The dimerization of the O2 sensor FNR is regulated by extremely fine-tuned interactions.
Keywords: biochemisty, biomolecules, Fumarate and Nitrate Reduction regulator (FNR), crystal structure
Abstract
The structure of the dimeric holo–fumarate and nitrate reduction regulator (FNR) from Aliivibrio fischeri has been solved at 2.65 Å resolution. FNR globally controls the transition between anaerobic and aerobic respiration in facultative anaerobes through the assembly/degradation of its oxygen-sensitive [4Fe-4S] cluster. In the absence of O2, FNR forms a dimer and specifically binds to DNA, whereas in its presence, the cluster is degraded causing FNR monomerization and DNA dissociation. We have used our crystal structure and the information previously gathered from numerous FNR variants to propose that this process is governed by extremely fine-tuned interactions, mediated by two salt bridges near the amino-terminal cluster-binding domain and an “imperfect” coiled-coil dimer interface. [4Fe-4S] to [2Fe-2S] cluster degradation propagates a conformational signal that indirectly causes monomerization by disrupting the first of these interactions and unleashing the “unzipping” of the FNR dimer in the direction of the carboxyl-terminal DNA binding domain.
The fumarate and nitrate reduction regulator (FNR) is the protein switch that controls the transition between anaerobic and aerobic respiration in Escherichia coli (Ec) and related facultative anaerobes (1). FNR is a member of the CRP [cyclic adenosine monophosphate (cAMP) receptor protein] superfamily of homodimeric transcription factors (2). Proteins belonging to this superfamily consist of a C-terminal DNA binding domain that recognizes specific sequences within numerous promoters, an extensive helical coiled-coil dimer interface, and an N-terminal sensory domain (2). In FNR, the latter domain contains residues of Cys20, Cys23, and Cys29, which, along with Cys122 (E. coli numbering), can coordinate an iron-sulfur cluster (3). In the absence of O2, FNR coordinates a [4Fe-4S]2+ center, forms a dimer, and specifically binds to DNA (4, 5). Conversely, in the presence of O2, the cluster is rapidly degraded in a second-order reaction to a [3Fe-4S]+ intermediate and then spontaneously to a [2Fe-2S]2+ center, causing FNR monomerization (6). Monomeric FNR does not bind DNA and consequently is unable to interact with RNA polymerase and regulate gene expression (7). After prolonged O2 exposure, FNR completely loses its iron-sulfur cluster (8). Transcriptional control through an iron-sulfur cluster-modulated monomer-dimer transition is unique to the FNR family of regulators and has been the subject of intense scrutiny (1, 9–11). However, and in spite of long-term efforts, no FNR three-dimensional structure is yet available. Accordingly, the numerous site-directed mutagenesis studies and their interpretation have been based on homology models built from the known CRP structure (12). Here, we report the 2.65 Å resolution dimeric structure of [4Fe-4S]–containing FNR from the marine bacterium Aliivibrio fischeri (Af) (table S1) (13). In addition, we briefly discuss the structure of a form of AfFNR with a partially degraded cluster. AfFNR has the same length as EcFNR and shares 84% of amino acid sequence identity with it. It can also complement an fnr− E. coli mutant (13), and in our hands, it behaves very much like the E. coli regulator (Fig. 1). Although CRP-based models provided a reasonable basis for mutagenesis studies of EcFNR, we will show that many of the interpretations derived from these efforts need to be revised now that our closely related AfFNR structure is available.
Fig. 1. AfFNR characterization.
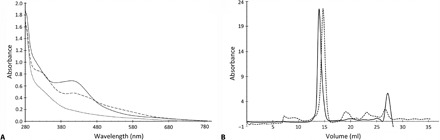
(A) Ultraviolet-visible spectra of AfFNR. The spectra of anaerobic (solid line), 15-min air-exposed (dashed line), and 330-min air-exposed (dotted line) AfFNR correspond to the [4Fe-4S], [2Fe-2S], and apo forms of the protein, respectively. (B) Size-exclusion chromatography elution profiles of anaerobically prepared (dimeric; solid line) and aerobically treated (monomeric; dashed line) AfFNR. These figures are to be compared to those obtained for EcFNR under similar conditions (15, 17, 23).
