Abstract
Mutations in the parkin or PINK1 genes are the leading cause of the autosomal recessive form of Parkinson’s disease. The gene products, the E3 ubiquitin ligase parkin and the serine/threonine kinase PINK1, are neuroprotective proteins, which act together in a mitochondrial quality control pathway. Here, we review the structure of parkin and mechanisms of its autoinhibition and function as a ubiquitin ligase. We present a model for the recruitment and activation of parkin as a key regulatory step in the clearance of depolarized or damaged mitochondria by autophagy (mitophagy). We conclude with a brief overview of other functions of parkin and considerations for drug discovery in the mitochondrial quality control pathway.
Keywords: mitophagy, parkin, PINK1, ubiquitin, ubiquitination
Introduction
Parkinson’s disease (PD) is a degenerative movement disorder caused by the progressive death of dopamine-producing neurons in the substantia nigra pars compacta of the mid-brain. Studies linking PD to defects in the electron transport chain suggest that damaged mitochondria may play a central role in PD pathology 1. While most PD cases occur sporadically, research on inherited forms of PD in the past decade have shed new light on the disease and pathogenesis. Studies of two recessive PD genes, PINK1 (PTEN-induced putative kinase protein 1 or PARK6) and parkin (PARK2), have provided direct evidence for the contribution of damaged mitochondria in PD pathology 2. Parkin is a cytosolic E3 ubiquitin ligase and PINK1 is the only known protein kinase with a mitochondrial targeting domain. These two proteins are involved in a common pathway regulating mitochondrial quality control and promoting the selective autophagy of depolarized mitochondria (mitophagy) 3. Under basal conditions, parkin E3 ligase activity is repressed in the cytoplasm and PINK1 is imported into mitochondria via TOM40 and TOM20 core containing complexes 4 and cleaved sequentially by mitochondrial proteases such as MPP and PARL 5–8. Cleaved PINK1 then degrades rapidly in the cytosol via the N-end rule pathway so that levels of PINK1 are low in healthy cells 9. However, when the organelle loses its inner membrane electrochemical gradient, import of PINK1 to the mitochondria is inhibited and the protein is stabilized on the mitochondrial outer membrane with its kinase domain on the cytosolic face 10,11. The accumulation of PINK1 kinase activity on the mitochondria triggers parkin recruitment and activation. Activated parkin produces ubiquitin chains on various outer mitochondrial membrane proteins leading to autophagic elimination of the damaged organelle 12–15. Pathogenic mutations in either of these genes lead to loss of this quality control pathway and accumulation of impaired mitochondria, which are thought to be a source of toxic reactive oxygen species (ROS) and contribute to neuronal cell death and PD.
Parkin is an RBR E3 ubiquitin ligase
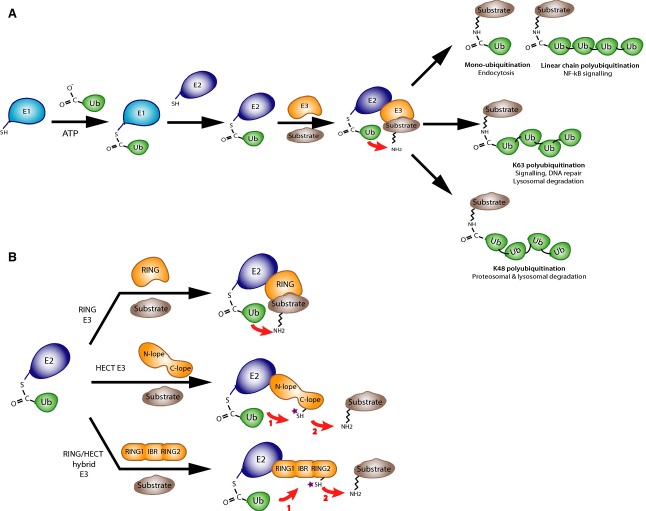
Ubiquitination is a post-translational modification that typically marks proteins for degradation through the covalent attachment of ubiquitin and ubiquitin chains to lysine residues or the N-terminal amino group of a substrate protein 16. In addition to degradation via the proteasome, ubiquitination can act as a signal for autophagy 17 – degradation via lysosomes – as well as alter substrate protein activity or location 18. Ubiquitination is carried out through the sequential action of three enzymes: E1 ubiquitin-activating enzymes, E2 ubiquitin-conjugating enzymes and E3 ubiquitin ligases (Fig.1A). In the pathway, E1 first uses ATP to activate ubiquitin for conjugation by forming a thioester between its catalytic cysteine and the C-terminal carboxyl group of ubiquitin. The ubiquitin is then passed to a second cysteine of an E2 ubiquitin conjugating enzyme. In the final step, the ubiquitin-charged E2 enzyme interacts with a specific E3 ubiquitin ligase and transfers the ubiquitin to the amino group of a substrate protein. Ubiquitin contains seven lysine residues as well as an N-terminal amino group that can be used to build chains of polyubiquitin. The most common chain types are K48 and K63 chains in which multiple ubiquitin molecules are linked in a linear arrangement with the C terminus of one molecule attached to lysine 48 (or 63) of the next. In the ubiquitination pathway, the E3 ubiquitin ligases typically provide the majority of the specificity and regulation in recognition of substrates and control of activity 19. Based on considerations of structure and chemistry, three classes of E3 ligases are distinguished: RING-type (including U-box ligases), HECT-type and RING-HECT hybrids 20 (Fig.1B). RING-type E3s are characterized by the presence of a canonical C3HC4-type RING (really interesting new gene) domain that binds the E2 enzyme but does not participate directly in catalysis. This class of E3s functions as inert scaffolding ligases that facilitate the direct transfer of ubiquitin from the E2 onto the substrate. HECT-type E3s contain a HECT (homologous to the E6-AP carboxyl terminus) domain with an active site cysteine, which accepts ubiquitin from an E2 enzyme in the form of a thioester intermediate and then transfers it to substrates. The third class consists of the RBR (RING-between-RING) family of E3 ubiquitin ligases that combine the chemistry of HECT-type ligases with structural similarity to RING-type ligases 21,22. RBRs contain a canonical C3HC4-type RING (named RING1), followed by two conserved Cys/His-rich Zn-binding domains, in-between-RING (IBR) and RING2 domains, that contain an active site cysteine residue. This cysteine accepts ubiquitin from the E2 enzyme and transfers it onto substrates; hence, ligases in this class are sometimes referred to as RING-HECT hybrids 22.
Figure 1.
