Abstract
Domain swapping that contributes to the stability of biologically crucial multisubunit complexes has been implicated in protein oligomerization. In the case of membrane protein assemblies, domain swapping of the iron–sulfur protein (ISP) subunit occurs in the hetero-oligomeric cytochrome b6f and bc1 complexes, which are organized as symmetric dimers that generate the transmembrane proton electrochemical gradient utilized for ATP synthesis. In these complexes, the ISP C-terminal predominantly β-sheet extrinsic domain containing the redox-active [2Fe-2S] cluster resides on the electrochemically positive side of each monomer in the dimeric complex. This domain is bound to the membrane sector of the complex through an N-terminal transmembrane α-helix that is “swapped’ to the other monomer of the complex where it spans the complex and the membrane. Detailed analysis of the function and structure of the b6f complex isolated from the cyanobacterium Fremyella diplosiphon SF33 shows that the domain-swapped ISP structure is necessary for function but is not necessarily essential for maintenance of the dimeric structure of the complex. On the basis of crystal structures of the cytochrome complex, the stability of the cytochrome dimer is attributed to specific intermonomer protein–protein and protein–lipid hydrophobic interactions. The geometry of the domain-swapped ISP structure is proposed to be a consequence of the requirement that the anchoring helix of the ISP not perturb the heme organization or quinone channel in the conserved core of each monomer.
Graphical abstract
Protein oligomerization under in vivo conditions is crucial for cellular survival1 as it leads to stability, enhanced allosteric control, increased availability of active sites, and formation of novel sites at intersubunit interfaces.1–3 Despite the significance of oligomerization to cellular physiology, the mechanism by which individual proteins assemble and interact to form a multisubunit complex remains enigmatic. Domain swapping provides a mechanism for achieving oligomerization and involves exchange of identical domains, or secondary and tertiary structure elements, between monomeric units to generate a higher-order assembly.2–4 Domain swapping can provide a physical connection between monomeric subunits, leading to the stabilization of a multisubunit oligomer. Domain swapping has also been implicated in disease development, especially in neurodegenerative disorders.5,6 Elucidation of the mechanism and effects of domain swapping is central to an understanding of the relation of structure to function in heterooligomeric membrane protein complexes.
Oligomerization of polypeptides into higher-order assemblies has been extensively reported for membrane protein complexes, including those involved in ion conductance,7,8 nutrient transport,9 cellular signaling,10–12 photosynthetic electron transfer,13–16 respiratory electron transfer,17–19 and ATP synthesis.20,21 Oligomerization is achieved mostly through limited exchange of structural elements between the monomeric subunits of membrane protein complexes. A unique case of oligomerization is presented by the membrane-associated dimeric cytochrome bc complexes (b6f of oxygenic photosynthesis and bc1 of anoxygenic photosynthesis and respiration) that catalyze quinone redox reactions during photosynthesis and respiration. These complexes consist of multiple subunits that are organized into the monomeric unit of the dimeric complex.22–31 In the case of the cytochrome b6f complex, eight distinct polypeptides (Figure 1A) and seven prosthetic groups (Figure 1B) have been resolved crystallographically in the monomer, for which a number of crystal structures have been published, the latest described in Protein Data Bank (PDB) entry 4OGQ.32 The complex contains four relatively large subunits, cytochrome b6 [cyt b subunit containing four transmembrane helices (TMH), “A–D”], subunit IV (subIV with three TMH, “E–G”), cytochrome f, and the iron–sulfur protein (cyt f and ISP, respectively, each with a single TMH and a large extrinsic domain). The complex also contains four small peripheral subunits, PetG, L, M, and N, each spanning the membrane as a single TMH. The dimer encloses an intermonomer cavity, which has been proposed to be involved in quinone/quinol capture from the lipid bilayer, and subsequent transfer to the quinol deprotonation–oxidation (Qp) site through a narrow, partially occluded channel.33,34 The intermonomer cavity is occupied by lipid molecules,32 which have been inferred to influence electron transfer through dielectric constant modulation within the transmembrane domain.35
Figure 1.
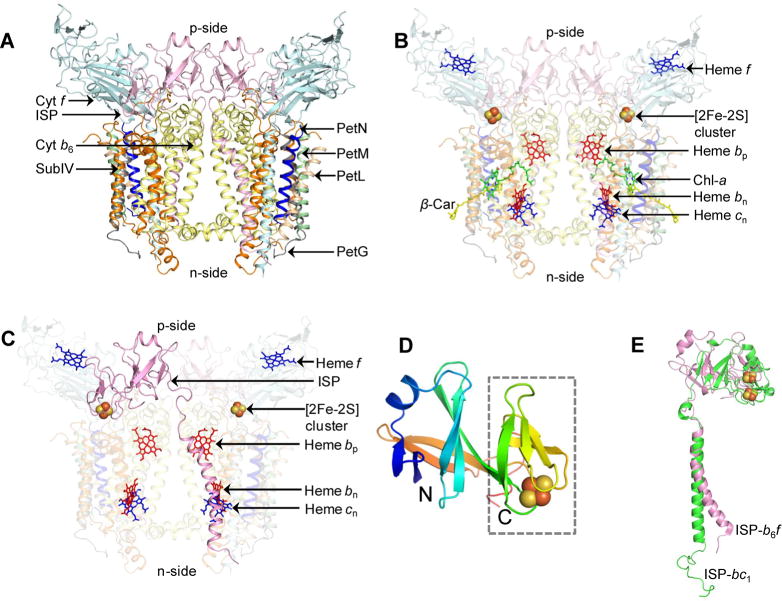
Cyt b6f complex of oxygenic photosynthesis (PDB entry 2E74). (A) Polypeptide composition of the cyt b6f complex. Polypeptide subunits are shown as ribbons. Color code: cytochrome b6 (cyt b6), yellow; subunit IV (subIV), orange; cytochrome f (cyt f), cyan; iron–sulfur protein (ISP), pink; PetG, gray; PetL, wheat; PetM, green; PetN, blue. (B) Prosthetic groups involved in electron transfer (hemes f, bp, bn, and cn and the [2Fe-2S] cluster). The photosynthetic pigments chlorophyll a (chl-a) and β-carotene (β-car) are colored green and yellow, respectively. (C) Domain swapping of the iron–sulfur protein (ISP). The ISP subunit (colored pink) provides a connection between the monomers of the dimeric cyt b6f complex (PDB entry 2E74). The extrinsic domain is associated with one monomer, while the transmembrane helix (TMH) interacts with the other monomer, to stabilize the dimer. For the sake of simplicity, the cyt b6f monomers are shown as semitransparent surfaces. (D) Extrinsic domain of the ISP subunit from spinach (PDB entry 1RFS). The polypeptide sequence is colored in a rainbow pattern, blue at the N-terminus to red at the C-terminus. The [2Fe-2S] cluster is shown as spheres (Fe, dark brown; S, light brown). The [2Fe-2S] cluster binding “small” subunit is outlined with a dashed box. (E) Difference in ISP TMH geometry. The TMH of cyt b6f ISP (pink) is bent, while the cyt bc1 ISP TMH (green) does not show any apparent bending.
In the b6f and bc1 complexes, the ISP subunit is organized in a transmonomer conformation by “domain swapping”, through which the ISP p-side extrinsic domain, which must undergo significant rotation and translation to accomplish competent electron transfer,36 is located proximal to one monomer, while its single TMH is associated with the other (Figure 1C). In the cyt b6f complex, electron transfer from plastoquinol to the ISP subunit leads to generation of a neutral semiplastoquinone and reactive oxygen species, which have been implicated in cellular signaling.37 The first structure of the ISP extrinsic domain in the cyt b6f complex [PDB entry 1RFS (Figure 1D)] was isolated from spinach thylakoids, crystallized, and resolved to a resolution of 1.83 Å.38 The C-terminal ISP extrinsic domain consists primarily of β-sheet and has a bipartite structure consisting of two distinct subdomains (Figure 1D), a [2Fe-2S] cluster binding “small” subdomain and a large subdomain that are distal and proximal, respectively, to the domain-swapped transmembrane α-helical domain (Figure 1C). The small subdomain, which provides ligation to the [2Fe-2S] cluster, shares structural homology with the corresponding subdomain in the related cyt bc1 ISP subunit and has a rubredoxin-like fold.38 The large subdomain, which connects to the TMH in the adjacent monomer, is not similar in structure to the ISP subunit of the bc1 complex.
The N-terminal ISP subunit TMH shows limited conservation of structure between the cyt b6f and bc1 complexes (Figure 1E, PDB entry 1Q90). In the cyt b6f complex, the ISP TMH shows a distinct bend induced by the presence of Pro37, as seen in the structure of the cyanobacterium Mastigocladus laminosus (PDB entry 2E74), and Pro56 in the Chlamydomonas reinhardtii complex. It is of interest to note that Pro37 in the cyanobacterial ISP structure is replaced by Gly63 in the C. reinhardtii structure, while Pro56 in the C. reinhardtii ISP structure is replaced by Gly28-Val29 in the M. laminosus structure. The functional significance of the bent ISP TMH remains unknown, although in the 2.5 Å structure of the cyt b6f complex (PDB entry 4OGQ),32 the ISP TMH is found to be connected to the TMH of the PetL subunit via an n-side lipidic molecule. PetL has previously been implicated in the stability and assembly of the b6f complex in the eukaryotic C. reinhardtii.39–42 The ISP-encoding petC gene is nuclear-encoded in eukaryotes, and the insertion and assembly of the petC gene product into a stable and functional complex may require additional factors, such as interaction with PetL. The bend in the ISP TMH would prevent potential steric clashes with the PetL helix and generates an n-side interhelix lipidic site, occupied by the detergent UDM, between the transmembrane helices of the ISP and PetL subunits (Supporting Information, Figure S1).