The [4Fe-4S]–holo-AfFNR structure appears to be increasingly agitated or disordered in going from the C-terminal DNA binding motif to the N-terminal iron-cluster binding region and the α-B helix, which packs against its α-A counterpart (Fig. 2, A and B). As in the related FixK2 (14), some N-terminal residues of AfFNR (in our case, 18) and its Strep-tag are not visible in the electron density maps. Several side chains also lack matching electron density at the N-terminal domain, but most of it is relatively well defined (fig. S1), including the [4Fe-4S] cluster ligands (Fig. 2C). Conversely, in the AfFNR containing a partially degraded cluster, the N-terminal region up to residue 42 is not resolved.
Fig. 2. Structure of holo-AfFNR.
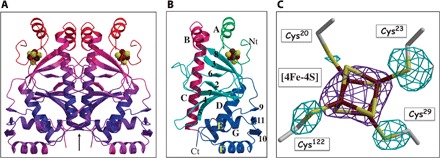
(A) Ribbon depiction of the AfFNR dimer color-coded according to increasing temperature factor values (blue to red). The arrow represents the twofold axis of the AfFNR dimer. (B) The AfFNR monomer domains; crimson, dimerization α-C helix; blue, C-terminal (Ct) DNA binding domain; light blue, β-roll domain; green, N-terminal (Nt) iron-sulfur cluster-binding domain. Helices are sequentially named with letters as in other CRP superfamily members (12); β-strands are denoted with numbers. (C) Composite electron density at the AfFNR [4Fe-4S] cluster; purple, 2.65 Å resolution anomalous scattering peak from the data collected at the iron edge and contoured at 7σ (see table S1); light blue, (Fobs − Fcalc) map with the sulfur atoms of Cys20, Cys23, Cys29, and Cys122 omitted from the calculated structure factors and phases (3.6σ).
Of the large number of EcFNR variants studied (11, 15, 16), S24F, L28H, R140A, L151A, and D154A deserve special attention. The first two mutations increase iron-sulfur cluster stability toward O2-induced degradation (17–19). In the case of the S24F variant, using a partially homologous [4Fe-4S]–containing amino acid sequence from the known structure of endonuclease III, it was postulated that the Phe side chain could shield the [4Fe-4S]2+ cluster from O2 attack (17). However, from our crystal structure, it is unclear how much protection Phe24 could afford against O2 because the Cα atom of Ser24 is >7 Å from the nearest cluster atom and its Cβ atom points away from the cluster (Fig. 3A)
Fig. 3. Analysis of the [4Fe-4S] cluster environment.
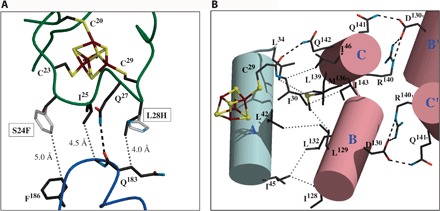
(A) Likely positions of mutated residues (gray) in the S24F and L28H AfFNR variants. Both mutations should influence the interaction of the Cys23-Cys29 loop (green) with the top of the DNA binding domain (blue). (B) Intramolecular connections between the [4Fe-4S] cluster and the Asp130-Arg140 salt bridges. Helices A, B, and C (Fig. 2B) are represented by cylinders.
