Significance
Our results suggest that particular ferredoxins in photosynthetic organisms are tailored to serve as electron carriers that sustain day-time and night-time metabolism and that the chloroplast-localized ferredoxin-5 (FDX5) appears to function in the desaturation of fatty acids required for maintaining the correct ratio of the dominant lipids in the thylakoid membranes and the integration of chloroplast and mitochondrial metabolism, which is absolutely required for growth in the dark. The most important messages from this work are that redox components associated with critical activities in photosynthetic organisms must be tuned to the redox conditions of the cells and the overall carbon budget of photosynthetic cells requires an understanding of metabolic features that accompany the movement of cells between light and dark conditions.
Keywords: ferredoxin, dark growth, thylakoid lipids, triacylglycerol, redox regulation
Abstract
Photosynthetic microorganisms typically have multiple isoforms of the electron transfer protein ferredoxin, although we know little about their exact functions. Surprisingly, a Chlamydomonas reinhardtii mutant null for the ferredoxin-5 gene (FDX5) completely ceased growth in the dark, with both photosynthetic and respiratory functions severely compromised; growth in the light was unaffected. Thylakoid membranes in dark-maintained fdx5 mutant cells became severely disorganized concomitant with a marked decrease in the ratio of monogalactosyldiacylglycerol to digalactosyldiacylglycerol, major lipids in photosynthetic membranes, and the accumulation of triacylglycerol. Furthermore, FDX5 was shown to physically interact with the fatty acid desaturases CrΔ4FAD and CrFAD6, likely donating electrons for the desaturation of fatty acids that stabilize monogalactosyldiacylglycerol. Our results suggest that in photosynthetic organisms, specific redox reactions sustain dark metabolism, with little impact on daytime growth, likely reflecting the tailoring of electron carriers to unique intracellular metabolic circuits under these two very distinct redox conditions.
Ferredoxins (FDXs) are soluble, iron–sulfur proteins that mediate electron transfer in a variety of essential metabolic reactions (1–3) (SI Appendix, Fig. S1). The Chlamydomonas genome encodes 13 FDXs (SI Appendix, Table S1) with localization and redox properties that suggest involvement in specific redox reactions (4). Using a yeast two hybrid approach, a global FDX interaction network was established for Chlamydomonas, suggesting putative roles for FDX1 (originally designated Fd) in redox metabolism, carbohydrate modification, and fatty acid biosynthesis (5); this FDX was already known to function in both linear and cyclic photosynthetic electron flow (4, 6). FDX1 also accepts electrons from FDX–NADP oxidoreductase (FNR) (7) and donates electrons to HYDA hydrogenases (5, 8, 9). Other FDXs may be involved in state transitions, nitrogen metabolism, cellular responses to reactive oxygen species (ROS), and dark anoxia (4, 5, 10–13).
The major lipids in thylakoid membranes are monogalactosyldiacylglycerol (MGDG), digalactosyldiacylglycerol (DGDG), phosphatidylglycerol (PG), and sulfoquinovosyldiacylglycerol (SQDG) (14–17), with MGDG being the most abundant, followed by DGDG (14, 17). In Arabidopsis, three enzyme systems are involved in MGDG and DGDG synthesis (18). In contrast, there is only a single copy each of an MGDG and DGDG synthase gene in Chlamydomonas (19). Chlamydomonas uses the prokaryotic pathway for thylakoid lipid synthesis (20) in which the MGDG species synthesized in chloroplasts contain predominantly C18:3Δ9,12,15–sn-1 with the unusual hexadeca-4, 7, 10, 13-tetraenoic acid C16:4Δ4,7,10,13 at the sn-2 position (20). Desaturation of the sn-2 acyl group of Chlamydomonas MGDG requires the CrΔ4FAD desaturase, which is not present in Arabidopsis (21), and MGDG accumulation in Chlamydomonas is reduced in strains that make less CrΔ4FAD (21). Under conditions of nutrient deprivation, there are small amounts of C16:4Δ4,7,10,13 and C18:3Δ9,12,15 that are integrated into triacylglycerol (TAG) (22, 23), indicating that fatty acids within TAG can be at least partly recycled from membrane lipids such as MGDG.
In this study, we show that a fdx5 mutant (i) cannot grow in the dark despite having a growth rate similar to that of wild-type (WT) cells in the light; (ii) has altered photosynthetic properties, membrane structure, and lipid composition relative to WT cells; and (iii) accumulates higher levels of TAG. In addition, FDX5 appears to interact with the CrΔ4FAD and CrFAD6 desaturases. Together, these results suggest a critical role for FDX5 in fatty acid desaturation and maintaining thylakoid membrane composition and functionality specifically during growth in the dark.
Results
fdx5 Is a Null Mutant.
The Chlamydomonas FDX5 gene (SI Appendix, Fig. S2A) encodes a protein of 130 amino acids, with residues 35–107 forming the 2Fe–2S cluster binding domain (SI Appendix, Fig. S2B). We used random insertional mutagenesis (24, 25) and a PCR-based screen to isolate a mutant disrupted for FDX5 (FDX5-specific primers shown in SI Appendix, Table S2). Southern blot hybridizations indicated that the FDX5 gene was disrupted by the AphVIII cassette (SI Appendix, Fig. S2 C and D). Characterization of the site of insertion in FDX5 was performed by PCR and sequencing (SI Appendix, Fig. S2E). The cassette insertion resulted in a 46-bp deletion in the second exon that caused a shift in the ORF and a truncation of the C-terminal sequence (SI Appendix, Fig. S2 B and F). The truncated protein no longer contains the conserved cysteine residues that bind the 2Fe–2S cluster required for protein activity. Using immunological analyses, we were unable to detect FDX5 in the mutant (SI Appendix, Fig. S2 G, Left), whereas the FDX5 protein was restored in the rescued strains (SI Appendix, Fig. S2 G, Left and Right).
FDX5 Localizes to Chloroplasts.
Amino acids 1–18 of FDX5 are predicted to encode an N-terminal chloroplast transit peptide (26) (SI Appendix, Fig. S2B). Previous biochemical (13) and proteomic (27) studies demonstrated chloroplast localization of FDX5, which was confirmed in this study (SI Appendix, Fig. S3); the various mutant strains and constructs used for this and other analyses presented below are given in SI Appendix, Tables S3 and S4.