Ubiquitination pathway. (A) Cascade of ubiquitination enzymes. The ubiquitin-activating enzyme E1 uses ATP to conjugate the C-terminal carboxylic acid group of ubiquitin to an active site cysteine. This is then transferred to a cysteine on one of a number of E2 enzymes that work in concert with E3 enzymes to ubiquitinate substrate proteins on amino groups of lysine residues or protein N termini. The formations of mono-ubiquitin or polyubiquitin chains on substrates are signals for different downstream pathways. (B) Classes of E3 ubiquitin ligases. E3 ligases can be distinguished based on the presence of a RING domain, a catalytic cysteine or both. RING and U-box domain ligases act as scaffolds to bring the substrate and ubiquitin-conjugated E2 together. HECT ligases have a catalytic cysteine in their C-lobe that is transiently conjugated to ubiquitin as an intermediate in a two-step process of substrate ubiquitination. Parkin is a RING/HECT hybrid ligase that contains a RING domain that binds the E2 enzyme and a catalytic cysteine that transfers ubiquitin to the substrate.
Parkin, a member of RBR E3 ubiquitin ligases, ubiquitinates a wide variety of cytosolic and outer mitochondrial membrane proteins upon mitochondrial depolarization 23,24. It forms multiple types of ubiquitin chains, most frequently K63, K48, K11 and K6 linkages 25. Parkin also shows relatively lax substrate specificity 23,24. Accumulation of polyubiquitin chains on mitochondria signals recruitment of the autophagosome and proteasome machinery to initiate mitophagy 12–14,23. Parkin itself becomes ubiquitinated by the attachment of K6 ubiquitin chains, which may play a role in its own degradation 26. The activity of parkin is tightly regulated and normally repressed 27–30. In cells, parkin activity can be activated under a variety of conditions such as depolarization of mitochondria or epidermal growth factor signaling 31. A large number of treatments that disrupt its structure are also known to de-repress its ligase activity in vitro. These include heat treatment, N-terminal deletions and some point mutations 28–30,32.
Lessons from the structure
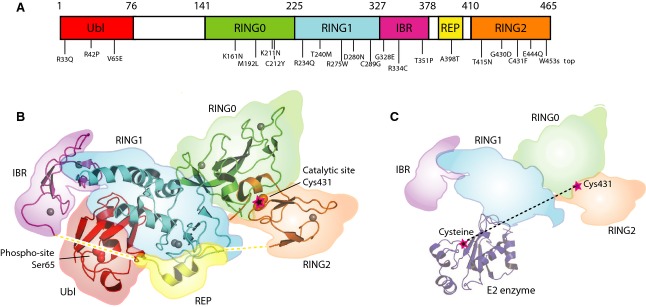
Recent structures of parkin have revealed much about its regulation and function (Fig.2A). Parkin consists of a ubiquitin-like domain (Ubl) at its N terminus and four zinc-coordinating RING-like domains: RING0, RING1, IBR and RING2. More than 120 pathogenic PD mutations are spread throughout parkin domains, attesting to critical functions for each of the individual domains 33. The Ubl domain is involved in substrate recognition, binding SH3 and ubiquitin interacting motif (UIM) domains, proteasome association, and regulation of cellular parkin levels and activity 27,31,34–37.
Figure 2.
The structure of parkin reveals the mechanism of its autoinhibition. (A) Domain architecture of the five parkin domains and identification of selected PD mutations. (B) Cartoon representation of parkin (PDB: 4K95) 29. Parkin activity depends on two functional sites: a binding site for ubiquitin-conjugated E2 enzyme on the RING1 of parkin and a catalytic site with a cysteine that forms a transient covalent linkage with ubiquitin on the RING2 domain. Both sites are occluded in the autoinhibited structure. The Ubl domain and REP linker between the IBR and RING2 domains prevent the E2 from binding to RING1. The RING0 domain partially covers the catalytic cysteine on the RING2 domain. Mutations at interdomain interfaces as well as phosphorylation of Ser65 in the Ubl domain increase parkin ubiquitin ligase activity. The eight structural zinc ions in parkin are shown as gray spheres. Dashed lines indicate portions of the IBR and RING2 linker that were not observed in the crystal structure. (C) Model of an E2 enzyme bound to parkin with Ubl and REP linker removed. An additional structural rearrangement must occur to allow ubiquitin on the E2 enzyme to transfer to the catalytic cysteine of parkin. The E2 and parkin catalytic cysteines are ∼ 50 Å apart in the model.
Last year, several groups reported high-resolution crystal structures of a parkin fragment consisting of the RING0, RING1, IBR and RING2 domains (PDB: 4K7D, 4I1H, 4I1F, 4BM9) along with a low-resolution structure of the full-length protein (PDB: 4K95) 28–30,38. In the structures, parkin adopts a compact arrangement, stabilized by multiple hydrophobic interactions, similar to a coiled snake 39 (Fig.2B). The N-terminal Ubl domain uses a hydrophobic surface centered around Ile44 to bind to the RING1 domain, while the C-terminal catalytic domain is tightly associated with the RING0 domain. Domains RING0 through RING2 (collectively referred to as R0–RBR) each coordinate two zinc ions through histidine and cysteine residues, confirming the stoichiometry of eight zinc ions per parkin 40. The RING0 domain binds zinc ions in a hairpin arrangement unique to parkin while the RING2 and IBR domains show a sequential arrangement of zinc-coordinating residues. RING0 and RING2 were originally identified as RING domains based on the primary amino acid sequence but their structural topology differs from a classical RING fold. The RING1 domain is the only RING domain with the cross-brace zinc coordination topology observed in other RING-type E3 ligases. The similarity of RING1 to other RING E3 ligases suggested that it is the E2 binding site on parkin (Fig.2C). NMR titrations, mutagenesis and molecular modeling confirmed this and identified an α-helix (residues 263–271) and two loops (residues 239–244 and 290–292) in RING1 as the E2 binding site 28–30. The RING2 domain is the catalytic module harboring the catalytic cysteine (Cys431). While RING0 is unique to parkin, the IBR domain is conserved among the RBR E3 family; the precise function of the domains is currently unknown. Parkin contains two interdomain linkers that are flexible and not observed in the crystal structures. The first consists of 70 poorly conserved residues of unknown function that follow the Ubl domain. The second, within the R0–RBR fragment, occurs between the IBR and RING2 domains and is composed of disordered segments and a conserved α-helix (residues 391–403) that binds to the RING1 domain. This helix has been termed the repressor element of parkin (REP) due to its role in the regulation of parkin activity.
Parkin is a difficult protein to work with unless certain precautions are taken 41. The ligase activity is normally repressed and surprisingly some mutations or heat treatment increase activity 27–30. The bound zinc ions are required for structural stability and properly regulated enzymatic activity 40,42. While supplemental zinc is not required during purification, metal-chelating agents such as ethylenediaminetetraacetic acid (EDTA) need to be avoided since they denature the protein. Unfortunately, some published co-immunoprecipitation experiments have used EDTA and need to be interpreted with caution. With a total of 35 cysteine residues, parkin requires a high concentration of a reducing agent such as dithiothreitol to maintain the cysteine thiols reduced and available for coordinating zinc ions.