In the bc1 complex, the ISP extrinsic domain has been shown to undergo a large-scale motion relevant to electron transfer.43 Motion of the ISP extrinsic domain in the cyt b6f complex has been inferred from the crystallographic disorder of the domain,44 projection map analysis of the isolated dimer in the presence of a quinol analogue,45 and mutagenesis of the flexible hinge that connects the ISP extrinsic domain to the TMH.46,47 It has been suggested that domain swapping of the ISP subunit contributes to dimer stability,22,23 as loss of this subunit during detergent extraction and purification of the b6f complex from thylakoid membranes has been associated with monomerization and inactivation of the complex.24,39,48–50 Co-crystal structures of the b6f complex with the quinol analogue TDS (tridecyl-stigmatellin) (PDB entry 4H13) show that the electron donor, the [2Fe-2S] cluster, is separated from the electron acceptor, a ring of the heme of cytochrome f, by ∼28 Å.51 To transfer electrons from the bound quinol to the heme of the cyt f subunit, the ISP extrinsic domain must undergo motion from a quinol-proximal position to a heme f-proximal position,52 as documented crystallographically for the mitochondrial bc1 complex.36 Although tight binding of the ISP extrinsic domain to the b6f monomer at the quinol-proximal or heme f-proximal site would enhance dimer stabilization, a strong interaction of the ISP extrinsic domain with either docking site would interfere with ISP extrinsic domain motion. Contrary to the view presented previously,48 it is inferred from these studies that dimer stability in cyt bc complexes does not require a connection between the two monomers mediated by an intact ISP subunit and must involve intermonomer hydrophobic interactions other than those mediated by ISP domain swapping. In the study presented here, isolation and biochemical characterization of the cyt b6f complex from the cyanobacterium Fremyella diplosiphon SF33 are reported. It is demonstrated that the ISP subunit contributes minimally to the stabilization of the dimeric state of the cytochrome b6f complex. Analysis of the interactions that are significant for stabilization of the cyt b6f dimer in the context of the recent structure data has identified several residues in the transmembrane helices near the intermonomer interface that are well positioned to stabilize the dimeric structure, along with internal lipids that can have a role in the stabilization.
MATERIALS AND METHODS
Genetic Modification of the petA Gene
A PetA-His strain of F. diplosiphon SF33 that contained a petA gene with six histidine codons added in-frame after the final codon at the C-terminus (see Table S1 of the Supporting Information for details of primers) was obtained by genetic modification. This strain was created using plasmid pPetA-His. The pPetA-His insert was obtained by overlap polymerase chain reaction amplification as previously described,53 using the primer pairs petA-F-NcoI and petA-His-R3 for one fragment and petA-R-EcoRI and petA-His-F3 for the second fragment. The final fusion product was amplified using the primer pair petA-F-NcoI and petA-R-EcoRI, and the 2.9 kb product was cleaved with EcoRI and NcoI and then cloned into EcoRI- and NcoI-cut pJCF276.54 The insert and junctions of pPetA-His were verified by sequencing prior to conjugation into the cpc3 P+L strain of F. diplosiphon SF3355 as previously described.54,56 Colonies with chromosomally integrated pPetA-His were selected on 1% agar-Medium D (pH 8.0) containing 10 mg/mL neomycin. Transformed colonies were grown in 50 mL of BG-11 medium (pH 8.0) without selection and bubbled with air containing 3% CO2 while being illuminated by 10–15 mmol of photons of red light m−2 s−1 from LEDs (Digikey.com model 160-1415-2-ND). The culture was grown to an absorbance of 0.7 at 750 nm and then plated. The resulting colonies were sucrose-resistant and screened for allelic replacement.54,56 Genomic DNA from these colonies was sequenced to confirm the presence of a six-histidine-tagged petA gene. Colonies containing six-histidine-tagged petA were restreaked four times and resequenced to verify the presence of the six-histidine tag.
Cell Growth
Mutant F. diplosiphon SF33 with a six-histidine tag at the C-terminus of cyt f was grown in two phases, under different illumination conditions. A starter culture of 1 L was grown under red-white light (~100 μmol m−2 s−1) at 30 °C for ~7 days. The culture was used as an inoculum for a 10 L carboy that was grown for an additional 7–8 days under blue-white light (~100 μmol m−2 s−1), to an optical density of ~0.5 at 730 nm.
Membrane Preparation
A 350 mL bead-beater chamber was filled with ~175 mL of sterile glass beads (0.1 mm diameter). Cells from three carboys (~25–30 g of wet cell mass) were suspended in ~165 mL of a cell breakage solution containing 25 mM HEPES buffer (pH 7.5), 400 mM sucrose, 10 mM MgCl2, 10 mM CaCl2, and protease inhibitors (2 mM benzamidine, 2 mM 6-aminohexanoic acid, and 0.2 mM phenylmethanesulfonyl fluoride) and packed in the bead-beater chamber. Cells were broken by a short (~10 s) agitation followed by cooling (~6 min). The cycle was repeated 15 times. Cell debris was removed by centrifugation of the cell lysate at 2500g (4 °C). Thylakoid membranes were harvested from the supernatant by ultracentrifugation (300000g, 40 min, 4 °C).
Protein Purification and Characterization
Membranes were resuspended in an extraction buffer containing Tris-HCl (20 mM, pH 8.0), NaCl (50–100 mM), and protease inhibitors and extracted with UDM detergent [0.5% (w/v)] for 30 min at 4 °C in darkness at a chlorophyll a concentration of 2 mg/mL. The extract was clarified by centrifugation (300000g, 40 min, 4 °C), and the pH of the debris-free supernatant was re-equilibrated to 8.0 with Tris-HCl. The extract was passed through a 0.45 μm syringe filter and loaded under gravity on a 15 mL Ni-NTA metal affinity chromatography column equilibrated with extraction buffer supplemented with 0.05% UDM. The column was washed with the same buffer, and the protein was eluted with 300 mM imidazole dissolved in equilibration buffer (pH 8.0). The eluted protein was loaded on a 10 to 32% linear sucrose gradient prepared in Tris-HCl (30 mM, pH 7.5), NaCl (50 mM), EDTA (1 mM), UDM (0.05%), and protease inhibitors and centrifuged at 160000g for 12–16 h 4 °C to separate the dimer from the monomer. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) analysis of the purified protein was performed on a 12% polyacrylamide gel using standard procedures. Purification of the b6f complex from spinach and Nostoc PCC 7120 has been described previously.24,57 Genetic modification and purification of the cyt b6f complex from Synechocystis sp. PCC 6803 were performed following protocols described for Synechococcus sp. PCC 7002.58 Clear native gel electrophoresis was performed according to published protocols59 on a 4 to 12% gradient gel. Gel densitometry was performed using AlphaView software from ProteinSimple.60 Briefly, the intensity of bands corresponding to cyt f, cyt b6, ISP, and subIV was determined, followed by scaling to the corresponding molecular masses to obtain the subunit stoichiometry relative to cyt f. Two-dimensional (2D) clear native-PAGE was performed according to a protocol described elsewhere.61
Assay of Electron Transfer Activity
Thylakoid membranes and cytochrome b6f were isolated from F. diplosiphon, as described in this section. Cytochrome b6f oxidoreductase activity was measured at room temperature in a 20 mM Tricine-NaOH buffer (pH 8.0) containing 0.05% UDM. The reaction mixture containing 25 μM decyl-plastoquinone, 2.5 μM horse heart cytochrome c, and 250 μM potassium ferricyanide was initiated by addition of ~10 nM cytochrome b6f complex or thylakoids containing ~5 μg of chlorophyll. The reaction was monitored through the change in ferricyanide absorbance at 420 nm, relative to the background rate, based on a millimolar extinction coefficient ɛmM of 1.02 mM−1 cm−1.
Mass Spectrometric Analysis
The polypeptide composition of the Fremyella cytochrome b6f preparation was analyzed by liquid chromatography and mass spectrometry (Supporting Information, Figures S2–S9) with fraction collection as described previously (LC−MS+).62 Selected polypeptide masses were further analyzed by high-resolution Fourier transform mass spectrometry using collisionally activated or electron capture/transfer dissociation (CAD/ETD) as described previously.63 The high-resolution data were collected using positive ion nanoelectrospray ionization on ion cyclotron resonance/orbitrap Fourier transform instruments (Thermo Scientific) externally calibrated to yield data at mass accuracy tolerances of <15 ppm. Data were interpreted using Prosight PC 2.0 (Thermo Scientific) to transform raw files in to peak lists and match the resulting monoisotopic masses to protein structures with various post-translational modifications.64
Structure Studies of Intermonomer Contacts
Intermonomer interactions were identified in the recently published 2.5 Å crystal structure of the b6f complex (PDB entry 4OGQ),32 and the 1.9 Å crystal structure of the cyt bc1 complex (PDB entry 3CX5).31 The crystallographic dimers of the b6f and bc1 complex were analyzed for intersubunit interactions and interfacial area calculations using COCOMAPS.65 Residue pairs separated by more than 4 Å were classified as noninteracting.
