Abstract
Minimally invasive approaches to detect/predict target organ toxicity have significant practical applications in occupational toxicology. The potential application of peripheral blood transcriptomics as a practical approach to study the mechanisms of silica-induced pulmonary toxicity was investigated. Rats were exposed by inhalation to crystalline silica (15 mg/m3, 6 h/day, 5 days) and pulmonary toxicity and global gene expression profiles of lungs and peripheral blood were determined at 32 weeks following termination of exposure. A significant elevation in bronchoalveolar lavage fluid lactate dehydrogenase activity and moderate histological changes in the lungs, including type II pneumocyte hyperplasia and fibrosis, indicated pulmonary toxicity in the rats. Similarly, significant infiltration of neutrophils and elevated monocyte chemotactic protein-1 levels in the lungs showed pulmonary inflammation in the rats. Microarray analysis of global gene expression profiles identified significant differential expression [>1.5-fold change and false discovery rate (FDR) p < 0.01] of 520 and 537 genes, respectively, in the lungs and blood of the exposed rats. Bioinformatics analysis of the differentially expressed genes demonstrated significant similarity in the biological processes, molecular networks, and canonical pathways enriched by silica exposure in the lungs and blood of the rats. Several genes involved in functions relevant to silica-induced pulmonary toxicity such as inflammation, respiratory diseases, cancer, cellular movement, fibrosis, etc, were found significantly differentially expressed in the lungs and blood of the silica-exposed rats. The results of this study suggested the potential application of peripheral blood gene expression profiling as a toxicologically relevant and minimally invasive surrogate approach to study the mechanisms underlying silica-induced pulmonary toxicity.
Keywords: Crystalline silica, pulmonary toxicity, gene expression profile, blood, mechanisms
Introduction
Occupational exposure to crystalline silica occurs in a variety of industries and occupations because of its extremely common natural occurrence and the wide range of products that contain it. In addition to sand blasting, silica milling, surface mining and tunneling, all activities that involve the movement of earth (e.g. mining, farming, construction, etc) are considered to be major sources of occupational exposure to crystalline silica. It has been estimated that approximately 2 million workers in USA and millions more worldwide are occupationally exposed to crystalline silica annually. In many cases, occupational exposure to crystalline silica takes place at levels much higher than the National Institute for Occupational Safety and Health (NIOSH) recommended exposure level (REL) of 0.05 mg/m3 (Linch et al., 1998).
Occupational exposure to crystalline silica is a major concern among healthcare providers primarily because of the various diseases that are associated with the exposure. Lungs, in addition to being the major route for occupational exposure to crystalline silica, are also the primary targets for its health effects. Silicosis, an incurable, irreversible but preventable progressive lung fibrosis condition, is the most serious health effect associated with occupational exposure to crystalline silica (Mundt et al., 2011; Nasrullah et al., 2011). In addition, silica exposure is associated with lung cancer, cardiopulmonary failure, mycobacterial infection, kidney failure and autoimmune diseases (IARC, 1997; Cooper et al., 2002).
A major constraint in the prevention of diseases associated with occupational exposure to crystalline silica, especially silicosis, is the lack of practical techniques that are sensitive enough to detect the early pulmonary effects of silica exposure. Chest X-ray and pulmonary function tests are the two techniques most commonly employed to detect silicosis depending on the structural and/or functional impairment of the lungs associated with the onset of clinical symptoms of the disease. As silicosis is an irreversible and potentially life-threatening disease, detection of the disease at a later stage following the onset of clinical symptoms is of no significant value in the prevention of silicosis. In view of this limitation, NIOSH has recommended developing practical (noninvasive or minimally invasive) and highly sensitive techniques capable of detecting silicosis preclinically (NIOSH, 2002). In compliance with the NIOSH recommendation, we have initiated a research project investigating the potential application of peripheral blood gene expression profiling as a minimally invasive and highly sensitive surrogate approach to detect silicosis prior to the onset of clinical symptoms of the disease. Recently, we have reported the potential application of peripheral blood gene expression profiling as a practical and highly sensitive approach to detect/predict the pulmonary effects of silica exposure in a rat model (Sellamuthu et al., 2011a). In that study, a gene expression signature, developed based on the differential gene expression profile in the blood of the silica-exposed rats, identified with significant accuracy, the rats which were exposed to a very low concentration of silica (1 mg/m3, 6 h/day, 5 days) that did not result in pulmonary toxicity detectable by the currently available biochemical and histological toxicity parameters.
In the present study, we have investigated the potential application of peripheral blood global gene expression profiling as a relevant surrogate approach to study the molecular mechanism(s) underlying the pulmonary effects of silica exposure. Data presented in this report demonstrated a remarkable similarity between the lungs and peripheral blood of the silica-exposed rats with respect to the biological functions and cellular pathways that are known to underlie the silica-induced pulmonary toxicity. Our results suggested the potential application of peripheral blood gene expression profiling as a mechanistically relevant, minimally invasive surrogate approach to study silica-induced pulmonary toxicity.
Materials and methods
Inhalation exposure of rats to crystalline silica
The entire animal experiment was conducted in an Association for Assessment and Accreditation of Laboratory Animal Care International approved animal facility (NIOSH, Morgantown, WV) following an animal protocol approved by the Institutional Animal Care and Use Committee. Male Fischer CDF (F344/DuCrl) rats from Charles River Laboratories Wilmington, MA) monitored free of viral pathogens, parasites, mycoplasmas, Helicobacter and CAR Bacillus, weighing approximately 150 g, were housed in ventilated isolator cages in specific pathogen-free and environmentally controlled conditions, on Alpha-Dri virgin cellulose chips and hardwood Beta-chips as bedding, and provided HEPA-filtered air, irradiated Teklad 2918 diet, and tap water ad libitum. Following an acclimatization period of approximately 2 weeks, the rats were exposed to crystalline silica (Min-U-Sil silica, US Silica, Berkley Springs, WV) by inhalation. Generation of an aerosol consisting of silica particles of respirable size and exposure of rats to the particles by inhalation were done exactly as reported previously from our laboratory (McKinney et al., 2009; Sellamuthu et al., 2011a). Stated briefly, eight rats were exposed to an aerosol containing silica at a final concentration of 15 mg/ m3, 6 h/day, for five consecutive days. Another group of eight rats exposed simultaneously to filtered air served as the control. Following termination of exposure to air or silica aerosol, the control and silica-exposed rats were maintained on a 12-hour light – dark schedule with free access to food and tap water for 32 weeks. Throughout the period of the experiment, the rats were monitored for overt symptoms of toxicity and weekly body weight fluctuations, if any.
Pulmonary toxicity determination
Following the 32-week post-exposure time period, the control and silica-exposed rats were euthanized with an intraperitoneal injection of ≥100 mg sodium pentobarbital/kg body weight (Fort Dodge Animal Health, Fort Dodge, IA). Blood collected directly from the abdominal aorta was transferred to Vacutainer tubes (Becton-Dickinson, Franklin Lakes, NJ) containing EDTA as the anticoagulant and used to determine global gene expression profile and hematological parameters as described below.
The right lungs of the rats were clamped off and bronchoalveolar lavage (BAL) was performed in the left lungs as described previously (Antonini & Roberts, 2007). The cellular fraction of BAL and a concentrated acellular fraction of BAL fluid (BALF) that was collected were used for further biochemical determination of pulmonary toxicity. The diaphragmatic lobe and cardiac lobe of the unlavaged right lung were inflated with 10% neutral-buffered formaldehyde and preserved in the same solution for histopathological analysis of damage. The apical lobe of the unlavaged right lung was stored in RNAlater (Invitrogen, Carlsbad, CA) and used in gene expression studies as described below.
BALF lactate dehydrogenase (LDH) assay
Activity of LDH, a general indicator of cytotoxicity, was assayed in the acellular fraction of BALF of the rats using a COBAS MIRA autoanalyzer (Roche Diagnostics Systems, Mount Clair, NJ) as previously reported (Antonini & Roberts, 2007).