The parkin structure provides rationales for many of the mutations associated with PD (Fig.2A). Certain mutations compromise the structural integrity of the protein. Others interfere with binding of substrates or directly affect the enzyme catalysis. Among the best understood, mutations C212Y, C289G and C441R affect zinc coordination, while R42P, K211N and T351P disrupt protein folding or stability. C431F and G430D are in the catalytic site, and T240R prevents E2 binding. The effects of other mutations such as R33Q, D280N, G328E or T415N are less clear. These may disrupt the interdomain interactions or affect binding of other proteins by parkin. A comprehensive list of known disease-causing parkin mutations together with the predicted structure rationale and biochemical effects is provided as supplementary material by Wauer and Komander 30.
Autoinhibition
The activity of parkin is tightly controlled by multiple mechanisms of autoinhibition. The first is access to the catalytic RING2 domain, which is blocked by RING0. Other RBR E3 ligases show similar mechanisms of autoinhibition 43–45. The Ariadne domain of the RBR E3 HHARI and the UBA domain of the RBR E3 HOIP both participate in inhibiting the catalytic domain. Deletion of the N terminus of parkin through to the RING0 domain leads to very high ubiquitin ligase activity in vitro 28–30.
A second mechanism is the control of binding of the upstream E2 enzyme to parkin. Modeling and mutagenesis have confirmed that the E2 binding site is on the RING1 domain but the site is occluded by the Ubl domain and REP linker (Fig.2C). Deletion of the Ubl domain and mutagenesis of the REP linker both increase the affinity of E2 binding and parkin activity 27,29. Finally, the large distance between the E2 binding site and catalytic site on RING2 prevents transfer of ubiquitin from the ubiquitin–E2 conjugate to the parkin catalytic cysteine (Fig.2C).
Catalytic mechanism
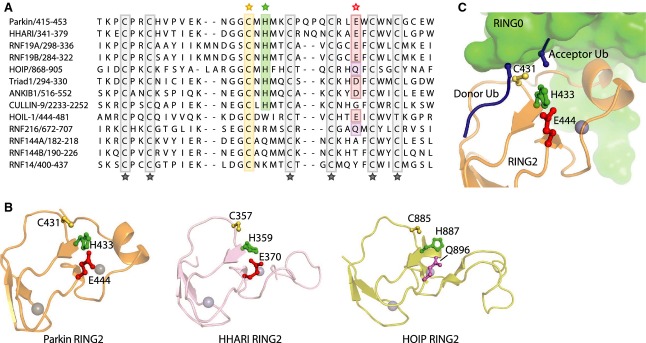
The detailed structure of parkin RING2 domain reveals conserved chemistry across the RBR family of E3 ligases for ubiquitin transfer to the target protein (Fig.3). Catalytically, ubiquitination proceeds through two steps: (a) formation of a ubiquitin–cysteine thioester intermediate where the C terminus of ubiquitin is covalently linked to the parkin catalytic cysteine, and (b) acyl transfer where the ubiquitin is transferred from parkin to an amino group of the substrate.
Figure 3.
Mechanism of parkin ubiquitin ligase activity. (A) Sequence alignment of RBR E3 ubiquitin ligases shows conservation in the catalytic RING2 domains. The catalytic cysteine (highlighted in yellow) is invariant across RBR proteins, whereas histidine (green) and glutamate residues (red) that play secondary roles in catalysis are less conserved. Gray indicates zinc-coordinating residues. (B) Structural comparison of catalytic domains of parkin, HHARI and HOIP (PDB: 4K7D 29; 4KBL 43; 4LJO 44). The alignment of the catalytic cysteine, histidine and glutamate/glutamine is suggestive of a catalytic triad where the histidine acts as a general base to promote transesterification of ubiquitin onto the substrate. (C) Model of the positions of the donor and acceptor ubiquitin molecules in the catalytic site of parkin based on the crystal structure of HOIP 44. The C terminus of the donor ubiquitin lies in a groove of the RING2 domain and terminates next to the catalytic cysteine, mimicking the thioester intermediate. The amino group of the acceptor ubiquitin approaches Cys431 from the opposite side but would be sterically blocked by the parkin RING0 domain in the absence of a conformational change.
The catalytic cysteine residue (in the RING2 domain) is conserved in all the members of the RBR family as are the residues involved in zinc coordination. Following the cysteine, there is partial conservation of a histidine residue and an acidic residue. In parkin, these residues adopt a linear arrangement that has been proposed to act as a catalytic triad where the histidine acts as a general base to promote catalysis 28–30. The structures of two other RBR ligases are known: HHARI, a member of the Ariadne family of E3 ligases 43, and HOIP, a component of the linear ubiquitination assembly complex (LUBAC) 44. The RING2 domains are highly conserved and share the linear arrangement of the three residues (Fig.3B). Nonetheless, the results of mutagenesis are ambiguous about the importance of the catalytic triad. The effect of the loss of the histidine is substrate dependent and can be suppressed in vitro by raising the pH 28,44. In cultured cells, mutation of the histidine only moderately slows parkin-mediated mitophagy. And, while the mutation E444Q reduces autoubiquitination 29 and may be implicated in PD 46, it has no effect on parkin activity in cells 28.
Additional insight into the catalytic mechanism comes from the crystal structure of HOIP, which contains two ubiquitin molecules in contact with catalytic RING2 domain (Fig.3C). The C terminus of one ubiquitin molecule is positioned to mimic the thioester linkage with the catalytic cysteine while the amino group of the second ubiquitin molecule approaches the cysteine from the other side and occupies the position of the acceptor molecule. Transposition of the two ubiquitin molecules onto the parkin crystal structure generates a hypothetical model of the active site with a thioester intermediate prior to acyl transfer. While the donor ubiquitin can be accommodated to fit in a groove on the surface of the RING2 domain, the position of the acceptor ubiquitin or substrate protein clashes with the RING0 domain, implying that this domain must move in order for a substrate amino group to access the active site.
Parkin activation
Recent studies have made tremendous progress in understanding how parkin is activated. In vitro studies of parkin have found that a wide variety of effectors can promote parkin activity. These include point mutations that disrupt inhibitory interdomain interactions as well as N-terminal deletions 27–30. The Ubl domain plays a special role in parkin activation and a number of binding partners that bind the Ubl domain are associated with parkin activation 27. The Ubl domain recruits these binding partners such as the SH3 domain and UIMs of endophilin A1, Eps15, proteasomal subunits and ataxin-3 31,35–37 through the same hydrophobic surface centered around Ile44 that it uses to interact with the RING1 domain, indicating that the Ubl dissociates from RING1 upon activation.
