Summary
CARD9 is a central component of anti-fungal innate immune signaling via C-type lectin receptors, and several immune-related disorders are associated with CARD9 mutations. Here we used a rare CARD9 variant that confers protection against inflammatory bowel disease as an entry point to investigate CARD9 regulation. We showed that the C-terminal truncated CARD9 protective variant acted in a dominant negative manner for CARD9-mediated cytokine production, indicating an important role for the C terminus in CARD9 signaling. We identified TRIM62 as a CARD9 binding partner and showed that TRIM62 facilitated K27-linked poly-ubiquitination of CARD9. We identified K125 as the ubiquitinated residue on CARD9 and demonstrated that this ubiquitination was essential for CARD9 activity. Furthermore, we showed that Trim62-deficient mice have increased susceptibility to fungal infection, similar to Card9-deficient mice. This study utilizes a rare protective allele to uncover a TRIM62-mediated mechanism for regulation of CARD9 activation.
Introduction
CARD9 is a central adaptor protein in innate immune signaling via C-type lectin receptors (CLRs) such as Dectin-1, Dectin-2, and Mincle (Goodridge et al., 2009; Gross et al., 2006; Ishikawa et al., 2009; Roth and Ruland, 2013; Saijo et al., 2010; Schoenen et al., 2010; Werninghaus et al., 2009), and has more recently been reported to regulate cytokine production induced by cytosolic nucleic acid sensors RIG-I and Rad50 in mouse models (Abdullah et al., 2012; Poeck et al., 2010; Roth et al., 2014). CLRs sense components of fungal and bacterial cell walls, linking signaling from these immune receptors to nuclear factor-κB (NF-κB) activation through a series of sequential phosphorylation events. CARD9 thereby mediates pro-inflammatory cytokine production, including TNFα, IL-6, and IL-1β, ultimately regulating the responses of Th1 and Th17 cells (Drummond et al., 2011; Glocker et al., 2009; LeibundGut-Landmann et al., 2007; Marakalala et al., 2010; Robinson et al., 2009; Saijo et al., 2010; Sokol et al., 2013).
CARD9 was recently identified as a gene associated with determining risk for inflammatory bowel disease (IBD) (Jostins et al., 2012; Rivas et al., 2011), ankylosing spondylitis (Pointon et al., 2010), primary sclerosing cholangitis (Janse et al., 2011), and IgA nephropathy (Kiryluk et al., 2014). Patients with early stop codons or point mutations in the N-terminal portion of CARD9 also show increased susceptibility to fungal infection (Drewniak et al., 2013; Gavino et al., 2014; Gazendam et al., 2014; Glocker et al., 2009; Grumach et al., 2015; Herbst et al., 2015; Jachiet et al., 2015; Lanternier et al., 2015a; Lanternier et al., 2015b; Lanternier et al., 2013; Wang et al., 2013, 2014). Unlike most genetic risk factors for complex diseases, CARD9 alleles exist in both predisposing and protective forms for IBD. The predisposing variant, CARD9 S12N, is a common coding single nucleotide polymorphism that was identified via genome-wide association studies (GWAS) and is associated with increased expression of CARD9 mRNA (Franke et al., 2010; Jostins et al., 2012; McGovern et al., 2010; Zhernakova et al., 2008). The protective variant, CARD9 S12NΔ11, is a rare splice variant in which exon 11 of CARD9 is deleted. This allele, identified by deep sequencing of GWAS loci, results in a protein with a C-terminal truncation and confers strong protection against disease (P < 10−16) (Beaudoin et al., 2013; Rivas et al., 2011).
The CARD9 signaling cascade is initiated following engagement of Dectin-1 by β-glucans, which results in the phosphorylation of Dectin-1 itself, or engagement of Dectin-2 or Mincle by fungal mannans, which results in phosphorylation of the ITAM-containing signaling adaptor FcRγ. These phosphorylation events activate Syk kinase, which subsequently activates PKCδ to phosphorylate CARD9 at T231 (Strasser et al., 2012). Phosphorylated CARD9 recruits BCL10 and MALT1 to form a CARD9-BCL10-MALT1 (CBM) complex, which activates the canonical NF-κB pathway (Roth and Ruland, 2013). Rubicon acts as a feedback inhibitor, displacing CARD9 from the CBM complex and thereby terminating CARD9-mediated signaling (Yang et al., 2012). Interestingly, a recent report showed that Dectin-1-CARD9 signaling induces neutrophilic myeloid-derived suppressor cells; these cells functionally suppress T and NK cell responses, suggesting that the CARD9 pathway may play an important role in balancing inflammation in response to pathogenic fungi (Rieber et al., 2015).
Aside from the kinase-dependent steps that contribute to the initiation of the CBM signalosome and Rubicon-dependent feedback inhibition, little is known regarding other mechanisms that may regulate the assembly, stability, or activity of CARD9 in this complex. CARD9 is a scaffold protein with an N-terminal domain composed of a CARD domain followed by two coiled-coil domains. However, CARD9 has no clear domain within its C terminus and its mode of regulation is not fully defined (Hara and Saito, 2009; Roth and Ruland, 2013).
In this study, we use disease-associated CARD9 alleles to uncover fundamental insights into the protein-protein interactions and post-translational modifications that regulate CARD9 function. We demonstrate that the C terminus of CARD9 is a critical regulatory module for CARD9 activity and identify TRIM62 as a novel interactor with the CARD9 C terminus. We show that TRIM62 ubiquitinates CARD9 at K125 and demonstrate that a CARD9 mutation at this residue (K125R) abrogates CLR-induced CARD9-mediated cytokine production. Furthermore, Trim62−/− mice show reduced CLR-CARD9-dependent cytokine production and increased susceptibility to fungal infection. In parallel, we show that the protective CARD9 Δ11 variant acts in a dominant negative fashion for CLR-CARD9-dependent cytokine signaling and that TRIM62-mediated ubiquitination does not occur in this variant.
Results
The C Terminus of CARD9 Is Essential for CARD9 Activation
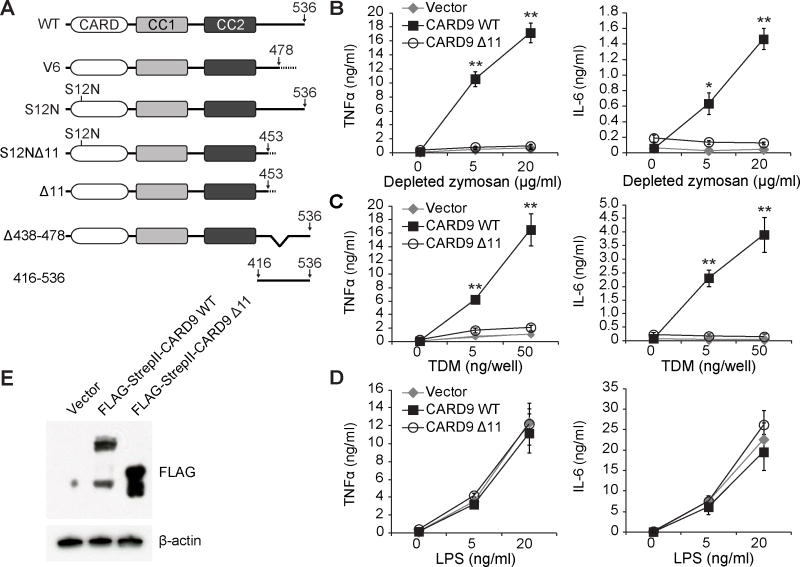
The association of CARD9 with both protective and predisposing alleles for IBD raises the need to characterize regulatory mechanisms for CARD9-dependent pathways. To this end, we employed naturally occurring truncated CARD9 variants to investigate CARD9 regulation. We first used a human immune cDNA panel to collect a library of naturally occurring human CARD9 alleles and screened this collection for effects on CARD9-mediated cytokine production. In addition to full-length CARD9 and the disease-associated S12N and S12NΔ11 alleles, we identified several novel variants of CARD9 (Figures 1A and S1A), noting that the majority of the allelic variations were located in the C-terminal portion of the protein.
Figure 1. C-terminal CARD9 Truncations Impair Depleted Zymosan- and TDM-induced Cytokine Production.
(A) Schematic of CARD9 variants used. All indicated variants were found in a human immune cDNA panel with the exception of CARD9 416-536, which was designed for experimental use. CC: coiled-coil domain. (B–D) Card9−/− murine BMDCs transduced with indicated CARD9 variants were stimulated with either depleted zymosan, TDM or LPS and cytokine levels were assessed by ELISA. Data were obtained from three independent experiments performed in duplicate (n = 3). (E) Expression of CARD9 variants in total lysates from (B–D) as detected by Western blot. Bars represent means ± s.d. *P < 0.05, **P < 0.01. Comparisons in (B–D) are relative to stimulated CARD9 WT. See also Figure S1.