Sequence Alignment
Multiple-sequence alignment was performed in Clustal-Omega, using default settings. NCBI accession numbers: cyt b6, Synechocystis sp. PCC 6803 (BAA10149.1), Synechococcus PCC 7002 (P28056.1), Nostoc PCC 7120 (BAB75120.1), M. laminosus (AAR26242.1), C. reinhardtii (CAA44690.1), Arabidopsis thaliana (NP_051088.1), and Spinacia oleracea (CAA30128.1); subIV, Synechocystis sp. PCC 6803 (BAA10150.1), Synechococcus PCC 7002 (YP_001734101.1), Nostoc PCC 7120 (NP_487462.1), M. laminosus (AAR26243.1), C. reinhardtii (CAA40030.1), A. thaliana (NP_051089.1), and S. oleracea (CAA30129.1); cyt b (bc1 complex), Rhodobacter sphaeroides (ABA80574.1), Rhodobacter capsulatus (ETE52795.1), Saccharomyces cerevisiae (NP_009315.1), Gallus gallus (BAE16037.1), and Bos taurus (AAZ95350.1). Sequence alignment figures were generated in ESPript 3.0 (http://espript.ibcp.fr/ESPript/ESPript/index.php).66 Assignment of secondary structure elements to cyt b6 and subIV polypeptide sequences utilized the cyt b6f complex structure (PDB entry 4OGQ) from Nostoc PCC 7120.32
Model Building of the ISP Extrinsic Domain
Structures of the cyt bc1 complex (PDB entries 1BE3 and 3CX5) were superimposed on a cyt b6f dimer structure (PDB entry 4H13) in PyMol (www.pymol.org) using the N-terminal domain of the cyt b polypeptide as a template. An additional finding is that the cyt b6f ISP extrinsic domain (PDB entry 4H13, chain D, Thr54–Val179) is more similar to the yeast ISP (PDB entry 3CX5, chain P, Val94–Gly215) than the bovine ISP (PDB entry 1BE3, chain E, Ile76–Gly196). The yeast ISP extrinsic domain was rotated and superposed onto the bovine ISP extrinsic domain in PyMol using the “align” command. The cyt b6f ISP extrinsic domain was then superposed on the rotated yeast cyt bc1 ISP extrinsic domain in PyMol. The angle of rotation of the cyt b6f ISP extrinsic domain was calculated in Chimera.67
RESULTS
Purification and Characterization of the F. diplosiphon SF33 Cyt b6f Complex
The petA gene, which encodes the cyt f polypeptide, was genetically modified in F. diplosiphon SF33, to include a C-terminal six-histidine affinity purification tag. Following mass cell culture and membrane preparation, detergent-mediated extraction of proteins was performed as described previously.24 Nonionic maltoside detergents have been demonstrated to keep the cyt b6f complex in a stable dimeric state.23,24 The detergent n-undecyl β-D-maltopyranoside (UDM), which was used for the successful purification and crystallization of the cyt b6f complex from Nostoc PCC 712024 and M. laminosus,22,49 was utilized for the extraction and purification of the b6f complex from F. diplosiphon SF33. A simple two-step procedure was followed for isolation of the complex. The clarified extract from UDM-solubilized thylakoid membranes was applied to a Ni affinity chromatography column to selectively purify the b6f complex, followed by sucrose density centrifugation of the eluted protein (Figure 2A) to separate the b6f dimer from the monomer. Approximately 65% of the purified b6f protein was found to be dimeric as quantitated from redox difference spectra.68 Fractions of purified monomer and dimer were resolved via clear native-PAGE to estimate their size. The dimer, presumably with bound detergent, migrated at ~300 kDa close to the spinach and Nostoc dimers (Figure 2B), whereas the migration of the monomer at ~150 kDa was similar to that of the b6f monomer of Synechocystis PCC 6803 (Figure 2B).
Figure 2.
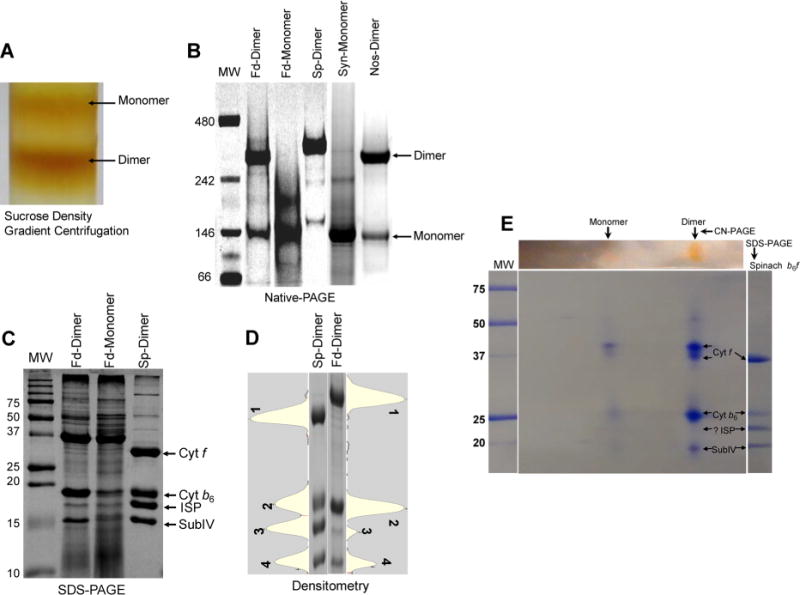
Biochemical characterization of the oligomeric state of the cyt b6f complex isolated from F. diplosiphon SF33. (A) Sucrose density gradient profile of the cyt b6f complex from F. diplosiphon SF33 eluted from a Ni affinity chromatography column, showing the presence of the monomer and dimer. (B) Native gel electrophoresis of the purified cyt b6f complex dimer (Fd-dimer) and monomer (Fd-monomer) from F. diplosiphon SF33. Purified cyt b6f from spinach (Sp-dimer), Synechocystis PCC 6803 (Syn-monomer), and Nostoc PCC 7120 (Nos-dimer) were used as controls for comparison. Molecular mass standards (MW, in kilodaltons) are also shown (lane 1). (C) Subunit composition of the cyt b6f complex analyzed by SDS–PAGE. F. diplosiphon SF33 dimer (Fd-dimer) and monomer (Fd-monomer) show prominent bands corresponding to three large subunits, i.e., cyt f, cyt b6, and subIV. The band corresponding to ISP was comparatively faint. Purified dimeric cyt b6f from spinach (Sp-dimer) was used as a standard for comparison. Molecular mass standards (MW, kilodaltons) are shown. (D) Densitometry profile of the four large cyt b6f subunits from spinach (Sp-dimer, lane 1) and F. diplosiphon SF33 dimer (Fd-dimer, lane 2). Cyt f, cyt b6, ISP, and subIV are labeled 1–4, respectively. The corresponding lane profiles are shown adjacent to the gel image. (E) Two-dimensional CN-PAGE of the cyt b6f dimer from F. diplosiphon SF33. Ten micrograms of the protein was resolved via 4 to 12% CN-PAGE in the first dimension followed by 12% SDS–PAGE in the second dimension. The gel was Coomassie-stained for visualization. Protein spots corresponding to cyt f, cyt b6, and subIV are visible. For reference, molecular mass standards (MW, kilodaltons) are shown in the left-most lane, and spinach cyt b6f is shown in the right-most lane.
The subunit composition of the two forms of the cyt b6f complex was analyzed by SDS–PAGE (Figure 2C). The purified dimeric b6f complex from spinach was utilized as a control for molecular mass estimation. All four “large” transmembrane subunits (cyt f, cyt b6, ISP, and subIV) were resolved by SDS–PAGE in both the dimer and monomer of the F. diplosiphon SF33 b6f complex (Figure 2C). However, the content of ISP in both dimer and monomer was substoichiometric in comparison to that of the spinach b6f complex (Figure 2C). A densitometric analysis of the purified spinach cyt b6f complex, normalized to the content of cyt f, showed an approximate unit stoichiometry (1:0.96:0.95:0.85 cyt f:cyt b6:ISP:subIV) (Figure 2D and Table 1). However, in the F. diplosiphon SF33 cyt b6f dimer, the ISP and subIV polypeptide subunits were approximately 17 and 45% of that of the cyt f content, respectively (Table1). The ISP subunit content of the purified dimeric and monomeric cyt b6f complex from F. diplosiphon SF33 membranes was further estimated by 2D-PAGE (Figure 2E). The 2D gel showed prominent bands corresponding to cyt f, cyt b6, and subIV, but the ISP band was found to have a significantly lower intensity (Figure 2E). This loss of ISP paralleled the decline in electron transport activity of the complex. The electron transport rate was 375 electrons monomer−1s −1 in the isolated thylakoids, whereas it was 94 electrons monomer−1 s−1 in the purified dimer, approximately 25% of the activity in thylakoids. This decrease in electron transport activity in the purified dimer is similar to the 17% reduction in ISP subunit stoichiometry relative to cytochrome f calculated via one-dimensional SDS–PAGE (Figure 2D and Table 1).
Table 1.
Subunit Stoichiometries in the Cyt b6f Complex from F. diplosiphon SF33 (dimer and monomer) and Spinacha
| cytochrome b6f complex subunit | Fremyella dimer | Fremyella monomer | spinach dimer |
|---|---|---|---|
| cyt f | 1.00 | 1.00 | 1.00 |
| cyt b6 | 1.09 | 0.69 | 0.96 |
| ISP | 0.17 | 0.21 | 0.95 |
| subIV | 0.45 | 0.29 | 0.85 |
The subunit stoichiometry was calculated from the intensities of the Coomassie-stained bands after SDS–PAGE. The values for cyt f were normalized to 1 for each analyzed lane.
Intact protein mass spectrometry of the F. diplosiphon SF33 cyt b6f complex eluted from the Ni affinity chromatography column was performed to determine polypeptide composition (Table 2). LC−MS+ analysis identified nine components (Table 2), including a 14579 Da species that was considerably more hydrophilic than the ISP subunit, which is the most hydrophilic member of the cyt b6f complex. The hydrophilic species was unequivocally identified as an N-terminally truncated extrinsic domain of the ISP subunit. The transmembrane helix of the ISP subunit was found to be absent from the identified species, as a consequence of cleavage between residues 41 and 42 (Supporting Information, Figure S9A,B), as reported previously for the cyt b6f complex isolated from M. laminosus.48 The truncated ISP extrinsic domain was found to have lost the [2Fe-2S] cluster during sample preparation, and consequently, the thiols became partially oxidized to disulfide bonds. The calculated mass of the truncated ISP extrinsic domain reflects a single disulfide, while the measured monoisotopic mass was lighter by 1 Da, presumably indicative of a mixture of one- and two-disulfide species. High-resolution PetG mass measurements were in full agreement with the F. diplosiphon SF33 gene sequences (Supporting Information, Figure S9C). As F. diplosiphon SF33 PetM and PetL sequences were unavailable, experimental data required manual assignment. For PetM, this was achieved by using Prosight PC version 2.0 (Thermo Scientific) to extract sequence tags from the data that were then screened against a database of cyanobacterial PetM sequences from the Uniprot protein database. Two sequence tags matched to Nostoc azollae PetM, and the high-resolution mass spectrometry data matched well to an N-formylated version of this sequence, confirming the 3578 Da small subunit as F. diplosiphon SF33 PetM (Supporting Information, Figure S9D). Two sequence tags were obtained using Prosight and by manual interpretation of the top-down data set for the 3255 Da species and were matched to the N. azollae PetL sequence, followed by manual sequence adjustment to achieve an optimal match of precursor and product ions, again with N-formylation of the initiating Met residue (Supporting Information, Figure S9E). It is significant to note the strong agreement of top-down mass spectrometry data with the sequence, which confirms the MS species as F. diplosiphon SF33 PetL, and rules out other assignments. PetN, the fourth peripheral transmembrane subunit, was also identified (Supporting Information, Figure S9F). In conclusion, the cyt b6f complex isolated from F. diplosiphon SF33 consists of eight subunits but shows evidence of the truncated ISP in a subpopulation of the isolated complex. Notably, because of the high sensitivity of the MS technique, the 14570 Da extrinsic domain of the ISP subunit could be detected in the mass spectrometry results of the isolated b6f complex after the Ni affinity chromatography purification.