PINK1 acts upstream of parkin and is required for parkin activation and recruitment to depolarized mitochondria 12–15,47. PINK1 phosphorylates parkin Ubl domain on residue Ser65 to activate parkin 48–50. Although the phospho-mimetic S65E mutation stimulates parkin ligase activity in vitro, this mutation is not able to bypass the PINK1 requirement for mitochondrial recruitment in cells and the non-phosphorylatable S65A mutation is not completely impaired 48. These observations led to the search for an additional PINK1 substrate involved in the parkin activation and the breakthrough discovery that ubiquitin can be phosphorylated by PINK1 51–53. Three groups concurrently observed that PINK1 can phosphorylate ubiquitin on Ser65, the same position that is phosphorylated in the parkin Ubl domain. Further, they showed that phosphorylated ubiquitin or phospho-mimetic ubiquitin mutants (S65D/E) directly activate parkin by enhancing the rate of E2–ubiquitin discharge 51–53. Parkin binds strongly to phospho-ubiquitin with an affinity of 400 nm. Phosphorylation of Ser65 of parkin further increases the affinity 20-fold. In cells, expression of the non-phosphorylatable S65A ubiquitin delays parkin recruitment to the depolarized mitochondria, and mutation of both parkin and ubiquitin at Ser65 abolishes parkin activation 53. Further studies have revealed that PINK1 is not only a ubiquitin kinase but is also capable of phosphorylating ubiquitin in ubiquitin chains 25,54,55. While phospho-ubiquitin can still become activated by E1 and charged onto E2 enzymes, its activity in ubiquitination assays is E3 dependent 55. Parkin shows somewhat less activity with phospho-ubiquitin–E2 than with ubiquitin–E2 51. Potentially more relevant is the observation that phosphorylated ubiquitin chains were more resistant to hydrolysis by 10 out of 12 deubiquitinases tested 55. One possible explanation is the presence of a minor conformation of phospho-ubiquitin that was detected by NMR 55. The minor conformation, corresponding to about 30% of molecules, shows a β-strand slippage that disrupts the Ile44 hydrophobic patch involved in many ubiquitin interactions.
The structural details of parkin activation are unknown but most of the evidence is compatible with a simple two-state system. Inputs that activate parkin shift the equilibrium between an inactive and active conformation. There is no evidence of multiple steps in the pathway: disruption of the RING0–RING2 interface increases ligase activity as does disruption of the REP–RING1 interaction. The conformational change between the two states has been suggested to be a ‘butterfly’ movement in which parkin folds along the axis between the RING0 and RING1 domains to bring the two functional sites together 28. Alternatively, the active conformation could consist of an ensemble of structures where the E2 binding and catalytic sites transiently interact like beads on a string. The release of the inhibitory interactions would suffice to activate the RING1 and RING2 domains to carry out the ubiquitin ligase activity.
The site of parkin phosphorylation is in close proximity to the REP linker between IBR and RING2 domain in the autoinhibited conformation (Fig.2B) and could promote displacement of the REP linker and Ubl domain to allow E2 binding. The site of phospho-ubiquitin binding to parkin is also unknown and it is not clear if the phospho-ubiquitin and phosphoUbl binding sites are the same or distinct. In one of the crystal structures, a pocket formed by three solvent exposed positively charged residues Lys161, Arg163 and Lys211 in the RING0 domain was occupied by a sulfate ion, suggesting a potential phosphate binding site 30. While the PD-associated mutants K161N and K211N fail to recruit to depolarized mitochondria 12–14, mutation of the basic patch did not affect the binding affinity towards phospho-ubiquitin, suggesting another role for this pocket 25.
Parkin recruitment to mitochondria
In cultured cells, PINK1/parkin pathways can be activated by depolarizing mitochondria with uncouplers, such as carbonyl cyanide m-chlorophenyl hydrazone (CCCP). Following CCCP treatment, parkin shows a very robust and complete recruitment to mitochondria within several hours, which is then followed by the clearance of mitochondria. Parkin recruitment to mitochondria requires PINK1 kinase activity; however, there is no fixed stoichiometry between PINK1 levels and parkin recruitment. This suggests that a molecule other than PINK1 is recognized by parkin. This contrasts with the surprising observation that PINK1 artificially targeted to peroxisomes is sufficient to recruit and activate parkin on peroxisomes 4. A further enigma is the requirement for parkin ubiquitin ligase activity for its recruitment. Catalytically inactive mutants of parkin do not show detectable recruitment following mitochondrial depolarization 12,21.
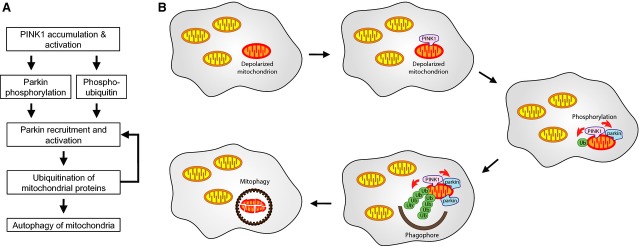
The solution to this puzzle is now emerging with a model of the events leading to the recruitment and activation of parkin by phospho-ubiquitin (Fig.4). In the first step, the selective accumulation of PINK1 on depolarized mitochondria leads to the phosphorylation of low, basal levels of ubiquitin or parkin present on mitochondria 25,51–53. The system exhibits feedforward control as both ubiquitin and parkin phosphorylation are positive effectors of parkin ubiquitin ligase activity 25,51–53. Phosphorylation of ubiquitin chains was recently shown to inhibit the action of deubiquitinases 55, which would act as an additional feedforward mechanism. Full activation of the system requires a positive feedback loop where activation of parkin increases the amount of mitochondrially conjugated ubiquitin, which is then phosphorylated by PINK1 to recruit more parkin. The positive feedback explains the requirement for both PINK1 and parkin catalytic activities as well as how high levels of parkin can be recruited and depleted from the cytosol by low, endogenous levels of PINK1. In agreement with the model, the requirement for parkin ligase activity can be bypassed by using overexpressed tetra-ubiquitin chains artificially targeted to mitochondria 56. The requirement for PINK1 activity can be further bypassed by the use of phospho-mimetic chains for S65E tetra-ubiquitin 54.
Figure 4.
Pathway of PINK1 activation of parkin leading to autophagy of depolarized mitochondria. (A) Flowchart of feedforward and feedback activation of parkin. (B) Schematic of the quality control pathway. PINK1 accumulates on depolarized or damaged mitochondria and phosphorylates ubiquitin and parkin present on the surface. Activated parkin produces additional ubiquitin chains on the mitochondria, which in turn are phosphorylated by PINK1 to promote the recruitment of more parkin. The autophagosome forms around the heavily ubiquitinated mitochondria, which are then eliminated by autophagy.
A number of questions remain unanswered. The relative importance of the feedforward pathways is unknown as is the order of the initial events. It is possible, although unlikely, that ubiquitin phosphorylation precedes chain formation and that parkin incorporates phospho-ubiquitin directly into chains 25,51,52. The relative importance of mono-ubiquitin and polyubiquitin chains is also unknown. Mono-ubiquitin tethered to mitochondria is not sufficient to recruit catalytically inactive parkin which suggests that ubiquitin chains may play a special role 56.