Lung histopathology
The diaphragmatic lobes of the unlavaged right lung of the control and silica-exposed rats fixed in formaldehyde solution were embedded in paraffin, sectioned at a thickness of 5 µm and stained with hematoxylin and eosin (H & E) or Mason’s trichrome stain. The slides were assessed for histological changes (H & E stains) or for fibrosis (trichrome stain) by a board-certified veterinary pathologist (Experimental Pathology Laboratory, Inc., Sterling, VA).
Determination of silica-induced pulmonary inflammation
Protein concentration of the inflammatory cytokine, monocyte chemoattractant protein-1 (MCP-1), was determined in the acellular fraction of the BALF using the Rat MCP-1 ELISA Kit (Invitrogen, Carlsbad, CA) and a Spectramax 250 plate spectrophotometer. The cellular fraction of BALF was resuspended in 1 mL of phosphate buffered saline (PBS), and the total number of alveolar macrophages (AMs) and polymorphonuclear leukocytes (PMNs) was determined using a Coulter Multisizer II and Accu Comp software (Coulter Electronics, Hialeah, FL). BALF cells (5 × 104) were spun onto microscope slides using a Cytospin 3 centrifuge (Shandon Life Sciences International, Cheshire, UK) and stained with a Leukostat stain (Fisher Scientific, Pittsburgh, PA) to differentiate AM and PMN. Two-hundred cells were counted per rat, and percentages of each cell type were multiplied back across the total cell count to obtain total AM and PMN numbers.
Hematology
The total and differential white blood cell (WBC) counts of the blood samples obtained from the air and silica-exposed rats were determined by the previously described flow cytometer procedure (Erdely et al., 2011) using rat antibodies (BD Pharmingen, San Diego, CA). The leukocytes were separated into three gates (lymphocytes, monocytes, and neutrophils) by forward and side scattering. After collecting 3500 counting beads, the data were analyzed using FlowJo software (Treestar, Costa Mesa, CA).
Gene expression analysis of lung and blood samples
RNA isolation from lung and blood samples
Total RNA was isolated from the apical lobe of the unlavaged right lung of the control and silica-exposed rats using an RNeasy Fibrous Tissue Mini Kit (Qiagen Inc., Valencia, CA). Approximately 30 mg of lung tissue were homogenized in buffer RLT containing β-mercaptoethanol and two 2.4 mm Zirconia beads (BioSpec Products Inc., Bartlesville, OK) using a mini beadbeater-8 (BioSpec Products Inc.) for 20 seconds. The tissue homogenate was centrifuged at 10,000×g for 10 min at room temperature, and the RNA present in the supernatant was extracted and purified using RNeasy columns as directed in the RNeasy Fibrous Tissue Mini Kit protocol.
Total RNA present in blood samples of the rats was isolated using the Mouse Ribopure Blood RNA Isolation Kit (Ambion Inc., Austin, TX) following the protocol provided by the manufacturer. The RNA isolated was digested with RNase-free DNase and further purified using the RNeasy Mini Kit (Qiagen Inc.). Globin mRNA present in abundance in the blood is known to interfere with global gene expression profiling using microarray (Raghavachari et al., 2009); and, therefore, depletion of globin mRNA from the blood RNA samples was performed using the Globinclear-Mouse/Rat Globin mRNA Removal Kit (Ambion Inc.).
The integrity of the purified lung and blood RNA samples was determined using the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA) and RNA was quantitated by UV spectrophotometry. Only RNA samples exhibiting an RNA Integrity Number (RIN) ≥8.0 were used in the microarray analysis of the global gene expression profile.
Microarray analysis of global gene expression profile
The global gene expression profiles of the lungs and blood RNA samples were determined using RatRef-12 V1.0 Expression BeadChip array (Illumina Inc., San Diego, CA). All microarray experiments were conducted following the Minimum Information About a Microarray Experiment (MIAME) guidelines. Biotin-labeled cRNA was synthesized from 375 ng RNA for each control and silica-exposed lung and blood sample using the Illumina Totalprep RNA Amplification Kit (Ambion Inc.). Details regarding chip hybridization and washing, scanning the chips, normalization of the microarray data and further analysis can be found in our recent publications (Sellamuthu et al., 2011a,b). With respect to the statistical analysis, Illumina BeadArray expression data were analyzed in Bioconductor (Gentleman et al., 2004) using the “lumi” and “limma” packages. The “lumi” Bioconductor package was specifically developed to process Illumina microarrays and covers data input, quality control, variance stabilization, normalization, and gene annotation. Normalized data were then analyzed using the “limma” package in R (Smyth, 2004). In short, limma fits a linear model for each gene, generates group means of expression, conducts pairwise comparisons between selected groups, and calculates p values using empirical Bayes methodology. The log fold changes of gene expression between the control and silica-exposed samples were converted to standard fold changes and raw p values were corrected for false discovery rate (FDR) using the Benjamini and Hochberg method (Benjamini & Hochberg, 1995). Only genes with FDR p value <0.01 and fold change in expression > 1.5 compared with the control samples were considered as significantly differentially expressed genes (SDEGs) and used as input for further bioinformatics analysis. The microarray data presented in this manuscript have been deposited in the Gene Expression Omnibus (GEO) Database (http://www.ncbi.nlm.nih.gov/geo) and are accessible through GEO accession number GSE36208.
Quantitative real-time polymerase chain reaction (QRT-PCR) analysis
Seven genes which exhibited a significant difference in expression (FDR p < 0.01 and fold change >1.5) in both the lung and blood samples of the silica-exposed rats compared with the control rats and belonging to the IPA biological function category inflammatory response (details about IPA analysis and findings are presented below) were selected for QRT-PCR analysis to confirm the microarray data. The nucleotide sequences of the primers used in the QRT-PCR analysis of the selected genes and the housekeeping gene, β-actin, are presented in Supplementary Table S1. The PCR amplification, detection of the PCR amplified gene products, and their quantitations were performed with a 7900 HT Fast Realtime PCR machine and SYBR Green PCR MasterMix (Applied Biosystems, Foster City, CA). The specificity and integrity of the PCR products were determined by analyzing the dissociation curves of all PCR amplified gene products. The expression levels of the genes were normalized to that of the housekeeping gene, and the fold changes in expression compared with the controls were calculated using the formula 2^(−(ΔCt_target gene − ΔCt_ housekeeping gene).
Bioinformatics analysis of SDEGs
Bioinformatics analysis of the SDEGs (both upregulated and downregulated in expression in the lungs and blood samples of silica-exposed rats compared with the controls) was conducted using Ingenuity Pathway Analysis software (IPA, Ingenuity Systems, www.ingenuity.com). IPA software is designed to map the biological relationship of the uploaded genes and classify them into categories of biological functions, molecular networks, and canonical pathways according to published literature in the database. Fisher’s exact test was conducted to calculate a p value to determine the significance of a particular biological function, molecular network, or canonical pathway enriched by silica exposure in the rat lungs and blood. The criteria required for a biological function, molecular network, or canonical pathway to be considered significantly enriched in response to silica exposure and toxicity was that the category within a gene set was represented with a significance level for enrichment of p < 0.05. The biological functions, molecular networks, and canonical pathways significantly enriched (p < 0.05) but represented by fewer than 5 genes were filtered out to identify the functions, networks, and pathways most robustly perturbed by silica exposure in the rat lung and blood samples.
Statistical analysis of the data
Non-microarray data between the silica-exposed and control rats were compared using the one-way ANOVA test. Post hoc comparisons were made with Fisher’s least significant difference (LSD) test. The level of statistical significance was set at p < 0.05.