To definitively evaluate whether the deletion of exon 11 (Δ11) imparts protection from disease, we analyzed Immunochip data sets with 33,311 IBD cases and 33,938 healthy controls from the International Inflammatory Bowel Disease Genetics Consortium (IIBDGC) and found that individuals with the Δ11 splice variant are less likely to develop IBD regardless of the presence of S12N mutation (Figure S1B), further suggesting an important functional role for the C terminus of CARD9.
To examine how C-terminal truncated variants affect CARD9 function, we stimulated CARD9-dependent pathways using either depleted zymosan, a glucan-enriched preparation derived from fungal cell walls that mainly activates Dectin-1 (Gross et al., 2006), or trehalose 6,6-dimycolate (TDM), a component of mycobacterial cell walls that activates two other CLRs, Mincle and macrophage C-type lectin (Miyake et al., 2013; Roth and Ruland, 2013; Werninghaus et al., 2009). Activation by these two ligands led to CARD9-dependent production of proinflammatory cytokines, including TNFα and IL-6 (Figure S1C) (Gross et al., 2006; Saijo et al., 2010; Werninghaus et al., 2009). We therefore re-expressed human CARD9 isoforms in murine Card9−/− bone marrow-derived dendritic cells (BMDCs) and assessed cytokine production in response to these ligands by ELISA (Figures 1B–1E and S1D–S1F). BMDCs expressing the predisposing variant CARD9 S12N showed increased TNFα and IL-6 production compared to BMDCs expressing wild-type CARD9. In contrast, CARD9 Δ11 and CARD9 S12NΔ11, as well as the C-terminal truncated variant CARD9 V6, showed significant impairment in TNFα and IL-6 production. This effect did not occur when cytokine production was stimulated by lipopolysaccharide (LPS), a ligand for Toll-like receptor 4 (TLR4), indicating that the impairment was specific to the CARD9 pathway (Figures 1D and S1E). These data suggest that the C terminus of CARD9 is required for CLR-CARD9-mediated cytokine production.
A Protective CARD9 Variant Acts as Dominant Negative
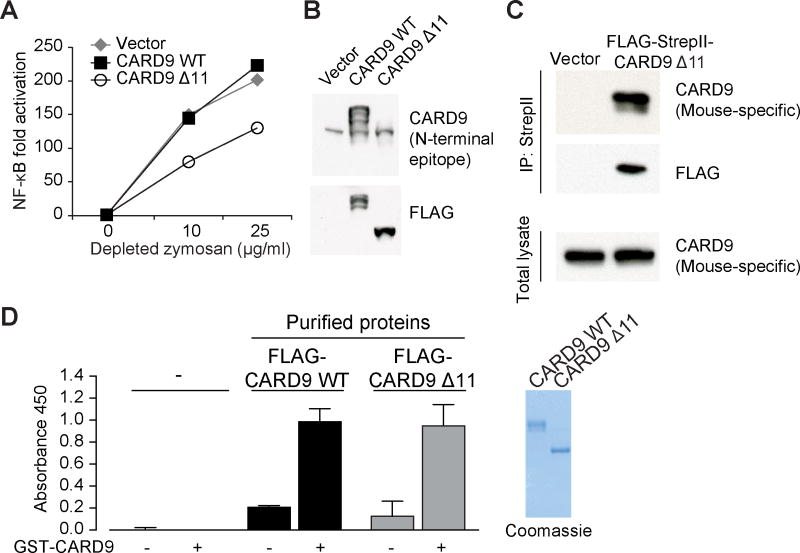
Pursuing our finding that CARD9 Δ11 demonstrates loss of CARD9 function (Figure 1), we next examined whether this variant would have a dominant negative effect on CARD9 function when co-expressed with wild-type CARD9 in human cells. To test this hypothesis, we expressed CARD9 Δ11 in the human monocytic cell line THP-1, as well as in primary human monocyte-derived dendritic cells (MDDCs) and found that CARD9 Δ11 suppressed depleted zymosan-induced NF-κB activation in THP-1 cells (Figures 2A and 2B) as well as depleted zymosan-induced TNFα production in MDDCs (Figures S2A and S2B). Consistent with these findings, we also expressed CARD9 Δ11 in wild-type murine BMDCs and found that CARD9 Δ11 inhibited TDM-induced TNFα and IL-6 production (Figures S2C and S2D). In contrast, LPS-induced cytokine production was unaltered under the same experimental conditions in both human MDDCs and murine BMDCs (Figures S2E and S2F). Collectively, the results indicate that CARD9 Δ11 has a dominant negative effect in both human and mouse dendritic cells. Furthermore, FLAG-tagged human CARD9 Δ11 co-immunoprecipitated with endogenous CARD9 in wild-type murine BMDCs (Figure 2C). This result demonstrates that CARD9 Δ11 can form a complex with endogenous full-length CARD9, suggesting that CARD9 Δ11 may inhibit CARD9 function via hetero-oligomerization with full-length CARD9. To pursue these findings, we developed an ELISA-based assay that detects the interaction between differentially tagged purified CARD9 proteins in vitro in a cell-free system. By using GST- and FLAG-tagged purified CARD9 protein constructs, we confirmed that both wild-type CARD9 and CARD9 Δ11 are capable of directly interacting with CARD9. Importantly, the ability of CARD9 Δ11 to form complexes with wild-type CARD9 was not impaired (Figure 2D).
Figure 2. CARD9 Δ11 Is a Dominant Negative Variant.
(A) THP-1 cells were transduced with FLAG-CARD9 WT or FLAG-CARD9 Δ11, stimulated with depleted zymosan, and NF-κB levels were assessed by NF-κB luciferase assay. (B) Western blot showing expression of CARD9 as detected by antibodies against endogenous CARD9 (top) or FLAG (bottom) in lysates from (A). Note that the antibody detecting endogenous CARD9 has an N-terminal epitope and therefore does not recognize CARD9 Δ11. (C) WT BMDCs were transduced with FLAG-StrepII empty vector or FLAG-StrepII-tagged CARD9 Δ11 followed by immunoprecipitation of tagged CARD9 Δ11. (D) Purified FLAG-tagged CARD9 WT or CARD9 Δ11 proteins were incubated with immobilized GST-CARD9; interactions were assessed by ELISA. Coomassie gel at right shows purified proteins used in ELISA. See also Figure S2.
TRIM62 Binds the C Terminus of CARD9
Because CARD9 functions as a scaffold protein, we next investigated whether loss of specific protein-protein interactions by C-terminal truncated variants might underlie their effects on CARD9 function. Since the details of how the C terminus of CARD9 delivers signals from receptors to effectors are largely unknown, we sought to identify novel CARD9 binding partners that might be responsible for the loss of signal transduction in the truncated variants. To this end, we re-expressed a tagged version of wild-type CARD9 in immortalized Card9−/− cells (Blasi et al., 1985) and employed tandem affinity purification to purify CARD9. Using mass spectrometry, we identified TRIM62 as a top hit co-purified with CARD9 based on based high peptide counts and a high percentage of protein coverage.
