k-space. Regressions were then performed on either the magnitude and phase (M/P) or the real and imaginary (R/I) k-space data. The corrected complex k-space data were then transformed back into image space to form the corrected images. The temporal variance in image space was used to assess the efficacy of the different correction strategies.
RetroICor and RetroKCor were also compared in single-slice 2D in-vivo data. These data were acquired for a previous study with a dynamic scan time of only 152 ms (Tijssen et al., 2009). The short volume scan time compared to physiological fluctuations makes it a reasonable approximation to instantaneous data, allowing the same regressors to be used for both RetroICor and RetroKCor. The time-series acquisition consisted of a single coronal slice through the brainstem and motor cortex with the following scan parameters: bSSFP with a segmented EPI readout, matrix = 128 × 128, resolution = 2 × 2 × 2.5 mm, α = 30°, TR/TE = 9/3.5 ms, 1860 Hz/pix, 8 lines per TR, and 2000 volumes. Heart and respiratory rates were recorded using a pulse oximeter and pneumatic bellows, respectively. RetroKCor corrections were performed on either the M/P channels or the R/I channels of the raw k-space data, whereas RetroICor was only performed on the magnitude data of the reconstructed images. The corrections were assessed on temporal variance and temporal SNR (tSNR).
The influence of regressor resolution and reference time-point
To investigate the effect of reference time point and regressor resolution (i.e., the number of unique physiological phases within each volume), another set of simulations was performed based on a 3D Shepp–Logan phantom with a similar spatial distribution of simulated physiological noise to the regions in the 2D phantom. To create a realistic 4D time-series, a multi-shot 3D EPI sequence with a matrix size of 64 × 64 in-plane and 64 kz-planes was simulated, in which each k-space segment was sampled at different time-points in the respiratory cycle. For this readout each kz-plane is acquired as a single shot with a simulated TR of 50 ms, which resulted in a volume acquisition time (Tvol) of 3.2 s. In order to create each 3D volume the respiratory trace, which served as the physiological input function, was sampled at 50 ms. For each time-point a 3D volume was generated and Fourier transformed to generate the instantaneous signal in k-space at 50 ms temporal resolution (which we will refer to as k-space frames). The 3D sampling of k-space then simply involves drawing a different kz-plane from each simulated time point to generate the “sampled” 3D volumes. In total 150 volumes were created to form the simulated time-series (SNR = 25, pCNR = 2.5).
The same respiratory trace served as input for the regressors, simulating a perfect physiological model as before. A series of regressors were generated ranging from segment-specific regressors (where each kz-plane is assigned a unique physiological phase) to volume-specific regressors (only one phase per volume). For intermediate resolutions groups of segments were assigned the same phase. A second test included varying the reference time-point of the volume-specific regressor starting with the first z-plane, at the start of the volume acquisition, up to the last acquired kz-plane. For RetroKCor, the regressions were performed on the real-imaginary data. The results were assessed based on the tSNR within the simulated noise regions.
Optimal physiological models for 3D FMRI
Twelve healthy volunteers were scanned under resting conditions on a 3 T Siemens TIM Trio system (Siemens, Erlangen, Germany) using a 12-channel head receive coil. All subjects were scanned with informed consent under a technical development protocol approved by the local ethics committee.
Data acquisition protocol
A range of six different FMRI acquisitions was scanned: 2D GRE-EPI, 3D GRE-EPI, highly segmented 3D SPGR, and highly segmented 3D passband bSSFP (Bowen et al., 2005; Miller et al., 2007) (scan parameters in Table 1). All 3D acquisitions used a stack-of-segmented EPI readout (Miller et al., 2006), which was based on the Siemens product code for EPI. This sequence includes ramp sampling and uses 1D navigator echoes (two forward lines, and one backward line at ky = 0), to correct for EPI Nyquist ghost. The SPGR and bSSFP acquisitions were acquired with and without cardiac synchronization (Tijssen et al., 2011), to assess the effects on cardiac corrections independent of overall performance. Cardiac synchronization prospectively cleans up the cardiac fluctuations, by determining the readout order in real-time based on the cardiac cycle. For each scan physiological monitoring was performed using pneumatic bellows and a pulse oximeter. The 3D GRE-EPI data were only acquired in eight subjects (this sequence was added at a later stage to include a 3D sequence with a more conventional long-TE BOLD contrast). In one subject the bSSFP data could not be analyzed, because the pulse ox signal was lost during the scan so these data were excluded.
Table 1.
Sequence parameters. The single-shot GRE-EPI data were acquired with an acceleration factor (R) of 2 in order to reduce the readout length and thus distortion.
| Parameter | 2D GRE-EPI | 3D GRE-EPI | 3D SPGR | 3D bSSFP |
|---|---|---|---|---|
| RF spoiling | N/A | 117° inc. | 117° inc. | N/A |
| Grad. spoiling | N/A | 2π phase | 2π phase | N/A |
| Excitation flip angle | 90° | 30° | 30° | 30° |
| TR | 3500 ms | 72 ms | 12 ms | 12 ms |
| TE | 30 ms | 30 ms | 6 ms | 6 ms |
| Segmentation | Single-shot | 48 lines per TR | 8 lines per TR | 8 lines per TR |
| Parallel acceleration | R = 2 | None | None | None |
| Imaging plane | Coronal |
|---|---|
| Readout bandwidth | 1860 Hz/pix |
| Field of view | 192 × 192 × 48 mm |
| Matrix size | 96 × 96 × 24 |
| Resolution | 2×2×2 mm |
| Tvol | 3500 ms |
| Number of volumes | 60 volumes |
| Total imaging time | 210 s |
In each subject, single volume whole-brain scans were collected with each imaging contrast for registration purposes using the same parameters as for the time-series acquisitions, but with an increased FOV of 192 × 192 × 160 mm (matrix size = 96 × 96 × 24) to cover the entire brain. For the anatomical scan, an inversion-prepared 3D MPRAGE sequence was used with TI = 1100 ms, kz-plane time = 3300 ms, and TE = 4.6 ms, matrix = 256 × 176 × 224, and 1 mm3 isotropic resolution.
Image reconstruction
Image reconstruction was performed offline using custom in-house code in Matlab (MathWorks, Natick, MA). The reconstruction steps included regridding and phase correction to correct for inconsistencies between the odd and even lines of the EPI trajectory. Mean phase offsets between volumes were not corrected. For the 2D GRE-EPI data, which were acquired with an acceleration factor of R = 2, GRAPPA was performed. The individual coil images were combined using a sum of squares combination.
Physiological noise model (regressor set)
The regressors used for this experiment were generated in Matlab. In total 22 regressors were generated: 14 phase terms, 4 interaction terms, 2 rate regressors and 2 rate derivatives. The generated regressors are listed in Table 2. The numbering and naming convention in this table will be used when the results are discussed. The cardiac, respiratory and interaction terms (regressors 1–18) were defined according to Glover et al. (2000) and Brooks et al. (2008). The cardiac rate (regressor 19) and respiratory rate (regressor 21) were calculated according to Chang et al. (2009), and Birn et al. (2006a), respectively. Temporal derivatives of the rate regressors were included to allow for temporal shifts (see Appendix A).
Table 2.