Table 2.
Intact Protein Mass Spectrometry of the F. diplosiphon SF33 Cyt b6f Complex after Ni Affinity Chromatography
| retention time (min) | measured average molecular mass (Da) | calculated average molecular mass (Da) | measured monoisotopic molecular mass (Da) | modifications | subunit identity |
|---|---|---|---|---|---|
| 35.7 | 14578.1 | 14579.3917 | 14567.0896 | 42–179 | ISP (truncated) |
| 46.1 | 18939.5 | 18940.3345 | NA | 2–179, N-acetyl | ISP (full length) |
| 50.9 | 32743.0 | 32718.4635 | NA | 36–333 + six His, heme | cyt f (PetA)a |
| 53.4 | 31768.0 | 31748.4403 | NA | 36–332, heme | cyt f (without N333, His tag) |
| 84.0 | 4026.1 | 4025.7869 | 4023.1578 | N-formyl | PetG |
| 95.9 | 3578.0 | 3578.2265b | 3575.8351 | N-formyl | PetMc |
| 97.8 | 3254.6 | 3256.0387d | 3252.8471 | N-formyl | PetLd |
| 100 | 24772.6 | 24771.6082 | NA | N-acetyl, heme | cyt b6 (PetB) |
| 109 | 3261.5 | 3261.9616 | 3259.7588 | N-formyl | PetN |
| 110 | 17409.3 | 17409.7298 | NA | Met1 removed | PetD |
During the course of this analysis, the results presented in our 2002 paper were reviewed. The current spinach PetA sequence (P16013; September, 6, 2014) now yields full mass match with the same modifications we originally reported, further supporting the YP motif at the N-terminus of this protein.
Based upon the PetM sequence of N. azollae (strain 0708) (D7DYS4).
BLAST searches of a pair of sequence tags (FGLIFV and GALLLK) match various cyanobacterial PetM sequences from the Nostocales. D7DYS4 provided a full sequence in agreement with the low- and high-resolution mass spectrometry data.
Based upon the PetL sequence of N. azollae (strain 0708) (D7E059).
Protein–Protein Intermonomer Contacts within the Cyt b6f Dimer
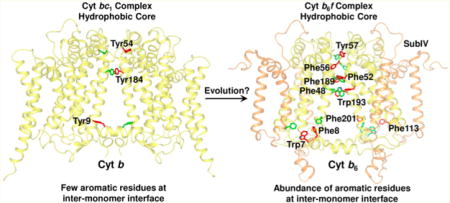
Crystal structures of the cyanobacterial cyt b6f complex have been obtained from M. laminosus (PDB entries 1VF5, 2D2C, 2E74, 2E75, 2E76, 4H13, and 4H0L)22,51,69,70 and Nostoc PCC 7120 (PDB entries 2ZT9 and 4H44),24,51 including a 2.5 Å structure (PDB entry 4OGQ).32 Multiple-sequence analysis of the core subunits, cyt b6 and subIV, of the b6f complex from M. laminosus, Nostoc PCC 7120, and F. diplosiphon SF33 showed sequence identities of 95.4 and 90.0% for cyt b6 and subIV, respectively (Supporting Information, Figures S10 and S11). From the high level of sequence identity, it is expected that structure features are conserved between the b6f complex of F. diplosiphon SF33 and of Nostoc PCC 7120 and M. laminosus. The cyt b6f crystal structure from Nostoc PCC 7120 (PDB entry 4OGQ) was analyzed to identify the major source of intermonomer contacts. The largest number of residues involved in intermonomer interactions is contributed by the cyt b6 polypeptide, whose four TM helices form a large fraction of the intermonomer interface, including 31 distinct cyt b6–cyt b6 interactions (Supporting Information, Table S2), and one pair of interacting residues between cyt b6 and subIV (Supporting Information, Table S2). It is significant to note that the intermonomer interface is enriched with aromatic residues (Figure 3A,B), which show significant conservation across prokaryotic and eukaryotic species (Supporting Information, Figures S10 and S11). In the cyt b6f complex from Nostoc PCC 7120 (PDB entry 4OGQ), 10 aromatic residues are inferred to provide stability to the dimer. In contrast, of a total of 21 residues of the cyt b polypeptide that constitute intermonomer interactions (Supporting Information, Table S3), only three aromatic residues (Tyr9, Tyr54, and Tyr184) are observed to provide intermonomer contacts in the related cyt bc1 complex (PDB entry 3CX5) (Figure 3C). Of the three aromatics involved in intermonomer interactions between the cyt b subunit of the bc1 complex, Tyr184 of the bc1 complex is substituted with Phe189 in the cyt b6 subunit of Nostoc PCC 7120 (Supporting Information, Figures S12 and S13).
Figure 3.
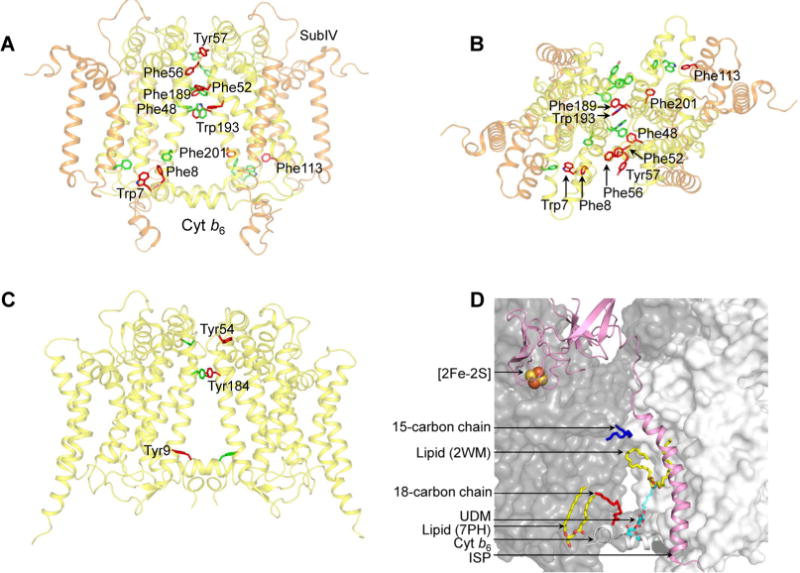
Cyt b6f complex (PDB entry 4OGQ) intermonomer interface. (A) Distribution of transmembrane aromatic residues that contribute to stabilization of the cyt b6f dimer core, which consists of cyt b6 (yellow) and subIV (orange). For the sake of simplicity, only the residues of one monomer (green) are labeled. Residues belonging to the other monomer (red) are not labeled. (B) View of the cyt b6f core along the axis of 2-fold rotational symmetry, normal to the membrane plane. (C) Distribution of transmembrane aromatic residues that contribute to stabilization of the cyt bc1 dimer core (PDB entry 3CX5), which consists of cyt b (yellow). For the sake of simplicity, only the residues of one monomer (green) are labeled. Residues belonging to the other monomer (red) are not labeled. (D) Lipid-mediated stabilization of the cyt b6f complex dimer core. In the 2.5 Å structure of the dimeric complex (PDB entry 4OGQ), four lipidic sites were identified at the intermonomer interface: a 15-carbon chain (dark blue), an 18-carbon chain (red), and two lipid sites (yellow and red). A fifth site, occupied by the detergent UDM (cyan and red), was previously reported in the M. laminosus cyt b6f complex (PDB entry 2E74). The ISP subunit is shown as a pink ribbon. The cyt b6f monomers are shown as white and gray surfaces.
The amino acid residues of the cyt b6 and subIV polypeptide that are inferred to provide stabilizing intermonomer contacts show strong conservation across species (Supporting Information, Figures S10 and S11), with three significant exceptions. In prokaryotic organisms, positions 48 and 193 of the cyt b6 subunit are occupied by aromatic residues Phe and Trp, respectively. However, in eukaryotic organisms, the residues are replaced with Val48 and Leu193, respectively. Position 201 of the cyt b6 polypeptide shows strong conservation only in eukaryotic organisms, where Met201 is conserved.
Protein–Lipid Interactions
The core of the cyt b6f complex dimer is composed of the cyt b6 polypeptide dimer.22–24 Apart from protein–protein interactions, the cyt b6 subunit also participates in protein–lipid interactions. In the 2.5 Å structure of the cyt b6f dimer (PDB entry 4OGQ), a total of four lipidic sites were located at the monomer–monomer interface (Figure 3D). On the n-side of the b6f complex (PDB entry 2E74),70 a crystallographically ordered UDM detergent molecule is found to provide cross-linking interactions between the two monomers (Figure 3D). It has been proposed that crystallographically ordered detergent molecules reside at physiologically relevant lipid binding sites71,72 and inferred that the dimer of the b6f complex is stabilized by lipidic sites. The lipid molecules within the intermonomer cavity have been inferred to provide directionality to transmembrane electron transfer.35
Analysis of the ISP Subunit Structure
Motion of the ISP extrinsic domain has been previously reported in the related cyt bc1 complex.43 In the heme c1-proximal position of the cyt bc1 ISP extrinsic domain (PDB entry 1BE3), the [2Fe-2S] cluster is located within 11 Å of the heme c1 ring. A rotation of ∼57° of the ISP extrinsic domain from the cyt b-proximal position orients the [2Fe-2S] cluster into a position to donate an electron to the heme of cytochrome c1.43 A continuous diffusion-based motion of the ISP extrinsic domain between positions proximal to cyt b and cyt c1 was inferred from a steered molecular dynamics study of the cyt bc1 complex.73 In the absence of structural data to define a heme f-proximal state of the ISP subunit in the cyt b6f complex, a model was generated using the heme c1-proximal orientation of the cyt bc1 ISP subunit. (PDB entry 1BE3, Figure 4A,B). The heme f-proximal state was achieved by a 74.5° rigid-body rotation of the ISP extrinsic domain from the cyt b-proximal position (PDB entry 4H13). In the heme f-proximal state, the [2Fe-2S] cluster is located within 14.5 Å of the heme f porphyrin ring and 20.5 Å from the quinol analogue TDS bound within the quinol oxidation site (Figure 4C).