Disruption of FDX5 Causes a Dark Growth Deficiency.
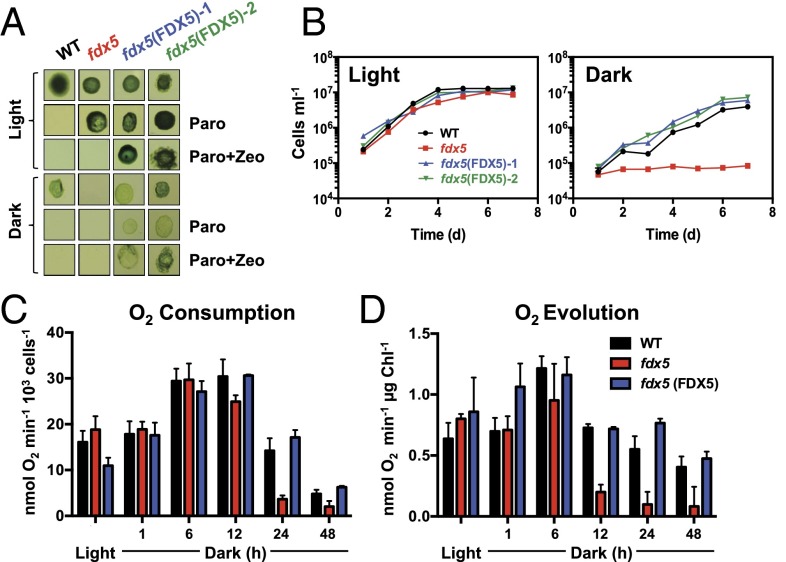
As shown in Fig. 1 A and B, the fdx5 mutant grows normally on Tris–acetate–phosphate (TAP) agar and liquid medium in the light but is unable to grow on the same medium in the dark. Very low light intensities (2 and 5 μmol photons·m−2·s−1) were sufficient for fdx5 to grow, albeit at a slower rate than WT and the complemented cells (SI Appendix, Fig. S4 A and B). With slight increases in light intensity (10 and 40 μmol photons·m−2·s−1), the rate of fdx5 growth became the same as that of WT and the complemented strains (SI Appendix, Fig. S4 C and D). Tetrad analyses demonstrated that the dark growth phenotype cosegregated with the insertion, conferring paromomycin resistance (SI Appendix, Fig. S5). To confirm that disruption of FDX5 was the cause of the phenotype, we introduced WT FDX5 cDNA and genomic DNA (gDNA) into fdx5 cells, which grew under both light and dark conditions on solid and in liquid medium, as shown for fdx5(FDX5)-1– and fdx5(FDX5)-2–rescued strains (Fig. 1 A and B); fdx5(FDX5)-1 was used for the remaining studies and is hereafter designated fdx5(FDX5). The growth-rescued transformants also regained their ability to accumulate FDX5 protein (SI Appendix, Fig. S2G). Overall, these results conclusively demonstrate that a strain unable to synthesize FDX5 cannot grow in the dark but readily resumes growth in the light.
Fig. 1.
The fdx5 mutant is unable to grow and has attenuated respiration and photosynthesis rates in the dark. (A) Colonies of WT, fdx5, and two rescued strains [fdx5(FDX5)-1 and fdx5(FDX5)-2] after 9 d of growth on solid TAP medium in the light and dark. The medium was unsupplemented, supplemented with paromomycin (Paro), or supplemented with paromomycin and Zeocin (Paro + Zeo). (B) Growth of WT, fdx5, and two rescued strains in liquid TAP medium over 7 d in the light and dark. (C) Respiratory O2 consumption after transferring WT cells, fdx5, and the fdx5(FDX5) complemented strain from the light to the dark for various times (1–48 h). (D) Photosynthetic O2 evolution in saturating actinic light after transferring WT cells, fdx5, and the fdx5(FDX5) complemented strain from the light to the dark for various times (1–48 h). The results for the various strains are given in different colors, as indicated. Error bars for B, C, and D represent SD (n = 3); experiments were performed in triplicate.
Photosynthesis and Respiration Decrease in Dark-Maintained fdx5.
The inability of fdx5 to grow in the dark suggested a loss of respiratory activity. As shown in Fig. 1C, respiratory O2 consumption is comparable in WT and mutant cells in the light and after transfer to the dark for up to 6 h. However, after 24 and 48 h in the dark, there was ∼twofold more respiratory O2 consumption in WT and the complemented strain than in fdx5. A similar trend was observed for light-mediated net photosynthetic O2 evolution, indicating a rapid decline in the ability of fdx5 to perform photosynthetic electron transport after dark incubation (Fig. 1D). Hence, both respiratory and photosynthetic processes were seriously compromised in the dark-maintained fdx5 mutant.
To determine the extent of absolute O2 consumption after dark-grown cells were placed in the light, we performed membrane inlet mass spectrometry (MIMS) with added gaseous 18O2 to distinguish between absolute O2 consumption and evolution (from H216O). As shown in SI Appendix, Fig. S6, the decline in net light-dependent O2 evolution in fdx5 is partly reflected by an increased rate of light-dependent O2 consumption, which is substantially higher in fdx5 than in WT cells.
Photosynthetic Electron Flow Is Altered in Dark-Maintained fdx5.
To examine photosynthetic electron transport rates (ETRs), we monitored the induction of chlorophyll (Chl) fluorescence. Induction curves of cultures transferred to the dark for 24 h showed that illumination caused an increase in steady-state fluorescence (Fs) for fdx5 relative to WT or complemented cells (SI Appendix, Fig. S7A), indicating that the plastoquinone (PQ) pool in the mutant was more reduced. Following the addition of the PSII inhibitor DCMU to the cultures (SI Appendix, Fig. S7A), Chl fluorescence was the same for all dark-grown strains, indicating normal functioning of PSII reaction centers. These results indicate that when maintained in the dark, fdx5 develops a partial block in photosynthetic electron flow downstream of PSII.