Members of the TRIM family of proteins are involved in multiple cellular processes, including innate immunity (Uchil et al., 2013; Versteeg et al., 2013), and mutations in multiple TRIM-encoding genes are associated with human disease (Marin, 2012; McNab et al., 2011). However, most studies of TRIM proteins have employed overexpression or knockdown approaches (Arimoto et al., 2010; Tsuchida et al., 2010; Uchil et al., 2013; Versteeg et al., 2013), and very few studies in knockout mice or human cells have examined ligand specificity (Gack et al., 2007; Zhang et al., 2013). We next examined whether we could detect a direct interaction between CARD9 and TRIM62. To demonstrate TRIM62 binding of CARD9, we developed an ELISA-based system that employs differentially tagged CARD9 and TRIM62. These experiments demonstrated that wild-type CARD9, but not CARD9 Δ11, interacted with TRIM62 as assessed by CARD9-TRIM62 ELISA. Addition of purified C-terminal domain (CTD) of CARD9 abrogated this interaction, supporting the hypothesis that TRIM62 binds the C-terminus of CARD9 (Figure S3A). To further study the molecular interaction between CARD9 and TRIM62 and the dominant negative effect of CARD9 Δ11, we developed a cell-based BRET (bioluminescence resonance energy transfer) system that enables detection of specific protein-protein interactions in the context of an intact cell. BRET has been used to characterize disease-related genetic variants of GNAL (Fuchs et al., 2013) as well as to investigate the function of cancer-associated PTEN mutations in a dominant-negative context (Papa et al., 2014). Notably, a BRET assay was recently used to discover a novel CARD9 interactor in a physiologic setting (Roth et al., 2014). In our BRET system, nano-luciferase-tagged CARD9 (donor) can interact with Halo-tagged TRIM62 (acceptor) to generate energy measured as a BRET signal (Figure S3B). We first confirmed that the assay robustly detects an interaction between CARD9 and TRIM62. In agreement with our findings that the interaction is mediated by the C terminus of CARD9, we found that the interaction can be competitively disrupted by co-expression of the CARD9 C terminus (aa 416-536) (Figure S3C). To more precisely examine the dominant negative effect of CARD9 Δ11, we assessed the interaction between CARD9 and TRIM62 in the presence of co-expressed CARD9 Δ11 or negative controls such as unrelated proteins SULT1 and LacZ (Figure S3D). The presence of CARD9 Δ11 disrupted the CARD9-TRIM62 interaction, whereas SULT1, LacZ, or vector alone showed no effect. These results demonstrate the inhibitory effect of the protective variant on the CARD9-TRIM62 interaction, and support the hypothesis that hetero-oligomerization of wild-type CARD9 and CARD9 Δ11 may functionally inhibit wild-type CARD9 signaling.
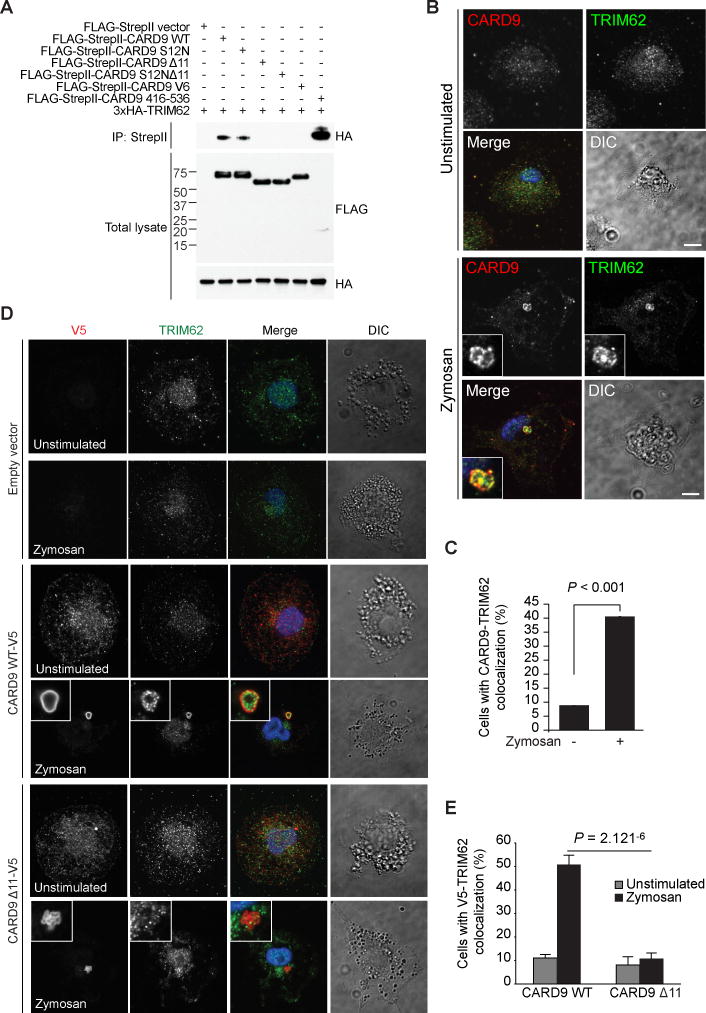
Consistent with these findings, co-immunoprecipitation studies of CARD9 and TRIM62 demonstrated that CARD9 Δ11, CARD9 S12NΔ11, and CARD9 V6 failed to bind TRIM62, whereas the CARD9 C terminus alone (aa 416-536) bound TRIM62 even more efficiently than full-length wild-type CARD9 (Figure 3A). We further confirmed these results by demonstrating that endogenous CARD9 interacts with endogenous TRIM62 in a zymosan-dependent manner in THP-1 cells (Figure S3E). These results demonstrate that these proteins interact endogenously in an activation-dependent manner.
Figure 3. TRIM62 Interacts with Full-length CARD9, but not CARD9 C-terminal Truncations.
(A) HEK293T cells were transfected with indicated constructs followed by immunoprecipitation of tagged CARD9. (B) WT BMDCs were stimulated with depleted zymosan and stained for endogenous CARD9 and TRIM62. Scale bars, 5 μm. (C) Quantification of results shown in (B). (D) CARD9−/− THP-1 cells were transduced with V5-tagged CARD9 WT or CARD9 Δ11 and stimulated with depleted zymosan. Cells were stained for endogenous TRIM62 and tagged CARD9. (E) Quantification of results shown in (D). See also Figure S3.
Having found a biochemical interaction between TRIM62 and CARD9, we next examined their colocalization by immunofluorescence. We found that TRIM62 co-localized with wild-type CARD9 and CARD9 S12N, but did not co-localize with CARD9 variants lacking the C terminus (CARD9 Δ11, CARD9 S12N Δ11, or CARD9 V6) in HeLa cells (Figures S3F–S3H); all isoforms of CARD9 showed relatively diffuse cytoplasmic localization when expressed alone, whereas TRIM62 showed a diffuse localization with some cytoplasmic bodies, consistent with localization patterns observed for TRIM5α (Campbell et al., 2007) and TRIM22 (Reymond et al., 2001) (Figures S3F–S3H). Consistent with these observations, endogenous CARD9 and TRIM62 co-localized in a zymosan-dependent manner in wild-type BMDCs (Figures 3B and 3C).
Furthermore, using THP-1 cells in which endogenous CARD9 was deleted using CRISPR, we re-expressed tagged wild-type CARD9 or CARD9 Δ11 and found that although wild-type CARD9 colocalized with endogenous TRIM62 in a stimulation-dependent manner, CARD9 Δ11 did not (Figures 3D and 3E). Notably, TRIM62 co-localized with BCL10, a well-characterized CARD9 interactor, in a zymosan-dependent manner in wild-type BMDCs, suggesting that TRIM62 may associate with CARD9 as part of the CBM complex (Figures S3I and S3J).
TRIM62 Induces K27-Linked CARD9 Ubiquitination
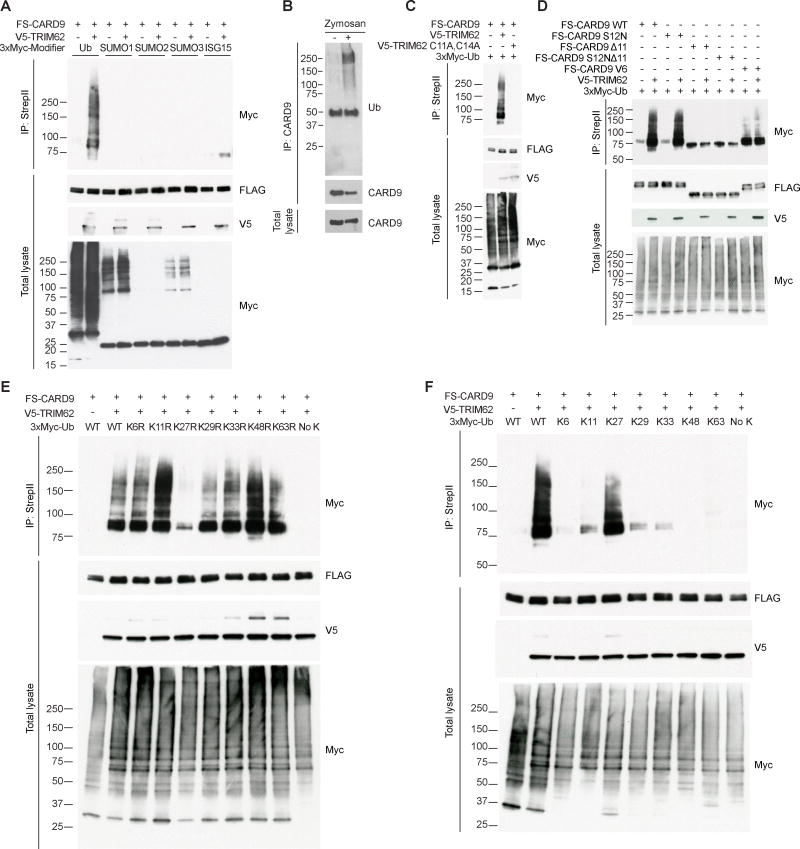
We next examined the functional consequences of the CARD9-TRIM62 interaction. TRIM family proteins generally modify their targets via E3 ligase activity, which can include ubiquitination, SUMOylation, or ISGylation (Chu and Yang, 2011; McNab et al., 2011). We found that TRIM62 specifically promotes CARD9 ubiquitination in HEK293T cells (Figure 4A) and confirmed that endogenous CARD9 is ubiquitinated in a stimulation-dependent manner in dendritic cells (Figure 4B). We next designed an E3 ligase-dead variant of TRIM62 by mutating two conserved catalytic cysteines in the RING domain to alanine (TRIM62 C11A,C14A) (Deshaies and Joazeiro, 2009), showing that CARD9 ubiquitination is dependent on E3 ligase activity in HEK293T cells (Figure 4C). Importantly, CARD9 Δ11 was not ubiquitinated by TRIM62 (Figure 4D). Similarly to wild-type TRIM62, TRIM62 C11A,C14A colocalized extensively with CARD9 (Figure S4).