Results
Pulmonary toxicity in the silica-exposed rats
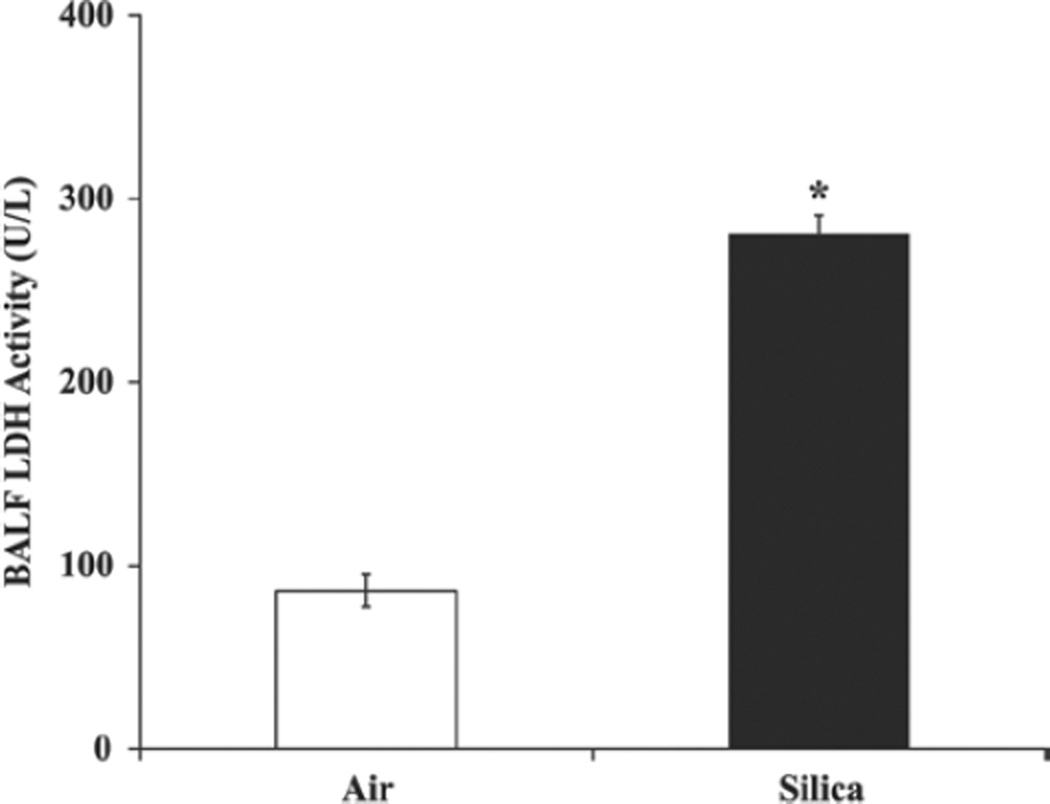
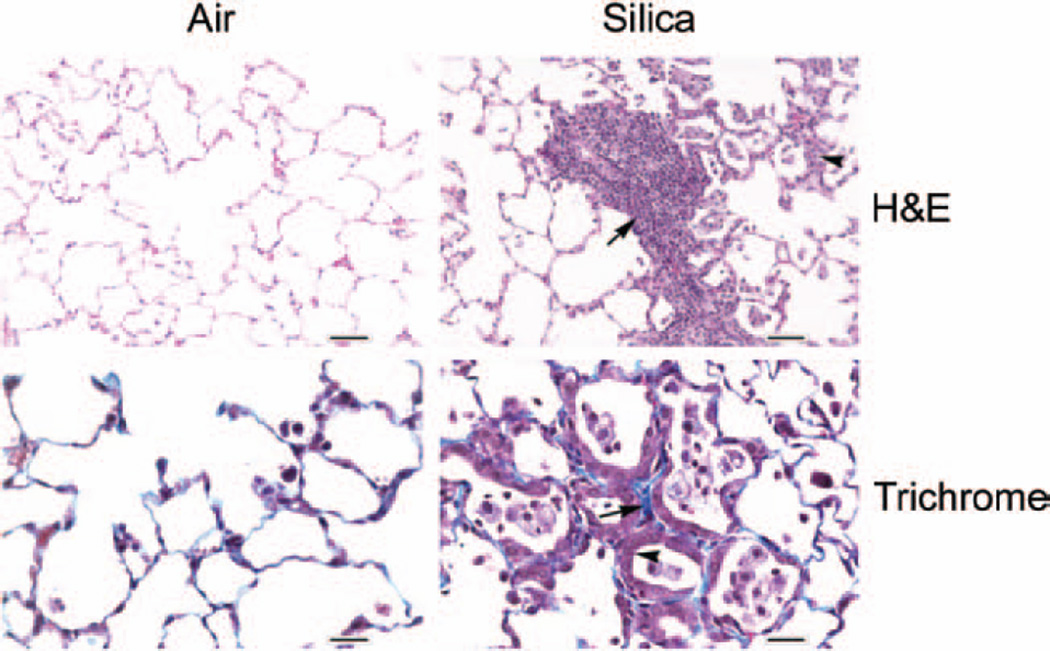
Compared with the control rats, the BALF LDH activity in the silica-exposed rat lungs was approximately three-fold higher (p< 0.05) at the 32-week post silica exposure time interval (Figure 1). Silica-induced pulmonary toxicity in the rats was further confirmed by histological analysis of the lungs (Figure 2). Compared with the control rats, significant histological changes suggesting silica-induced pulmonary toxicity were detected in the lungs of the silica-exposed rats. The major histological changes noticed in the rat lungs in response to silica exposure were multifocal areas of inflammation and type II pneumocyte hyperplasia. Inflammation ranged in degree with the majority of animals exhibiting sub-acute and the remaining animals exhibiting chronic or chronic-active inflammation. The affected areas of the lungs contained increased numbers of alveolar macrophages present in the alveolar space. These AMs were typically large and round with a foamy cytoplasm. In addition, accumulation of neutrophils, lymphocytes, and plasma cells was noticed in the lungs of the silica-exposed rats. When lungs were stained with trichrome, many of the areas affected by inflammation had increased fibrosis characterized by thickening of the alveolar septae with a blue stained cellular matrix, interpreted to be collagen (Figure 2). In general, the severity of silica-induced pulmonary damage in the rats was higher at the 32-week post-exposure time interval compared with the early post-exposure time intervals of 0–16 weeks (Sellamuthu et al., 2011a).
Figure 1.
Lactate dehydrogenase activity in rat lungs. Rats were exposed to crystalline silica or filtered air and lactate dehydrogenase activity was determined in bronchoalveolar lavage fluid (BALF) as described in the text. Values represent mean ± SE of eight rats per group. *Statistically significant (p < 0.05) compared to the control rats.
Figure 2.
Photomicrographs of lungs from control and silica-exposed rats. Lung samples obtained from control and crystalline silica-exposed rats were sectioned and stained with H & E (top panel) or Masson’s trichrome stain (bottom panel) as described in the text. The arrow and arrowhead in the H & E stained sections show type II pneumocyte hyperplasia and alveolar space filled with macrophages and neutrophils, respectively, in the silica-exposed rat lungs. The arrow and arrowhead in the trichrome stained sections show thickened alveolar septae and type II pneumocyte hyperplasia lining the alveolar septae, respectively, in the silica-exposed rat lungs Magnification: H & E = ×20; trichrome = ×40.