To assess the effect of different actinic light intensities on light- or dark-grown cultures, Chl fluorescence induction curves were assayed. For all growth conditions, Fv/Fm values were similar among the strains (SI Appendix, Fig. S7B; ΦPSII at time 0), suggesting that PSII is active and not responsible for the block in electron flow. The light curves also confirmed a reduction in ETR after the fdx5 mutant had been maintained in the dark (SI Appendix, Fig. S7 B, Middle). Furthermore, as shown in SI Appendix, Fig. S7 C, Bottom Right, the addition of methyl viologen (MV), which accepts electrons from PSI, to dark-grown cultures relieved elevated PQ pool reduction in fdx5 relative to WT cells, strongly suggesting that the block in electron transport was the result of a defect downstream of PSI, potentially caused by a lack of a suitable electron acceptor. Surprisingly, the rate of cyclic electron flow (CEF) was drastically reduced for fdx5 grown in the dark (SI Appendix, Fig. S7 D, Right) but not in the light (SI Appendix, Fig. S7 D, Left). The addition of MV, which competes with CEF for PSI electrons, had a similar effect for all strains (SI Appendix, Fig. S7D). Overall, fdx5 appears to be defective for both photosynthetic and respiratory processes after dark acclimation.
Photosynthetic Polypeptides Are Not Altered in fdx5.
The decrease in ETR and CEF in fdx5 grown in the dark could result from altered thylakoid membrane structure and/or aberrant accumulation of photosynthetic polypeptides or complexes. To explore the latter, we examined representative subunits of each photosynthetic complex by immunoblots. As shown in SI Appendix, Fig. S8, we immunologically detected the proteins of all of the major photosynthetic complexes; these include PsbA, OEE2, Cyt f, PSAD, both CF1-β and F1-β, LS, NDA2, and PGRL1 (full names provided in SI Appendix, Fig. S8). These proteins accumulated in fdx5 to approximately WT levels in both the light and dark (mitochondrial F1-β was somewhat lower in the “back to light” samples), suggesting no major alteration in the composition of membrane proteins associated with photosynthetic function in the mutant. These results oriented our further analyses toward examining membrane structure and lipid composition in dark-maintained fdx5.
Dark-Maintained fdx5 Has Altered Membrane Ultrastructure.
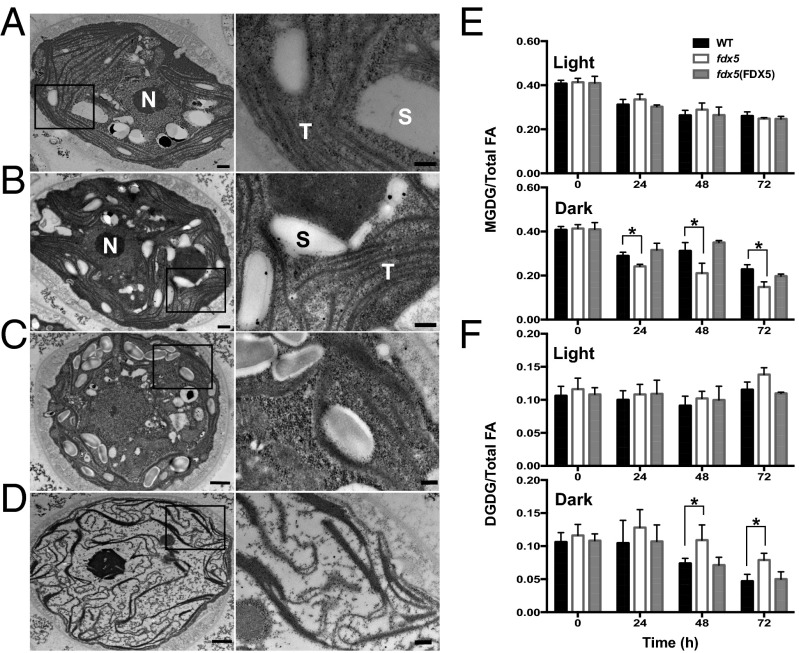
A potential explanation for the phenotypes described above is that the membrane structure/lipid composition is aberrant in dark-maintained fdx5, which in turn could retard efficient electron transport or other membrane-associated processes (e.g., ion transport). To evaluate this possibility, we performed transmission electron microscopy (TEM) of sectioned Chlamydomonas cells. In WT cells, thylakoid membranes are arranged as layers within chloroplasts, with some of the membranes appressed. Features in the light and dark were similar, although the membranes appeared to be more regularly arranged in light-grown cells (compare Fig. 2 A and C). Although the layered arrangement of thylakoids is observed in light-grown fdx5 (Fig. 2B), transfer of the mutant to the dark (Fig. 2D) caused the membrane structure to become highly aberrant. Dark-grown mutant membranes appear wavier, have more convoluted configurations, and seem to be more fragmented than membranes of WT cells. The cytoplasm and the chloroplast stroma in fdx5 had reduced granularity and did not accumulate large thylakoid-associated starch granules compared with WT (Fig. 2D).
Fig. 2.
Altered membrane morphologies and lipid compositions in dark-maintained fdx5. TEM of sectioned Chlamydomonas WT cells (A) and the fdx5 mutant (B) grown in the light. WT cells (C) and the fdx5 mutant (D) maintained in the dark for 48 h. Areas delineated by the rectangles are enlarged (Right) to reveal structural features of the membranes. N, nucleus; S, starch granules; T, thylakoid membranes; these structures are only noted for A and B. [Scale bar, 500 nm (A and B), 1,000 nm (C and D), and 200 nm (all enlargements at Right).] Relative levels of MGDG (E) and DGDG (F) in WT cells, fdx5, and the fdx5(FDX5) complemented strain in the light and following transfer to the dark for the indicated times. Error bars represent SD (n = 3); experiments were performed three times. “0” was the time just before transferring cells to the light or dark. *P < 0.05, by two-tailed Student’s t tests.
Membrane Lipids Are Altered in Dark-Maintained fdx5.