Figure 4. TRIM62 Promotes the Ubiquitination of Full-length CARD9, but not CARD9 Truncations.
(A) HEK293T cells were transfected with indicated constructs followed by immunoprecipitation of tagged CARD9, then blotted for tagged ubiquitin, SUMO1, SUMO2, SUMO3, and ISG15. (B) WT BDMCs were stimulated with depleted zymosan and immunoprecipitated for CARD9, then blotted for endogenous CARD9 and ubiquitin. (C) HEK293T cells were transfected with indicated constructs followed by immunoprecipitation of tagged CARD9, then blotted for ubiquitin in the presence of either WT or a ligase-dead (C11A,C14A) version of TRIM62. (D) HEK293T cells were transfected with indicated constructs followed by immunoprecipitation of tagged CARD9, then blotted for tagged ubiquitin. (E–F) HEK293T cells were transfected with indicated constructs followed by immunoprecipitation of tagged CARD9, then blotted for indicated ubiquitin mutants. In (E), transfected ubiquitin constructs have single point mutations to indicated lysine residues. In (F), transfected ubiquitin constructs have point mutations to multiple lysines, leaving only the indicated lysine residue intact. FS, FLAG-StrepII. See also Figure S4.
Next we identified which type of ubiquitin linkage was occurring on CARD9, showing that ubiquitination was dramatically reduced by a K27R mutation in ubiquitin (Figure 4E). In a reciprocal assay using ubiquitin mutants that each contained only one lysine, we found that K27 alone was sufficient for TRIM62-mediated ubiquitination of CARD9 (Figure 4F). Taken together, we conclude that TRIM62 mediates K27-linked ubiquitination of full-length CARD9, but not CARD9 Δ11.
CARD9 Ubiquitination by TRIM62 Plays a Critical Role in CLR-CARD9 Signaling
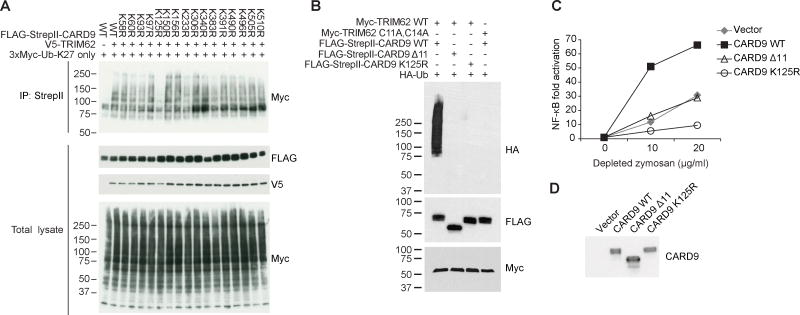
To identify the specific lysine target(s) on CARD9 that is ubiquitinated by TRIM62, we first examined all 40 lysines in human CARD9. We identified 17 of these lysines as being absolutely conserved across 17 mammalian species and elected to individually mutate each of these residues to arginine. We found that mutation of K125 significantly reduced TRIM62-mediated CARD9 poly-ubiquitination in HEK293T cells (Figure 5A). Importantly, using in vitro ubiquitination assays with purified proteins, we verified that CARD9 is directly ubiquitinated by TRIM62 at residue K125; this ubiquitination is dependent on the ligase activity of TRIM62 and does not occur in CARD9 Δ11 (Figure 5B).
Figure 5. TRIM62-mediated CARD9 Ubiquitination at K125 is Critical for CARD9 Activation.
(A) HEK293T cells were transfected with TRIM62, ubiquitin (K27 only), and indicated CARD9 point mutants, followed by immunoprecipitation of CARD9. (B) Indicated proteins were purified and incubated in vitro to assess ubiquitination of CARD9. (C) CARD9−/− THP-1 cells were reconstituted with CARD9 WT, CARD9 Δ11, or CARD9 K125R, stimulated with depleted zymosan, and NF-κB levels were assessed by NF-κB luciferase assay. (D) Western blot showing expression levels of CARD9 in cells used in (C). See also Figure S5.
To identify the functional consequence of this ubiquitination, we re-expressed this ubiquitination-deficient CARD9 in Card9−/− BMDCs. We found that K125R mutation abolished CARD9-mediated cytokine production upon depleted zymosan or TDM stimulation (Figures S5A–S5D), indicating that ubiquitination of CARD9 K125 is critical for CARD9 activation. Similar results were obtained in human CARD9−/− THP-1 cells reconstituted with wild-type CARD9 or CARD9 K125R. In this system, we found that stimulation-dependent NF-κB activity was dramatically reduced by the introduction of the K125R mutation (Figures 5C and 5D). Notably, using Card9−/− BMDCs reconstituted with wild-type CARD9 or CARD9 K125R, both the wild-type and mutant forms of CARD9 retained zymosan-dependent colocalization with BCL10 (Figures S5E and S5F). However, the CARD9-BCL10 interaction remains unproductive in the context of the K125R mutation, as shown by the disruption of NF-κB activity by this mutation. Taken together, these results demonstrate that K125-mediated ubiquitination is critical for CARD9-mediated NF-κB signaling.
TRIM62 Regulates Immune Responses and Susceptibility to Candida albicans Infection In Vivo
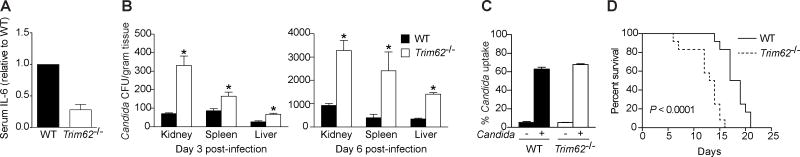
To directly assess whether the novel CARD9 interactor TRIM62 is important for immune responses elicited via the CARD9 signaling pathway, we generated Trim62−/− mice (Figures S6A and S6B). Intravenous injection of heat-killed C. albicans, a fungal pathogen that activates the CLR-CARD9 pathway, induces acute, systemic IL-6 production, which was significantly impaired in the absence of TRIM62 (Figure 6A).
Figure 6. TRIM62 Regulates Immune Responses and Susceptibility to Candida albicans Infection In Vivo.
(A) WT and Trim62−/− mice were injected i.v. with heat-killed C. albicans and serum IL-6 levels were quantified 4 h later by ELISA (n = 5). Bars display IL-6 relative to WT levels. (B) WT and Trim62−/− mice were injected with C. albicans and fungal loads in kidney, spleen, and liver were assessed at 3 and 6 days post-infection. (C) WT and Trim62−/− BMDMs were incubated with pHrodo-labeled C. albicans, and phagocytosed C. albicans was quantified by intracellular fluorescence via flow cytometry. (D) Survival curve showing percent survival over time in WT and Trim62−/− mice infected with C. albicans. Bars represent means ± s.d. *P < 0.05. See also Figure S6.
To examine pathways involved in pro-inflammatory cytokine production, we used intracellular staining and flow cytometry to measure activity of both NF-κB and mitogen-activated protein kinase (MAPK) pathways, as a recent study in macrophages identified a role for ERK signaling in susceptibility to Candida (Jia et al., 2014). Isolated wild-type and Trim62−/− splenic dendritic cells were stimulated ex vivo for ten minutes with LPS or heat-killed C. albicans. Consistent with the in vivo findings, Trim62−/− dendritic cells had less phosphorylated IκBα and ERK1/2 in response to heat-killed C. albicans stimulation, indicating impaired NF-κB and MAPK signaling, respectively (Figure S6C). This difference was not observed in response to LPS, indicating that the impaired signaling is specific to the CLR-CARD9 signaling pathway. These results are in agreement with a recent report showing that TRIM62 knockdown in human primary macrophages does not affect MyD88-dependent signaling at early time points (Uchil et al., 2013).