Induction of inflammation in silica-exposed rats
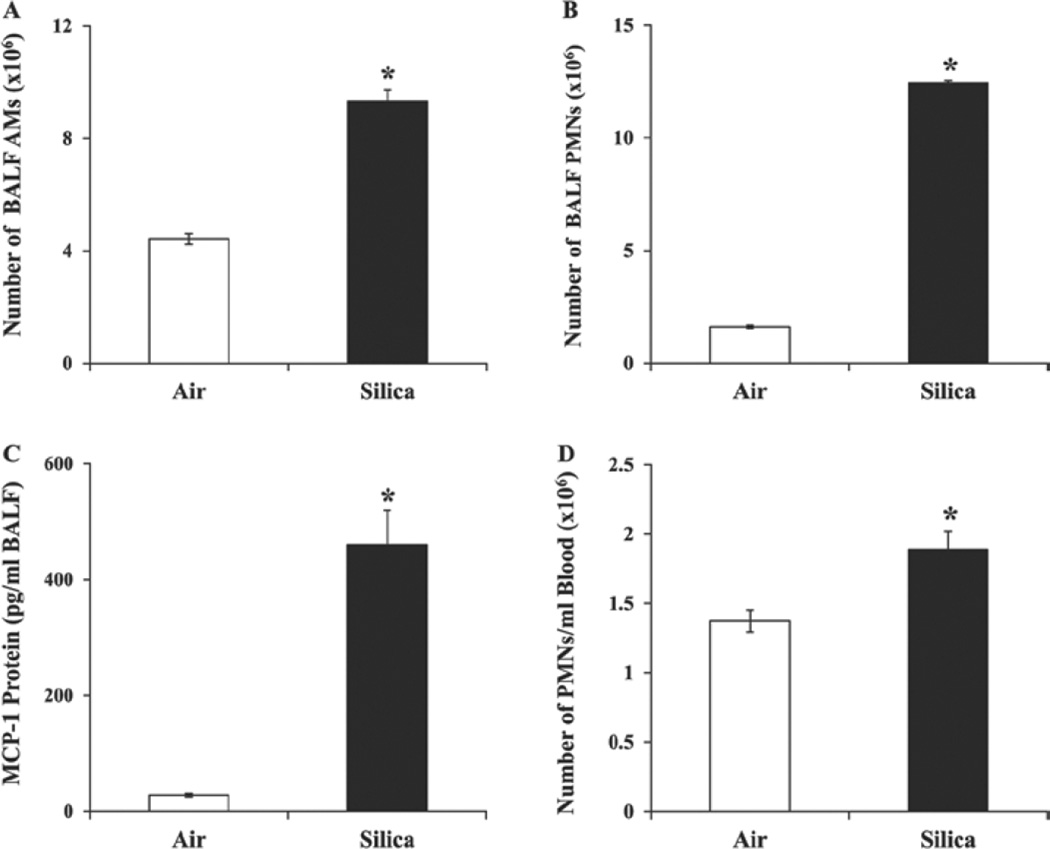
A significant increase in the number of AMs and PMNs and in the concentration of MCP-1 was noticed in the BALF of the silica-exposed rats compared with the control rats (Figure 3A, 3B and 3C), suggesting silica-induced pulmonary inflammation. As described above, histological analysis of the lung samples further supported silica-induced pulmonary inflammation in the rats. A significant increase in the number of neutrophils in the blood of the silica-exposed rats compared with the control rats was also noticed (Figure 3D), providing evidence for silica-induced systemic inflammation. Overall, a further progression in silica-induced pulmonary inflammation was seen in the rats at the 32-week post-exposure time interval compared to the earlier time intervals of 0–16 weeks (Sellamuthu et al., 2011a).
Figure 3.
Bronchoalveolar lavage fluid (BALF) and blood parameters of inflammation in the control and silica-exposed rats. Rats were exposed to crystalline silica or air and total number of alveolar macrophages (AMs) (A) and polymorphonuclear leukocytes (PMNs) (B) and concentration of MCP-1(C) were determined in the BALF as described in the text. Number of neutrophils (D) in the blood of the rats was also determined. Values represent mean ± SE of eight rats per group. *Statistically significant (p < 0.05) compared to the control rats.
Differential gene expression profiles in the lungs and blood of silica-exposed rats
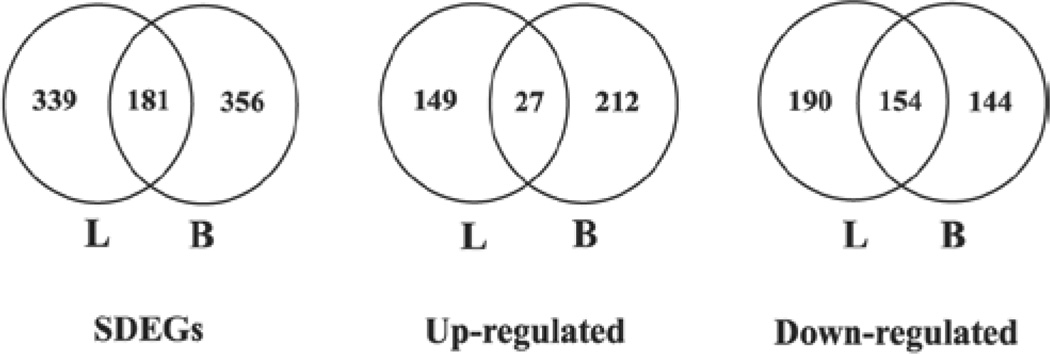
Microarray analysis of global gene expression profile identified a significant differential expression (> 1.5-fold change and FDR p < 0.01) of 520 and 537 genes, respectively, in the lungs and blood of the silica-exposed rats compared with the controls. A complete list of the SDEGs in the lungs and blood of the silica-exposed rats along with their fold changes in expression and FDR p values compared with the controls are presented in Supplementary Table S2 and Supplementary Table S3. Of the SDEGs, 181 genes were found to be common between lungs and blood. Details regarding the number of significantly differentially expressed genes including those which were overexpressed and underexpressed in the lungs and blood of the silica-exposed rats compared with the controls, are presented in Figure 4. In general, the number of SDEGs detected in the lungs and blood of the silica-exposed rats at the 32-week postexposure time interval was more than those detected at the earlier time intervals of 0–16 weeks (Sellamuthu et al., 2011a; 2012).
Figure 4.
Number of significantly differentially expressed genes (SDEGs) in the lungs (L) and blood (B) of silica-exposed rats. Rats were exposed to crystalline silica or air and gene expression profile was determined in the lungs and blood by microarray analysis. Genes with >1.5-fold change and a false discovery rate (FDR) p < 0.01 in the silica-exposed rat lungs and blood compared with the corresponding control samples were considered as significantly differentially expressed. SDEGs – Significantly differentially expressed genes in the lungs and blood of the silica-exposed rats compared with the control samples; Upregulated – genes which were significantly overexpressed in the lungs and blood of the silica-exposed rats compared with the control samples; and Downregulated – genes whose expressions were significantly downregulated in the lungs and blood of the silica-exposed rats compared with the control samples.
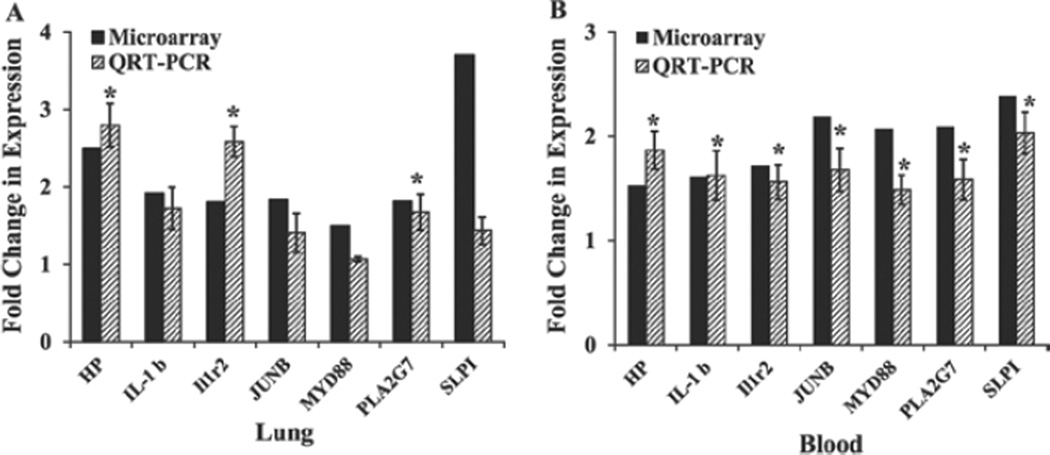
The RT-PCR analysis results confirmed the differential expression (> 1.5-fold change) of all the blood genes selected based on the microarray results (Figure 5). In the case of the lung tissue, three genes were confirmed to be differentially expressed at a level of > 1.5-fold change.