Figure 4.
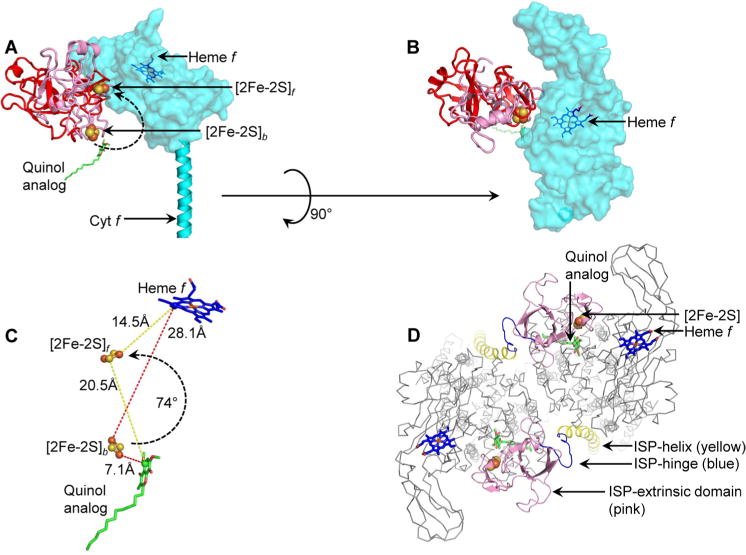
Model of ISP extrinsic domain movement in the cyt b6f complex. (A) The ISP extrinsic domain (pink, PDB entry 4H13) was observed crystallographically in a position proximal to the quinol oxidation site, where the [2Fe-2S] cluster is labeled [2Fe-2S]b. Model of the ISP extrinsic domain in the heme f-proximal position ([2Fe-2S]f). The model is colored red to distinguish it from the crystallographically observed [2Fe-2S]b position. The curved arrow shows the 74.5° rotation of the ISP domain to move the cluster from the [2Fe-2S]b position to the [2Fe-2S]f position. The quinol analogue TDS is shown for reference. The cyt f extrinsic domain is shown as a semitransparent surface for the sake of clarity of the ISP extrinsic domains. (B) View of ISP and cyt f extrinsic domains, rotated by 90° from the view in panel A. The ISP subunit extrinsic domain shows minimal overlap with the cyt f extrinsic domain, indicating limited steric clashes. (C) Distance of the ISP [2Fe-2S] cluster from the electron donor (represented by the quinol analogue TDS, green and red sticks) and electron acceptor heme f (blue and red sticks). The donor–acceptor distances of [2Fe-2S]b and [2Fe-2S]f positions are shown with red and yellow dashes, respectively. (D) Extrinsic packing of the ISP subunit in the cyt b6f complex (view along the normal to the membrane). The N-terminal ISP transmembrane helix (Asp9–Phe42) is colored yellow, the hinge blue (Ile43–Ala56), and the C-terminal extrinsic domain pink (Lys57–Ser179). As a reference for location of the p-side quinol binding site, a quinol analogue inhibitor (TDS) is shown as green and red sticks. The ISP [2Fe-2S] cluster is shown as brown and yellow spheres and heme f as blue and red sticks. For the sake of simplicity, other subunits are shown as thin gray wires (“ribbon” representation in PyMol).
It is significant to note that the flexible linker that connects the ISP TMH to the ISP extrinsic domain is a poly-Gly tract in the b6f complex, which is devoid of well-defined secondary structure. In the context of the cyt bc1 complex, it has been suggested that the ISP linker may undergo an order–disorder transition, which determines the position of the ISP extrinsic domain.74–76 Docking of the bc1 ISP extrinsic domain proximal to the Qp site has been suggested to involve loss of secondary structure of the linker. Similarly, the ISP extrinsic domain of the cyt b6f complex has been observed to be located proximal to the Qp site when the site is unoccupied (PDB entries 2E74, 2ZT9, 4H44, and 4OGQ), which may be attributed to the absence of defined secondary structure in the linker.
The cyt f extrinsic domain consists of an extended, 75 Å long β-sheet-based arrangement, which is closely wrapped around the ISP extrinsic domain.22,23,77 The model of the heme f-proximal state of the ISP subunit, achieved through a rigid-body rotation of 74.5°, does not show significant steric clashes with the cyt f extrinsic domain, indicating that an ISP extrinsic domain rotation similar to the domain motion reported in the cyt bc1 complex is plausible in the b6f complex. It is noted that in the cyt b6f complex, the propionate groups of the c-type heme of the cyt f extrinsic domain point away from the ISP binding interface (Figure 4B), which is different from the organization of the high-potential chain in the cyt bc1 complex, where the propionate groups of the c-type heme of the cyt c1 subunit are oriented toward the ISP binding interface.43,75
The view orthogonal to the membrane plane of the peripheral arrangement of the N-terminal TMH of the ISP in the second monomer (Figure 4D) shows that an anchor function is achieved with minimal perturbation of the functional domains of the ISP subunit.
DISCUSSION
The role of the ISP subunit in stabilization of the cyt b6f complex dimer in the context of the 2.5 Å structure (PDB entry 4OGQ) was investigated using a two-step purification scheme. The petA gene encoding the cyt f subunit was genetically manipulated in the cyanobacterium F. diplosiphon SF33, to introduce a C-terminal six-histidine purification tag. Extraction of protein from isolated thylakoid membranes was performed using the neutral maltoside detergent UDM, which has previously been used for the successful isolation of dimeric cyt b6f from Nostoc PCC 7120.68,78 The complex was purified using metal affinity Ni-NTA chromatography, and the dimer fraction was separated from the monomer fraction by sucrose density gradient centrifugation. It was observed that the isolated dimeric b6f complex from F. diplosiphon SF33 contains a substoichiometric amount of ISP, as inferred from SDS–PAGE analysis. Therefore, the loss of the ISP subunit may not be the primary determinant of dimer to monomer conversion of the cyt b6f complex. In this context, it is significant to note the isolation of a stable cytochrome b6f dimer devoid of ISP.39,79
Structural Basis of Dimer Stability: Role of Aromatic Residues
Analysis of the cyt b6f structure (PDB entry 4OGQ) provides a basis for the structural stability of the ISP-devoid cyt b6f dimer. The cyt b6f structure shows that the intermonomer interface consists primarily of the cyt b6 polypeptide within the transmembrane domain (Figure 3A,B). A total of 31 cyt b6 residues from TMH A, B, and D and the ab and cd loops contribute to intermonomer interactions. It is significant that the cyt b6 polypeptide, which not only shows strong conservation of sequence in organisms that perform oxygenic photosynthesis but also shares conserved residues and structure-related features with the cyt b subunit N-terminal domain of the bc1 complex,80 shows limited conservation of critical aromatic amino acids that are inferred to provide intermonomer contacts (Supporting Information, Figure S12). From the analysis presented here, it is inferred that evolution from the cyt bc1 complex of anoxygenic photosynthesis and respiration to the cyt b6f complex involved a transition in the sequence of the core cyt b subunit to favor aromatic residues for dimer stabilization in the cyt b6f complex.
Structural Basis of Dimer Stability: Role of Lipids
The intermonomer contacts within the transmembrane domain of the cyt b6f complex are further strengthened by lipid–protein interactions at the intermonomer interface (Figure 3D). As discussed elsewhere,81,82 the cytochrome b6f complex structure consists of an inner polytopic core domain, and a peripheral monotopic domain. Lipidic sites between the core and peripheral domains have been noted, as well as lipidic sites at the intermonomer interface. Hence, lipids may be involved in the assembly of the core domain of the cyt b6f complex dimer, and in stabilization of the assembled dimer.
Formation of the polytopic core during initial steps of assembly may be a feature that is conserved within the cyt bc complexes.82 The isolation of a cyt b–cyt c1 subcomplex of the homologous cyt bc1 complex has been reported from the photosynthetic bacterium R. capsulatus.83 The absence of the ISP does not cause destabilization of the subcomplex, and augmentation with purified ISP restores the activity of the complex, providing evidence of association of the single helix peripheral subunits after the formation of a polytopic nucleus.
Evolutionary Optimization of the ISP Position
The ISP extrinsic domain must undergo motion to transfer an electron from the bound quinol to a high-potential p-side heme (of cyt f in b6f, and of cyt c1 in the bc1 complex). Key requirements for the positioning of the ISP subunit within the cyt bc complex must be (i) the proximity of the ISP extrinsic domain to the Qp site and the high-potential heme (of cyt f and cyt c1) and (ii) the lack of occlusion of the Qp channel by the ISP TMH. In this study, it is inferred that the ISP extrinsic domain may undergo a simple rotation from the quinol-proximal position to the heme f-proximal position (Figure 4A,C). Such a rigid-body motion would involve minimal steric clashes with the surrounding p-side protein environment of the cyt b6 cd loop and the cyt f extrinsic domain, thereby reducing the energetic cost of ISP motion during catalysis. Moreover, the ISP TMH is distal in location from the p-side quinol channel, which prevents steric hindrance for quinol diffusion. In view of requirements (i) and (ii) discussed above, an analysis (points a–d below) of the cyt bc complex structures shows that the position of the ISP subunit may have been optimized during evolution.
It has been suggested that the intermonomer cavity may function in quinone/quinol exchange.33 If the ISP subunit TMH were located within the intermonomer cavity, it would interfere with the capture of quinone/quinol (Figure 5A).