In addition to impaired proinflammatory pathway activation and cytokine production in response to heat-killed C. albicans, we found using live C. albicans infection that Trim62−/− mice had significantly more C. albicans CFU present in kidney, spleen, and liver (Figure 6B), suggesting impaired pathogen clearance. To determine whether the increased fungal burden in Trim62−/− mice following in vivo infection was due to defective phagocytosis, BMDMs were exposed to fluorescently labeled C. albicans and uptake was determined via flow cytometry. Uptake of C. albicans was comparable in wild-type and Trim62−/− mice, indicating that the enhanced fungal burden in Trim62−/− mice is not due to differences in phagocytosis efficiency (Figure 6C). Additionally, expression of cytokines and chemokines important during C. albicans infection was significantly lower in Trim62−/− mice than in wild-type mice, suggesting that downstream inflammatory signaling events (through the TRIM62-CARD9 pathway) are blunted (Figure S6D). Lastly, to determine whether the decreased cytokine responses and increased fungal burden in Trim62−/− mice following infection confer enhanced susceptibility to C. albicans, mice were monitored over time following in vivo infection. Strikingly, and consistent with the above data, Trim62−/− mice were more susceptible to in vivo C. albicans infection, with 100% mortality by 15 days post-infection (Figure 6D).
Given the association of this pathway with IBD, we also assessed Trim62−/− mice in the dextran sulfate sodium (DSS) model of intestinal inflammation. Similar to previously published studies in Card9−/− mice, Trim62−/− mice show increased weight loss, greater inflammation, and impaired cytokine responses compared to wild-type mice treated with DSS (Figure S6E–S6G). Taken together, these results demonstrate that TRIM62 is important for immunity in in vivo models of C. albicans infection and DSS-induced colitis.
Discussion
CARD9 coding mutations are associated with multiple immune-related diseases, underscoring the physiologic importance of CARD9 signaling in immunity. In the current study, we used a CARD9 allele strongly protective against IBD as an entry point to uncover basic insights into the biology of CARD9 regulation, as well as a potential mechanism for the protective effect of the Δ11 variant. Through systematic pathway mapping, we found a CARD9 C-terminal interacting partner that regulates CARD9 function in a ligand-specific manner; this interactor (TRIM62), which does not interact with the CARD9 protective variant, acts as an E3 ubiquitin ligase to activate CARD9 function. Given our findings that TRIM62 acts as a restriction factor for fungal infections, our results expand the list of E3 ligases that function as restriction factors, and extend the range of pathogens implicated in TRIM family signaling. The TRIM family of proteins expanded relatively recently in evolution (Ozato et al., 2008) and play important roles in the crosstalk between the innate and adaptive immune systems (Rajsbaum et al., 2014a). The majority of studies of TRIM proteins have focused on their roles as antiviral restriction factors (Ozato et al., 2008), with recent screens reporting that mRNA and localization of most known TRIM proteins are altered during viral infection (Uchil et al., 2013; Versteeg et al., 2013). Extending these studies, a recent report identified TRIM5 as a regulator of autophagy that acts as a restriction factor against HIV-1 (Mandell et al., 2014). TRIM family members largely function in negative regulatory circuits, as was recently demonstrated for TRIM9, a negative regulator of NF-κB pro-inflammatory cytokine signaling (Shi et al., 2014). An increasing number of TRIM proteins have also been described as enhancers of innate immune pathways, as in the case of TRIM44 stabilizing MAVS to enhance signaling (Yang et al., 2013). TRIM proteins mediate several types of ubiquitin linkages, with most studies focusing on K48- and K63-based linkages and their respective roles in degradation and stabilization of protein complexes (Davis and Gack, 2015; Flannick et al., 2014; Gack et al., 2007; Rajsbaum et al., 2014b; Tsuchida et al., 2010; Zhao et al., 2012), although K27-linked ubiquitination has been reported for TRIM23 (Arimoto et al., 2010). Our findings of TRIM62-mediated K27-linked ubiquitination of CARD9 suggest that this type of linkage promotes an important mechanism for protein activation, although notably the K125R mutation does not affect colocalization of CARD9 and BCL10.
To date, all clinically reported homozygous or compound heterozygous carriers with either no CARD9 expression or CARD9 loss-of-function alleles are more susceptible to fungal infection (Drewniak et al., 2013; Gavino et al., 2014; Gazendam et al., 2014; Glocker et al., 2009; Grumach et al., 2015; Herbst et al., 2015; Jachiet et al., 2015; Lanternier et al., 2015a; Lanternier et al., 2015b; Lanternier et al., 2013; Wang et al., 2014). Our results from Trim62−/− mice suggest that TRIM62-mediated regulation of CARD9 activation is critical in the context of fungal infection. These data complement previous studies reporting critical roles for CARD9 and PKCδ during C. albicans infection and suggest TRIM62 as a novel member of this pathway important for anti-fungal immunity (Gross et al., 2006; Strasser et al., 2012). Furthermore, we found that Trim62−/− mice, similar to Card9−/− mice, are more susceptible to DSS colitis, underscoring the importance of this pathway in intestinal homeostasis and fortifying previous reports on the relationship between fungi and IBD (Chehoud et al., 2015; Richard et al., 2015; Romani, 2011).
CARD9 has both a common predisposing allele and rare protective splice variant identified using next-generation sequencing (Rivas et al., 2011). The presence of both common and rare risk variants in a single gene has been now increasingly observed in multiple studies, advancing the concept that when multiple common and rare variants are discovered, the resulting allelic series can serve to anchor a dose-response relationship between gene and disease. For example, the connection between rare genetic variants and therapeutic advances has been well demonstrated by PCSK9 in LDL, where protective variation can be therapeutically mimicked (Cohen et al., 2006), and a recent finding of loss-of-function mutations in SLC30A8 that protect against type 2 diabetes (Flannick et al., 2014). Our findings demonstrate that the CARD9 protective variant identified by genetic sequencing of patients with IBD does not undergo ubiquitination by TRIM62 and propose that the protective effect of the C-terminal truncation may be mediated by loss of TRIM62 interaction, thereby limiting proinflammatory cytokine responses. Since TRIM62-dependent ubiquitination and subsequent activation of CARD9 may be potentially targetable, these findings suggest a model in which a naturally occurring protective allele may be used as a guide for rational design of therapeutics. Indeed, evolution has “validated” this approach as a safe and effective strategy for decreasing the likelihood of developing IBD.
Experimental Procedures
Plasmids
For lentiviral vectors, FLAG-StrepII fusion human CARD9 variants were constructed by PCR-based subcloning of CARD9 coding sequences into a lentivirus-based CSGW vector backbone (gift from Christian Münz, University of Zurich). For other CARD9 constructs, CARD9 was either cloned into a pCMV vector (see section describing CARD9 variant cloning below) or pcDNA4/TO-FLAG-StrepII. Ubiquitin cDNA was kindly provided by Dr. M. Scheffner (University of Konstanz, Germany). All other genes were originally obtained from either OpenBiosystems or Origene and subcloned into indicated tagged vectors by PCR. WT and mutant ubiquitin, SUMO1, SUMO2, SUMO3, and ISG15 were subcloned into pCMV-3xMyc. pCMV-3xHA and pCMV-3xMyc were derived from pCMV-Myc (Clontech). TRIM62 constructs were subcloned into either pCMV-3xHA or pcDNA4/TO-V5. pcDNA4/TO-FLAG-StrepII and pcDNA4/TO-V5 were derived from pcDNA4/TO (Invitrogen). FLAG-StrepII, HA, and Myc are all N-terminal tags. All cDNAs were confirmed by DNA sequencing.
Chemical Reagents
Depleted zymosan was purchased from Invivogen. Trehalose 6,6′-dimycolate (TDM) was obtained from Enzo Life Sciences. TDM was dissolved in chloroform:methanol:water (90:10:1, 1 mg/ml) and further diluted with isopropanol. LPS was purchased from Sigma or Invivogen.