The very similar phenotype of the L28H variant has been explained using a model based on a ferredoxin structure, which suggests that the imidazole ring of His could form a hydrogen bond with a cluster sulfide ion (20). However, given the orientation of the Cys23-Cys29 loop relative to the [4Fe-4S]2+ cluster, such H-bond is not plausible (Fig. 3A). Instead, both mutations could hinder the conformational flexibility of this region expectedly required for both iron-sulfur cluster conversion/degradation from the [3Fe-4S]+ species to the [2Fe-2S]2+ cluster and the ensuing monomerization (Fig. 3A) (21). Phe24 and other bulky residues at that position (17) are likely to prevent this process by contacting the (183-186) stretch at the top of the DNA binding domain (Fig. 3A). Conversely, His28 could establish polar/electrostatic interactions with the same region or, like the L28F variant, also cause steric hindrance (17). The weak electron density at the Cys20 ligand (Fig. 2C) suggests that the oxygen-induced formation of the [3Fe-4S]+ species results from the dissociation of the Cys20S-Fe bond and subsequent loss of the metal as a ferrous ion, a first step in the [4Fe-4S]2+ to [2Fe-2S]2+ cluster transition (6). The cluster-stabilizing effect of the S24F and L28H mutations, located in the Cys23-Cys29 loop, also favors Cys20 as the most likely candidate for initial iron loss (Cys122 belongs to a different more ordered domain). Indeed, a double D154A-L28H variant has been reported to stabilize the [3Fe-4S]+ species (4).
Model-based predictions located the invariant Arg140, Leu151, and Asp154 at the long coiled-coil dimerization interface (15). Although this is indeed the case for Leu151 and Asp154, Arg140 is near the N terminus of the dimerization α-C helix where our AfFNR structure shows no coiled-coil interface contacts (the Cα-Cα distance for the twofold symmetry-related Arg140 residues is 8.7 Å). Instead, the guanidinium group of Arg140 forms a salt bridge with Asp130 from the opposite α-B helix (Figs. 3B and 4, A and B). This is a bond that, as discussed below, may be a determinant in modulating monomer-dimer equilibrium in FNRs. Mutation of Asp154 to Ala resulted in a dimeric EcFNR variant that did not dissociate into monomers after O2-induced iron-sulfur cluster degradation (15, 16). It was thus concluded from CRP-based models that the dimer twofold axis-related Asp154 side chains should inhibit FNR dimerization through charge repulsion. Indeed, removal of their carboxylate group enhanced dimer stability (16). A similar line of reasoning was used in the case of the less drastic E150A mutation. These conclusions may now be reanalyzed using our AfFNR structure. We note that charge repulsion between the two twofold axis-related Asp154 residues is somewhat minimized by the opposite orientation of their carboxylate groups, in a d-d′ helical interaction (Fig. 4, C and D); their closest O-O distance is about 4.6 Å. Conversely, these groups sit in protein pockets that, although polar in nature, will not compensate their expected negative charges. Furthermore, the carboxylate group of Glu150, which is across the dimer interface (Fig. 4, C and D), is located about 5.5 Å from its counterpart in Asp154. Replacement of either Asp154 or Glu150 by Lys resulted in variants with unexpectedly high activity under aerobic conditions (16), an observation that can now be explained by charge neutralization through the formation of salt bridges between Asp154 and Lys150 or Glu150 and Lys154 from the same monomer. In conclusion, our structure-based observations indicate that dimer interface stabilization through removal of the carboxylate groups in the D154A variant may not depend only on direct Asp154-Asp154′ repulsion. The proximity of Asp154 to Ile151 and Ile158 may also play a role. Li et al. (22) have reached comparable conclusions when analyzing the kinase VPS34 complex component Beclin 1 antiparallel coiled-coil domain. In their crystal structure, several a-d′ helical pairings were defined as “imperfect,” resulting in a metastable homodimer that readily dissociates to establish metabolically relevant heterodimeric interactions with other partners (22). Imperfect pairings were characterized, among others, by two a-d′ Glu-Val interactions; their Glu-to-Leu mutation enhanced homodimeric Beclin 1 thermostability (22).
Fig. 4. Structural determinants of the monomer-dimer equilibrium in FNRs.