The location of the ISP subunit within the same monomer would require positioning of the ISP TMH between helices F and G of the subIV polypeptide of cyt b6f (Figure 5B). However, in such an orientation, the ISP TMH would be almost normal to the membrane plane. Electron transfer from the bound quinol to the [2Fe-2S] cluster would require extensive bending of the ISP extrinsic domain upon the flexible hinge toward the bound p-side quinol. It has been suggested that interaction with the cd loop is essential for ISP function in quinol oxidation.84 However, if the ISP TMH were located between TMH F and G, then significant steric hindrance would be imposed on ISP extrinsic domain motion, especially because of clashes with the cd loop of the cyt b6 subunit (Figure 5B). Moreover, the space between TMH F and G is occupied by helix H of the cyt b subunit in the cyt bc1 complex.23,81,85
To avoid the need for domain swapping, the ISP subunit might be placed within the same monomer of cyt bc complexes (Figure 5C). Rotation of the ISP TMH along the axis normal to the membrane places the ISP extrinsic domain within the same monomer (Figure 5C). In such a situation, the ISP TMH still does not cause steric interference with quinol diffusion. However, the ISP extrinsic domain would require a significantly elongated hinge to reach the Qp site on the same monomer. Moreover, extensive steric clashes would occur between the ISP extrinsic domain and the cd loop of cyt b6 (Figure 5C).
The final possibility may be to place the ISP subunit proximal to the PetG-L-M-N peripheral four-helix bundle on the cyt b6f complex (Figure 5D). However, the p-side extramembrane space proximal to the PetG-L-M-N bundle is occupied by the extrinsic domain of the cyt f polypeptide. Insertion and motion of the ISP subunit in the peripheral position would involve extensive clashes of the N-terminal portion of the ISP extrinsic domain with the cyt f subunit, and of the [2Fe-2S] cluster containing the C-terminal ISP extrinsic domain with the ab and cd loops (cyt b6) and ef loops (subIV) (Figure 5D).
Figure 5.
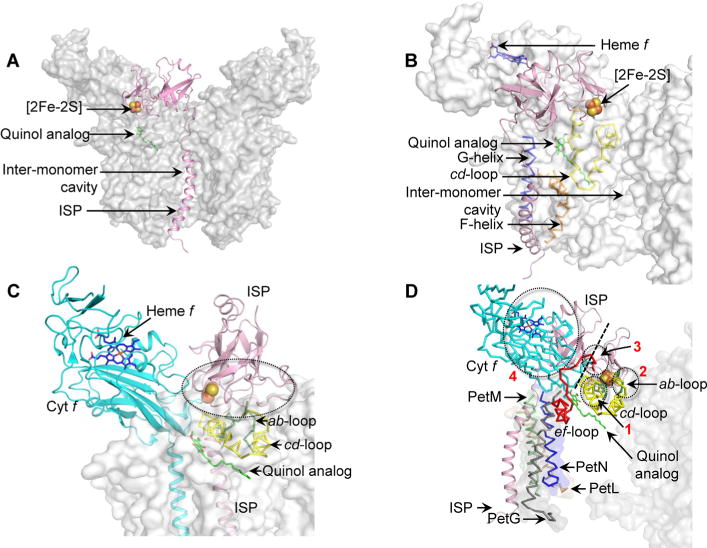
Models of steric hindrance in ISP interactions when ISP is inserted in a non-native position within the cyt b6f complex (PDB entry 4OGQ). (A) Location of the ISP TMH (pink) within the intermonomer cavity partially occludes the quinone/quinol exchange function of the cavity. (B) Insertion of the ISP TMH (pink) between the TMH F (orange ribbon) and G (blue ribbon) leads to extensive clashes between the ISP extrinsic domain and the cd loop (yellow ribbon) of cyt b6. (C) The ISP subunit within the same monomer of the cyt b6f complex orients the ISP extrinsic domain within unfavorable proximity of the cd loop (yellow), which leads to extensive clashes between the extrinsic domain and the cd loop. Regions of steric clashes are highlighted with a dotted oval. (D) Placement of the ISP TMH proximal to the PetG-L-M-N subunits located on the periphery of the cyt b6f complex. In such a location, the TMH-proximal N-terminal ISP subdomain undergoes extensive clashes with the cyt f extrinsic domain (cyan), while the distal, [2Fe-2S] binding C-terminal ISP subdomain clashes with the ab loop (green), the cd loop (yellow), and the ef loop (red). Regions of steric clashes are highlighted by three dotted ovals (1, clash with cd loop; 2, clash with ab and cd loops; 3, clash with ef loop; 4, clash with cyt f). A dotted black line separates the N-terminal, helix-proximal subdomain and the C-terminal cluster binding subdomain. For the sake of simplicity, the cyt b6f complex is shown as a gray surface. The Qp site is marked by the quinol analogue TDS molecule (green and red sticks, superposed from PDB entry 2E76). Loops ab (green, Phe56–Asn77), cd (yellow, Ser137–Gln177), and ef (red, Pro65–Asn93) are shown in the PyMol “ribbon” format, while the ISP and cyt f subunits are depicted in the PyMol “cartoon” format.
Hence, the orientation of the ISP subunit presented here may be optimal for functioning in electron transfer and minimal interference with quinol/quinone diffusion.
Supplementary Material
Acknowledgments
Funding: This study was supported by National Institutes of Health (NIH) Grant GM-038323 and the Henry Koffler Professorship (W.A.C.), core support from the UCSD/UCLA National Institute of Diabetes and Digestive and Kidney Diseases Diabetes Research Center Grant P30 DK063491 (J.P.W.), and a graduate research fellowship from Purdue University (S.S.H.). Infrastructure support was facilitated by NIH Center Grant P30 CA023168.
ABBREVIATIONS
- cyt
cytochrome
- ISP
iron–sulfur protein
- PET
photosynthetic electron transport
- TMH
transmembrane helix
- UDM
n-undecyl β-D-maltopyranoside
Footnotes
Supporting Information
Primer sequences for genetic manipulation (Table S1), intermonomer interactions within the cyt b6f dimer (Table S2), intermonomer interactions within the cyt bc1 dimer (Table S3), bend in the ISP subunit TMH (Figure S1), results of mass spectroscopy (low resolution) of truncated ISP (Figure S2), results of mass spectroscopy (high resolution) of truncated ISP (Figure S3), results of mass spectroscopy (low resolution) of full-length ISP (Figure S4), results of mass spectroscopy (high resolution) of PetG (Figure S5), results of mass spectroscopy (high resolution) of PetM (Figure S6), results of mass spectroscopy (high resolution) of PetL (Figure S7), results of mass spectroscopy (high resolution) of PetN (Figure S8), results of mass spectroscopy (high resolution) of small subunits and the ISP proteolysis site (Figure S9), sequence alignment of the cyt b6 subunit (Figure S10), sequence alignment of the subIV polypeptide (Figure S11), sequence alignment of cyt b6 (b6f) and cyt b (bc1) subunits (Figure S12), and sequence alignment of subIV (b6f) and cyt b (bc1) subunits (Figure S13). The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.5b00279.
Notes
The authors declare no competing financial interest.
References
- 1.Ali MH, Imperiali B. Protein oligomerization: How and why. Bioorg Med Chem. 2005;13:5013–5020. doi: 10.1016/j.bmc.2005.05.037. [DOI] [PubMed] [Google Scholar]
- 2.Liu Y, Eisenberg D. 3D domain swapping: As domains continue to swap. Protein Sci. 2002;11:1285–1299. doi: 10.1110/ps.0201402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett MJ, Schlunegger MP, Eisenberg D. 3D domain swapping: A mechanism for oligomer assembly. Protein Sci. 1995;4:2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett MJ, Choe S, Eisenberg D. Domain swapping: Entangling alliances between proteins. Proc Natl Acad Sci USA. 1994;91:3127–3131. doi: 10.1073/pnas.91.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Wel PC. Domain swapping and amyloid fibril conformation. Prion. 2012;6:211–216. doi: 10.4161/pri.18987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Hoop CL, Kodali R, Sivanandam VN, van der Wel PC. Amyloid-like fibrils from a domain-swapping protein feature a parallel. J Biol Chem. 2011;286:28988–28995. doi: 10.1074/jbc.M111.261750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.MacKinnon R. Determination of the subunit stoichiometry of a voltage-activated potassium channel. Nature. 1991;350:232–235. doi: 10.1038/350232a0. [DOI] [PubMed] [Google Scholar]
- 8.Dementieva IS, Tereshko V, McCrossan ZA, Solomaha E, Araki D, Xu C, Grigorieff N, Goldstein SA. Pentameric assembly of potassium channel tetramerization domain-containing protein 5. J Mol Biol. 2009;387:175–191. doi: 10.1016/j.jmb.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veenhoff LM, Heuberger EH, Poolman B. Quaternary structure and function of transport proteins. Trends Biochem Sci. 2002;27:242–249. doi: 10.1016/s0968-0004(02)02077-7. [DOI] [PubMed] [Google Scholar]
- 10.Bouvier M. Oligomerization of G-protein-coupled transmitter receptors. Nat Rev Neurosci. 2001;2:274–286. doi: 10.1038/35067575. [DOI] [PubMed] [Google Scholar]
- 11.Clayton AH, Walker F, Orchard SG, Henderson C, Fuchs D, Rothacker J, Nice EC, Burgess AW. Ligand-induced dimer-tetramer transition during the activation of the cell surface epidermal growth factor receptor: A multidimensional microscopy analysis. J Biol Chem. 2005;280:30392–30399. doi: 10.1074/jbc.M504770200. [DOI] [PubMed] [Google Scholar]
- 12.Ullrich A, Schlessinger J. Signal transduction by receptors with tyrosine kinase activity. Cell. 1990;61:203–212. doi: 10.1016/0092-8674(90)90801-k. [DOI] [PubMed] [Google Scholar]
- 13.Newman PJ, Sherman LA. Isolation and characterization of photosystem I and II membrane particles from the blue-green alga, Synechococcus cedrorum. Biochim Biophys Acta. 1978;503:343–361. doi: 10.1016/0005-2728(78)90193-7. [DOI] [PubMed] [Google Scholar]
- 14.Ford RC, Holzenburg A. Investigation of the structure of trimeric and monomeric photosystem I reaction centre complexes. EMBO J. 1988;7:2287–2293. doi: 10.1002/j.1460-2075.1988.tb03071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogner M, Dekker JP, Boekema EJ, Witt HT. Size, shape and mass of the oxygen-evolving photosystem-II complex from the thermophilic cyanobacterium Synechococcus sp. FEBS Lett. 1987;219:207–211. [Google Scholar]