Parkin/PINK1-mediated mitophagy in neurons
Although parkin recruitment to depolarized mitochondria is a robust phenomenon in diverse mammalian cell lines, it has been controversial whether mitophagy is applicable in neurons and in PD pathogenesis. Concerns are that most of these experiments use overexpressed parkin and that CCCP treatment leads to rapid depolarization of the entire mitochondrial network and non-physiological levels of damage to mitochondria, which probably is never the case in PD. In experiments in primary neurons, parkin recruitment to depolarized mitochondria is modest and only happens after prolonged CCCP treatment or in the presence of lysosomal or apoptosis inhibitors or special culture conditions 57,58. In part, this could arise from the fact that most cell lines are glycolytic while neurons are strictly dependent on oxidative phosphorylation for mitochondrial ATP production; neuronal mitophagy studies may require more physiological methods for induction of depolarized mitochondria. In a recent paper, the short mitochondrial ARF protein was shown to induce mitochondrial depolarization and parkin/PINK1 autophagy both in cell lines and in neurons 59. Moreover, light activated ROS-induced mitochondrial depolarization was also shown to initiate parkin- and PINK1-dependent mitochondrial degradation by autophagy in axonal mitochondria 60. This confirms that parkin recruitment and activation on mitochondria is relevant for PD.
Other functions of parkin
Parkin plays a number of roles outside of the induction of mitophagy. Parkin ubiquitination of outer mitochondrial membrane proteins such as mitofusins, optic atrophy 1 (OPA1) and Miro alters the balance of fission to fusion and mitochondrial motility, facilitating the isolation of dysfunctional mitochondria from the mitochondrial network for mitophagy 61. Parkin and PINK1 also work together to repair mildly damaged mitochondria in response to mild oxidative stress through the formation of mitochondria derived vesicles (MDVs) enriched in oxidized proteins 62. These MDVs carry damaged cargo to lysosomes for degradation 63. Complementary to mitophagy, parkin and PINK1 balance the turnover of mitochondria by promoting the synthesis of new mitochondria 64. Overexpressing parkin in proliferating cells and SH-SY5 cells increases mitochondrial transcription through interaction with TFAM 65,66.
Parkin has been reported to promote cell survival through various mechanisms although how parkin becomes activated in these pathways is not always clear. Parkin has been ascribed as preventing cell death through proteosomal degradation of several proteins such as parkin interacting substrate (PARIS), aminoacyl-tRNA synthetase complex-interacting multifunctional protein-2 (AIMP2) and Fbw7b, a substrate-binding adaptor protein subunit of SCF E3 ubiquitin ligase complex 67–69. Parkin may activate prosurvival pathways by increasing nuclear factor κB (NF-κB) signaling 70 or decreasing activation of c-jun N-terminal kinase 71–73. A number of recent papers describe interactions between parkin and Bcl proteins in the mitochondrial apoptotic pathway 69,74–77. Parkin transcription is under the control of p53, and reportedly mediates the Warburg effect of p53 on glucose metabolism 78. Parkin has also been implicated in cell surface signaling by controlling epidermal growth factor receptor internalization and Akt signaling by ubiquitination of the endocytic scaffold protein Eps15 31. Parkin is a tumor suppressor and its inactivation has been reported in various human cancers. Parkin is deleted in 30% of human tumors and parkin-deficient mice are more susceptible to tumorigenesis 78–82.
Parkin also has a key role in pathogen defense through xenophagy, a pathway related to mitophagy. In xenophagy, bacteria are marked with ubiquitin chains that recruit ubiquitin-binding autophagy adaptors, leading to autophagosome formation and eventually fusion with the lysosome. The ubiquitinated substrates and ligases involved in this pathway are poorly understood. Genomic studies identified parkin as a susceptibility factor for the intracellular bacterial pathogen Mycobacterium leprae 83. In a recent paper, parkin has been shown to be required for resistance to intracellular pathogens such as Mycobacterium tuberculosis and Salmonella enterica through an autophagy-dependent mechanism 84. The shared ancestry between mitochondria and bacteria points to a common mechanism of parkin-mediated autophagy, but whether PINK1 or a related kinase is required for xenophagy is unknown.
Is parkin a good therapeutic target?
The recent progress in understanding the regulation of parkin activity may seem appealing for new routes for treating diseases with mitochondrial dysfunction. Parkin displays low basal activity and a small increase in the activation of parkin could be sufficient to slow the progression of PD in sporadic forms of the disease where the wild-type protein is present 41. Although simplistic, small molecules that mimic phospho-ubiquitin or disrupt autoinhibitory interactions might enhance its neuroprotective action. In cultured cells, mutation of Trp403 or Phe463 speeds recruitment of parkin to mitochondria in a regulated process that remains dependent on PINK1 and mitochondrial depolarization 29. A small molecule that binds tightly to the pocket occupied by the amino acid side chains would be expected to have the same effect. Alternatively, the deubiquitinating (DUB) enzymes USP30 and USP15 were recently found to oppose parkin/PINK1 in mitophagy, making inhibitors of these DUBs prime candidates for drug design 85,86. In contrast, USP8 promotes parkin-mediated mitophagy and thus agonists of this DUB could be developed 26. While it remains speculative with many challenges, the quality control pathway mediated by PINK1 and parkin appears to offer multiple therapeutic targets for the treatment of PD and other diseases caused by dysfunctional mitochondria.
Acknowledgments
We thank Jean-François Trempe and Xinlu Li for reading the manuscript and constructive comments. This work was supported by the Canadian Institutes of Health Research grant MOP-125924 and studentships from the Fonds de la Recherche en Santé du Québec in partnership with la Société Parkinson du Québec and the NSERC CREATE Training Program in Bionanomachines.
Glossary
- CCCP
carbonyl cyanide m-chlorophenyl hydrazone
- HECT
homologous to the E6-AP carboxyl terminus
- IBR
in-between-RING
- PD
Parkinson’s disease
- RBR
RING-between-RING
- REP
repressor element of parkin
- RING
really interesting new gene
- ROS
reactive oxygen species
- Ubl
ubiquitin-like domain
- UIM
ubiquitin interacting motif
Author contributions
MS, GK, and KG conceived and wrote the review.