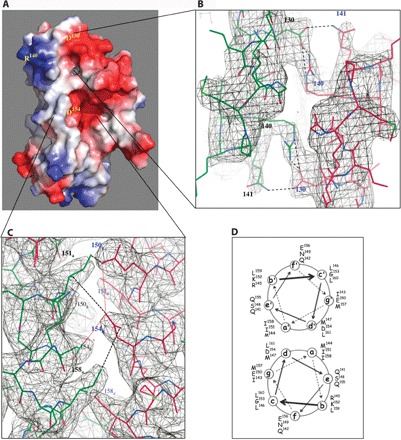
(A) Space-filling model depicting electrostatic surfaces at the monomer-monomer interface. (B) View down the dimer twofold axis at the “top” salt bridges between Asp130 and Arg140 in their (2Fo − Fc) electron density map contoured at 1σ. (C) Coiled-coil dimer interface. The positions of E150, I151, D154, and I158 are highlighted in their (2Fo − Fc) electron density contoured at 1σ (see the text). (D) Helical wheel projection of the distribution of amino acid residues in the two α-C helices at the AfFNR dimer interface. Stereo images of (B) and (C) are shown in figs. S3 and S4.
The phenotype of the FNR I151A variant, which remains mostly monomeric even when coordinating a [4Fe-4S]2+ cluster (15), is intriguing. The postulated screening effect of Ile151 putatively placed between the carboxylate groups of the Asp154 pair (15) is not corroborated by our structure (Fig. 4C). Instead, the Ile151 pair establishes van der Waals contacts through an a-a′ helical interaction, just above the Asp154 pair. This hydrophobic contact appears to be a determinant in keeping the dimeric structure because of its proximity to the destabilizing Asp154 residues. In the case of Beclin 1, double Leu-to-Ala variants also generated monomeric species, underscoring the stabilizing effect of Leu-Leu interactions in its coiled-coil domain (22). Several other variants of residues belonging to the coiled-coil domain of FNR favor the monomeric state (15). However, in most cases, dimerization could be reestablished either by higher in vivo expression of the EcFNR variant or by making the corresponding double mutant with D154A (15). In conclusion, the numerous mutagenesis studies indicate that because of its imperfect interface caused by the Asp154 pair and apparently aided by the presence of Glu150 (16), the FNR dimer is metastable, being significantly affected by the removal of any of the hydrophobic side chains not only at the a and d coiled-coil positions but also elsewhere in that domain (15).
Although, as mentioned above, it is well established that in wild-type FNRs the degradation of the [4Fe-4S] cluster results in monomerization (3, 6, 23), so far, no well-founded models have been postulated to explain the mechanism of such process. The fact that D154A-EcFNR dimers can exist without a cluster (18) and that I151A-EcFNR monomers can coordinate a [4Fe-4S] center (15) indicates that iron-sulfur cluster disassembly is not directly related to monomerization. Available data from other members of the CRP-FNR superfamily are not particularly helpful in this respect because they do not depend on dimer-monomer equilibrium for DNA binding. Rather, they rely on effector-driven modulation of the orientation and distance of the two DNA binding helix-turn-helix motifs in the dimer, involving the α-E and α-F helices (Fig. 2B) (12), or direct chemical modification at the regulator-DNA interface (14). Notably, by acquiring a [4Fe-4S] cluster, EcFNR increases its DNA binding affinity by only sevenfold, whereas when CRP binds its cAMP effector, its affinity augments by a factor of 104 (5). Clearly, very different mechanisms are at play. The FNR dimer is markedly different from other members of the homodimeric CPR superfamily with known structures not only functionally, as just described, but also structurally. Besides the expected dissimilarities at the [4Fe-4S] binding region, the N-terminal ends of the two long α-C helices are more agitated and kept further apart than in other members of the superfamily by interactions with the rest of the structure (fig. S2). In addition, the imperfect nature of the helical dimer interface and the salt bridges at the top of the structure described above are also unique to FNRs (Fig. 4A). Although some side chains in this latter region are not clearly visible in the electron density map, the α-B helices between Tyr126 and Gly134 and the α-A helices between Asp36 and Ile45 are rather well defined. There are three stabilizing hydrophobic interactions between these two antiparallel helices: Leu42a-Leu129e′, Ile45d-Ile128d′, and Ile45d-Ile132a′ (Fig. 3B).