- 16.Huang D, Everly RM, Cheng RH, Heymann JB, Schagger H, Sled V, Ohnishi T, Baker TS, Cramer WA. Characterization of the chloroplast cytochrome b6f complex as a structural and functional dimer. Biochemistry. 1994;33:4401–4409. doi: 10.1021/bi00180a038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sone N, Takagi T. Monomer-dimer structure of cytochrome-c oxidase and cytochrome bc1 complex from the thermophilic bacterium PS3. Biochim Biophys Acta. 1990;1020:207–212. doi: 10.1016/0005-2728(90)90052-6. [DOI] [PubMed] [Google Scholar]
- 18.Leonard K, Wingfield P, Arad T, Weiss H. Three-dimensional structure of ubiquinol:cytochrome c reductase from Neurospora mitochondria determined by electron microscopy of membrane crystals. J Mol Biol. 1981;149:259–274. doi: 10.1016/0022-2836(81)90301-6. [DOI] [PubMed] [Google Scholar]
- 19.Montoya G, te Kaat K, Rodgers S, Nitschke W, Sinning I. The cytochrome bc1 complex from Rhodovulum sulfidophilum is a dimer with six quinones per monomer and an additional 6-kDa component. Eur J Biochem. 1999;259:709–718. doi: 10.1046/j.1432-1327.1999.00094.x. [DOI] [PubMed] [Google Scholar]
- 20.Arnold I, Pfeiffer K, Neupert W, Stuart RA, Schagger H. Yeast mitochondrial F1F0-ATP synthase exists as a dimer: Identification of three dimer-specific subunits. EMBO J. 1998;17:7170–7178. doi: 10.1093/emboj/17.24.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies KM, Anselmi C, Wittig I, Faraldo-Gomez JD, Kuhlbrandt W. Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc Natl Acad Sci USA. 2012;109:13602–13607. doi: 10.1073/pnas.1204593109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurisu G, Zhang H, Smith JL, Cramer WA. Structure of the cytochrome b6f complex of oxygenic photosynthesis: Tuning the cavity. Science. 2003;302:1009–1014. doi: 10.1126/science.1090165. [DOI] [PubMed] [Google Scholar]
- 23.Stroebel D, Choquet Y, Popot JL, Picot D. An atypical haem in the cytochrome b6f complex. Nature. 2003;426:413–418. doi: 10.1038/nature02155. [DOI] [PubMed] [Google Scholar]
- 24.Baniulis D, Yamashita E, Whitelegge JP, Zatsman AI, Hendrich MP, Hasan SS, Ryan CM, Cramer WA. Structure-function, stability, and chemical modification of the cyanobacterial cytochrome b6f complex from Nostoc sp. PCC 7120. J Biol Chem. 2009;284:9861–9869. doi: 10.1074/jbc.M809196200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia D, Yu CA, Kim H, Xia JZ, Kachurin AM, Zhang L, Yu L, Deisenhofer J. Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science. 1997;277:60–66. doi: 10.1126/science.277.5322.60. [DOI] [PubMed] [Google Scholar]
- 26.Iwata S, Lee JW, Okada K, Lee JK, Iwata M, Rasmussen B, Link TA, Ramaswamy S, Jap BK. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science. 1998;281:64–71. doi: 10.1126/science.281.5373.64. [DOI] [PubMed] [Google Scholar]
- 27.Hunte C, Koepke J, Lange C, Rossmanith T, Michel H. Structure at 2.3 Å resolution of the cytochrome bc1 complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment. Structure. 2000;8:669–684. doi: 10.1016/s0969-2126(00)00152-0. [DOI] [PubMed] [Google Scholar]
- 28.Kleinschroth T, Castellani M, Trinh CH, Morgner N, Brutschy B, Ludwig B, Hunte C. X-ray structure of the dimeric cytochrome bc1 complex from the soil bacterium Paracoccus denitrificans at 2.7-Å resolution. Biochim Biophys Acta. 2011;1807:1606–1615. doi: 10.1016/j.bbabio.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 29.Berry EA, Huang LS, Saechao LK, Pon NG, Valkova-Valchanova M, Daldal F. X-ray structure of Rhodobacter capsulatus cytochrome bc1: Comparison with its mitochondrial and chloroplast counterparts. Photosynth Res. 2004;81:251–275. doi: 10.1023/B:PRES.0000036888.18223.0e. [DOI] [PubMed] [Google Scholar]
- 30.Berry EA, Guergova-Kuras M, Huang LS, Crofts AR. Structure and function of cytochrome bc complexes. Annu Rev Biochem. 2000;69:1005–1075. doi: 10.1146/annurev.biochem.69.1.1005. [DOI] [PubMed] [Google Scholar]
- 31.Solmaz SR, Hunte C. Structure of complex III with bound cytochrome c in reduced state and definition of a minimal core interface for electron transfer. J Biol Chem. 2008;283:17542–17549. doi: 10.1074/jbc.M710126200. [DOI] [PubMed] [Google Scholar]
- 32.Hasan SS, Cramer WA. Internal lipid architecture of the hetero-oligomeric cytochrome b6f complex. Structure. 2014;22:1008–1015. doi: 10.1016/j.str.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cramer WA, Zhang H, Yan J, Kurisu G, Smith JL. Trans-membrane traffic in the cytochrome b6f complex. Annu Rev Biochem. 2006;75:769–790. doi: 10.1146/annurev.biochem.75.103004.142756. [DOI] [PubMed] [Google Scholar]
- 34.Hasan SS, Proctor EA, Yamashita E, Dokholyan NV, Cramer WA. Traffic within the cytochrome b6f lipoprotein complex: Gating of the quinone. Biophys J. 2014;107:1620–1628. doi: 10.1016/j.bpj.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hasan SS, Zakharov SD, Chauvet A, Stadnytskyi V, Savikhin S, Cramer WA. A map of dielectric heterogeneity in a membrane protein: The hetero-oligomeric cytochrome b6f complex. J Phys Chem B. 2014;118:6614–6625. doi: 10.1021/jp501165k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z, Huang L, Shulmeister VM, Chi YI, Kim KK, Hung LW, Crofts AR, Berry EA, Kim SH. Electron transfer by domain movement in cytochrome bc1. Nature. 1998;392:677–684. doi: 10.1038/33612. [DOI] [PubMed] [Google Scholar]
- 37.Baniulis D, Hasan SS, Stofleth JT, Cramer WA. Mechanism of enhanced superoxide production in the cytochrome b6f complex of oxygenic photosynthesis. Biochemistry. 2013;52:8975–8983. doi: 10.1021/bi4013534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carrell CJ, Zhang H, Cramer WA, Smith JL. Biological identity and diversity in photosynthesis and respiration: Structure of the lumen-side domain of the chloroplast Rieske protein. Structure. 1997;5:1613–1625. doi: 10.1016/s0969-2126(97)00309-2. [DOI] [PubMed] [Google Scholar]
- 39.Breyton C, Tribet C, Olive J, Dubacq JP, Popot JL. Dimer to monomer conversion of the cytochrome b6f complex. Causes and consequences. J Biol Chem. 1997;272:21892–21900. doi: 10.1074/jbc.272.35.21892. [DOI] [PubMed] [Google Scholar]
- 40.Schwenkert S, Legen J, Takami T, Shikanai T, Herrmann RG, Meurer J. Role of the low-molecular-weight subunits PetL, PetG, and PetN in assembly, stability, and dimerization of the cytochrome b6f complex in tobacco. Plant Physiol. 2007;144:1924–1935. doi: 10.1104/pp.107.100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schottler MA, Flugel C, Thiele W, Bock R. Knock-out of the plastid-encoded PetL subunit results in reduced stability and accelerated leaf age-dependent loss of the cytochrome b6f complex. J Biol Chem. 2007;282:976–985. doi: 10.1074/jbc.M606436200. [DOI] [PubMed] [Google Scholar]
- 42.Schneider D, Volkmer T, Rogner M. PetG and PetN, but not PetL, are essential subunits of the cytochrome b6f complex from Synechocystis PCC 6803. Res Microbiol. 2007;158:45–50. doi: 10.1016/j.resmic.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Z, Huang L, Shulmeister VM, Chi YI, Kim KK, Hung LW, Crofts AR, Berry EA, Kim SH. Electron transfer by domain movement in cytochrome bc1. Nature. 1998;392:677–684. doi: 10.1038/33612. [DOI] [PubMed] [Google Scholar]
- 44.Hasan SS, Stofleth JT, Yamashita E, Cramer WA. Lipid-induced conformational changes within the cytochrome b6f complex of oxygenic photosynthesis. Biochemistry. 2013;52:2649–2654. doi: 10.1021/bi301638h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Breyton C. Conformational changes in the cytochrome b6f complex induced by inhibitor binding. J Biol Chem. 2000;275:13195–13201. doi: 10.1074/jbc.275.18.13195. [DOI] [PubMed] [Google Scholar]
- 46.de Vitry C, Ouyang Y, Finazzi G, Wollman FA, Kallas T. The chloroplast Rieske iron-sulfur protein. At the crossroad of electron transport and signal transduction. J Biol Chem. 2004;279:44621–44627. doi: 10.1074/jbc.M406955200. [DOI] [PubMed] [Google Scholar]
- 47.Yan J, Cramer WA. Functional insensitivity of the cytochrome b6f complex to structure changes in the hinge region of the Rieske iron-sulfur protein. J Biol Chem. 2003;278:20925–20933. doi: 10.1074/jbc.M212616200. [DOI] [PubMed] [Google Scholar]
- 48.Zhang H, Cramer WA. Problems in obtaining diffraction-quality crystals of hetero-oligomeric integral membrane proteins. J Struct Funct Genomics. 2005;6:219–223. doi: 10.1007/s10969-005-1912-y. [DOI] [PubMed] [Google Scholar]
- 49.Zhang H, Kurisu G, Smith JL, Cramer WA. A defined protein-detergent-lipid complex for crystallization of integral membrane proteins: The cytochrome b6f complex of oxygenic photosynthesis. Proc Natl Acad Sci USA. 2003;100:5160–5163. doi: 10.1073/pnas.0931431100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baniulis D, Yamashita E, Zhang H, Hasan SS, Cramer WA. Structure-function of the cytochrome b6f complex. Photochem Photobiol. 2008;84:1349–1358. doi: 10.1111/j.1751-1097.2008.00444.x. [DOI] [PubMed] [Google Scholar]