References
- Venderova K. Park DS. Programmed cell death in Parkinson’s disease. Cold Spring Harb Perspect Med. 2012;2:pii: a009365. doi: 10.1101/cshperspect.a009365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- Narendra D, Walker JE. Youle R. Mitochondrial quality control mediated by PINK1 and parkin: links to parkinsonism. Cold Spring Harb Perspect Biol. 2012;4:pii: a011338. doi: 10.1101/cshperspect.a011338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M, Jin SM, Kane LA. Youle RJ. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase parkin. Dev Cell. 2012;22:320–333. doi: 10.1016/j.devcel.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deas E, Plun-Favreau H, Gandhi S, Desmond H, Kjaer S, Loh SHY, Renton AEM, Harvey RJ, Whitworth AJ, Martins LM, et al. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum Mol Genet. 2011;20:867–879. doi: 10.1093/hmg/ddq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene AW, Grenier K, Aguileta MA, Muise S, Farazifard R, Haque ME, McBride HM, Park DS. Fon EA. Mitochondrial processing peptidase regulates PINK1 processing, import and parkin recruitment. EMBO Rep. 2012;13:378–385. doi: 10.1038/embor.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SM, Lazarou M, Wang CX, Kane LA, Narendra DP. Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191:933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth AJ, Lee JR, Ho VMW, Flick R, Chowdhury R. McQuibban GA. Rhomboid-7 and HtrA2/Omi act in a common pathway with the Parkinson’s disease factors Pink1 and parkin. Dis Model Mech. 2008;1:168–174. doi: 10.1242/dmm.000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K. Youle RJ. PINK1 is degraded through the N-end rule pathway. Autophagy. 2013;9:1758–1769. doi: 10.4161/auto.24633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W. Kang UJ. Characterization of PINK1 processing, stability, and subcellular localization. J Neurochem. 2008;106:464–474. doi: 10.1111/j.1471-4159.2008.05398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Huang Y, Shao YF, May J, Prou D, Perier C, Dauer W, Schon EA. Przedborski S. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc Natl Acad Sci USA. 2008;105:12022–12027. doi: 10.1073/pnas.0802814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, Cookson MR. Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Holmstrom KM, Skujat D, Fiesel FC, Rothfuss OC, Kahle PJ. Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, Sou Y, Saiki S, Kawajiri S, Sato F, et al. PINK1 stabilized by mitochondrial depolarization recruits parkin to damaged mitochondria and activates latent parkin for mitophagy. J Cell Biol. 2010;189:211–221. doi: 10.1083/jcb.200910140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vives-Bauza C, Zhou C, Huang Y, Cui M, de Vries RLA, Kim J, May J, Tocilescu MA, Liu WC, Ko HS, et al. PINK1-dependent recruitment of parkin to mitochondria in mitophagy. Proc Natl Acad Sci USA. 2010;107:378–383. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A. Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Randow F. Youle RJ. Self and nonself: how autophagy targets mitochondria and bacteria. Cell Host Microbe. 2014;15:403–411. doi: 10.1016/j.chom.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D. Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- Dye BT. Schulman BA. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu Rev Biophys Biomol Struct. 2007;36:131–150. doi: 10.1146/annurev.biophys.36.040306.132820. [DOI] [PubMed] [Google Scholar]
- Berndsen CE. Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol. 2014;21:301–307. doi: 10.1038/nsmb.2780. [DOI] [PubMed] [Google Scholar]
- Lazarou M, Narendra DP, Jin SM, Tekle E, Banerjee S. Youle RJ. PINK1 drives parkin self-association and HECT-like E3 activity upstream of mitochondrial binding. J Cell Biol. 2013;200:163–172. doi: 10.1083/jcb.201210111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel DM, Lissounov A, Brzovic PS. Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474:105–108. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan NC, Salazar AM, Pham AH, Sweredoski MJ, Kolawa NJ, Graham RL, Hess S. Chan DC. Broad activation of the ubiquitin-proteasome system by parkin is critical for mitophagy. Hum Mol Genet. 2011;20:1726–1737. doi: 10.1093/hmg/ddr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarraf SA, Raman M, Guarani-Pereira V, Sowa ME, Huttlin EL, Gygi SP. Harper JW. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013;496:372–376. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordureau A, Sarraf SA, Duda DM, Heo JM, Jedrychowski MP, Sviderskiy VO, Olszewski JL, Koerber JT, Xie T, Beausoleil SA, et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol Cell. 2014;56:360–375. doi: 10.1016/j.molcel.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durcan TM, Tang MY, Perusse JR, Dashti EA, Aguileta MA, McLelland GL, Gros P, Shaler TA, Faubert D, Coulombe B, et al. USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin. EMBO J. 2014;33:2473–2491. doi: 10.15252/embj.201489729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaugule VK, Burchell L, Barber KR, Sidhu A, Leslie SJ, Shaw GS. Walden H. Autoregulation of parkin activity through its ubiquitin-like domain. EMBO J. 2011;30:2853–2867. doi: 10.1038/emboj.2011.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley BE, Lougheed JC, Callaway K, Velasquez M, Brecht E, Nguyen L, Shaler T, Walker D, Yang Y, Regnstrom K, et al. Structure and function of parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat Commun. 2013;4:1982. doi: 10.1038/ncomms2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe JF, Sauve V, Grenier K, Seirafi M, Tang MY, Menade M, Al-Abdul-Wahid S, Krett J, Wong K, Kozlov G, et al. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 2013;340:1451–1455. doi: 10.1126/science.1237908. [DOI] [PubMed] [Google Scholar]
- Wauer T. Komander D. Structure of the human parkin ligase domain in an autoinhibited state. EMBO J. 2013;32:2099–2112. doi: 10.1038/emboj.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon L, Belanger CML, Corera AT, Kontogiannea M, Regan-Klapisz E, Moreau F, Voortman J, Haber M, Rouleau G, Thorarinsdottir T, et al. A regulated interaction with the UIM protein Eps15 implicates parkin in EGF receptor trafficking and PI(3) K-Akt signalling. Nat Cell Biol. 2006;8:834–842. doi: 10.1038/ncb1441. [DOI] [PubMed] [Google Scholar]
- Spratt DE, Walden H. Shaw GS. RBR E3 ubiquitin ligases: new structures, new insights, new questions. Biochem J. 2014;458:421–437. doi: 10.1042/BJ20140006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruts M, Theuns J. Van Broeckhoven C. Locus-specific mutation databases for neurodegenerative brain diseases. Hum Mutat. 2012;33:1340–1344. doi: 10.1002/humu.22117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney N, Walther F, Mantel PY, Stauffer D, Rovelli G. Dev KK. The cellular protein level of parkin is regulated by its ubiquitin-like domain. J Biol Chem. 2003;278:16054–16058. doi: 10.1074/jbc.C300051200. [DOI] [PubMed] [Google Scholar]