The full set of regressors that are tested in this study. The numbering and naming convention shown here will be used throughout the paper.
| No. | Name | Description |
|---|---|---|
| 1 | ev01_cardcos_01 | Cardiac: First order cosine |
| 2 | ev02_cardsin_01 | Cardiac: First order sine |
| 3 | ev03_cardcos_02 | Cardiac: Second order cosine |
| 4 | ev04_cardsin_02 | Cardiac: Second order sine |
| 5 | ev05_cardcos_03 | Cardiac: Third order cosine |
| 6 | ev06_cardsin_03 | Cardiac: Third order sine |
| 7 | ev07_respcos_01 | Respiratory: First order cosine |
| 8 | ev08_respsin_01 | Respiratory: First order sine |
| 9 | ev09_respcos_02 | Respiratory: Second order cosine |
| 10 | ev10_respsin_02 | Respiratory: Second order sine |
| 11 | ev11_respcos_03 | Respiratory: Third order cosine |
| 12 | ev12_respsin_03 | Respiratory: Third order sine |
| 13 | ev13_respcos_04 | Respiratory: Fourth order cosine |
| 14 | ev14_respsin_04 | Respiratory: Fourth order sine |
| 15 | ev15_cosadd | Interaction: (card + resp) cosine |
| 16 | ev16_cossub | Interaction: (card − resp) cosine |
| 17 | ev17_sinadd | Interaction: (card + resp) sine |
| 18 | ev18_sinsub | Interaction: (card − resp) sine |
| 19 | ev19_cr | Cardiac rate |
| 20 | ev20_dcr | Cardiac rate derivative |
| 21 | ev21_rvt | Respiratory rate |
| 22 | ev22_drvt | Respiratory rate derivative |
In the case of RetroKCor, each k-space segment was assigned a cardiac phase and a respiratory phase. For the RetroICor corrections on 3D data, the cardiac and respiratory phases are sampled at one reference time point during the acquisition. This reference time point was chosen to be the halfway point of the readout, which for conventionally ordered 3D acquisitions coincides with the acquisition of the center kz-plane.
All regressions, both in k-space and image space were performed using the core regression function within FSL (film_gls), to exclude additional processing steps, such as temporal and spatial filtering. Motion correction was also not performed, to avoid interference between motion correction and the physiological noise regression (Jones et al., 2008). Visual inspection of the data showed no extensive movement in any of the data sets.
Model evaluation
In order to determine the optimal physiological noise model for each of the noise correction methods, a Bayesian model selection method was implemented based on the method proposed by Jenkinson et al. (2009). The Bayesian Information Criterion (BIC) (Kass and Raftery, 1993) is defined as:
| (1) |
where N is the number of samples (time points), k is the number of regressors, and RSS is the residual sum of squares. The BIC is used to compare different models, particularly models with a different number of parameters (here, regressors), with low BIC indicating a favorable model. BIC selection favors models that explain a large portion of the variance, which is weighted against a penalty for the total number of regressors, since this reduces the degrees of freedom (DOF) of the GLM analysis.
The selection procedure aims to find the optimal set of regressors iteratively by expanding the model to encapsulate one additional regressor at a time and comparing the resulting BIC score. If the BIC is smaller, we include it in our optimal set; otherwise, we consider the penultimate model (without the most recently-added regressor) to be optimal. The order in which the regressors are added to the model is determined based on the performance of each regressor when tested individually. So, the procedure followed is:
Step 1 Regressions are run using each of the candidate regressors (22 in total, Table 2) as a single regressor. Temporal variance maps are calculated for each of the data sets.
Step 2 The regressors are sorted based on how much variance each regressor explains within a given region of interest (ROI). The variance reduction is assessed based on the mean variance across the brainstem.
Step 3 An optimal regressor set is created iteratively by adding one regressor at a time in the order determined in step 2. After each model expansion step the BIC value is calculated using the mean RSS within the brainstem ROI. The set of regressors with the lowest BIC is taken as the optimal model for the correction.
ROI analysis
Time-series images were registered to the MNI template with FLIRT (Jenkinson et al., 2002) using the whole brain acquisitions and the anatomical scan for each subject as intermediate steps. The transformations obtained from this registration were used to transform the brainstem ROI, which we defined in the MNI space, to the native spaces of each of the acquired scans. This way spatial interpolation of the data, which can bias the temporal variance, was avoided. The Harvard–Oxford probabilistic atlas for subcortical structures (Desikan et al., 2006; Smith et al., 2004) was used to define the brainstem ROI by setting the threshold of the probability map to 0.95 (i.e., only voxels with a probability greater than 95% were included). The regions of CSF surrounding the brainstem are known to demonstrate high signal fluctuations and are likely to dominate the optimization if included. Therefore a conservative mask was defined in order to restrict the optimization only to areas of tissue and avoid partial voluming with CSF. To be certain that the masks did not include regions of CSF the masks were visually inspected and manually adjusted if needed.
Results
Optimization of the regression on 2D data
Fig. 2a shows the simulation pipeline of the simulations on a single, well-defined respiratory phase (corresponding to a single-shot acquisition). Example magnitude time-courses of two voxels with and without added physiological noise (SNR = 100, pCNR = 10) are given in Fig. 2b. The performance of the standard RetroICor and RetroKCor corrections were assessed on temporal signal-to-noise ratio (tSNR), shown in Figs. 2c and d (using the original RetroKCor method performed on M/P data by Hu et al. (1995)). Although both methods remove a similar amount of signal fluctuations in the large ROI with simulated physiological noise, RetroKCor also alters the tSNR in voxels without simulated physiological noise, indicated by the inhomogeneous tSNR in voxels that only simulated thermal noise.
The noise-like spatial distribution of the M/P RetroKCor correction was found to scale inversely with SNR (high SNR data not shown). This may indicate that the regression fails at the edges of k-space where low signal magnitude translates to high uncertainty in the phase (since phase noise scales non-linearly with signal magnitude). Limiting the phase regressions to only those pixels with sufficient signal magnitude improves the stability of the correction, as shown in Figs. 2e and f. By excluding k-space voxels corresponding to the lowest 5% signal magnitude, the ΔtSNR map becomes more homogeneous in regions of purely thermal noise; while a 20% threshold results in homogeneous tSNR comparable to that of RetroICor. Confining the regressions to the center of k-space, however, also reduces the spatial resolution of the correction. This causes the correction to blur into the voxels directly adjacent to regions of physiological noise (i.e., the blue rings around the red regions denote a reduction in tSNR).
The original paper by Hu et al. (1995) argued that the regressions were to be performed on the magnitude and phase of the signal, as respiration was expected to mainly have an effect on the phase of the signal. However, there is no real reason why the regressions cannot be performed on the real and imaginary parts of the signal instead. This avoids the non-linear scaling of phase noise in the outer k-space pixels and also gets around practical issues such as phase unwrapping (Hagberg et al., 2008) and the arbitrary nature of k-space masking, which could bias the results. Fig. 2g shows the results when the corrections are performed on the real and imaginary data instead of the magnitude and phase. The results are similar, but not identical to the RetroICor results (Panel c). Some background voxels still show a small reduction in tSNR. Nevertheless, a strong improvement is observed compared to the standard M/P correction in Fig. 2d, but without the loss in spatial resolution involved with masking the phase corrections (Fig. 2f).