The phenotypes described above raised the possibility that fdx5 is defective in generating normal membranes in the dark, which may explain the aberrant membrane structures and disrupted activities that require specific lipid environments. Therefore, we analyzed classes of lipids and their acyl groups in membranes of fdx5 and WT cells. The relative levels of MGDG and DGDG in light-grown fdx5 and WT cells exhibited minor differences (Fig. 2 E and F, light). However, after 24, 48, and 72 h in the dark (Fig. 2 E and F, dark), the ratio of MGDG to DGDG in fdx5 strongly declined in comparison with WT cells. By 48 and 72 h in the dark, the mutant had a ∼2:1 ratio of MGDG to DGDG, whereas the ratio in WT cells was ∼5:1 (Fig. 2 E and F, dark).
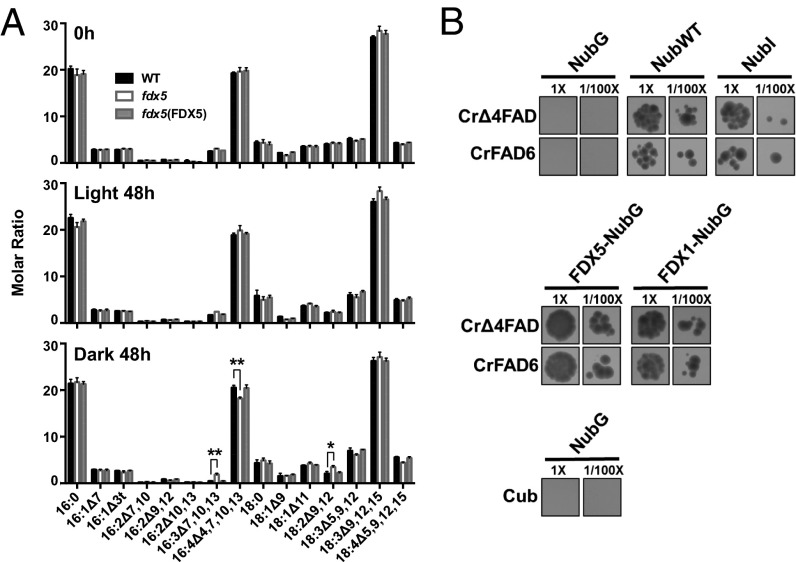
We also examined total fatty acid profiles under the same experimental conditions (Fig. 3A). The fraction of specific fatty acids that contain four double bonds (C16:4Δ4,7,10,13) was decreased in fdx5 maintained in the dark for 48 h but not in the light. This difference could be a consequence of a decrease in the synthesis of C16:4Δ4,7,10,13 (in accord with this finding, more C16:3Δ7,10,13 accumulated in fdx5 than in WT cells), an increase in degradation of MGDG containing C16:0 relative to C16:4Δ4,7,10,13 on the sn-2 position of MGDG, an increase in conversion of MGDG to DGDG (before desaturation of C16 on sn-2 of MGDG), or a combination of these events. We note that the change in the level of C16:4Δ4,7,10,13 in fdx5 in the dark was not as extensive as the change in the relative MGDG levels, which is consistent with previous results (21) and suggests (i) that a decline in the synthesis of C16:4Δ4,7,10,13 is also accompanied by conversion of MGDG containing C16:4Δ4,7,10,13 to DGDG and (ii) a proportion of the MGDG is degraded and the C16:4Δ4,7,10,13 is converted into TAG and potentially other lipids. The latter possibility is consistent with additional findings. The MGDG fatty acid composition showed no major difference in the fraction of C16:4Δ4,7,10,13 (relative to total fatty acids) in fdx5 in the dark relative to WT cells (SI Appendix, Fig. S9), with only a small change in the level of C16:4Δ4,7,10,13 in DGDG (SI Appendix, Fig. S10). There were also changes in the levels of C16:0, C16:1Δ7, 18:1Δ9, and 18:3Δ9,12,15 in DGDG (SI Appendix, Fig. S10) of fdx5, but these changes are independent of light/dark conditions and do not appear to impact the MGDG:DGDG ratio (in the light, the ratio of these lipids in fdx5 is similar to that of WT cells). Overall, aberrations in lipid composition (most notably the MGDG:DGDG ratio) and an altered ability to maintain the proper desaturation state of the fatty acids in dark-maintained fdx5 could explain the defects in photosynthesis, distorted membrane structure, and the overall appearance of mutant chloroplasts (membranes and stroma), which could also impact other cellular processes including respiration. Therefore, we sought to obtain mechanistic insights into the ways in which the fdx5 mutation impacts membrane lipid and fatty acid composition.
Fig. 3.
C16:4Δ4,7,10,13 fatty acid is decreased in dark-grown fdx5 and FDX5 interacts with fatty acid desaturases. (A) Fatty acid profiles in WT, fdx5, and the fdx5(FDX5) complemented strain in the light and dark at 0 and 48 h. Error bars represent SD (n = 3). Experiments were performed three times. *P < 0.05 and **P < 0.01, by two-tailed Student’s t tests. (B) Mating-based split-ubiquitin system showing interactions of FDX5 and FDX1 (Fd) with CrΔ4FAD and CrFAD6 in the presence of 500 µM methionine (Met500). The NubG (empty) vector combined with Cub or vectors containing the desaturase genes served as negative controls, whereas NubWT and NubI vectors served as positive controls. The assay was performed on eight separate occasions, yielding similar results.
FDX5 Interacts with CrΔ4FAD and CrFAD6 Desaturases.
Previous yeast two hybrid experiments demonstrated that FDX5 might associate with two fatty acid desaturases (5): CrFAD6 (Cre13.g590500), which is involved in desaturation of C18:1 in plastidic lipids (MGDG, DGDG, PG, SQDG) to C18:2, and CrΔ4FAD (Cre01.g037700), which is specific to Chlamydomonas MGDG and is involved in desaturation of C16:3Δ7,10,13 to C16:4Δ4,7,10,13. Venus-tagged CrΔ4FAD and CrFAD6 desaturases, like FDX5 (SI Appendix, Fig. S3), localized to chloroplasts (SI Appendix, Fig. S11).
As shown in Fig. 3B, Central, the mating-based split ubiquitin system (28) clearly demonstrated an interaction between FDX5 and both CrΔ4FAD and CrFAD6. No interaction was detected between NubG and Cub or NubG and the FDX5–Cub fusion protein (negative controls), whereas NubWT and NubI interacted with the fused protein FDX5–Cub (Fig. 3B) (positive control). These studies show that FDX1 also interacts with CrΔ4FAD and CrFAD6 (Fig. 3B, Central).