Cell Culture and Lentiviral Production
HEK293T and HeLa cells were maintained at 37°C and 5% CO2 in DMEM supplemented with 10% fetal calf serum and 15 μg/ml gentamycin sulfate. THP-1 cells were maintained at 37°C and 5% CO2 in RPMI1640 supplemented with 10% fetal calf serum, 5×10−4M beta-mercaptoethanol and 15 μg/ml gentamycin sulfate. To prepare lentivirus for infection, protocols from the Broad Institute’s RNAi Consortium shRNA Library were used (http://www.broadinstitute.org/rnai/trc/lib). Lentivirus qPCR titer kit (Applied Biological Materials) was used for lentiviral titration. pSIV3+ plasmid for generating Vpx-VLP was a gift from Dr. Andrea Cimarelli (ENS de Lyon, France) (Berger et al., 2011).
Trim62−/− Mice Strain
Trim62 heterozygote mice on 129/SvEv genetic background were purchased from Taconic (Cat. No: TF2743). Trim62 heterozygote mice, derived from a F1 generation of the Trim62 heterozygotes mice from Taconic and Trim62 wild type C57BL/6, were bred to generate Trim62 WT and knockout littermates for the functional assay. The targeted locus is located at the junction of the first exon and first intron of Trim62, resulting in deletion of a portion of the exon 1 coding region and intron 1 of Trim62. See also Figure S6. Trim62−/− mice are viable and born at Mendelian ratios.
Preparation of Heat-Killed Candida albicans
C. albicans strain SC5314 (ATCC: MYA-2876 was obtained from Eleftherios Mylonakis (Massachusetts General Hospital, Boston). C. albicans were harvested from an overnight culture in YPD medium (Y1375, Sigma) at 30°C. C. albicans were washed with PBS twice and resuspended into PBS in 2 × 108/ml. Resuspended C. albicans were then incubated for 2 h at 68°C followed by cooling on ice. Heat-killed cell death was verified by plating cells on YPD agar plates.
In Vivo Injection of Heat-Killed C. albicans
1 × 107 heat-killed C. albicans in PBS were injected i.v. After 4 h, serum was harvested and IL-6 levels were determined via ELISA.
Ex Vivo Splenic DC Stimulations and Intracellular Phosphoprotein Staining
CD11c+ cells were positively selected from the spleen using MACS technology per the manufacturer’s protocol (Miltenyi Biotec). 1 × 106 cells were resuspended in complete RPMI (10% FBS and 15 μg/mL gentamicin) and incubated for 10 min at 37°C with 5% CO2 in one of three conditions: (1) no stimulation, (2) 100 ng/mL LPS stimulation, or (3) 1 × 106 heat-killed C. albicans stimulation. Cells were immediately fixed using BD Cytofix Fixation Buffer and permeabilized using BD Phosflow Permeabilization Buffer 3, per the manufacturer’s protocol (BD Biosciences). After 20 min of Fc block on ice, cells were stained as indicated with the following antibodies: CD11c-PECy7 (BD Biosciences), phospho-ERK1/2-PE (BD Biosciences), phospho-IκBα (Cell Signaling Technologies), and donkey anti-rabbit IgG DyLight 488 (BioLegend). Cells were acquired on the BD FACSVerse (BD Biosciences) and analyzed using FlowJo Software.
C. albicans for In Vivo Infection
C. albicans (strain ATCC 90028) were grown on yeast-peptone-dextrose agar (Difco) at 30°C. For each infection, a fresh culture of C. albicans was started from −80°C stocks. For experiments, a fresh colony was isolated from an agar plate and grown in YPD media for 24 h at 30°C. Prior to inoculation, yeast cells were washed three times in PBS, counted, and adjusted to the appropriate concentration.
Viable C. albicans Counts
Mice were infected i.v. with 105 C. albicans and sacrificed at days 3 and 5 post infection. The CFU of C. albicans in the homogenized kidney, spleen, and liver were determined by plate counts. Data are expressed as CFU per mg of tissue.
Mice
Mice were maintained in specific-pathogen-free facilities at Massachusetts General Hospital. All animal studies were conducted under protocols approved by the Institutional Animal Care and Use Committee (IACUC) at Massachusetts General Hospital.
Statistical Analysis
Unpaired two-tailed Student’s t-tests were used for comparisons for ELISA. All statistical comparisons were made between multiple independent experiments performed in parallel.
Supplementary Material
Acknowledgments
We thank Drs. Hiromitsu Hara (Saga Medical School, Japan) for providing Card9−/− mice (Hara et al., 2007), Kara Lassen for guidance with manuscript preparation, and Petric Kuballa, Jakob Begun, and David Fei for assistance with experiments. This work was supported by funding from the Helmsley Trust and N.I.H. grants DK097485, DK062432, and DK086502 to R.J.X. M.G.N. was supported by an ERC Consolidator Grant (nr.310372). T.K.M. was supported by N.I.H. grant AI084884. Z.G.R.-O. was supported by N.I.H. grant AR066716.
Footnotes
Author Contributions
R.J.X. and Z.C. conceived the project. Z.C., K.L.C., R.J.H, J.S.R., E.S.L., Z.G.R.-O., A.N., A.G., S.C.C., and T.K.M. performed experiments and H.H analyzed Immunochip data. R.J.X., M.J.D., and T.K.M. supervised the project. A.F.S., J.D.R., C.W., and M.G.N. provided intellectual contributions throughout the project. R.J.X., N.N., and Z.C. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdullah Z, Schlee M, Roth S, Mraheil MA, Barchet W, Bottcher J, Hain T, Geiger S, Hayakawa Y, Fritz JH, et al. RIG-I detects infection with live Listeria by sensing secreted bacterial nucleic acids. EMBO J. 2012;31:4153–4164. doi: 10.1038/emboj.2012.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimoto K, Funami K, Saeki Y, Tanaka K, Okawa K, Takeuchi O, Akira S, Murakami Y, Shimotohno K. Polyubiquitin conjugation to NEMO by triparite motif protein 23 (TRIM23) is critical in antiviral defense. Proc Natl Acad Sci U S A. 2010;107:15856–15861. doi: 10.1073/pnas.1004621107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin M, Goyette P, Boucher G, Lo KS, Rivas MA, Stevens C, Alikashani A, Ladouceur M, Ellinghaus D, Torkvist L, et al. Deep resequencing of GWAS loci identifies rare variants in CARD9, IL23R and RNF186 that are associated with ulcerative colitis. PLoS Genet. 2013;9:e1003723. doi: 10.1371/journal.pgen.1003723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger G, Durand S, Goujon C, Nguyen XN, Cordeil S, Darlix JL, Cimarelli A. A simple, versatile and efficient method to genetically modify human monocyte-derived dendritic cells with HIV-1-derived lentiviral vectors. Nat Protoc. 2011;6:806–816. doi: 10.1038/nprot.2011.327. [DOI] [PubMed] [Google Scholar]
- Blasi E, Mathieson BJ, Varesio L, Cleveland JL, Borchert PA, Rapp UR. Selective immortalization of murine macrophages from fresh bone marrow by a raf/myc recombinant murine retrovirus. Nature. 1985;318:667–670. doi: 10.1038/318667a0. [DOI] [PubMed] [Google Scholar]
- Campbell EM, Dodding MP, Yap MW, Wu X, Gallois-Montbrun S, Malim MH, Stoye JP, Hope TJ. TRIM5 alpha cytoplasmic bodies are highly dynamic structures. Mol Biol Cell. 2007;18:2102–2111. doi: 10.1091/mbc.E06-12-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehoud C, Albenberg LG, Judge C, Hoffmann C, Grunberg S, Bittinger K, Baldassano RN, Lewis JD, Bushman FD, Wu GD. Fungal Signature in the Gut Microbiota of Pediatric Patients With Inflammatory Bowel Disease. Inflamm Bowel Dis. 2015 doi: 10.1097/MIB.0000000000000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y, Yang X. SUMO E3 ligase activity of TRIM proteins. Oncogene. 2011;30:1108–1116. doi: 10.1038/onc.2010.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- Davis ME, Gack MU. Ubiquitination in the antiviral immune response. Virology. 2015 doi: 10.1016/j.virol.2015.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Drewniak A, Gazendam RP, Tool AT, van Houdt M, Jansen MH, van Hamme JL, van Leeuwen EM, Roos D, Scalais E, de Beaufort C, et al. Invasive fungal infection and impaired neutrophil killing in human CARD9 deficiency. Blood. 2013;121:2385–2392. doi: 10.1182/blood-2012-08-450551. [DOI] [PubMed] [Google Scholar]