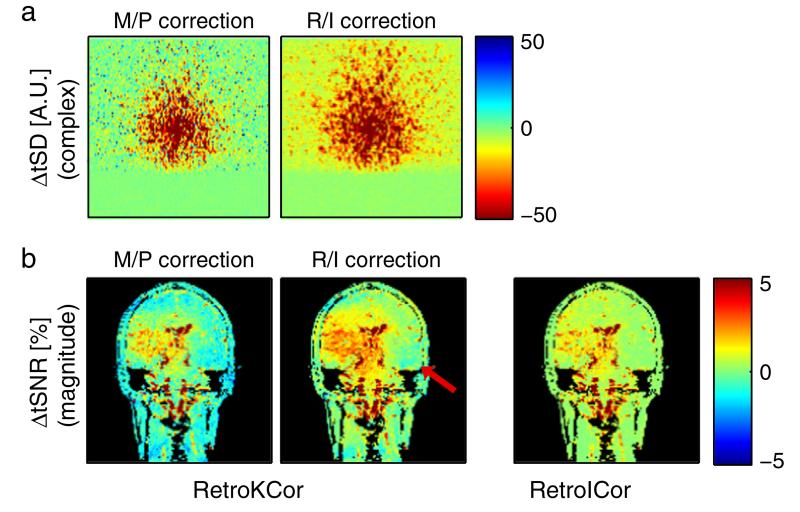
Fig. 3 compares the performance of RetroICor and RetroKCor corrections on 2D, single-slice in-vivo data for which k-space acquisitions are acquired over a brief (150 ms) window. These data have the important properties that each image can be reasonably considered to be well represented by an instantaneous physiological phase. The constructed regressors are therefore the same for both image-space and k-space based regressions. Fig. 3a shows the effect of RetroKCor on temporal fluctuations in k-space. When RetroKCor corrections are performed on the M/P channels the reduction in temporal SD (tSD) is somewhat confined to the center of k-space. Some k-space positions in the periphery of k-space even show an increase tSD after correction. This effect is not observed when the regressions are performed on the R/I channels instead. The improvement of RetroKCor corrections is also seen in image space (Fig. 3b). Across the image the tSNR increase is larger for the R/I corrected image than the M/P corrected image. When the corrections are performed on the R/I channels, the RetroKCor results are comparable to the RetroICor results. However, small differences between the two methods are still observed. In areas where a tSNR increase was obtained RetroKCor showed a slightly bigger improvement, whereas areas adjacent to this (red arrow) showed a marginal drop in tSNR of up to 0.5% compared to the RetroICor corrected data.
Fig. 3.
2D in-vivo results comparing RetroKCor and RetroICor. a: the change in complex temporal standard deviation (tSD) after RetroKCor regressions on the M/P (left) and R/I channels (right). Here, we show absolute tSD to provide a depiction of the signal power (since percent change in tSNR at outer k-space is misleading). b: RetroKCor vs. RetroICor. Performing the regressions on the R/I channels has a clear advantage for RetroKCor, although small reductions in tSNR (up to 0.5%) can still be observed (denoted by the red arrow). For both cases, red (blue) indicates increased (decreased) temporal stability.
The influence of regressor resolution and reference time-point
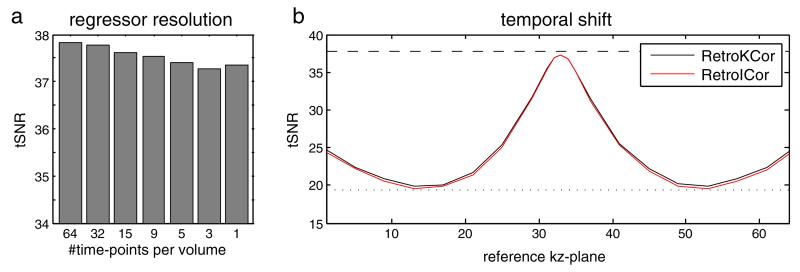
Fig. 4 shows the effect of regressor resolution and shift in reference time-point on the tSNR within the simulated noise region after regression. With only thermal noise simulated the tSNR in this region is 37.8. With simulated physiological fluctuations the tSNR drops to 19.5. As the regressor is directly derived from the input for the physiological fluctuations (simulating a perfect physiological model), we expect the tSNR for regressions with sufficient resolution to lie close to 37.8. Fig. 4a demonstrates that the best results are achieved when segment-specific regressors are used (i.e., 64 time-points per volume; tSNR = 37.8). However, lowering the temporal resolution only has a marginal effect on the resulting tSNR. When only 1 time-point per volume is used a tSNR of 37.4 is still achieved (1.3% difference).
Fig. 4.
3D simulation results showing the tSNR within the simulated noise region after regression. a: the effect of regressor resolution. b: the effect of temporal shift. The tSNR when only thermal noise is simulated is 37.8 whereas the tSNR with simulated physiological fluctuations is 19.5 denoted by the striped and dotted line in (b), respectively. The best results are achieved when 64 regressors are used (segment-specific regression). The tSNR drops with decreasing temporal resolution, although the effect is small. When only 1 time-point is used per volume, the optimal reference time-point is the time at which the center kz-plane is acquired. A shift in either positive or negative direction causes the regression to become less affective.
Fig. 4b shows that, when only one reference time-point per volume is used, the physiological fluctuations are almost completely removed as long as the reference point is set to the center kz-plane (i.e., kz-plane 33). When the reference time-point is shifted away from the center kz-plane the efficacy drops and is minimal when the shift is around 1 s, which corresponds to roughly 1/4 respiratory cycle. For this shift the constructed regressor is orthogonal to the simulated fluctuation and thus the correction is almost completely ineffective.
The results presented in Fig. 4 indicate that the temporal resolution does not affect the correction much as long as the reference time-point is set correctly to the time at which the center of k-space is acquired.
Optimal physiological models for 3D FMRI
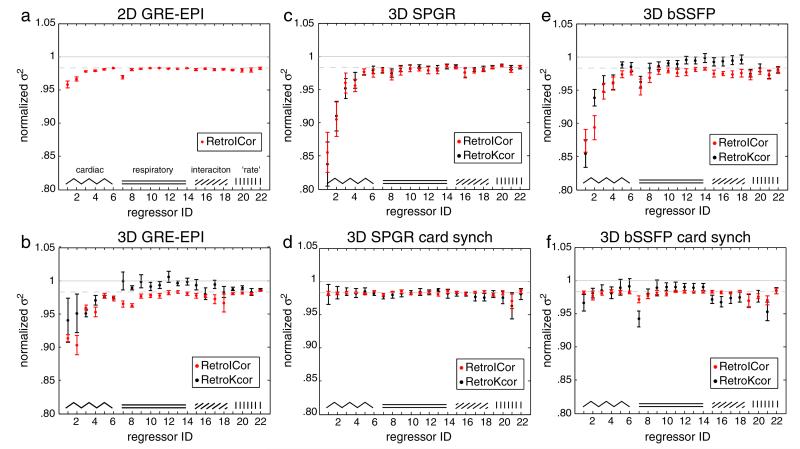
Fig. 5 compares the efficacy of image-space and k-space regressions on 2D GRE-EPI, 3D GRE-EPI, non-synchronized and cardiac-synchronized 3D bSSFP and SPGR. The plots display the residual variance in the brainstem ROI after regression with individual regressors. The residual variance is normalized against the non-corrected data set. The regressors are plotted in the order as listed in Table 2. The gray dashed line denotes the average amount of residual variance obtained when a random regressor would be used for the regression. On average a randomly constructed regressor would explain 1/Nth of the variance in a data set, where N is the number of time points (Jenkinson et al., 2009). This value can thus be used as a reference to assess the effectiveness of each individual regressor.
Fig. 5.