Figure 5.
Validation of microarray results by QRT-PCR. A set of seven genes whose expressions were significantly different in the lungs and blood of the silica-exposed rats as indicated by the microarray findings were analyzed by QRT-PCR as described in the “Materials and methods” section. The microarray data presented is the mean fold change (n= 6) and the QRT-PCR data presented is mean fold change ± SE (n= 6) of silica-exposed rats compared with the control. A. Lung results, B. Blood results. *Statistically significant (p < 0.05) compared to the control rats.
The expressions of JunB and SLP1 were slightly less than but close to 1.5-fold – the criteria employed to select the SDEGs on the basis of the microarray data.
Bioinformatics analysis of SDEGs in the lungs and blood of silica-exposed rats
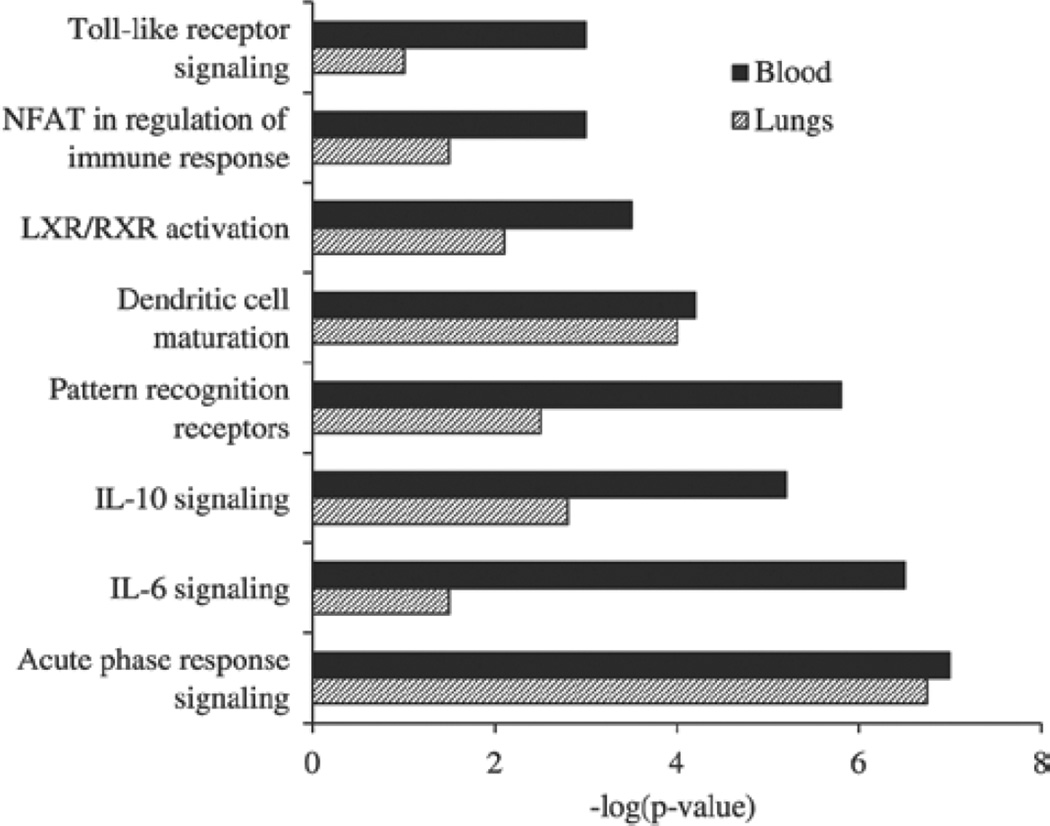
Bioinformatics analysis by IPA of the SDEGs in the lungs of the silica-exposed rats provided molecular insights into the mechanisms potentially underlying the pulmonary effects of silica. Inflammatory response and cancer were among the highly ranked IPA biological function categories significantly enriched in the lungs and blood of the silica-exposed rats (Figure 6 and Supplementary Table S4–Supplementary Table S7). Inflammatory response ranked second among the significantly enriched IPA biological function categories in the lungs of the silica-exposed rats further confirming the central prominent role played by inflammation in silica-induced pulmonary toxicity (DiMatteo & Reasor, 1997; Barbarin et al., 2005). Results of the IPA bioinformatics analysis of the canonical pathways further suggested the prominent role played by inflammation in silica-induced pulmonary toxicity (Figure 7). A marked similarity was noticed between the lungs and blood of the rats with respect to the IPA biological functions and canonical pathways enriched by their pulmonary exposure to crystalline silica (Figures 6 and 7). A similar observation was also made with respect to the IPA molecular networks which were significantly enriched in the lungs and blood of the silica-exposed rats (data not presented).
Figure 6.
Enrichment of IPA biological functions in the lungs and blood of silica-exposed rats. Bioinformatics analysis of the significantly differentially expressed genes identified in the silica-exposed rat lungs and blood was done using IPA software. The top 10 significantly enriched biological functions in the lungs of the silica-exposed rat lungs compared with the control rat lungs and the same biological functions in the blood samples are presented to demonstrate the similarity in gene expression profile between lungs and blood of the silica-exposed rats. Data represents the mean of six rats per group.
Figure 7.
Enrichment of IPA canonical pathways in the lungs and blood of silica-exposed rats. Bioinformatics analysis of the significantly differentially expressed genes in the silica-exposed rat lungs and blood was done using IPA software. The top eight significantly enriched canonical pathways in the silica-exposed rat lungs compared with the control rat lungs and the same pathways in the blood samples of the rats are presented to demonstrate the similarity in the gene expression profile between lungs and blood of the silica-exposed rats. Data represents the mean of six rats per group.
Discussion
A significant increase in mining, refining, and manufacturing, as well as the increased use of products containing potentially toxic agents over the past several decades, has resulted in a corresponding increase in human exposure to toxic agents capable of resulting in adverse health effects including potentially life-threatening diseases. It has been reported that millions of workers in the USA and elsewhere are occupationally exposed to crystalline silica (Sanderson, 1986), and in many cases the exposure takes place at levels as high as 10-fold higher than the NIOSH recommended exposure level of 0.05 mg/m3 (Linch et al., 1998). In addition to having an association with silicosis, a life-threatening but preventable pulmonary disease, occupational exposure to crystalline silica is associated with the development of autoimmune diseases, rheumatoid arthritis, chronic renal diseases, lupus, and cancer (Parks et al., 1999; Steenland et al., 2001). A better understanding of the molecular mechanisms underlying the pulmonary effects of silica exposure as well as developing the capability to detect the adverse pulmonary effects at an early stage (prior to the appearance of clinical symptoms of toxicity or disease) are critical in preventing diseases and mortality associated with occupational exposure to crystalline silica.
Laboratory animals have been used extensively in the past to study the pulmonary effects of silica exposure (Carter & Driscoll, 2001; Porter et al., 2001; Castranova et al., 2002), and the rat model employed in the current study appears to be consistent with the progression of human silicosis. Based on the findings of epidemiological studies, it has been demonstrated that silicosis may develop and/or progress in workers even after cessation of their occupational exposure to crystalline silica (Kreiss & Zhen, 1996; Miller et al., 1998). Compared to our related study (Sellamuthu et al., 2011a), which examined rats 0–16 weeks after a 1 week crystalline silica exposure, inflammation progressed and was more severe at 32-weeks. In addition, type II cell hyperplasia and fibrosis were prominent histological findings indicating the progression of lung disease due to silica exposure. Analogous to the human situation, the pulmonary toxicity induced by inhalation exposure of rats to silica for 1 week further progressed through the 32-week postexposure time interval. The mechanisms potentially underlying silica-induced pulmonary toxicity include the induction of oxidative stress (Fubini & Hubbard, 2003), apoptosis (Porter et al., 2002), and chronic inflammation (Sato et al., 2008). Based on the results obtained from a large number of in vitro and in vivo studies, it has been fairly well established that the induction of inflammation plays a predominant and central role in the pulmonary effects of silica exposure (Castranova, 2004; Hornung et al., 2008; Thakur et al., 2009). The current study also shows significant chronic inflammation in the lungs as well as systemic inflammation detected in the blood.