- Drummond RA, Saijo S, Iwakura Y, Brown GD. The role of Syk/CARD9 coupled C-type lectins in antifungal immunity. Eur J Immunol. 2011;41:276–281. doi: 10.1002/eji.201041252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannick J, Thorleifsson G, Beer NL, Jacobs SB, Grarup N, Burtt NP, Mahajan A, Fuchsberger C, Atzmon G, Benediktsson R, et al. Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nat Genet. 2014;46:357–363. doi: 10.1038/ng.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs T, Saunders-Pullman R, Masuho I, Luciano MS, Raymond D, Factor S, Lang AE, Liang TW, Trosch RM, White S, et al. Mutations in GNAL cause primary torsion dystonia. Nat Genet. 2013;45:88–92. doi: 10.1038/ng.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- Gavino C, Cotter A, Lichtenstein D, Lejtenyi D, Fortin C, Legault C, Alirezaie N, Majewski J, Sheppard DC, Behr MA, et al. CARD9 deficiency and spontaneous central nervous system candidiasis: complete clinical remission with GM-CSF therapy. Clin Infect Dis. 2014;59:81–84. doi: 10.1093/cid/ciu215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazendam RP, van Hamme JL, Tool AT, van Houdt M, Verkuijlen PJ, Herbst M, Liese JG, van de Veerdonk FL, Roos D, van den Berg TK, Kuijpers TW. Two independent killing mechanisms of Candida albicans by human neutrophils: evidence from innate immunity defects. Blood. 2014;124:590–597. doi: 10.1182/blood-2014-01-551473. [DOI] [PubMed] [Google Scholar]
- Glocker EO, Hennigs A, Nabavi M, Schaffer AA, Woellner C, Salzer U, Pfeifer D, Veelken H, Warnatz K, Tahami F, et al. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N Engl J Med. 2009;361:1727–1735. doi: 10.1056/NEJMoa0810719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodridge HS, Shimada T, Wolf AJ, Hsu YM, Becker CA, Lin X, Underhill DM. Differential use of CARD9 by dectin-1 in macrophages and dendritic cells. J Immunol. 2009;182:1146–1154. doi: 10.4049/jimmunol.182.2.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross O, Gewies A, Finger K, Schafer M, Sparwasser T, Peschel C, Forster I, Ruland J. Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity. Nature. 2006;442:651–656. doi: 10.1038/nature04926. [DOI] [PubMed] [Google Scholar]
- Grumach AS, de Queiroz-Telles F, Migaud M, Lanternier F, Filho NR, Palma SM, Constantino-Silva RN, Casanova JL, Puel A. A Homozygous CARD9 Mutation in a Brazilian Patient with Deep Dermatophytosis. J Clin Immunol. 2015 doi: 10.1007/s10875-015-0170-4. [DOI] [PubMed] [Google Scholar]
- Hara H, Ishihara C, Takeuchi A, Imanishi T, Xue L, Morris SW, Inui M, Takai T, Shibuya A, Saijo S, et al. The adaptor protein CARD9 is essential for the activation of myeloid cells through ITAM-associated and Toll-like receptors. Nat Immunol. 2007;8:619–629. doi: 10.1038/ni1466. [DOI] [PubMed] [Google Scholar]
- Hara H, Saito T. CARD9 versus CARMA1 in innate and adaptive immunity. Trends Immunol. 2009;30:234–242. doi: 10.1016/j.it.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Herbst M, Gazendam R, Reimnitz D, Sawalle-Belohradsky J, Groll A, Schlegel PG, Belohradsky B, Renner E, Klepper J, Grimbacher B, et al. Chronic Candida albicans Meningitis in a 4-Year-Old Girl with a Homozygous Mutation in the CARD9 Gene (Q295X) Pediatr Infect Dis J. 2015 doi: 10.1097/INF.0000000000000736. [DOI] [PubMed] [Google Scholar]
- Ishikawa E, Ishikawa T, Morita YS, Toyonaga K, Yamada H, Takeuchi O, Kinoshita T, Akira S, Yoshikai Y, Yamasaki S. Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. J Exp Med. 2009;206:2879–2888. doi: 10.1084/jem.20091750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jachiet M, Lanternier F, Rybojad M, Bagot M, Ibrahim L, Casanova JL, Puel A, Bouaziz JD. Posaconazole Treatment of Extensive Skin and Nail Dermatophytosis Due to Autosomal Recessive Deficiency of CARD9. JAMA Dermatol. 2015;151:192–194. doi: 10.1001/jamadermatol.2014.2154. [DOI] [PubMed] [Google Scholar]
- Janse M, Lamberts LE, Franke L, Raychaudhuri S, Ellinghaus E, Muri Boberg K, Melum E, Folseraas T, Schrumpf E, Bergquist A, et al. Three ulcerative colitis susceptibility loci are associated with primary sclerosing cholangitis and indicate a role for IL2, REL, and CARD9. Hepatology. 2011;53:1977–1985. doi: 10.1002/hep.24307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia XM, Tang B, Zhu LL, Liu YH, Zhao XQ, Gorjestani S, Hsu YM, Yang L, Guan JH, Xu GT, Lin X. CARD9 mediates Dectin-1-induced ERK activation by linking Ras-GRF1 to H-Ras for antifungal immunity. J Exp Med. 2014;211:2307–2321. doi: 10.1084/jem.20132349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiryluk K, Li Y, Scolari F, Sanna-Cherchi S, Choi M, Verbitsky M, Fasel D, Lata S, Prakash S, Shapiro S, et al. Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat Genet. 2014;46:1187–1196. doi: 10.1038/ng.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanternier F, Barbati E, Meinzer U, Liu L, Pedergnana V, Migaud M, Heritier S, Chomton M, Fremond ML, Gonzales E, et al. Inherited CARD9 Deficiency in 2 Unrelated Patients With Invasive Exophiala Infection. J Infect Dis. 2015a;211:1241–1250. doi: 10.1093/infdis/jiu412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanternier F, Mahdaviani SA, Barbati E, Chaussade H, Koumar Y, Levy R, Denis B, Brunel AS, Martin S, Loop M, et al. Inherited CARD9 deficiency in otherwise healthy children and adults with Candida species-induced meningoencephalitis, colitis, or both. J Allergy Clin Immunol. 2015b doi: 10.1016/j.jaci.2014.12.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanternier F, Pathan S, Vincent QB, Liu L, Cypowyj S, Prando C, Migaud M, Taibi L, Ammar-Khodja A, Boudghene Stambouli O, et al. Deep dermatophytosis and inherited CARD9 deficiency. N Engl J Med. 2013;369:1704–1714. doi: 10.1056/NEJMoa1208487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeibundGut-Landmann S, Gross O, Robinson MJ, Osorio F, Slack EC, Tsoni SV, Schweighoffer E, Tybulewicz V, Brown GD, Ruland J, Reis e Sousa C. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat Immunol. 2007;8:630–638. doi: 10.1038/ni1460. [DOI] [PubMed] [Google Scholar]
- Mandell MA, Kimura T, Jain A, Johansen T, Deretic V. TRIM proteins regulate autophagy: TRIM5 is a selective autophagy receptor mediating HIV-1 restriction. Autophagy. 2014;10:2387–2388. doi: 10.4161/15548627.2014.984278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marakalala MJ, Graham LM, Brown GD. The role of Syk/CARD9-coupled C-type lectin receptors in immunity to Mycobacterium tuberculosis infections. Clin Dev Immunol. 2010;2010:567571. doi: 10.1155/2010/567571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin I. Origin and Diversification of TRIM Ubiquitin Ligases. PLoS ONE. 2012;7:e50030. doi: 10.1371/journal.pone.0050030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern DP, Gardet A, Torkvist L, Goyette P, Essers J, Taylor KD, Neale BM, Ong RT, Lagace C, Li C, et al. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet. 2010;42:332–337. doi: 10.1038/ng.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab FW, Rajsbaum R, Stoye JP, O’Garra A. Tripartite-motif proteins and innate immune regulation. Curr Opin Immunol. 2011;23:46–56. doi: 10.1016/j.coi.2010.10.021. [DOI] [PubMed] [Google Scholar]
- Miyake Y, Toyonaga K, Mori D, Kakuta S, Hoshino Y, Oyamada A, Yamada H, Ono K, Suyama M, Iwakura Y, et al. C-type lectin MCL is an FcRgamma-coupled receptor that mediates the adjuvanticity of mycobacterial cord factor. Immunity. 2013;38:1050–1062. doi: 10.1016/j.immuni.2013.03.010. [DOI] [PubMed] [Google Scholar]