- Sakata E, Yamaguchi Y, Kurimoto E, Kikuchi J, Yokoyama S, Yamada S, Kawahara H, Yokosawa H, Hattori N, Mizuno Y, et al. Parkin binds the Rpn10 subunit of 26S proteasomes through its ubiquitin-like domain. EMBO Rep. 2003;4:301–306. doi: 10.1038/sj.embor.embor764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe JF, Chen CXQ, Grenier K, Camacho EM, Kozlov G, McPherson PS, Gehring K. Fon EA. SH3 domains from a subset of BAR proteins define a Ubl-binding domain and implicate parkin in synaptic ubiquitination. Mol Cell. 2009;36:1034–1047. doi: 10.1016/j.molcel.2009.11.021. [DOI] [PubMed] [Google Scholar]
- Bai JJ, Safadi SS, Mercier P, Barber KR. Shaw GS. Ataxin-3 is a multivalent ligand for the parkin Ubl domain. Biochemistry. 2013;52:7369–7376. doi: 10.1021/bi400780v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt DE, Martinez-Torres RJ, Noh YJ, Mercier P, Manczyk N, Barber KR, Aguirre JD, Burchell L, Purkiss A, Walden H, et al. A molecular explanation for the recessive nature of parkin-linked Parkinson’s disease. Nat Commun. 2013;4:1983. doi: 10.1038/ncomms2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove KK. Klevit RE. Structural biology: parkin’s serpentine shape revealed in the year of the snake. Curr Biol. 2013;23:R691–R693. doi: 10.1016/j.cub.2013.07.039. [DOI] [PubMed] [Google Scholar]
- Hristova VA, Beasley SA, Rylett RJ. Shaw GS. Identification of a novel Zn2+-binding domain in the autosomal recessive juvenile parkinson-related E3 ligase parkin. J Biol Chem. 2009;284:14978–14986. doi: 10.1074/jbc.M808700200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnstrom K, Yan J, Nguyen L, Callaway K, Yang Y, Diep L, Xing W, Adhikari A, Beroza P, Hom RK, et al. Label free fragment screening using surface plasmon resonance as a tool for fragment finding - analyzing parkin, a difficult CNS target. PLoS ONE. 2013;8:e66879. doi: 10.1371/journal.pone.0066879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley SA, Hristova VA. Shaw GS. Structure of the parkin in-between-ring domain provides insights for E3-ligase dysfunction in autosomal recessive Parkinson’s disease. Proc Natl Acad Sci USA. 2007;104:3095–3100. doi: 10.1073/pnas.0610548104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda DM, Olszewski JL, Schuermann JP, Kurinov I, Miller DJ, Nourse A, Alpi AF. Schulman BA. Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an ariadne-family E3 and insights into ligation mechanism. Structure. 2013;21:1030–1041. doi: 10.1016/j.str.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieglitz B, Rana RR, Koliopoulos MG, Morris-Davies AC, Schaeffer V, Christodoulou E, Howell S, Brown NR, Dikic I. Rittinger K. Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP. Nature. 2013;503:422–426. doi: 10.1038/nature12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieglitz B, Morris-Davies AC, Koliopoulos MG, Christodoulou E. Rittinger K. LUBAC synthesizes linear ubiquitin chains via a thioester intermediate. EMBO Rep. 2012;13:840–846. doi: 10.1038/embor.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madegowda RH, Kishore A. Anand A. Mutational screening of the parkin gene among South Indians with early onset Parkinson’s disease. J Neurol Neurosurg Psychiatry. 2005;76:1588–1590. doi: 10.1136/jnnp.2004.046888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziviani E, Tao RN. Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates Mitofusin. Proc Natl Acad Sci USA. 2010;107:5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iguchi M, Kujuro Y, Okatsu K, Koyano F, Kosako H, Kimura M, Suzuki N, Uchiyama S, Tanaka K. Matsuda N. Parkin-catalyzed ubiquitin-ester transfer is triggered by PINK1-dependent phosphorylation. J Biol Chem. 2013;288:22019–22032. doi: 10.1074/jbc.M113.467530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okatsu K, Oka T, Iguchi M, Imamura K, Kosako H, Tani N, Kimura M, Go E, Koyano F, Funayama M, et al. PINK1 autophosphorylation upon membrane potential dissipation is essential for parkin recruitment to damaged mitochondria. Nat Commun. 2012;3:1016. doi: 10.1038/ncomms2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, Gourlay R, Burchell L, Walden H, Macartney TJ, Deak M, et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012;2:120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyano F, Okatsu K, Kosako H, Tamura Y, Go E, Kimura M, Kimura Y, Tsuchiya H, Yoshihara H, Hirokawa T, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014;510:162–166. doi: 10.1038/nature13392. [DOI] [PubMed] [Google Scholar]
- Kazlauskaite A, Kondapalli C, Gourlay R, Campbell DG, Ritorto MS, Hofmann K, Alessi DR, Knebel A, Trost M. Muqit MMK. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser(65) Biochem J. 2014;460:127–139. doi: 10.1042/BJ20140334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane LA, Lazarou M, Fogel AI, Li Y, Yamano K, Sarraf SA, Banerjee S. Youle RJ. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba-Fukushima K, Arano T, Matsumoto G, Inoshita T, Yoshida S, Ishihama Y, Ryu KY, Nukina N, Hattori N. Imai Y. Phosphorylation of mitochondrial polyubiquitin by PINK1 promotes parkin mitochondrial tethering. PLoS Genet. 2014;10:e1004861. doi: 10.1371/journal.pgen.1004861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wauer T, Swatek KN, Wagstaff JL, Gladkova C, Pruneda JN, Michel MA, Gersch M, Johnson CM, Freund SM. Komander D. Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. EMBO J. 2015;34:307–325. doi: 10.15252/embj.201489847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XD. Hunter T. Parkin mitochondrial translocation is achieved through a novel catalytic activity coupled mechanism. Cell Res. 2013;23:886–897. doi: 10.1038/cr.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier K, McLelland GL. Fon EA. Parkin- and PINK1-dependent mitophagy in neurons: will the real pathway please stand up? Front Neurol. 2013;4:100. doi: 10.3389/fneur.2013.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Zakaria HM, Simone A. Sheng ZH. Spatial parkin translocation and degradation of damaged mitochondria via mitophagy in live cortical neurons. Curr Biol. 2012;22:545–552. doi: 10.1016/j.cub.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier K, Kontogiannea M. Fon EA. Short mitochondrial ARF triggers parkin/PINK1-dependent mitophagy. J Biol Chem. 2014;289:29519–29530. doi: 10.1074/jbc.M114.607150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi G, Schlehe JS, LaVoie MJ. Schwarz TL. Mitophagy of damaged mitochondria occurs locally in distal neuronal axons and requires PINK1 and Parkin. J Cell Biol. 2014;206:655–670. doi: 10.1083/jcb.201401070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarffe LA, Stevens DA, Dawson VL. Dawson TM. Parkin and PINK1: much more than mitophagy. Trends Neurosci. 2014;37:315–324. doi: 10.1016/j.tins.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLelland GL, Soubannier V, Chen CX, McBride HM. Fon EA. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 2014;33:282–295. doi: 10.1002/embj.201385902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubannier V, McLelland GL, Zunino R, Braschi E, Rippstein P, Fon EA. McBride HM. A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr Biol. 2012;22:135–141. doi: 10.1016/j.cub.2011.11.057. [DOI] [PubMed] [Google Scholar]