In the current study, we employed microarray technique for detecting the differentially expressed genes in the lungs and blood of the rats to provide further insight into the mechanisms of silica-induced pulmonary toxicity. Bioinformatics analysis of the significantly differentially expressed genes by IPA identified the biological functions, molecular networks, and canonical pathways that were significantly enriched in response to pulmonary exposure to silica and, therefore, provided molecular insights into the mechanisms potentially underlying silica-induced pulmonary toxicity. It has been fairly well established that inhaled silica particles interact with AMs and alveolar epithelium to result in the generation of reactive oxygen species (ROS) (Porter et al., 2002), induction of inflammation (Porter et al., 2004) and fibrosis (Porter et al., 2001) culminating in silicosis. In agreement with our previous reports (Sellamuthu et al., 2011a,b, 2012), several genes involved in the generation of ROS as well as in the cellular response to oxidative stress, viz: superoxide dismutase 2 (SOD2), hemeoxygenase 1 (HMOX1), metallothionein 1A (MT1A), neutrophil cytosolic factor 1 (NCF1), and NADPH oxidase organizer 1 (NOX01), were found significantly overexpressed in the lungs of the silica-exposed rats suggesting the potential involvement of oxidative stress in the induction of pulmonary toxicity detected in our rat model. As presented in Figures 6 and 7 and in agreement with our previous report (Sellamuthu et al., 2011a) as well as those of other investigators (Porter et al., 2004; Castranova, 2004), induction of inflammation appeared to be the major mechanism underlying the silica-induced pulmonary toxicity. Many of the canonical pathways that were significantly enriched in the silica-exposed rat lungs were those involved in an inflammatory response (Figure 7). Endogenous danger signals or alarmins that are released from AMs following cell death (Chen & Nuñez, 2010) may interact with pattern recognition receptors (PRRs) to result in pulmonary inflammation. The transcript for one such alarmin, SA100A8, was significantly overexpressed in our rat model. Transcript for MYD88, an adaptor molecule for TLR (Xiang & Fan, 2010) and CASP1, an adaptor molecule for NLRP3 inflammasome complex (Cassel et al., 2008) were also significantly overexpressed in our rat model. A definite role for NLRP3 inflammasome complex in silicosis has been previously demonstrated (Cassel et al., 2008). Activation of the PRRs and other inflammatory response receptors along with their pathways results in the release of proinflammatory cytokines and chemokines resulting in the induction of inflammation. The transcript for IL1β, a proinflammatory cytokine that plays a major role in silica-induced pulmonary inflammation and damage (Franchi et al., 2009), was significantly overexpressed in our rat model. The significant overexpression of the transcripts for genes involved in tissue remodeling and fibrosis viz: haptoglobin (HP), arginase 1 (ARG1), members of the metalloproteinase (MMP) family, secreted phosphoprotein 1 (SPP1), and JUNB in the silica-exposed rat lungs may account for the fibrosis detected in the lungs of the silica-exposed rats in our study (Figure 2). It is noteworthy that many of the genes whose expressions were significantly different in the lungs (Supplementary Table S2) and are mechanistically relevant to silica-induced pulmonary toxicity were also significantly differentially expressed in the blood of the silica-exposed rats (Supplementary Table S3 and Sellamuthu et al., 2011a), and a remarkable similarity was seen in the IPA biological functions and canonical pathways that were significantly enriched in the lungs and blood of the silica-exposed rats (Figures 6 and 7). These results, therefore, suggested the potential application of peripheral blood gene expression profiling as a mechanistically relevant and practical surrogate approach to study the pulmonary toxicity induced by crystalline silica.
The data presented in this report have clearly demonstrated the ability of silica particles deposited in the rat lungs to result in global gene expression changes in peripheral blood. Even though the exact mechanism(s) underlying the blood gene expression changes taking place in the rats exposed to crystalline silica by inhalation are currently unknown, several possibilities are worthy of consideration. Mainly because of poor solubility and the relatively large size (mass median aerodynamic diameter =1.6 µm), it is unlikely that the silica particles employed in our study would have crossed the lung-blood barrier to reach the peripheral blood. This argument is supported by the results of a previous study (Absher et al., 1992) which reported undetectable level of silica in the blood of rats at 60-days following a 1-week exposure (a much shorter post-exposure time interval compared to the present study) to a higher concentration of cristobalite (silicon dioxide) particles of an even smaller size (mass median aerodynamic diameter = 0.9 µm) than that was employed in our study. The most logical factor potentially contributing to the changes seen in the blood gene expression profile of the rats in response to their pulmonary exposure to silica, therefore, seems to be an indirect effect. Pulmonary exposure to crystalline silica results in the generation of a wide array of mediators of toxicity such as reactive oxygen and nitrogen species (Fubini & Hubbard, 2003), inflammatory mediators (Porter et al., 2004), danger signals (Chen & Nuñez, 2010) and fibrogenic signals (O’Reilly et al., 2005). One such toxicity mediator that deserves special attention with respect to the differential gene expression profile seen in the blood of the silica-exposed rats is hyaluronan, a low molecular mass extracellular membrane fragment, released into the BALF and peripheral blood following silica and other particulate material-induced pulmonary toxicity (Cantin et al., 1992; Eklund et al., 1991; Fraser et al., 1997). Hyaluronan functions as a chemoattractant and its chemotactic activity is mediated through its interaction with CD44 (Tzircotis et al., 2005). Interaction of hyaluronan with its receptors, TLR and CD44, is known to result in an inflammatory response (Campo et al., 2012) through activation of macrophages and recruitment of neutrophils into the lungs (Jiang et al., 2005). The proinflammatory cytokine, MCP-1, plays a prominent role in the hyaluronan-mediated inflammatory response (Haslinger et al., 2001). Significant overexpression of TLR2, TLR4 and CD44 detected in the blood (Sellamuthu et al., 2011a and Supplementary Table S3) and activation of MCP-1 seen in the BALF of the silica-exposed rats (Figure 3C) further support the potential role of hyaluronan in the pulmonary and systemic inflammation as well as in the differential expression of blood genes noticed in the silica-exposed rats.
The superior sensitivity of gene expression changes taking place in a target organ as an indicator of toxicity compared to the traditional biochemical and histological toxicity markers has been demonstrated previously (Heinloth et al., 2004). However, as summarized in Table 1, the results presented in this study as well as those presented in recent publications (Bushel et al., 2007; Lobenhofer et al., 2008; Umbright et al., 2010; Sellamuthu et al., 2011a) suggest that peripheral blood gene expression profiling may be an even more sensitive approach to detect target organ toxicity than gene expression profiling in target organs. In addition to the superior sensitivity, the present study results demonstrated that the gene expression changes taking place in the peripheral blood of the silica-exposed rats are mechanistically relevant to the pulmonary toxicity induced by the silica particles deposited in the lungs following inhalation exposure. Therefore, peripheral blood gene expression profiling appears to provide an opportunity to develop markers of target organ toxicity that are not only superior in sensitivity compared with the existing biochemical and histological toxicity markers (Sellamuthu et al., 2011a) but mechanistically relevant to the target organ toxicity. The capability to determine the mechanisms underlying the onset and progression of target organ toxicity by employing peripheral blood gene expression profiling, a minimally invasive surrogate approach, has implications in developing as well as applying effective strategies potentially capable of preventing target organ toxicity. This would represent a significant advancement considering silicosis, the principal pulmonary effect of occupational exposure to silica, is an irreversible but preventable, life-threatening pulmonary condition. While the findings of the present study are encouraging, additional studies are warranted, especially, those designed to determine whether peripheral blood gene expression changes can detect toxicity that is specific to both the target organ and a given toxicant, especially under conditions of human exposure to toxic agents.