Increased TAG in fdx5 Mutant in the Dark.
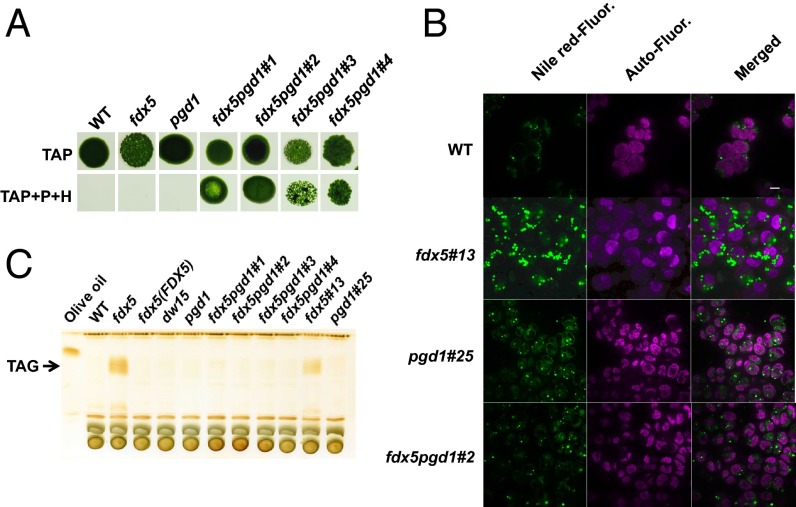
Mature MGDG synthesis in chloroplasts requires specific desaturation of fatty acids. The inability to properly desaturate fatty acids (e.g., generate C16:4Δ4,7,10,13 in dark) may destabilize newly synthesized MGDG, causing it to be recycled and its fatty acids to be stored as TAG. This possibility was supported by the finding that fdx5 accumulates TAG in the dark (Fig. 4 B and C and SI Appendix, Fig. S12). Sustained TAG accumulation was observed by TLC in the mutant maintained for 72 h in the dark, and much of the TAG was retained even 24 h after fdx5 was transferred back to the light (SI Appendix, Fig. S12). It was previously shown that the lipase PGD1 could cleave C18:1Δ9 from nascent MGDG and transfer it to TAG during nitrogen deprivation (29). To determine if TAG accumulation was mediated by PGD1, fdx5pgd1 double mutants were generated by crossing fdx5 with pgd1 (Fig. 4A); progeny with both lesions were identified by PCR (SI Appendix, Fig. S13). To visualize TAG production in fdx5, Nile-Red was used to stain lipid droplets (LDs). Fluorescence from stained LDs was observed in dark-maintained fdx5 cells, with LD content significantly lower in WT and the other strains (Fig. 4B). TLC (Fig. 4C) analysis also demonstrated TAG accumulation in fdx5 but not in any of the fdx5pgd1 double mutants, suggesting that PGD1 mediates the transfer of fatty acids from membrane lipids such as MGDG to TAG in the fdx5 mutant. The analysis of TAG in fdx5 and fdx5#13 (segregant from a cross between fdx5 and pgd1) showed that the TAG that accumulated in the mutant contained C16:0, C16:4Δ4,7,10,13, and C18:3Δ9,12,15 (SI Appendix, Fig. S14), suggesting that fatty acids of membrane-associated MGDG were transferred to TAG. These results suggest that fatty acid reshuffling and TAG accumulation in the dark depend on PGD1.
Fig. 4.
PGD1-mediated TAG accumulation in fdx5 in the dark. (A) Growth of WT, fdx5, and pgd1 single mutants and 4 fdx5pgd1 double mutants for 9 d on solid TAP medium in the light. The medium was unsupplemented or supplemented with both paromomycin (P) and hygromycin (H), as indicated. (B) Imaging of LDs in WT, fdx5#13, and pgd1#25 single mutants and the fdx5pgd1#2 double mutant. (C) TLC analysis of TAG in various strains grown in the dark. dw15 was the parental strain of the pgd1 mutant. Different isolates of the fdx5pgd1 double mutants, noted as “#,” are also shown.
Discussion
For photosynthetic organisms, metabolism under dark oxic conditions is impacted by a different cellular redox environment than metabolism in the light. Unlike WT cells, the fdx5 mutant is unable to grow in the dark. Dark-maintained fdx5 has low MGDG and high DGDG content (Fig. 2 E and F), has less C16:4Δ4,7,10,13 relative to total fatty acids (Fig. 3A), and accumulates TAG, which may at least in part be a consequence of a decrease in C16 desaturation by CrΔ4FAD to generate C16:4Δ4,7,10,13, as depicted in SI Appendix, Figs. S15 and S16. FDX5 also interacts with and may donate electrons to CrFAD6. However, CrFAD6 acts on fatty acids of both MGDG and DGDG (19, 30), and although the fdx5 mutant exhibits alterations in the ratio of the various C18 fatty acids (different desaturation states), the changes observed occur in both the light and dark; in the light, the mutant shows no defect in growth or in its MGDG:DGDG ratio. The donation of electrons from FDX5 (in the dark) and potentially FDX1 (in the light) to CrFAD6 (Cre13.g590500) (Fig. 3B) may be responsible for conversion of C18:1Δ9 on the sn-1 position to C18:2Δ9,12, and C16:1Δ7 on the sn-2 position to C16:2Δ7,10 in MGDG and also for conversion of C18:1Δ9 on the sn-1 position to C18:2Δ9,12 in DGDG (19). In the dark-maintained fdx5 mutant, the C18:3Δ9,12,15 level was considerably elevated in both total fatty acids and fatty acids in DGDG, whereas C18:2Δ9,12 showed only a modest increase (Fig. 3A and SI Appendix, Fig. S10). Plausible explanations for this finding are that another desaturase can function in place of CrFAD6 (Cre13.g590500) and/or that the CrFAD6 desaturase can accept electrons from another FDX or a cytochrome b5 (b-type cytochromes can also donate electrons to desaturases) that is reduced in the dark. Indeed, although most Arabidopsis desaturases have a single homolog, the Chlamydomonas genome encodes two plastidic forms of ω-6 desaturases (19), Cre06.g288650 and Cre13.g590500. Therefore, it is reasonable to suggest that Cre06.g288650 may function in the fdx5 mutant to synthesize C18:2Δ9,12 in the dark.