The residual variance after single regressor regressions for 2D GRE-EPI (a), 3D GRE-EPI (b), non-synchronized SPGR (c), synchronized SPGR (d), non-synchronized bSSFP (e), and synchronized bSSFP (f). RetroICor results are shown in red and RetroKCor results are shown in black. The mean across subjects is plotted with the error bars representing the standard deviation. The dashed gray line represents the expected residual variance obtained by a regression with a randomly constructed regressor.
Comparing the different plots, we found that the amount of variance explained by each of the regressors varies considerably between the acquisitions. For 2D GRE-EPI, only the first order cardiac regressors (i.e., ev01_cardcos_01 and ev02_cardsin_01) and one first order respiratory regressor (i.e., ev07_respcos_01) explain a considerable amount of variance. The other regressors do not remove more variance than a randomly constructed regressor. For non-synchronized bSSFP and SPGR, which are more prone to cardiac fluctuations, up to the second order cardiac regressors explain a considerable amount of variance as well as the first order respiratory regressors. For cardiac synchronized bSSFP and SPGR, the effect of the cardiac regressors is minute, as these fluctuations are prospectively corrected for by the adaptive readout.
The RetroICor regressions (red) appear as effective or, in some cases, even more effective than RetroKCor regressions (black) for 3D data. Only the cardiac synchronized bSSFP data shows an improved performance of RetroKCor for a few of the tested regressors. On the other hand, the RetroKCor results also show a slightly increased variance in the brainstem for some respiratory regressors in the 3D GRE-EPI and synchronized bSSFP data (Figs. 5b and f), which is not seen for the regressions that are performed in image space.
Physiological model selection
Finally, we consider BIC selection of a set of regressors to form a final model for each acquisition. Table 3 lists the 10 candidate regressors that showed the largest individual effect per data set. For each acquisition type, the regressors were added in this unique, acquisition specific order (from top to bottom) to find the model that provides the best tradeoff between variance reduction and loss in DOF. The optimal models (i.e., the regressors that passed the BIC selection) for each data set are listed in bold. The BIC metric indicates that the data supported far fewer than 10 regressors in every case, which is notable since this metric was computed in the brainstem, one of the areas most affected by physiological noise. It is further seen that, although generally the models include cardiac regressors, the number and type of regressors vary between the acquisitions. For both cardiac synchronized readouts no cardiac regressors pass the BIC, which is in good agreement with the results shown in Figs. 5d, f. However, note that one could not trivially predict the results of Table 3 from Fig. 5 since: (a) the full model is a combined regression rather than the variance explained by a single regressor, and (b) the BIC metric is not identical to the 1/Nth variance metric.
Table 3.
This table lists the order of the candidate regressors for the BIC selection. The regressors were ordered based on the amount of variance explained each regressor individually. The first 10 regressors are listed. The regressors in bold typeface are the regressors that ultimately passed the BIC selection procedure.
| Seq. | Without cardiac synch |
With cardiac synch |
||
|---|---|---|---|---|
| RetroICor | RetroKCor | RetroICor | RetroKCor | |
| 2D GRE | ev01_cardcos_01 | - | - | - |
| ev02_cardsin_01 | - | - | - | |
| ev07_respcos_01 | - | - | - | |
| ev03_cardcos_02 | - | - | - | |
| ev04_cardsin_02 | - | - | - | |
| ev20_dcr | - | - | - | |
| ev21_rvt | - | - | - | |
| ev19_cr | - | - | - | |
| ev15_cosadd_01c01r | - | - | - | |
| ev05_cardcos_03 | - | - | - | |
| 3D GRE | ev02_cardsin_01 | ev03_cardcos_02 | - | - |
| ev01_cardcos_01 | ev02_cardsin_01 | - | - | |
| ev04_cardsin_02 | ev01_cardcos_01 | - | - | |
| ev03_cardcos_02 | ev04_cardsin_02 | - | - | |
| ev08_respsin_01 | ev06_cardsin_03 | - | - | |
| ev07_respcos_01 | ev05_cardcos_03 | - | - | |
| ev18_sinsub_01c01r | ev21_rvt | - | - | |
| ev06_cardsin_03 | ev19_cr | - | - | |
| ev17_sinadd_01c01r | ev08_respsin_01 | - | - | |
| ev15_cosadd_01c01r | ev18_sinsub_01c01 | - | - | |
| SPGR | ev01_cardcos_01 | ev01_cardcos_01 | ev21_rvt | ev21_rvt |
| ev02_cardsin_01 | ev02_cardsin_01 | ev19_cr | ev20_dcr | |
| ev04_cardsin_02 | ev03_cardcos_02 | ev07_respcos_01 | ev18_sinsub_01c01r | |
| ev03_cardcos_02 | ev04_cardsin_02 | ev13_respcos_04 | ev17_sinadd_01 c01r | |
| ev08_respsin_01 | ev08_respsin_01 | ev05_cardcos_03 | ev07_respcos_01 | |
| ev06_cardsin_03 | ev16_cossub_01c01r | ev10_respsin_02 | ev08_respsin_01 | |
| ev16_cossub_01c01r | ev05_cardcos_03 | ev16_cossub_01c01r | ev01_cardcos_01 | |
| ev05_cardcos_03 | ev12_respsin_03 | ev02_cardsin_01 | ev16_cossub_01c01r | |
| ev09_respcos_02 | ev07_respcos_01 | ev01_cardcos_01 | ev19_cr | |
| ev17_sinadd_01c01r | ev06_cardsin_03 | ev04_cardsin_02 | ev22_drvt | |
| bSSFP | ev01_cardcos_01 | ev01_cardcos_01 | ev19_cr | ev07_respcos_01 |
| ev02_cardsin_01 | ev02_cardsin_01 | ev21_rvt | ev21_rvt | |
| ev03_cardcos_02 | ev07_respcos_01 | ev07_respcos_01 | ev01_cardcos_01 | |
| ev07_respcos_01 | ev03_cardcos_02 | ev20_dcr | ev16_cossub_01c01r | |
| ev04_cardsin_02 | ev04_cardsin_02 | ev08_respsin_01 | ev19_cr | |
| ev08_respsin_01 | ev21_rvt | ev02_cardsin_01 | ev15_cosadd_01c01r | |
| ev19_cr | ev19_cr | ev10_respsin_02 | ev17_sinadd_01c01r | |
| ev21_rvt | ev22_drvt | ev03_cardcos_02 | ev18_sinsub_01c01r | |
| ev05_cardcos_03 | ev08_respsin_01 | ev17_sinadd_01c01r | ev20_dcr | |
| ev16_cossub_01c01r | ev20_dcr | ev06_cardsin_03 | ev08_respsin_01 | |
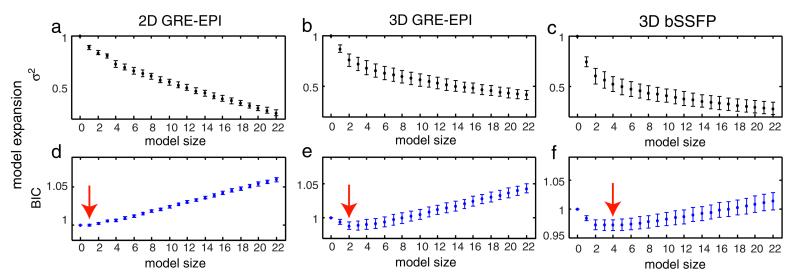
Figs. 6a–c show examples of the residual variance for each model expansion step during the BIC selection procedure. Results are shown for RetroICor on 2D GRE-EPI, 3D GRE-EPI, and non-synchronized 3D bSSFP data. The results are averaged across all subjects with error bars denoting the standard deviation. The largest effect is seen in bSSFP (Fig. 6c). A large variance reduction is achieved by the first few regressors. After the first few regressors the effect flattens to a constant reduction per added regressor, similar to the curve shown in Fig. 1 of Beall (2010). Note that the shape of this curve resembles the shape of the Eigenspectra curve common in PCA analysis, where the “knee” of the plot splits the significant regressors from those that simply capture additional variance (i.e., overfit the data) (Beckmann and Smith, 2004). A similar curve, although less pronounced is observed for 3D GRE-EPI, while 2D GRE-EPI shows an even smaller effect.