Table 1.
Correlation co-efficients (r2 values) for the relationship between silica-induced pulmonary toxicity (BALF LDH, PMN and MCP-1) and the number of significantly differentially expressed genes in the lungs and blood of silica exposed rats. The toxicity measurements and the number of differentially expressed genes in the silica exposed rats at postexposure time intervals of 0, 1, 2, 4, 8, 16, and 32 weeks following a one week exposure were used to determine the correlation co-efficients (r2 values). The 32-week data were obtained from the present study while data for all other post-exposure time intervals were obtained from our previous publications (Sellamuthu et al., 2011a, 2012).
| BALF LDH | BALF PMN | BALF MCP-1 | |
|---|---|---|---|
| Lung DEG | 0.776 | 0.879 | 0.927 |
| Blood DEG | 0.831 | 0.923 | 0.958 |
BALF, bronchoalveolar lavage fluid; LDH, lactate dehydrogenase, PMN, polymorphonuclear leukocytes; DEG, differentially expressed gene.
Supplementary Material
Acknowledgements
The authors thank Amy Cumpston and Howard Leonard (NIOSH, Morgantown, WV) for assistance with inhalation exposure of rats to crystalline silica.
Footnotes
Declaration of interest
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of NIOSH.
References
- Absher MP, Hemenway DR, Leslie KO, Trombley L, Vacek P. Intrathoracic distribution and transport of aerosolized silica in the rat. Exp Lung Res. 1992;18:743–757. doi: 10.3109/01902149209031705. [DOI] [PubMed] [Google Scholar]
- Antonini JM, Roberts JR. Chromium in stainless steel welding fume suppresses lung defense responses against bacterial infection in rats. J Immunotoxicol. 2007;4:117–127. doi: 10.1080/15476910701336953. [DOI] [PubMed] [Google Scholar]
- Barbarin V, Nihoul A, Misson P, Arras M, Delos M, Leclercq I, Lison D, Huaux F. The role of pro- and anti-inflammatory responses in silica-induced lung fibrosis. Respir Res. 2005;6:112. doi: 10.1186/1465-9921-6-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser. 1995;57:289–300. [Google Scholar]
- Bushel PR, Heinloth AN, Li J, Huang L, Chou JW, Boorman GA, Malarkey DE, Houle CD, Ward SM, Wilson RE, Fannin RD, Russo MW, Watkins PB, Tennant RW, Paules RS. Blood gene expression signatures predict exposure levels. Proc Natl Acad Sci USA. 2007;104:18211–18216. doi: 10.1073/pnas.0706987104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo GM, Avenoso A, D’Ascola A, Scuruchi M, Prestipino V, Nastasi G, Calatroni A, Campo S. The inhibition of hyaluronan degradation reduced pro-inflammatory cytokines in mouse synovial fibroblasts subjected to collagen-induced arthritis. J Cell Biochem. 2012;113:1852–1867. doi: 10.1002/jcb.24054. [DOI] [PubMed] [Google Scholar]
- Cantin AM, Larivée P, Martel M, Bégin R. Hyaluronan (hyaluronic acid) in lung lavage of asbestos-exposed humans and sheep. Lung. 1992;170:211–220. doi: 10.1007/BF00174118. [DOI] [PubMed] [Google Scholar]
- Carter JM, Driscoll KE. The role of inflammation, oxidative stress, and proliferation in silica-induced lung disease: a species comparison. J Environ Pathol Toxicol Oncol. 2001;20(Suppl 1):33–43. [PubMed] [Google Scholar]
- Cassel SL, Eisenbarth SC, Iyer SS, Sadler JJ, Colegio OR, Tephly LA, Carter AB, Rothman PB, Flavell RA, Sutterwala FS. The Nalp3 inflammasome is essential for the development of silicosis. Proc Natl Acad Sci USA. 2008;105:9035–9040. doi: 10.1073/pnas.0803933105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castranova V, Porter D, Millecchia L, Ma JY, Hubbs AF, Teass A. Effect of inhaled crystalline silica in a rat model: time course of pulmonary reactions. Mol Cell Biochem. 2002:234–235. 177–184. [PubMed] [Google Scholar]
- Castranova V. Signaling pathways controlling the production of inflammatory mediators in response to crystalline silica exposure: role of reactive oxygen/nitrogen species. Free Radic Biol Med. 2004;37:916–925. doi: 10.1016/j.freeradbiomed.2004.05.032. [DOI] [PubMed] [Google Scholar]
- Chen GY, Nuñez G. Sterile inflammation: sensing and reacting to damage. Nat Rev Immunol. 2010;10:826–837. doi: 10.1038/nri2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GS, Miller FW, Germolec DR. Occupational exposures and autoimmune diseases. Int Immunopharmacol. 2002;2:303–313. doi: 10.1016/s1567-5769(01)00181-3. [DOI] [PubMed] [Google Scholar]
- DiMatteo M, Reasor MJ. Modulation of silica-induced pulmonary toxicity by dexamethasone-containing liposomes. Toxicol Appl Pharmacol. 1997;142:411–421. doi: 10.1006/taap.1996.8057. [DOI] [PubMed] [Google Scholar]
- Eklund A, Tornling G, Blaschke E, Curstedt T. Extracellular matrix components in bronchoalveolar lavage fluid in quartz exposed rats. Br J Ind Med. 1991;48:776–782. doi: 10.1136/oem.48.11.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdely A, Liston A, Salmen-Muniz R, Hulderman T, Young SH, Zeidler-Erdely PC, Castranova V, Simeonova PP. Identification of systemic markers from a pulmonary carbon nanotube exposure. J Occup Environ Med. 2011;53:S80–S86. doi: 10.1097/JOM.0b013e31821ad724. [DOI] [PubMed] [Google Scholar]
- Franchi L, Eigenbrod T, Núñez G. Cutting edge: TNF-alpha mediates sensitization to ATP and silica via the NLRP3 inflammasome in the absence of microbial stimulation. J Immunol. 2009;183:792–796. doi: 10.4049/jimmunol.0900173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser JR, Laurent TC, Laurent UB. Hyaluronan: its nature, distribution, functions and turnover. J Intern Med. 1997;242:27–33. doi: 10.1046/j.1365-2796.1997.00170.x. [DOI] [PubMed] [Google Scholar]
- Fubini B, Hubbard A. Reactive oxygen species (ROS) and reactive nitrogen species (RNS) generation by silica in inflammation and fibrosis. Free Radic Biol Med. 2003;34:1507–1516. doi: 10.1016/s0891-5849(03)00149-7. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslinger B, Mandl-Weber S, Sellmayer A, Sitter T. Hyaluronan fragments induce the synthesis of MCP-1 and IL-8 in cultured human peritoneal mesothelial cells. Cell Tissue Res. 2001;305:79–86. doi: 10.1007/s004410100409. [DOI] [PubMed] [Google Scholar]
- Heinloth AN, Irwin RD, Boorman GA, Nettesheim P, Fannin RD, Sieber SO, Snell ML, Tucker CJ, Li L, Travlos GS, Vansant G, Blackshear PE, Tennant RW, Cunningham ML, Paules RS. Gene expression profiling of rat livers reveals indicators of potential adverse effects. Toxicol Sci. 2004;80:193–202. doi: 10.1093/toxsci/kfh145. [DOI] [PubMed] [Google Scholar]
- Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, Latz E. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC. International Agency for Research on Cancer, Monograph on the evaluation of carcinogenic risk to human. 1997;68:1–475. [Google Scholar]
- Jiang D, Liang J, Fan J, Yu S, Chen S, Luo Y, Prestwich GD, Mascarenhas MM, Garg HG, Quinn DA, Homer RJ, Goldstein DR, Bucala R, Lee PJ, Medzhitov R, Noble PW. Regulation of lung injury and repair by Toll-like receptors and hyaluronan. Nat Med. 2005;11:1173–1179. doi: 10.1038/nm1315. [DOI] [PubMed] [Google Scholar]