C18:3Δ9,12,15 is typically synthesized through the activity of an ω-3 desaturase, which uses C18:2Δ9,12 as its substrate. CrFAD7 (Cre01.g038600), likely localized to chloroplasts, was shown to be the only ω-3 desaturase in Chlamydomonas and is responsible for ω-3 desaturation of plastid- and ER-synthesized lipids (31). Because CrFAD7 does not appear to interact with FDX5 (5), it likely requires a different electron donor (e.g., another FDX isoform). The presence of multiple CrFAD6 desaturases and a chloroplast CrFAD7 enzyme may explain why there is no reduction of DGDG C18:3Δ9,12,15 in the fdx5 mutant in the dark. It is also likely that fatty acids from other lipids serve as substrates that are used in the synthesis of DGDG. This possibility is supported by the finding that there are over 100 mostly uncharacterized lipases encoded on the Chlamydomonas genome and that some of these lipases function in conjunction with acyl-transferases to modify or edit the acyl chains on membrane glycerolipids (29). Thus, certain lipases may release C18:2Δ9,12 and C18:3Δ9,12,15 from diacylglyceryl trimethyl homoserine and phosphatidylethanolamine in the ER or PG and SQDG in the chloroplast, promoting the recycling of these fatty acids.
In Chlamydomonas, “immature” MGDG (C18:1Δ9/C16:0) is known to be used in three ways (SI Appendix, Fig. S15): (i) desaturation to make mature MGDG (C18:3Δ9,12,15/C16:4Δ4,7,10,13) by multiple fatty acid desaturations, (ii) galactosylation to synthesize DGDG by the enzyme DGD1, and (iii) hydrolysis of the fatty acids from the glycerol backbone by PGD1, with the export of C18:1Δ9 and its incorporation into TAG (19–21, 29, 32). This last fate of MGDG may in part explain the reduced levels of C18:1Δ9 in DGDG, although this is observed in both the light and dark. Some MGDG would also be converted to DGDG (Fig. 2 E and F).
Finally, an inability to generate C16:4Δ4,7,10,13 at the sn-2 position of newly synthesized MGDG in dark-maintained fdx5 would prevent the maturation of MGDG, resulting in its destabilization and decreased accumulation of both MGDG and total C18:3Δ9,12,15 in MGDG. The C18:1Δ9 fatty acid present at the sn-1 position of immature MGDG could then be further desaturated (or remain C18:1Δ9) and become a constituent of DGDG (through conversion of MGDG to DGDG) or be released from MGDG through a PGD1-dependent cleavage (29) and used for the production of TAG. Mature MGDG present in fdx5 after growth in the light also contributes to TAG production through its turnover and recycling of its fatty acids; a similar MGDG recycling process is observed in Chlamdyomonas cells deprived of nitrogen (22, 29).
In the fdx5 mutant, desaturation of the C16 fatty acid to C16:4Δ4,7,10,13 may be decreased in the dark, as evidenced by the lower MGDG:DGDG ratio and a reduced fraction of C16:4Δ4,7,10,13 among all fatty acids relative to WT cells (the level of C16:4Δ4,7,10,13 remains the same in MGDG relative to WT cells, as expected), probably due to the lack of FDX5 protein as an electron donor for the CrΔ4FAD desaturase. Generation of C16:4Δ4,7,10,13 through the activity of CrΔ4FAD was shown to be a crucial limiting factor in the conversion of immature MGDG into mature MGDG, which would limit the overall MGDG content (21). Consequently, we observed an elevated level of DGDG and accumulation of TAG (probably caused by greater conversion of immature MGDG into these products) as well as a lower overall amount of MGDG in dark-maintained fdx5 (Fig. 2 E and F and SI Appendix, Figs. S15 and S16).
Together, our results suggest that FDX5 is a redox carrier that donates electrons to at least two fatty acid desaturases (CrΔ4FAD and CrFAD6) involved in converting C16:3Δ7,10,13 to C16:4Δ4,7,10,13and C18:1Δ9 to C18:2Δ9,12. We confirmed physical interactions of FDX5 with these desaturases (Fig. 3B). Because the mutant is unable to synthesize the proper fatty acids in the dark, there is an increase in DGDG production relative to MGDG as well as an increase in TAG synthesis; elevated TAG accumulation can be prevented if the pgd1 lesion is introduced into the fdx5 strain (Fig. 4). The altered ratio of MGDG to DGDG (Fig. 2) in fdx5 causes aberrant membrane structure (Fig. 2), which impacts the activity of many cellular processes including photosynthetic electron flow and causes other abnormalities potentially associated with altered membrane composition. These results raised several questions about TAG production in fdx5: (i) How are the enzymes involved in TAG production regulated in the dark? (ii) How does PGD1 affect TAG production in fdx5 in the dark? (iii) What is the contribution of other lipases to the accumulation of TAG? TAG production is low in WT cells and the pgd1 mutant, and these two strains have a similar complement of MGDG, DGDG, and TAG in the dark (SI Appendix, Fig. S16). However, TAG accumulation in fdx5 in the dark is blocked in the fdx5pgd1 double mutant (SI Appendix, Fig. S16). The incompletely processed MGDG in this double mutant cannot provide fatty acids for TAG biosynthesis, although it can still be converted to DGDG (SI Appendix, Fig. S16).
The dramatic dark-specific defect in lipid biosynthesis in fdx5 may limit its growth in the dark, although we cannot eliminate the possibility that the growth phenotype may be the consequence of an inability to perform other FDX5-mediated reactions in the dark (33). Preliminary pull-down assays have revealed several potential FDX5-interacting proteins (SI Appendix, Table S5). Furthermore, rescue of the fdx5 growth phenotype occurs in very low light, suggesting that the rescue may require only low-level production of reductant by the photosynthetic apparatus but may also involve light-associated regulatory processes. What seems most apparent from these studies is that some redox carriers have light- or dark-specific activities and play a major role in the diel metabolism of photosynthetic organisms.