- Vincow ES, Merrihew G, Thomas RE, Shulman NJ, Beyer RP, MacCoss MJ. Pallanck LJ. The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc Natl Acad Sci USA. 2013;110:6400–6405. doi: 10.1073/pnas.1221132110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda Y, Mitsui T, Kunishige M, Shono M, Akaike M, Azuma H. Matsumoto T. Parkin enhances mitochondrial biogenesis in proliferating cells. Hum Mol Genet. 2006;15:883–895. doi: 10.1093/hmg/ddl006. [DOI] [PubMed] [Google Scholar]
- Rothfuss O, Fischer H, Hasegawa T, Maisel M, Leitner P, Miesel F, Sharma M, Bornemann A, Berg D, Gasser T, et al. Parkin protects mitochondrial genome integrity and supports mitochondrial DNA repair. Hum Mol Genet. 2009;18:3832–3850. doi: 10.1093/hmg/ddp327. [DOI] [PubMed] [Google Scholar]
- Shin JH, Ko HS, Kang H, Lee Y, Lee YI, Pletinkova O, Troconso JC, Dawson VL. Dawson TM. PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson’s disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Karuppagounder SS, Shin JH, Lee YI, Ko HS, Swing D, Jiang H, Kang SU, Lee BD, Kang HC, et al. Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss. Nat Neurosci. 2013;16:1392–1400. doi: 10.1038/nn.3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekholm-Reed S, Goldberg MS, Schlossmacher MG. Reed SI. Parkin-dependent degradation of the F-box protein Fbw7beta promotes neuronal survival in response to oxidative stress by stabilizing Mcl-1. Mol Cell Biol. 2013;33:3627–3643. doi: 10.1128/MCB.00535-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Rischart AK, Pilsl A, Beaudette P, Patra M, Hadian K, Funke M, Peis R, Deinlein A, Schweimer C, Kuhn PH, et al. The E3 ligase parkin maintains mitochondrial integrity by increasing linear ubiquitination of NEMO. Mol Cell. 2013;49:908–921. doi: 10.1016/j.molcel.2013.01.036. [DOI] [PubMed] [Google Scholar]
- Hwang S, Kim D, Choi G, An SW, Hong YK, Suh YS, Lee MJ. Cho KS. Parkin suppresses c-Jun N-Terminal kinase-induced cell death via transriptional regulation in drosophila. Mol Cells. 2010;29:575–580. doi: 10.1007/s10059-010-0068-1. [DOI] [PubMed] [Google Scholar]
- Jiang HB, Ren Y, Zhao JH. Feng J. Parkin protects human dopaminergic neuroblastoma cells against dopamine-induced apoptosis. Hum Mol Genet. 2004;13:1745–1754. doi: 10.1093/hmg/ddh180. [DOI] [PubMed] [Google Scholar]
- Ren Y, Jiang H, Yang F, Nakaso K. Feng J. Parkin protects dopaminergic neurons against microtubule-depolymerizing toxins by attenuating microtubule-associated protein kinase activation. J Biol Chem. 2009;284:4009–4017. doi: 10.1074/jbc.M806245200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollville E, Carroll RG, Cullen SP. Martin SJ. Bcl-2 family proteins participate in mitochondrial quality control by regulating Parkin/PINK1-dependent mitophagy. Mol Cell. 2014;55:451–466. doi: 10.1016/j.molcel.2014.06.001. [DOI] [PubMed] [Google Scholar]
- Johnson BN, Berger AK, Cortese GP. LaVoie MJ. The ubiquitin E3 ligase parkin regulates the proapoptotic function of Bax. Proc Natl Acad Sci USA. 2012;109:6283–6288. doi: 10.1073/pnas.1113248109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger AK, Cortese GP, Amodeo KD, Weihofen A, Letai A. LaVoie MJ. Parkin selectively alters the intrinsic threshold for mitochondrial cytochrome c release. Hum Mol Genet. 2009;18:4317–4328. doi: 10.1093/hmg/ddp384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Lee S, Peng Y, Bunker E, Giaime E, Shen J, Zhou Z. Liu X. PINK1 triggers autocatalytic activation of parkin to specify cell fate decisions. Curr Biol. 2014;24:1854–1865. doi: 10.1016/j.cub.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Lin M, Wu R, Wang X, Yang B, Levine AJ, Hu W. Feng Z. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc Natl Acad Sci USA. 2011;108:16259–16264. doi: 10.1073/pnas.1113884108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari R, Martin ES, Calin GA, Pentimalli F, Bichi R, McAdams H, Trapasso F, Drusco A, Shimizu M, Masciullo V, et al. Parkin, a gene implicated in autosomal recessive juvenile parkinsonism, is a candidate tumor suppressor gene on chromosome 6q25-q27. Proc Natl Acad Sci U S A. 2003;100:5956–5961. doi: 10.1073/pnas.0931262100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M, Marusawa H, Wang HQ, Iwai A, Ikeuchi K, Imai Y, Kataoka A, Nukina N, Takahashi R. Chiba T. Parkin as a tumor suppressor gene for hepatocellular carcinoma. Oncogene. 2008;27:6002–6011. doi: 10.1038/onc.2008.199. [DOI] [PubMed] [Google Scholar]
- Gong Y, Zack TI, Morris LG, Lin K, Hukkelhoven E, Raheja R, Tan IL, Turcan S, Veeriah S, Meng S, et al. Pan-cancer genetic analysis identifies PARK2 as a master regulator of G1/S cyclins. Nat Genet. 2014;46:588–594. doi: 10.1038/ng.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeriah S, Taylor BS, Meng S, Fang F, Yilmaz E, Vivanco I, Janakiraman M, Schultz N, Hanrahan AJ, Pao W, et al. Somatic mutations of the Parkinson’s disease-associated gene PARK2 in glioblastoma and other human malignancies. Nat Genet. 2010;42:77–82. doi: 10.1038/ng.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira MT, Alcais A, Nguyen VT, Moraes MO, Di Flumeri C, Vu HT, Mai CP, Nguyen TH, Nguyen NB, Pham XK, et al. Susceptibility to leprosy is associated with PARK2 and PACRG. Nature. 2004;427:636–640. doi: 10.1038/nature02326. [DOI] [PubMed] [Google Scholar]
- Manzanillo PS, Ayres JS, Watson RO, Collins AC, Souza G, Rae CS, Schneider DS, Nakamura K, Shiloh MU. Cox JS. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature. 2013;501:512–516. doi: 10.1038/nature12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingol B, Tea JS, Phu L, Reichelt M, Bakalarski CE, Song Q, Foreman O, Kirkpatrick DS. Sheng M. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature. 2014;510:370–375. doi: 10.1038/nature13418. [DOI] [PubMed] [Google Scholar]
- Cornelissen T, Haddad D, Wauters F, Van Humbeeck C, Mandemakers W, Koentjoro B, Sue C, Gevaert K, De Strooper B, Verstreken P, et al. The deubiquitinase USP15 antagonizes parkin-mediated mitochondrial ubiquitination and mitophagy. Hum Mol Genet. 2014;23:5227–5242. doi: 10.1093/hmg/ddu244. [DOI] [PMC free article] [PubMed] [Google Scholar]