- Kreiss K, Zhen B. Risk of silicosis in a Colorado mining community. Am J Ind Med. 1996;30:529–539. doi: 10.1002/(SICI)1097-0274(199611)30:5<529::AID-AJIM2>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Linch KD, Miller WE, Althouse RB, Groce DW, Hale JM. Surveillance of respirable crystalline silica dust using OSHA compliance data (1979–1995) Am J Ind Med. 1998;34:547–558. doi: 10.1002/(sici)1097-0274(199812)34:6<547::aid-ajim2>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Lobenhofer EK, Auman JT, Blackshear PE, Boorman GA, Bushel PR, Cunningham ML, Fostel JM, Gerrish K, Heinloth AN, Irwin RD, Malarkey DE, Merrick BA, Sieber SO, Tucker CJ, Ward SM, Wilson RE, Hurban P, Tennant RW, Paules RS. Gene expression response in target organ and whole blood varies as a function of target organ injury phenotype. Genome Biol. 2008;9:R100. doi: 10.1186/gb-2008-9-6-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney W, Chen B, Frazer D. Computer controlled multi-walled carbon nanotube inhalation exposure system. Inhal Toxicol. 2009;21:1053–1061. doi: 10.1080/08958370802712713. [DOI] [PubMed] [Google Scholar]
- Miller BG, Hagen S, Love RG, Soutar CA, Cowie HA, Kidd MW, Robertson A. Risks of silicosis in coalworkers exposed to unusual concentrations of respirable quartz. Occup Environ Med. 1998;55:52–58. doi: 10.1136/oem.55.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundt KA, Birk T, Parsons W, Borsch-Galetke E, Siegmund K, Heavner K, Guldner K. Respirable crystalline silica exposure-response evaluation of silicosis morbidity and lung cancer mortality in the German porcelain industry cohort. J Occup Environ Med. 2011;53:282–289. doi: 10.1097/JOM.0b013e31820c2bff. [DOI] [PubMed] [Google Scholar]
- Nasrullah M, Mazurek JM, Wood JM, Bang KM, Kreiss K. Silicosis mortality with respiratory tuberculosis in the United States, 1968–2006. Am J Epidemiol. 2011;174:839–848. doi: 10.1093/aje/kwr159. [DOI] [PubMed] [Google Scholar]
- NIOSH. Hazard Review: Health Effects of Occupational Exposures to Respirable Crystalline Silica. US Department of Health and Human Services (NIOSH) Publication No. 2002–129. 2002 Available at: http://www.cdc.gov/niosh/docs/2002-129/02-129a.html.
- O’Reilly KM, Phipps RP, Thatcher TH, Graf BA, Van Kirk J, Sime PJ. Crystalline and amorphous silica differentially regulate the cyclooxygenase-prostaglandin pathway in pulmonary fibroblasts: implications for pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2005;288:L1010–L1016. doi: 10.1152/ajplung.00024.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks CG, Conrad K, Cooper GS. Occupational exposure to crystalline silica and autoimmune disease. Environ Health Perspect. 1999;107(Suppl 5):793–802. doi: 10.1289/ehp.99107s5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter DW, Ramsey D, Hubbs AF, Battelli L, Ma J, Barger M, Landsittel D, Robinson VA, McLaurin J, Khan A, Jones W, Teass A, Castranova V. Time course of pulmonary response of rats to inhalation of crystalline silica: histological results and biochemical indices of damage, lipidosis, and fibrosis. J Environ Pathol Toxicol Oncol. 2001;20(Suppl 1):1–14. [PubMed] [Google Scholar]
- Porter DW, Millecchia L, Robinson VA, Hubbs A, Willard P, Pack D, Ramsey D, McLaurin J, Khan A, Landsittel D, Teass A, Castranova V. Enhanced nitric oxide and reactive oxygen species production and damage after inhalation of silica. Am J Physiol Lung Cell Mol Physiol. 2002;283:L485–L493. doi: 10.1152/ajplung.00427.2001. [DOI] [PubMed] [Google Scholar]
- Porter DW, Hubbs AF, Mercer R, Robinson VA, Ramsey D, McLaurin J, Khan A, Battelli L, Brumbaugh K, Teass A, Castranova V. Progression of lung inflammation and damage in rats after cessation of silica inhalation. Toxicol Sci. 2004;79:370–380. doi: 10.1093/toxsci/kfh110. [DOI] [PubMed] [Google Scholar]
- Raghavachari N, Xu X, Munson PJ, Gladwin MT. Characterization of whole blood gene expression profiles as a sequel to globin mRNA reduction in patients with sickle cell disease. PLoS ONE. 2009;4:e6484. doi: 10.1371/journal.pone.0006484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson W. The US population-at-risk to occupational respiratory diseases. In: Merchant JA, editor. Occupational Respiratory Diseases. Washington, DC: DHHS (NIOSH) publication; 1986. pp. 86–102. [Google Scholar]
- Sato T, Shimosato T, Alvord WG, Klinman DM. Suppressive oligodeoxynucleotides inhibit silica-induced pulmonary inflammation. J Immunol. 2008;180:7648–7654. doi: 10.4049/jimmunol.180.11.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellamuthu R, Umbright C, Roberts JR, Chapman R, Young SH, Richardson D, Leonard H, McKinney W, Chen B, Frazer D, Li S, Kashon M, Joseph P. Blood gene expression profiling detects silica exposure and toxicity. Toxicol Sci. 2011a;122:253–264. doi: 10.1093/toxsci/kfr125. [DOI] [PubMed] [Google Scholar]
- Sellamuthu R, Umbright C, Li S, Kashon M, Joseph P. Mechanisms of crystalline silica-induced pulmonary toxicity revealed by global gene expression profiling. Inhal Toxicol. 2011b;23:927–937. doi: 10.3109/08958378.2011.625995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellamuthu R, Umbright C, Roberts JR, Cumpston A, McKinney W, Chen BT, Frazer D, Li S, Kashon M, Joseph P. Molecular insights into the progression of crystalline silica-induced pulmonary toxicity in rats. J Appl Toxicol. in press doi: 10.1002/jat.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Steenland K, Mannetje A, Boffetta P, Stayner L, Attfield M, Chen J, Dosemeci M, DeKlerk N, Hnizdo E, Koskela R, Checkoway H. International Agency for Research on Cancer Pooled exposure-response analyses and risk assessment for lung cancer in 10 cohorts of silica-exposed workers: an IARC multicentre study. Cancer Causes Control. 2001;12:773–784. doi: 10.1023/a:1012214102061. [DOI] [PubMed] [Google Scholar]
- Thakur SA, Beamer CA, Migliaccio CT, Holian A. Critical role of MARCO in crystalline silica-induced pulmonary inflammation. Toxicol Sci. 2009;108:462–471. doi: 10.1093/toxsci/kfp011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzircotis G, Thorne RF, Isacke CM. Chemotaxis towards hyaluronan is dependent on CD44 expression and modulated by cell type variation in CD44-hyaluronan binding. J Cell Sci. 2005;118:5119–5128. doi: 10.1242/jcs.02629. [DOI] [PubMed] [Google Scholar]
- Umbright C, Sellamuthu R, Li S, Kashon M, Luster M, Joseph P. Blood gene expression markers to detect and distinguish target organ toxicity. Mol Cell Biochem. 2010;335:223–234. doi: 10.1007/s11010-009-0272-5. [DOI] [PubMed] [Google Scholar]
- Xiang M, Fan J. Pattern recognition receptor-dependent mechanisms of acute lung injury. Mol Med. 2010;16:69–82. doi: 10.2119/molmed.2009.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.