Fig. 6.
a–c: the residual variance after each model expansion step for 3D GRE-EPI, conventional bSSFP, and cardiac-synchronized bSSFP, respectively. d–f: The corresponding BIC values for each model expansion. The minimum BIC value for each sequence is marked by the red arrow. All plots represent the group mean with error bars representing standard deviation.
Figs. 6d–f show the calculated BIC values for each model expansion step, with the minima marked with red arrows. Like the variances, the BIC values are normalized against the non-corrected data set. The figures show that for 3D GRE-EPI and non-synchronized 3D bSSFP the minima of the BIC curves agree well with the location of the “knees” in the variance plots. For 3D GRE-EPI the minimum is obtained when two regressors are included, while for bSSFP the optimal model contains four regressors. For 2D GRE-EPI only one regressor is selected.
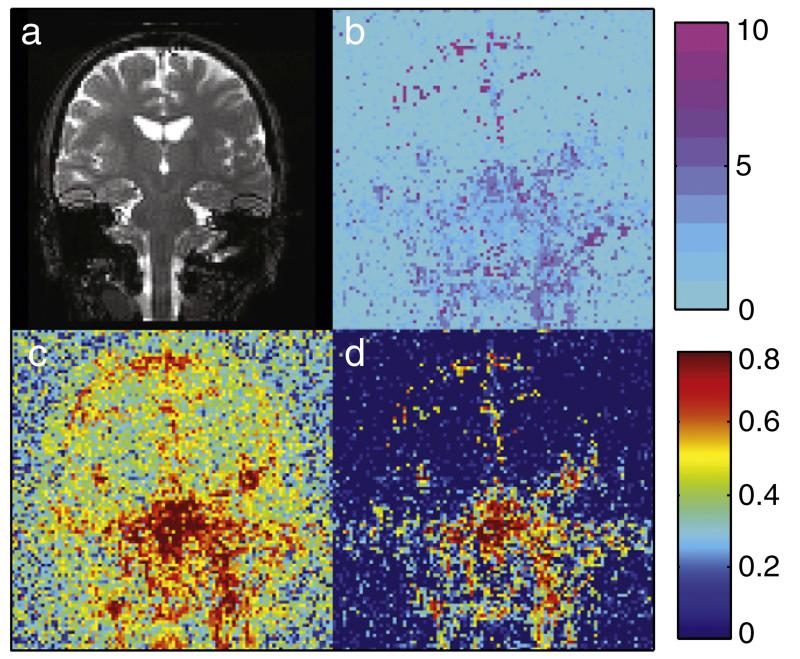
Fig. 7 shows an example of the spatial distribution of the BIC results as obtained in non-synchronized bSSFP data. An example image is shown in Fig. 7a, with the number of selected RetroICor regressors per voxel shown in Fig. 7b. It is seen that in the inferior regions of the brain the number of selected regressors is higher than in the cortical and white matter regions. In a few CSF voxels up to 10 regressors are selected by the BIC. For the majority of the brainstem voxels, however, the optimal model rarely contains more than six regressors. The bottom two images compare the relative variance reduction obtained by the full model of 22 regressors (Fig. 7c) and the optimal model of only four regressors (Fig. 7d). It is seen that, in areas of high physiological noise, a large amount of variance can be explained with the reduced set of only four regressors. When this model is used, an increase in tSNR of 18% within the brainstem ROI is achieved for non-synchronized bSSFP. For non-synchronized SPGR a tSNR increase of 15% is achieved. The synchronized versions of the sequences, on the other hand, show identical tSNR before and after, since no regressors were selected by the BIC selection. 2D GRE-EPI shows an improvement of 3.5%.
Fig. 7.
RetroICor BIC results for non-synchronized 3D bSSFP. a: Example image for anatomical reference. b: The number of BIC selected regressors calculated voxelwise. Inferior brain regions and regions of CSF show the largest number of selected voxels. c: the amount of variance reduction when the full set of 22 regressors is used for the regression and d: when the BIC determined model containing four regressors is used. Even with only a few regressors a large amount of variance can be removed.
Discussion
RetroKCor vs. RetroICor
The primary goal of this work was to optimize and compare image based and k-space based corrections for physiological noise in 3D data. RetroICor and RetroKCor were first compared under conditions in which they are equivalent: simulations in which k-space acquisition can be considered to be instantaneous and with high SNR (SNR = 100). In the case of a more realistic SNR (SNR = 25), however, the non-linear scaling of phase noise in the outer k-space pixels results in poor estimates of the weights of the confound regressors. Our results demonstrate that the problems associated with the correction of the higher spatial frequencies in RetroKCor could be largely mitigated by performing the correction on the real and imaginary channels of the data. The noise on the R/I scales linearly with the signal thereby improving the regressions in outer k-space regions.
Next, RetroKCor and RetroICor were assessed on rapidly acquired single-slice in-vivo data. For these 152 ms acquisitions, the entire image can be assumed to be acquired instantaneously and thus the regressors are the same for both RetroICor and RetroKCor. Differences can therefore be attributed to differences in the SNR distribution between image- and k-space. The RetroKCor corrections performed on the R/I channels showed improvements over the RetroKCor corrections on the M/P channels, similar to what was found by our simulations. Small differences with RetroICor were also observed in-vivo, which can be attributed to the fact that in RetroKCor the low spatial frequency components of respiration effects are more readily compensated for than higher frequencies because of the uneven distribution of signal (and therefore SNR) across k-space. This can introduce an “overcompensation” effect in areas adjacent to respiratory fluctuations, which could also explain the increased variance in the brainstem induced by the respiratory regressors in the 3D GRE-EPI data (Fig. 5b).