Materials and Methods
Strains, Mutant Isolation, and Growth Conditions.
The parental, WT Chlamydomonas strains used were CC-124 (nit2−, mt−) and CC-125 (nit2−, mt+) (SI Appendix, Table S3). The fdx5 mutant was generated in the D66 (CC-4425; nit2−, cw15, mt+) genetic background (34) and identified using a PCR-based screen (24, 25), whereas the pgd1 strain (nit1−, cw15, mt+) was previously characterized (29). Strain cMJ030 (CC-4533, nit2−, cw15, mt−) was used for protein localization. Primers used for the mutant screen and genotyping are listed in SI Appendix, Table S2. The fdx5 mutant was backcrossed 5 times to CC-124 or CC-125, and fdx5pgd1 double mutants were generated by crossing fdx5 and pgd1 single mutants. Cells were grown on TAP agar and liquid medium, minimal medium, or high-salt medium at various light intensities. For TEM, cells were grown in Hepes–Acetate–Phosphate buffered medium.
Complementation and Transformation.
Complementation was performed with either FDX5 cDNA or gDNA under the control of the PSAD promoter in the pGEM T-easy vector (Promega). Transformation was performed by introducing 1 µg DNA of plasmids pJM43Ble-FDX5cDNA and pJM43Ble-FDX5gDNA (both linearized with NotI) into fdx5 (in CC-124 background) by electroporation (0.8 kv, 25 uF) using a BioRad GenePulser II Electroporator (Bio-Rad), as previously described (35). Transformants were selected on solid TAP medium containing 100 μg/mL ampicillin, 5 μg/mL paromomycin (for original fdx5 mutation), and 6 μg/mL Zeocin (for the introduced FDX5 gene) and then selected for growth in the dark. Transformants that grew in the dark were assayed by PCR for insertion of the different drug resistance cassettes.
Phenotyping and Growth Rates.
Growth was analyzed in liquid and solid medium, with or without various antibiotics. Samples from liquid cultures were collected and cells counted and/or quantified based on Chl levels (36, 37).
Photosynthetic O2 Evolution and Respiratory O2 Consumption.
The evolution and consumption of O2 were measured using a Clark electrode (CBID; Hansatech). Respiratory O2 consumption was measured in the dark, whereas photosynthetic O2 evolution was measured using white actinic light (∼500 µmol photons·m−2·s−1). Experiments were the average of three biological replicates.
TEM.
For TEM, cells were grown in modified TAP medium (Tris replaced by 20 mM Hepes), fixed in growth medium containing glutaraldehyde and OsO4, negatively stained with uranyl acetate, dehydrated in ethanol, and embedded in London resin (LR) white. Ultrathin sections (∼60 nm) were mounted onto Formvar-coated copper grids, contrasted with lead citrate (38), and micrographs were obtained using a Jeol JEM-1400 transmission electron microscope.
Analysis of Total and Membrane Lipids.
Total lipids were extracted from samples frozen in liquid nitrogen (39), dried under a N2 stream, and stored at –80 °C. Dried lipids were dissolved in 55 μL of chloroform and separated by TLC using a mixture of chloroform:methanol:acetic acid:water (75:13:9:3 vol/vol, respectively). After separation, the MGDG and DGDG bands and a spot containing total lipid (derived from 20 μL of the extract) were scraped from the TLC plate, converted into fatty acid methyl esters (40), and the amount of each of the fatty acid methyl esters quantified by gas chromatography on an HP6890 system as described previously (21).
TAG Analysis and Staining of LDs.
Lipid extraction and TAG analysis by TLC were conducted as described previously (29). To visualize LDs, cells were suspended in PBS-0.011% Triton X-100, stained with 1 µg/mL Nile-Red, immobilized on a 0.5% TAP-agarose pad, and imaged by spinning disk confocal microscopy; excitation was at 512 nm and emission at 570–590 nm.
Mating-Based Split Ubiquitin Assay.
FDX1, FDX5, CrΔ4FAD, and CrFAD6 genes were amplified from cDNAs using primers listed in SI Appendix, Table S2, and cloned into pENTR/D_TOPO (Invitrogen). The FDX1 and FDX5 genes from pENTR/d-TOPO_FD and pENTR/d-TOPO_FDX5 were introduced into pNX22-DEST by LR cloning, whereas CrΔ4FAD and CrFAD6 genes from pENTR/d-TOPO_CrΔ4FAD and pENTR/d-TOPO_CrFAD6 were introduced into pMetYC-DEST by LR cloning (SI Appendix, Table S4). Interactions between FDX1, FDX5, and CrΔ4FAD and CrFAD6 were assessed by the mating-based split ubiquitin system (28) (https://associomics.dpb.carnegiescience.edu/Associomics/Protocols.html).
Supplementary Material
Acknowledgments
We thank Dr. Gilles Peltier for PGRL1 antibodies and Dr. Fabrice Franck for Nda2 antibodies. The plasmids of pMetYC-DEST and pNX22-DEST and the yeast strains THY.AP4 and THY.AP5 were kindly provided by Dr. Wolf Frommer. MIMS experiments (performed by J.A., X.J., and P.R.) were supported by HélioBiotec, EU Grant 1944-32670, Provence Alpes Cote d'Azur (PACA) DEB 09-621, and Commissariat à l'Énergie Atomique. This work was supported by the Office of Biological and Environmental Research, Genomic Science Program, Office of Science, US Department of Energy Grants DE-FG02-07ER64427 and DE-FG02-12ER16338 (to A.R.G.) and DE-FG02-12ER16339 (to M.C.P.); National Science Foundation (NSF) Grants IOS-1359682 (to M.C.J.) and MCB 1157231 (to C.B.); NIH Grant GM42143 (to S.S.M.); Michigan State University AgBioResearch (to C.B.); as well as funds from the Carnegie Institution for Science (to A.R.G. and M.C.J.). J.W. is supported by the Royal Thai Government Scholarship. We acknowledge NIH Grant 1s10RR02678001 for the Jeol TEM 1400.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515240112/-/DCSupplemental.