- 51.Hasan SS, Yamashita E, Baniulis D, Cramer WA. Quinone-dependent proton transfer pathways in the photosynthetic cytochrome b6f complex. Proc Natl Acad Sci USA. 2013;110:4297–4302. doi: 10.1073/pnas.1222248110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hasan SS, Cramer WA. On rate limitations of electron transfer in the photosynthetic cytochrome b6f complex. Phys Chem Chem Phys. 2012;14:13853–13860. doi: 10.1039/c2cp41386h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alvey RM, Bezy RP, Frankenberg-Dinkel N, Kehoe DM. A light regulated OmpR-class promoter element coordinates light-harvesting protein and chromophore biosynthetic enzyme gene expression. Mol Microbiol. 2007;64:319–332. doi: 10.1111/j.1365-2958.2007.05656.x. [DOI] [PubMed] [Google Scholar]
- 54.Cobley JG, Clark AC, Weerasurya S, Queseda FA, Xiao JY, Bandrapali N, D’Silva I, Thounaojam M, Oda JF, Sumiyoshi T, Chu MH. CpeR is an activator required for expression of the phycoerythrin operon (cpeBA) in the cyanobacterium Fremyella diplosiphon and is encoded in the phycoerythrin linker-polypeptide operon (cpeCDESTR) Mol Microbiol. 2002;44:1517–1531. doi: 10.1046/j.1365-2958.2002.02966.x. [DOI] [PubMed] [Google Scholar]
- 55.Gutu A, Alvey RM, Bashour S, Zingg D, Kehoe DM. Sulfate-driven elemental sparing is regulated at the transcriptional and posttranscriptional levels in a filamentous cyanobacterium. J Bacteriol. 2011;193:1449–1460. doi: 10.1128/JB.00885-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Noubir S, Luque I, Ochoa de Alda JA, Perewoska I, Tandeau de Marsac N, Cobley JG, Houmard J. Coordinated expression of phycobiliprotein operons in the chromatically adapting cyanobacterium Calothrix PCC 7601: A role for RcaD and RcaG. Mol Microbiol. 2002;43:749–762. doi: 10.1046/j.1365-2958.2002.02783.x. [DOI] [PubMed] [Google Scholar]
- 57.Zhang H, Whitelegge JP, Cramer WA. Ferredoxin:NADP+ oxidoreductase is a subunit of the chloroplast cytochrome b6f complex. J Biol Chem. 2001;276:38159–38165. doi: 10.1074/jbc.M105454200. [DOI] [PubMed] [Google Scholar]
- 58.Yan J, Dashdorj N, Baniulis D, Yamashita E, Savikhin S, Cramer WA. On the structural role of the aromatic residue environment of the chlorophyll-a in the cytochrome b6f complex. Biochemistry. 2008;47:3654–3661. doi: 10.1021/bi702299b. [DOI] [PubMed] [Google Scholar]
- 59.Schagger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 60.Chain RK, Malkin R. Functional activities of monomeric and dimeric forms of the chloroplast cytochrome b6f complex. Photosynth Res. 1995;46:419–426. doi: 10.1007/BF00032296. [DOI] [PubMed] [Google Scholar]
- 61.Wittig I, Braun HP, Schagger H. Blue native PAGE. Nat Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 62.Whitelegge JP, Zhang H, Taylor R, Cramer WA. Full subunit coverage liquid chromatography electrospray-ionization mass spectrometry (LCMS+) of an oligomeric membrane protein complex: The cytochrome b6f complex from spinach and the cyanobacterium M. laminosus. Mol Cell Proteomics. 2002;1:816–827. doi: 10.1074/mcp.m200045-mcp200. [DOI] [PubMed] [Google Scholar]
- 63.Thangaraj B, Ryan CM, Souda P, Krause K, Faull KF, Weber AP, Fromme P, Whitelegge JP. Data-directed top-down Fourier-transform mass spectrometry of a large integral membrane protein complex: Photosystem II from Galdieria sulphuraria. Proteomics. 2010;10:3644–3656. doi: 10.1002/pmic.201000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ryan CM, Souda P, Bassilian S, Ujwal R, Zhang J, Abramson J, Ping P, Durazo A, Bowie JU, Hasan SS, Baniulis D, Cramer WA, Faull KF, Whitelegge JP. Post-translational modifications of integral membrane proteins resolved by top-down Fourier transform mass spectrometry with collisionally activated dissociation. Mol Cell Proteomics. 2010;9:791–803. doi: 10.1074/mcp.M900516-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vangone A, Spinelli R, Scarano V, Cavallo L, Oliva R. COCOMAPS: A web application to analyze and visualize contacts at the interface of biomolecular complexes. Bioinformatics. 2011;27:2915–2916. doi: 10.1093/bioinformatics/btr484. [DOI] [PubMed] [Google Scholar]
- 66.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera: A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 68.Hasan SS, Baniulis D, Yamashita E, Zhalnina MV, Zakharov SD, Stofleth JT, Cramer WA. Current Protocols in Protein Science. Vol. 74. Wiley; New York: 2013. Methods for studying interactions of detergents and lipids with α-helical and β-barrel integral membrane proteins; pp. 29.7.1–29.7.30. Unit 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yan J, Kurisu G, Cramer WA. Intraprotein transfer of the quinone analogue inhibitor 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone in the cytochrome b6f complex. Proc Natl Acad Sci USA. 2006;103:69–74. doi: 10.1073/pnas.0504909102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yamashita E, Zhang H, Cramer WA. Structure of the cytochrome b6f complex: Quinone analogue inhibitors as ligands of heme cn. J Mol Biol. 2007;370:39–52. doi: 10.1016/j.jmb.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qin L, Hiser C, Mulichak A, Garavito RM, Ferguson-Miller S. Identification of conserved lipid/detergent-binding sites in a high-resolution structure of the membrane protein cytochrome c oxidase. Proc Natl Acad Sci USA. 2006;103:16117–16122. doi: 10.1073/pnas.0606149103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qin L, Sharpe MA, Garavito RM, Ferguson-Miller S. Conserved lipid-binding sites in membrane proteins: A focus on cytochrome c oxidase. Curr Opin Struct Biol. 2007;17:444–450. doi: 10.1016/j.sbi.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Izrailev S, Crofts AR, Berry EA, Schulten K. Steered molecular dynamics simulation of the Rieske subunit motion in the cytochrome bc1 complex. Biophys J. 1999;77:1753–1768. doi: 10.1016/S0006-3495(99)77022-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Berry EA, Guergova-Kuras M, Huang LS, Crofts AR. Structure and function of cytochrome bc complexes. Annu Rev Biochem. 2000;69:1005–1075. doi: 10.1146/annurev.biochem.69.1.1005. [DOI] [PubMed] [Google Scholar]
- 75.Berry EA, De Bari H, Huang LS. Unanswered questions about the structure of cytochrome bc1 complexes. Biochim Biophys Acta. 2013;1827:1258–1277. doi: 10.1016/j.bbabio.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 76.Crofts AR, Shinkarev VP, Dikanov SA, Samoilova RI, Kolling D. Interactions of quinone with the iron-sulfur protein of the bc1 complex: Is the mechanism spring-loaded? Biochim Biophys Acta. 2002;1555:48–53. doi: 10.1016/s0005-2728(02)00253-0. [DOI] [PubMed] [Google Scholar]
- 77.Martinez SE, Huang D, Szczepaniak A, Cramer WA, Smith JL. Crystal structure of chloroplast cytochrome f reveals a novel cytochrome fold and unexpected heme ligation. Structure. 1994;2:95–105. doi: 10.1016/s0969-2126(00)00012-5. [DOI] [PubMed] [Google Scholar]
- 78.Baniulis D, Zhang H, Zakharova T, Hasan SS, Cramer WA. Purification and crystallization of the cyanobacterial cytochrome b6f complex. Methods Mol Biol. 2011;684:65–77. doi: 10.1007/978-1-60761-925-3_7. [DOI] [PubMed] [Google Scholar]
- 79.de Vitry C, Finazzi G, Baymann F, Kallas T. Analysis of the nucleus-encoded and chloroplast-targeted Rieske protein by classic and site-directed mutagenesis of Chlamydomonas. Plant Cell. 1999;11:2031–2044. doi: 10.1105/tpc.11.10.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Widger WR, Cramer WA, Herrmann RG, Trebst A. Sequence homology and structural similarity between cytochrome b of mitochondrial complex III and the chloroplast b6f complex: Position of the cytochrome b hemes in the membrane. Proc Natl Acad Sci USA. 1984;81:674–678. doi: 10.1073/pnas.81.3.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hasan SS, Yamashita E, Ryan CM, Whitelegge JP, Cramer WA. Conservation of lipid functions in cytochrome bc complexes. J Mol Biol. 2011;414:145–162. doi: 10.1016/j.jmb.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hasan SS, Yamashita E, Cramer WA. Transmembrane signaling and assembly of the cytochrome b6f-lipidic charge transfer complex. Biochim Biophys Acta. 2013;1827:1295–1308. doi: 10.1016/j.bbabio.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Davidson E, Ohnishi T, Tokito M, Daldal F. Rhodobacter capsulatus mutants lacking the Rieske FeS protein form a stable cytochrome bc1 subcomplex with an intact quinone reduction site. Biochemistry. 1992;31:3351–3358. doi: 10.1021/bi00128a007. [DOI] [PubMed] [Google Scholar]
- 84.Finazzi G, Büschlen S, de Vitry C, Rappaport F, Joliot P, Wollman FA. Function-directed mutagenesis of the cytochrome b6f complex in Chlamydomonas reinhardtii: Involvement of the cd loop of cytochrome b6 in quinol binding to the Qo site. Biochemistry. 1997;36:2867–2874. doi: 10.1021/bi962717y. [DOI] [PubMed] [Google Scholar]
- 85.Hasan SS, Cramer WA. Lipid functions in cytochrome bc complexes: An odd evolutionary transition in a membrane protein structure. Philos Trans R Soc, B. 2012;367:3406–3411. doi: 10.1098/rstb.2012.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