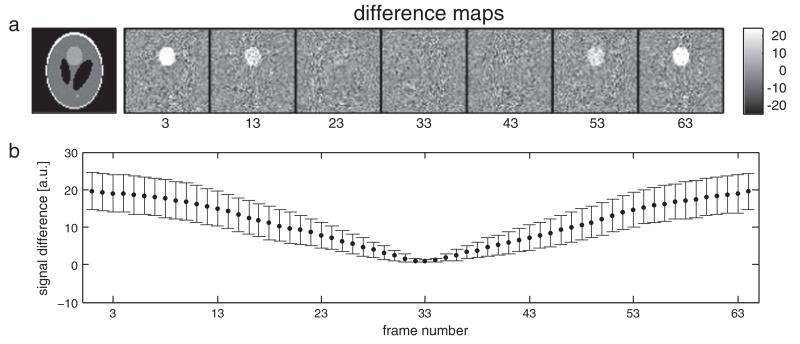
Simulations of RetroKCor on 3D data showed that the corrections are not sensitive to the temporal resolution of the regressors (i.e., the number of time-points per volume) as long as the time at which the center of k-space is acquired is well matched to the regressor (Fig. 4). In our simulations, which used perfect regressors (i.e., identical to the injected physiological signal), the physiological fluctuations are almost completely removed with only a single physiological phase per volume provided that the center k-plane is used as a reference time-point. However, the efficacy of the regression rapidly decreases when the reference time-point is shifted away from the center kz-plane. This result indicates that the physiological fluctuations are largely dominated by the center of k-space. This property is illustrated in Fig. 8. Fig. 8a displays the deviation of a simulated multi-shot 3D volume, acquired over 64 frames sampled throughout the physiological cycle, from the instantaneous magnetization at each time point. The multi-shot 3D volume most closely resembles the magnetization at time points where the center of k-space is sampled (segments 23–43), such that the deviation images are largely thermal noise. For time points that sampled outer k-space, the multi-shot volume deviates from the instantaneous magnetization in regions of physiological noise. Fig. 8b plots this absolute signal deviation for repeated simulations with different realizations of the physiological noise, demonstrating that this behavior holds regardless of the synchronization of physiological noise to the multi-shot acquisition (i.e., even if the signal fluctuations are strongest when central k-space is acquired).
Fig. 8.
Demonstration of the dominance of central k-space in the manifestation of physiological noise in 3D multi-shot volumes. a: Absolute deviation of the simulated multi-shot 3D volume from the instantaneous magnetization during each segment acquisition. The multi-shot volume deviates strongly from the instantaneous magnetization corresponding to outer k-space volumes, but closely matches the instantaneous magnetization at central k-space. This result demonstrates that physiological noise in the multi-shot 3D volume is dominated by the state of magnetization when central k-space is acquired, which is most likely the reason for the success of volume specific regressors like those used in RetroICor. b: the mean signal difference in the area of injected physiological noise averaged across all 150 simulated volumes. Error bars denote one standard deviation.
Our results on 3D in-vivo data confirm our findings that the temporal resolution of the regressors has little influence on the regression as RetroICor showed similar performance compared to RetroKCor. The fact that RetroICor and RetroKCor have similar performance is welcome, as it justifies the use of the simpler RetroICor corrections. RetroICor removes the need to collect the raw k-space data from the scanner and requires far fewer regressions to be performed. Where RetroKCor requires regressions on the complex data of each individual coil, only one regression (on the image magnitude data) is needed for RetroICor. RetroICor further allows the confound regressors to be included in the GLM together with the stimulus regressor, which statistically is a better approach (Lund et al., 2006). For example, if confound regressors are correlated to the stimulus paradigm, a separate regression would always result in an underestimation of the functional activation as the overlapping signal would always be fully attributed to the confound regressor.
Model evaluation
Secondary to the comparison of RetroICor and RetroKCor, we investigated what the optimal set of regressors would be for different types of 3D FMRI acquisitions. As shown by Hutton et al. (2011), the inclusion of more confound regressors does not necessarily introduce improvements in BOLD sensitivity. For example, the loss in DOF due to inclusion of a large number of regressors or correlations of the confound regressor with the regressor of interest (i.e., the stimulus) can actually degrade statistical significance. Most of the earlier studies, however, do not specifically address this and only report reductions in variance (which would be observed also in the presence of these issues) (Harvey et al., 2008; Jones et al., 2008).
The order in which the regressors were added to the BIC selection process was based on the amount of variance explained by independent, single regressions for each regressor, which was necessary to make the model selection compatible with RetroKCor. This approach, however, requires the regressors to be orthogonal. Inspection of the regressor correlation matrices showed that the regressors used in this study were largely orthogonal (with the highest correlation between two regressors being 0.2). When RetroICor is performed, the better approach is to determine the order of the regressors iteratively (i.e., the next candidate regressor is chosen by re-calculating how much of the remaining variance is explained by each of the remaining candidate regressors for every iteration). This would ensure an optimal ordering even if the candidate regressors are non-orthogonal.
Inspection of the selected regressors (Table 3 and Fig. 5) showed that the BIC selection robustly separates significant from insignificant regressors, but does not simply select all regressors that show a variance reduction larger than 1/N. The selection cut-off is dependent on the number of time-points, N, of the time-series. Including more time-points can therefore enable more regressors, and cause regressors that are on the border of significance (e.g., regressor 4 and 8 for RetroICor on non-synchronized bSSFP) to be included. A longer scan time, however, also means that regressors such as motion descriptors (Friston et al., 1996; Jones et al., 2008) become more relevant, which would then have to be added to the pool of potential regressors.
After regression with the optimal model as determined by the BIC, the increase in tSNR was 18% for bSSFP, 15% for SPGR, but only 3.5% for 2D GRE-EPI. This modest increase for 2D GRE-EPI deviates from previous studies that report tSNR improvements of up to 25% (Birn et al., 2006b; Hutton et al., 2011). We believe that the reason for this is two-fold: (1) the optimal model, determined by the BIC selection, only contained very few regressors compared to previous literature, and (2) we defined a very conservative brainstem ROI in order to deliberately exclude regions of CSF surrounding the brainstem. Harvey et al. (2008) reported a reduction in tSD of up to 15% in the brainstem, although most of the improvement was achieved near the boundaries with CSF. In the center of the brainstem the reduction was approximately 5%, which is roughly in line with our findings given the reduced number of regressors used in this study. When we perform a regression on the 2D GRE-EPI data using the full model of 22 regressors (something which would seriously reduce the DOF in this data set) we observe a tSNR increase of 29%. This is of the same order of magnitude as values previously reported in literature. Hutton et al. (2011) found a 35% increase in the lateral geniculate nucleus at 7 Tesla when 14 regressors were used (12 cardiac and respiratory phase regressors and two rate regressors).
The optimal regressor sets found in this study are not intended to serve as a general model for 3D FMRI corrections. As our results demonstrate the optimal model varies between the various acquisitions types tested. Although the acquisitions tested here varied considerably in terms of signal contrast or readout strategy, more subtle differences could also have a pronounced effect on the significance of certain regressors. For example, navigators that correct for global frequency drifts could prospectively remove respiratory fluctuations reducing the need to include respiratory regressors (Pfeuffer et al., 2002). It is therefore important to interrogate the data and assess which of the regressors are important before committing to any specific model. The BIC is an elegant approach for this purpose. For example, one could perform an exploratory BIC selection on a subset of the data, in order to find the optimal physiological noise model for a given experimental set-up (i.e., acquisition type, number of time-points, etc.). However, as shown in Fig. 6, it can occur that several models have a very similar BIC value (Fig. 6e; models 2 and 3). It is up to the investigator to make an informed decision as to which regressors to include, rather than just opting for the model that shows the lowest BIC. Although the BIC selection takes into account the loss in DOF by applying a penalty term for the number of regressors used, the weight of this penalty term can be varied. Other closely related selection criterion methods could be used such as the Akaike information criterion (AIC) (Akaike, 1974). The AIC differs from BIC in that the penalty term for the number of regressors is smaller. Therefore a larger regressor set would be preferred. In order to investigate the optimal weight that should be given to the penalty term, an assessment of the functional sensitivity should be included. In this study, however, such an investigation was not included.
Limitations and applicability
In this paper the effect of motion correction on physiological noise regression was not investigated. It has been shown that through-plane motion in 2D multi-slice acquisitions can have a significant effect on the effectiveness of physiological regressions (Jones et al., 2008). These effects occur when motion registration intervenes with the slice timing correction. Although such investigations are outside the scope of this paper we anticipate that these effects will be less pronounced for 3D acquisitions, as the z-direction in 3D imaging is not sampled at discreet time points like in 2D multi-slice imaging and volume-specific regressions showed similar results compared to regressions with high temporal resolution regressors.