The stretch between the cluster ligand Cys29 and Asn35 contains Ile30 and Leu34, which, along with Ile46, Ile124, Leu139, and Ile143 and residues from the two helices mentioned above, fill a cavity also lined by the [4Fe-4S] cluster. Thus, there is a conglomerate of interacting hydrophobic residues connecting the iron-sulfur cluster to the α-B helix (Fig. 3B). It seems then reasonable to conclude that conformational changes resulting from iron-sulfur cluster degradation will propagate through this hydrophobic network breaking the Arg140-Asp130 salt bridge and thereby causing the top-to-bottom directional disruption of the coiled-coil dimer interface (Fig. 1). This hypothesis is supported by the observation that the double D154A-I151A variant displays diminished activity (that is, it partially monomerizes), whereas the equivalent double D154A-I158A mutation does not have a significant effect on dimerization (15, 16). In this respect, and as indicated by the phenotypes of the R140A, R140L, and R140E EcFNR variants, Arg140 is essential for FNR dimer integrity (15).
Our crystal form of AfFNR with a partially degraded cluster shows that the loss of iron ion(s) disorganizes the Cys20-Cys29 cluster-binding loop to the point that it is no longer visible in the electron density map. We have already shown that a [4Fe-4S]2+ to [2Fe-2S]2+ structural transition in the model protein HydE involves major conformational changes (21). Equivalent changes cannot be accommodated in this AfFNR structure because of crystal packing constraints. Nevertheless, the structure shows that cluster loss leads to a small increase in both the disorder and the separation of the N-terminal regions of the α-C helices (fig. S2), as could be expected for a decrease in dimer stability. A comparison with the FixK2-DNA complex (14) shows that, in our structures, the α-F helices should be in a conformation close to the one required for DNA binding (fig. S5). In this respect, the AfFNR dimer does not differ from other members of the CRP superfamily. Although DNA binding is not required for O2-mediated cluster degradation, it appears that bound FNR is twice as prone to undergo this process as the free form (11). This might be related to the expected increased rigidity of FNR induced by its binding to DNA.
In summary, our analysis of the AfFNR crystal structure, along with the considerable functional information about EcFNR gathered during several decades, configures a picture of tunable monomer-dimer equilibrium that is mainly determined by two interactions: the top salt bridges and the imperfect interface of the two α-C helices. On the basis of these observations, we propose that FNR monomerization involves a dimer “unzipping” process that starts with the dissociation of the Asp130-Arg140 salt bridge near the N-terminal domain and propagates toward the C-terminal end of the interfacial α-C helices. We also put forward the idea that O2-modulated [4Fe-4S] to [2Fe-2S] cluster degradation indirectly initiates this process through a very significant local conformational change that disrupts the first of these two interactions. The work reported here will serve as a reliable source for new structural and functional studies aimed at further understanding the fascinating mechanism of O2-based regulation by FNRs.
Supplementary Material
Acknowledgments
We thank the staff from beamlines BM30A, ID23eh1, and ID23eh2 of the European Synchrotron Radiation Facility (ESRF, Grenoble, France). We are grateful to E. Barbier for the initial work with the FNR from A. fischeri. Funding: We thank the CEA and the CNRS for institutional funding. This work used the platforms of the Grenoble Instruct center (ISBG; UMS 3518 CNRS-CEA-UJF-EMBL) with support from FRISBI (ANR-10-INSB-05-02) within the Grenoble Partnership for Structural Biology (PSB). Author contributions: A.V. did the crystallography work, analyzed the structure, and wrote the paper; C.D. and O.R. performed the purification, characterization, and crystallization of FNR; Y.N. supervised the work by C.D. and O.R.; J.C.F.-C. supervised the work by C.D. and O.R., analyzed the structure and previous bibliography, and wrote the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. Atomic coordinates and structure factors have been deposited with the Protein Data Bank codes 5E44 (holo-AfFNR) and 5CVR (for partially degraded AfFNR).