- Ozato K, Shin DM, Chang TH, Morse HC., 3rd TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8:849–860. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa A, Wan L, Bonora M, Salmena L, Song MS, Hobbs RM, Lunardi A, Webster K, Ng C, Newton RH, et al. Cancer-associated PTEN mutants act in a dominant-negative manner to suppress PTEN protein function. Cell. 2014;157:595–610. doi: 10.1016/j.cell.2014.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poeck H, Bscheider M, Gross O, Finger K, Roth S, Rebsamen M, Hannesschlager N, Schlee M, Rothenfusser S, Barchet W, et al. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1 beta production. Nat Immunol. 2010;11:63–69. doi: 10.1038/ni.1824. [DOI] [PubMed] [Google Scholar]
- Pointon JJ, Harvey D, Karaderi T, Appleton LH, Farrar C, Stone MA, Sturrock RD, Brown MA, Wordsworth BP. Elucidating the chromosome 9 association with AS; CARD9 is a candidate gene. Genes Immun. 2010;11:490–496. doi: 10.1038/gene.2010.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajsbaum R, Garcia-Sastre A, Versteeg GA. TRIMmunity: the roles of the TRIM E3-ubiquitin ligase family in innate antiviral immunity. J Mol Biol. 2014a;426:1265–1284. doi: 10.1016/j.jmb.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajsbaum R, Versteeg GA, Schmid S, Maestre AM, Belicha-Villanueva A, Martinez-Romero C, Patel JR, Morrison J, Pisanelli G, Miorin L, et al. Unanchored K48-linked polyubiquitin synthesized by the E3-ubiquitin ligase TRIM6 stimulates the interferon-IKKepsilon kinase-mediated antiviral response. Immunity. 2014b;40:880–895. doi: 10.1016/j.immuni.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard ML, Lamas B, Liguori G, Hoffmann TW, Sokol H. Gut fungal microbiota: the Yin and Yang of inflammatory bowel disease. Inflamm Bowel Dis. 2015;21:656–665. doi: 10.1097/MIB.0000000000000261. [DOI] [PubMed] [Google Scholar]
- Rieber N, Singh A, Oz H, Carevic M, Bouzani M, Amich J, Ost M, Ye Z, Ballbach M, Schafer I, et al. Pathogenic Fungi Regulate Immunity by Inducing Neutrophilic Myeloid-Derived Suppressor Cells. Cell Host Microbe. 2015 doi: 10.1016/j.chom.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas MA, Beaudoin M, Gardet A, Stevens C, Sharma Y, Zhang CK, Boucher G, Ripke S, Ellinghaus D, Burtt N, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43:1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MJ, Osorio F, Rosas M, Freitas RP, Schweighoffer E, Gross O, Verbeek JS, Ruland J, Tybulewicz V, Brown GD, et al. Dectin-2 is a Syk-coupled pattern recognition receptor crucial for Th17 responses to fungal infection. J Exp Med. 2009;206:2037–2051. doi: 10.1084/jem.20082818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani L. Immunity to fungal infections. Nat Rev Immunol. 2011;11:275–288. doi: 10.1038/nri2939. [DOI] [PubMed] [Google Scholar]
- Roth S, Rottach A, Lotz-Havla AS, Laux V, Muschaweckh A, Gersting SW, Muntau AC, Hopfner KP, Jin L, Vanness K, et al. Rad50-CARD9 interactions link cytosolic DNA sensing to IL-1beta production. Nat Immunol. 2014;15:538–545. doi: 10.1038/ni.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S, Ruland J. Caspase recruitment domain-containing protein 9 signaling in innate immunity and inflammation. Trends Immunol. 2013;34:243–250. doi: 10.1016/j.it.2013.02.006. [DOI] [PubMed] [Google Scholar]
- Saijo S, Ikeda S, Yamabe K, Kakuta S, Ishigame H, Akitsu A, Fujikado N, Kusaka T, Kubo S, Chung SH, et al. Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity. 2010;32:681–691. doi: 10.1016/j.immuni.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Schoenen H, Bodendorfer B, Hitchens K, Manzanero S, Werninghaus K, Nimmerjahn F, Agger EM, Stenger S, Andersen P, Ruland J, et al. Cutting edge: Mincle is essential for recognition and adjuvanticity of the mycobacterial cord factor and its synthetic analog trehalose-dibehenate. J Immunol. 2010;184:2756–2760. doi: 10.4049/jimmunol.0904013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Cho H, Inn KS, Yang A, Zhao Z, Liang Q, Versteeg GA, Amini-Bavil-Olyaee S, Wong LY, Zlokovic BV, et al. Negative regulation of NF-kappaB activity by brain-specific TRIpartite Motif protein 9. Nat Commun. 2014;5:4820. doi: 10.1038/ncomms5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Conway KL, Zhang M, Choi M, Morin B, Cao Z, Villablanca EJ, Li C, Wijmenga C, Yun SH, et al. Card9 mediates intestinal epithelial cell restitution, T-helper 17 responses, and control of bacterial infection in mice. Gastroenterology. 2013;145:591–601. e593. doi: 10.1053/j.gastro.2013.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser D, Neumann K, Bergmann H, Marakalala MJ, Guler R, Rojowska A, Hopfner KP, Brombacher F, Urlaub H, Baier G, et al. Syk kinase-coupled C-type lectin receptors engage protein kinase C-sigma to elicit Card9 adaptor-mediated innate immunity. Immunity. 2012;36:32–42. doi: 10.1016/j.immuni.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida T, Zou J, Saitoh T, Kumar H, Abe T, Matsuura Y, Kawai T, Akira S. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity. 2010;33:765–776. doi: 10.1016/j.immuni.2010.10.013. [DOI] [PubMed] [Google Scholar]
- Uchil PD, Hinz A, Siegel S, Coenen-Stass A, Pertel T, Luban J, Mothes W. TRIM protein-mediated regulation of inflammatory and innate immune signaling and its association with antiretroviral activity. J Virol. 2013;87:257–272. doi: 10.1128/JVI.01804-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg GA, Rajsbaum R, Sanchez-Aparicio MT, Maestre AM, Valdiviezo J, Shi M, Inn KS, Fernandez-Sesma A, Jung J, Garcia-Sastre A. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity. 2013;38:384–398. doi: 10.1016/j.immuni.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang W, Lin Z, Wang X, Li T, Yu J, Liu W, Tong Z, Xu Y, Zhang J, et al. CARD9 mutations linked to subcutaneous phaeohyphomycosis and T17 cell deficiencies. J Allergy Clin Immunol. 2013 doi: 10.1016/j.jaci.2013.09.033. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang W, Lin Z, Wang X, Li T, Yu J, Liu W, Tong Z, Xu Y, Zhang J, et al. CARD9 mutations linked to subcutaneous phaeohyphomycosis and TH17 cell deficiencies. J Allergy Clin Immunol. 2014;133:905–908. e903. doi: 10.1016/j.jaci.2013.09.033. [DOI] [PubMed] [Google Scholar]
- Werninghaus K, Babiak A, Gross O, Holscher C, Dietrich H, Agger EM, Mages J, Mocsai A, Schoenen H, Finger K, et al. Adjuvanticity of a synthetic cord factor analogue for subunit Mycobacterium tuberculosis vaccination requires FcRgamma-Syk-Card9-dependent innate immune activation. J Exp Med. 2009;206:89–97. doi: 10.1084/jem.20081445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Wang J, Wang Y, Zhou H, Wu X, Tian Z, Sun B. Novel function of Trim44 promotes an antiviral response by stabilizing VISA. J Immunol. 2013;190:3613–3619. doi: 10.4049/jimmunol.1202507. [DOI] [PubMed] [Google Scholar]
- Yang CS, Rodgers M, Min CK, Lee JS, Kingeter L, Lee JY, Jong A, Kramnik I, Lin X, Jung JU. The autophagy regulator Rubicon is a feedback inhibitor of CARD9-mediated host innate immunity. Cell Host Microbe. 2012;11:277–289. doi: 10.1016/j.chom.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Bao M, Lu N, Weng L, Yuan B, Liu YJ. The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat Immunol. 2013;14:172–178. doi: 10.1038/ni.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Wang L, Zhang M, Wang P, Yuan C, Qi J, Meng H, Gao C. Tripartite motif-containing protein 38 negatively regulates TLR3/4- and RIG-I-mediated IFN-beta production and antiviral response by targeting NAP1. J Immunol. 2012;188:5311–5318. doi: 10.4049/jimmunol.1103506. [DOI] [PubMed] [Google Scholar]
- Zhernakova A, Festen EM, Franke L, Trynka G, van Diemen CC, Monsuur AJ, Bevova M, Nijmeijer RM, van ‘t Slot R, Heijmans R, et al. Genetic analysis of innate immunity in Crohn’s disease and ulcerative colitis identifies two susceptibility loci harboring CARD9 and IL18RAP. Am J Hum Genet. 2008;82:1202–1210. doi: 10.1016/j.ajhg.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.