Second, this study was performed at 3 Tesla. Although 3D FMRI is also performed at 3 T, the primary platform for these techniques may well be at 7 T. At higher field strength 2D GRE-EPI becomes more challenging (e.g., increased distortion and poor slice profile), whereas the reduced T2 and will boost the contrast in short-TE segmented acquisitions. In the present study the brainstem could be considered to mimic the increased physiological noise at high fields (Hutton et al., 2011; Triantafyllou et al., 2005; Van de Moortele et al., 2002). Although the noise levels in the brainstem at 3 T are greater than in cortical regions, and thus more comparable to the levels expected at 7 T, the relative contributions of cardiac and respiratory related fluctuations are likely to be different.
Conclusions
We have found that RetroKCor regressions can be significant improved when the regressions are performed on the real and imaginary channels instead of the phase and magnitude. However, the hypothesis that segment-specific regressions in k-space are preferable over volume-specific regressions (i.e., one physiological phase assigned to the entire volume) was rejected. Simulations showed that, when correcting 3D data, volume-specific regressors have similar performance compared to segment-specific regressors as long as the center of k-space is accurately corrected. Physiological regressions on in-vivo data showed that for a wide range of 3D acquisitions RetroICor performed as good as RetroKCor. It was found that the optimal model varies considerably between the different 3D acquisitions. The BIC-selection, which takes into account the loss in DOF, generally returned a small regressor set for both (with a maximum number of five regressors for our protocol). The exact regressor set varied between the acquisitions and depends on the contrast, readout, number of time points and region of interest.
Appendix A. Definition of the confound regressors
The cardiac phase, needed for the phase and interaction terms, was determined as follows:
| (A.1) |
where ttrig is the time since the last trigger and TRR the duration of the current RR interval.
The respiratory phase was calculated using the histogram-equalized transfer function as described in Glover et al. (2000). This approach takes into account the depth of breathing and distinguishes between inhalation and exhalation. The continuous respiratory waveform, R(t), is normalized to the maximum amplitude of inhalation, Rmax such that the range is (0,Rmax). From this normalized waveform a histogram, H(b), is calculated. For a given value of R(t) the respiratory phase is defined by the area under the histogram to the left of R(t) divided by the area under the entire histogram, and multiplied by the sign of dR/dt:
| (A.2) |
Using this approach, the end-expiration is assigned a phase of 0, whereas peak inspiration is assigned a value of ±π depending on whether we are at the end of inspiration or at the start of expiration.
Frequency regressors
The cardiac and respiratory frequency regressors are defined by the low order Fourier terms. The first order is the primary frequency, whereas the higher orders are also called the harmonics. Per order two regressors are generated, which are calculated by taking the sine and the cosine value for the current cardiac or respiratory phase. The signal that describes the cardiac (C[t]) and respiratory (R[t]) fluctuations can be written as the sum of the regressors:
| (A.3) |
Here, m and n are the order for the cardiac and respiratory waveforms, respectively. Here we have used: M ≤ 3 and N ≤ 4. The β’s are the individual amplitude of each of the regressors that are estimated by the linear regression.
Interaction regressors
The interaction terms describe the interactions between the cardiac and respiratory cycles (for example the effect of respiratory-induced pressure changes on the cardiac rhythm). Per order four regressors are created. The regressors are given by:
| (A.4) |
Again, β denotes the amplitude for each regressor that is estimated by the linear regression, and M and N represent the Fourier orders that are generated.
Cardiac and respiratory rate
Using the approach described Shmueli et al. (2007) the cardiac rate was calculated by taking the inverse of the period of each RR interval. This waveform was then smoothed with a moving average filter that spanned 10 s. The resulting signal was re-sampled at the acquisition rate of each k-space segment (in the case of RetroKCor) or each slice (in the case of RetroICor).
For respiratory, a regressor that takes into account the respiratory volume per time (RVT) was generated. The same procedure as described in Birn et al. (2006a) was followed. First, the amount of air inspired by each breath was calculated by taking the difference between the minimum and maximum peaks of the respiratory waveform. This difference is then divided by the duration of the respiratory cycle (i.e., the time between the peaks). The resulting waveform was smoothed with a moving average filter that spanned 10 s. For both these regressors the derivative was included to allow for temporal shifts of the regressors.
References
- Akaike H. A new look at the statistical model identification. IEEE Trans. Autom. Control. 1974;19(6):716–723. [Google Scholar]
- Barry RL, Strother SC, Gore JC. Complex and magnitude-only preprocessing of 2D and 3D BOLD FMRI data at 7 T. Magn. Reson. Med. 2012 Mar;67(3):867–871. doi: 10.1002/mrm.23072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth M, Meyer H, Kannengiesser SAR, Polimeni JR, Wald LL, Norris DG. T2-weighted 3D FMRI using S2-SSFP at 7 Tesla. Magn. Reson. Med. 2010 Apr;63(4):1015–1020. doi: 10.1002/mrm.22283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall EB. Adaptive cyclic physiologic noise modeling and correction in functional MRI. J. Neurosci. Methods. 2010 Mar;187(2):216–228. doi: 10.1016/j.jneumeth.2010.01.013. [DOI] [PubMed] [Google Scholar]
- Beckmann CF, Smith SM. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans. Med. Imaging. 2004 Feb;23(2):137–152. doi: 10.1109/TMI.2003.822821. [DOI] [PubMed] [Google Scholar]
- Bianciardi M, Fukunaga M, van Gelderen P, Horovitz SG, de Zwart JA, Shmueli K, Duyn JH. Sources of functional magnetic resonance imaging signal fluctuations in the human brain at rest: a 7 T study. Magn. Reson. Imaging. 2009 Oct;27(8):1019–1029. doi: 10.1016/j.mri.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birn RM, Diamond JB, Smith MA, Bandettini PA. Separating respiratory-variation-related fluctuations from neuronal-activity-related fluctuations in FMRI. NeuroImage. 2006a;31(4):1536–1548. doi: 10.1016/j.neuroimage.2006.02.048. [DOI] [PubMed] [Google Scholar]
- Birn RM, Murphy K, Bandettini P, Bodurka J. The use of multiple physiologic parameter regression increases gray matter temporal signal to noise by up to 50%; Proceedings 14th Scientific Meeting, International Society for Magnetic Resonance in Medicine; Seattle. 2006b. [Google Scholar]
- Bowen CV, Menon RS, Gati JS. High field balanced-SSFP FMRI: a BOLD technique with excellent tissue sensitivity and superior large vessel suppression; Proceedings of the 13th Annual Meeting of ISMRM; Miami. 2005.p. 119. [Google Scholar]