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/1/11/e1501086/DC1
Materials and Methods
Fig. S1. Stereo view of the electron density of the N-terminal region of holo-FNR (starting at His19), shown as a black mesh contoured at 1σ.
Fig. S2. Comparison of the α-C helical dimerization interfaces of FNR and other selected CRP-family members, using deposited structures at the Protein Data Bank.
Fig. S3. Stereo image viewed down the dimer twofold axis.
Fig. S4. Stereo image of the electron density near the center of the dimer interface.
Fig. S5. Stereo image of the superposition of FNR (purple) to FixK2 (green) showing the similar position and orientation of the DNA binding α-F helices at the bottom of the figure.
Table S1. X-ray data and refinement statistics.
REFERENCES AND NOTES
- 1.Kiley P. J., Beinert H., Oxygen sensing by the global regulator, FNR: The role of the iron-sulfur cluster. FEMS Microbiol. Rev. 22, 341–352 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Körner H., Sofia H. J., Zumft W. G., Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: Exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 27, 559–592 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Lazazzera B. A., Beinert H., Khoroshilova N., Kennedy M. C., Kiley P. J., DNA binding and dimerization of the Fe−S-containing FNR protein from Escherichia coli are regulated by oxygen. J. Biol. Chem. 271, 2762–2768 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Khoroshilova N., Beinert H., Kiley P. J., Association of a polynuclear iron-sulfur center with a mutant FNR protein enhances DNA binding. Proc. Natl. Acad. Sci. U.S.A. 92, 2499–2503 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green J., Bennett B., Jordan P., Ralph E. T., Thomson A. J., Guest J. R., Reconstitution of the [4Fe-4S] cluster in FNR and demonstration of the aerobic-anaerobic transcription switch in vitro. Biochem. J. 316, 887–892 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crack J. C., Green J., Cheesman M. R., Le Brun N. E., Thomson A. J., Superoxide-mediated amplification of the oxygen-induced switch from [4Fe-4S] to [2Fe-2S] clusters in the transcriptional regulator FNR. Proc. Natl. Acad. Sci. U.S.A. 104, 2092–2097 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott C., Green J., Miscoordination of the iron-sulfur clusters of the anaerobic transcription factor, FNR, allows simple repression but not activation. J. Biol. Chem. 277, 1749–1754 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Sutton V. R., Stubna A., Patschkowski T., Münck E., Beinert H., Kiley P. J., Superoxide destroys the [2Fe-2S]2+ cluster of FNR from Escherichia coli. Biochemistry 43, 791–798 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Lazazzera B. A., Bates D. M., Kiley P. J., The activity of the Escherichia coli transcription factor FNR is regulated by a change in oligomeric state. Genes Dev. 7, 1993–2005 (1993). [DOI] [PubMed] [Google Scholar]
- 10.Green J., Rolfe M. D., Smith L. J., Transcriptional regulation of bacterial virulence gene expression by molecular oxygen and nitric oxide. Virulence 5, 794–809 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crack J. C., Green J., Thomson A. J., Le Brun N. E., Iron-sulfur clusters as biological sensors: The chemistry of reactions with molecular oxygen and nitric oxide. Acc. Chem. Res. 47, 3196–3205 (2014). [DOI] [PubMed] [Google Scholar]
- 12.McKay D. B., Steitz T. A., Structure of catabolite gene activator protein at 2.9 Å resolution suggests binding to left-handed B-DNA. Nature 290, 744–749 (1981). [DOI] [PubMed] [Google Scholar]