- Brooks JCW, Beckmann CF, Miller KL, Wise RG, Porro CA, Tracey I, Jenkinson M. Physiological noise modelling for spinal functional magnetic resonance imaging studies. NeuroImage. 2008;39(2):680–692. doi: 10.1016/j.neuroimage.2007.09.018. [DOI] [PubMed] [Google Scholar]
- Chang C, Cunningham JP, Glover GH. Influence of heart rate on the BOLD signal: the cardiac response function. NeuroImage. 2009 Feb;44(3):857–869. doi: 10.1016/j.neuroimage.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS, Killiany RJ. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage. 2006 Jul;31(3):968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Friese S, Hamhaber U, Erb M, Klose U. B-waves in cerebral and spinal cerebro-spinal fluid pulsation measurement by magnetic resonance imaging. J. Comput. Assist. Tomogr. 2004;28(2):255–262. doi: 10.1097/00004728-200403000-00016. [DOI] [PubMed] [Google Scholar]
- Friston K, Williams S, Howard R, Frackowiak R, Turner R. Movement-related effects in FMRI time-series. Magn. Reson. Med. 1996;35:346–355. doi: 10.1002/mrm.1910350312. [DOI] [PubMed] [Google Scholar]
- Glover GH, Li TQ, Ress D. Image-based method for retrospective correction of physiological motion effects in FMRI: RETROICOR. Magn. Reson. Med. 2000;44(1):162–167. doi: 10.1002/1522-2594(200007)44:1<162::aid-mrm23>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Golay X, Pruessmann KP, Weiger M, Crelier GR, Folkers PJ, Kollias SS, Boesiger P. PRESTO-SENSE: an ultrafast whole-brain FMRI technique. Magn. Reson. Med. 2000;43(6):779–786. doi: 10.1002/1522-2594(200006)43:6<779::aid-mrm1>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Hagberg GE, Bianciardi M, Brainovich V, Cassara AM, Maraviglia B. The effect of physiological noise in phase functional magnetic resonance imaging: from blood oxygen level-dependent effects to direct detection of neuronal currents. Magn. Reson. Imaging. 2008;26(7):1026–1040. doi: 10.1016/j.mri.2008.01.010. [DOI] [PubMed] [Google Scholar]
- Harvey AK, Pattinson KTS, Brooks JCW, Mayhew SD, Jenkinson M, Wise RG. Brainstem functional magnetic resonance imaging: disentangling signal from physiological noise. J. Magn. Reson. Imaging. 2008;28(6):1337–1344. doi: 10.1002/jmri.21623. [DOI] [PubMed] [Google Scholar]
- Hu X, Le TH, Parrish T, Erhard P. Retrospective estimation and correction of physiological fluctuation in functional MRI. Magn. Reson. Imaging. 1995;34(2):201–212. doi: 10.1002/mrm.1910340211. [DOI] [PubMed] [Google Scholar]
- Hutton C, Josephs O, Stadler J, Featherstone E, Reid A, Speck O, Bernarding J, Weiskopf N. The impact of physiological noise correction on FMRI at 7 T. NeuroImage. 2011 Jul;57(1):101–112. doi: 10.1016/j.neuroimage.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002 Oct;17(2):825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Tijssen R, Brooks J, Miller K. Voxel-wise regressor selection for physiological noise correction with RETROICOR; Proceedings 17th Scientific Meeting, International Society for Magnetic Resonance in Medicine; Honolulu. 2009.p. 1582. [Google Scholar]
- Jones TB, Bandettini PA, Birn RM. Integration of motion correction and physiological noise regression in FMRI. NeuroImage. 2008 Aug;42(2):582–590. doi: 10.1016/j.neuroimage.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorge J, Figueiredo P, van der Zwaag W, Marques JP. Signal fluctuations in FMRI data acquired with 2D-EPI and 3D-EPI at 7 Tesla. Magn. Reson. Imaging. 2013;31(2):212–220. doi: 10.1016/j.mri.2012.07.001. [DOI] [PubMed] [Google Scholar]
- Kass R, Raftery A. Tech. Rep. Unversity of Washington; 1993. Bayes factors and model uncertainty; p. 254. [Google Scholar]
- Lund TE, Madsen KH, Sidaros K, Luo W-L, Nichols TE. Non-white noise in FMRI: does modelling have an impact? NeuroImage. 2006;29(1):54–66. doi: 10.1016/j.neuroimage.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Lutti A, Thomas DL, Hutton C, Weiskopf N. High-resolution functional MRI at 3 T: 3D/2D echo-planar imaging with optimized physiological noise correction. Magn. Reson. Med. 2013;69(6):1657–1664. doi: 10.1002/mrm.24398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KL, Smith SM, Jezzard P, Pauly JM. High-resolution FMRI at 1.5 T using balanced SSFP. Magn. Reson. Med. 2006;55(1):161–170. doi: 10.1002/mrm.20753. [DOI] [PubMed] [Google Scholar]
- Miller KL, Smith SM, Jezzard P, Wiggins GC, Wiggins CJ. Signal and noise characteristics of SSFP FMRI: a comparison with GRE at multiple field strengths. NeuroImage. 2007;37(4):1227–1236. doi: 10.1016/j.neuroimage.2007.06.024. [DOI] [PubMed] [Google Scholar]
- Pfeuffer J, Van de Moortele P-F, Ugurbil K, Hu X, Glover GH. Correction of physiologically induced global off-resonance effects in dynamic echo-planar and spiral functional imaging. Magn. Reson. Med. 2002;47(2):344–353. doi: 10.1002/mrm.10065. [DOI] [PubMed] [Google Scholar]
- Shmueli K, van Gelderen P, de Zwart JA, Horovitz SG, Fukunaga M, Jansma JM, Duyn JH. Low-frequency fluctuations in the cardiac rate as a source of variance in the resting-state FMRI BOLD signal. NeuroImage. 2007 Nov;38(2):306–320. doi: 10.1016/j.neuroimage.2007.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, Johansen-Berg H, Bannister PR, Luca MD, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, Stefano ND, Brady JM, Matthews PM. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage. 2004;23(Suppl. 1) doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Stenger VA, Peltier S, Boada FE, Noll DC. 3D spiral cardiac/respiratory ordered FMRI data acquisition at 3 Tesla. Magn. Reson. Med. 1999 May;41(5):983–991. doi: 10.1002/(sici)1522-2594(199905)41:5<983::aid-mrm18>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Tijssen R, Jenkinson M, Jezzard P, Miller K. Characterizing physiological noise in the brainstem: passband SSFP vs. GRE-EPI; Proceedings 17th Scientific Meeting. International Society for Magnetic Resonance in Medicine; Honolulu. 2009.p. 1591. [Google Scholar]
- Tijssen RHN, Okell TW, Miller KL. Real-time cardiac synchronization with fixed volume frame rate for reducing physiological instabilities in 3D FMRI. NeuroImage. 2011 Jun;57(4):1364–1375. doi: 10.1016/j.neuroimage.2011.05.070. [DOI] [PubMed] [Google Scholar]
- Triantafyllou C, Hoge RD, Krueger G, Wiggins CJ, Potthast A, Wiggins GC, Wald LL. Comparison of physiological noise at 1.5 T, 3 T and 7 T and optimization of FMRI acquisition parameters. NeuroImage. 2005;26(1):243–250. doi: 10.1016/j.neuroimage.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Van de Moortele P, Pfeuffer J, Glover GH, Ugurbil K, Hu X. Respiration-induced B0 fluctuations and their spatial distribution in the human brain at 7 Tesla. Magn. Reson. Med. 2002;47(5):888–895. doi: 10.1002/mrm.10145. [DOI] [PubMed] [Google Scholar]
- Wise RG, Ide K, Poulin MJ, Tracey I. Resting fluctuations in arterial carbon dioxide induce significant low frequency variations in BOLD signal. NeuroImage. 2004 Apr;21(4):1652–1664. doi: 10.1016/j.neuroimage.2003.11.025. [DOI] [PubMed] [Google Scholar]