- 13.Septer A. N., Bose J. L., Dunn A. K., Stabb E. V., FNR-mediated regulation of bioluminescence and anaerobic respiration in the light-organ symbiont Vibrio fischeri. FEMS Microbiol. Lett. 306, 72–81 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonnet M., Kurz M., Mesa S., Briand C., Hennecke H., Grütter M. G., The structure of Bradyrhizobium japonicum transcription factor FixK2 unveils sites of DNA binding and oxidation. J. Biol. Chem. 288, 14238–14246 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore L. J., Kiley P. J., Characterization of the dimerization domain in the FNR transcription factor. J. Biol. Chem. 276, 45744–45750 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Moore L. J., Mettert E. L., Kiley P. J., Regulation of FNR dimerization by subunit charge repulsion. J. Biol. Chem. 281, 33268–33275 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Jervis A. J., Crack J. C., White G., Artymiuk P. J., Cheesman M. R., Thomson A. J., Le Brun N. E., Green J., The O2 sensitivity of the transcription factor FNR is controlled by Ser24 modulating the kinetics of [4Fe-4S] to [2Fe-2S] conversion. Proc. Natl. Acad. Sci. U.S.A. 106, 4659–4664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bates D. M., Lazazzera B. A., Kiley P. J., Characterization of FNR* mutant proteins indicates two distinct mechanisms for altering oxygen regulation of the Escherichia coli transcription factor FNR. J. Bacteriol. 177, 3972–3978 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popescu C. V., Bates D. M., Beinert H., Münck E., Kiley P. J., Mössbauer spectroscopy as a tool for the study of activation/inactivation of the transcription regulator FNR in whole cells of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 95, 13431–13435 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bates D. M., Popescu C. V., Khoroshilova N., Vogt K., Beinert H., Münck E., Kiley P. J., Substitution of leucine 28 with histidine in the Escherichia coli transcription factor FNR results in increased stability of the [4Fe-4S]2+ cluster to oxygen. J. Biol. Chem. 275, 6234–6240 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Nicolet Y., Rohac R., Martin L., Fontecilla-Camps J. C., X-ray snapshots of possible intermediates in the time course of synthesis and degradation of protein-bound Fe4S4 clusters. Proc. Natl. Acad. Sci. U.S.A. 110, 7188–7192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X., He L., Che K. H., Funderburk S. F., Pan L., Pan N., Zhang M., Yue Z., Zhao Y., Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat. Commun. 3, 662 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khoroshilova N., Popescu C., Münck E., Beinert H., Kiley P. J., Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity. Proc. Natl. Acad. Sci. U.S.A. 94, 6087–6092 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicolet Y., Zeppieri L., Amara P., Fontecilla-Camps J. C., Crystal structure of tryptophan lyase (NosL): Evidence for radical formation at the amino group of tryptophan. Angew. Chem. Int. Ed. Engl. 53, 11840–11844 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Elliott J. I., Brewer J. M., The inactivation of yeast enolase by 2,3-butanedione. Arch. Biochem. Biophys. 190, 351–357 (1978). [DOI] [PubMed] [Google Scholar]
- 26.Evans G., Pettifer R. F., CHOOCH: A program for deriving anomalous-scattering factors from X-ray fluorescence spectra. J. Appl. Crystallogr. 34, 82–86 (2001). [Google Scholar]
- 27.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans P. R., Murshudov G. N., How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G. W., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S., Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J., Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A., REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/1/11/e1501086/DC1
Materials and Methods
Fig. S1. Stereo view of the electron density of the N-terminal region of holo-FNR (starting at His19), shown as a black mesh contoured at 1σ.
Fig. S2. Comparison of the α-C helical dimerization interfaces of FNR and other selected CRP-family members, using deposited structures at the Protein Data Bank.
Fig. S3. Stereo image viewed down the dimer twofold axis.
Fig. S4. Stereo image of the electron density near the center of the dimer interface.
Fig. S5. Stereo image of the superposition of FNR (purple) to FixK2 (green) showing the similar position and orientation of the DNA binding α-F helices at the bottom of the figure.
Table S1. X-ray data and refinement statistics.