References
- 1.Valentine RC. Bacterial ferredoxin. Bacteriol Rev. 1964;28:497–517. doi: 10.1128/br.28.4.497-517.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanke GT, Kimata-Ariga Y, Taniguchi I, Hase T. A post genomic characterization of Arabidopsis ferredoxins. Plant Physiol. 2004;134(1):255–264. doi: 10.1104/pp.103.032755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanke G, Mulo P. Plant type ferredoxins and ferredoxin-dependent metabolism. Plant Cell Environ. 2013;36(6):1071–1084. doi: 10.1111/pce.12046. [DOI] [PubMed] [Google Scholar]
- 4.Terauchi AM, et al. Pattern of expression and substrate specificity of chloroplast ferredoxins from Chlamydomonas reinhardtii. J Biol Chem. 2009;284(38):25867–25878. doi: 10.1074/jbc.M109.023622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peden EA, et al. Identification of global ferredoxin interaction networks in Chlamydomonas reinhardtii. J Biol Chem. 2013;288(49):35192–35209. doi: 10.1074/jbc.M113.483727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmitter JM, et al. Purification, properties and complete amino acid sequence of the ferredoxin from a green alga, Chlamydomonas reinhardtii. Eur J Biochem. 1988;172(2):405–412. doi: 10.1111/j.1432-1033.1988.tb13901.x. [DOI] [PubMed] [Google Scholar]
- 7.van Lis R, Baffert C, Couté Y, Nitschke W, Atteia A. Chlamydomonas reinhardtii chloroplasts contain a homodimeric pyruvate:ferredoxin oxidoreductase that functions with FDX1. Plant Physiol. 2013;161(1):57–71. doi: 10.1104/pp.112.208181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Winkler M, Hemschemeier A, Jacobs J, Stripp S, Happe T. Multiple ferredoxin isoforms in Chlamydomonas reinhardtii—Their role under stress conditions and biotechnological implications. Eur J Cell Biol. 2010;89(12):998–1004. doi: 10.1016/j.ejcb.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 9.Noth J, Krawietz D, Hemschemeier A, Happe T. Pyruvate:ferredoxin oxidoreductase is coupled to light-independent hydrogen production in Chlamydomonas reinhardtii. J Biol Chem. 2013;288(6):4368–4377. doi: 10.1074/jbc.M112.429985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hemschemeier A, et al. Copper response regulator1-dependent and -independent responses of the Chlamydomonas reinhardtii transcriptome to dark anoxia. Plant Cell. 2013;25(9):3186–3211. doi: 10.1105/tpc.113.115741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambertz C, Hemschemeier A, Happe T. Anaerobic expression of the ferredoxin-encoding FDX5 gene of Chlamydomonas reinhardtii is regulated by the Crr1 transcription factor. Eukaryot Cell. 2010;9(11):1747–1754. doi: 10.1128/EC.00127-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mus F, Dubini A, Seibert M, Posewitz MC, Grossman AR. Anaerobic acclimation in Chlamydomonas reinhardtii: Anoxic gene expression, hydrogenase induction, and metabolic pathways. J Biol Chem. 2007;282(35):25475–25486. doi: 10.1074/jbc.M701415200. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs J, Pudollek S, Hemschemeier A, Happe T. A novel, anaerobically induced ferredoxin in Chlamydomonas reinhardtii. FEBS Lett. 2009;583(2):325–329. doi: 10.1016/j.febslet.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 14.Dörmann P, Benning C. Galactolipids rule in seed plants. Trends Plant Sci. 2002;7(3):112–118. doi: 10.1016/s1360-1385(01)02216-6. [DOI] [PubMed] [Google Scholar]
- 15.Kelly AA, Dörmann P. Green light for galactolipid trafficking. Curr Opin Plant Biol. 2004;7(3):262–269. doi: 10.1016/j.pbi.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Liu B, Benning C. Lipid metabolism in microalgae distinguishes itself. Curr Opin Biotechnol. 2013;24(2):300–309. doi: 10.1016/j.copbio.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 17.Dörmann P, Hoffmann-Benning S, Balbo I, Benning C. Isolation and characterization of an Arabidopsis mutant deficient in the thylakoid lipid digalactosyl diacylglycerol. Plant Cell. 1995;7(11):1801–1810. doi: 10.1105/tpc.7.11.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benning C, Ohta H. Three enzyme systems for galactoglycerolipid biosynthesis are coordinately regulated in plants. J Biol Chem. 2005;280(4):2397–2400. doi: 10.1074/jbc.R400032200. [DOI] [PubMed] [Google Scholar]
- 19.Riekhof WR, Sears BB, Benning C. Annotation of genes involved in glycerolipid biosynthesis in Chlamydomonas reinhardtii: Discovery of the betaine lipid synthase BTA1Cr. Eukaryot Cell. 2005;4(2):242–252. doi: 10.1128/EC.4.2.242-252.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giroud C, Gerber A, Eichenberger W. Lipids of Chlamydomonas reinhardtii: Analysis of molecular species and intracellular site(s) of biosynthesis. Plant Cell Physiol. 1988;29(4):587–595. [Google Scholar]
- 21.Zäuner S, Jochum W, Bigorowski T, Benning C. A cytochrome b5-containing plastid-located fatty acid desaturase from Chlamydomonas reinhardtii. Eukaryot Cell. 2012;11(7):856–863. doi: 10.1128/EC.00079-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moellering ER, Benning C. RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot Cell. 2010;9(1):97–106. doi: 10.1128/EC.00203-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan J, Andre C, Xu C. A chloroplast pathway for the de novo biosynthesis of triacylglycerol in Chlamydomonas reinhardtii. FEBS Lett. 2011;585(12):1985–1991. doi: 10.1016/j.febslet.2011.05.018. [DOI] [PubMed] [Google Scholar]
- 24.Pootakham W, Gonzalez-Ballester D, Grossman AR. Identification and regulation of plasma membrane sulfate transporters in Chlamydomonas. Plant Physiol. 2010;153(4):1653–1668. doi: 10.1104/pp.110.157875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez-Ballester D, et al. Reverse genetics in Chlamydomonas: A platform for isolating insertional mutants. Plant Methods. 2011;7:24. doi: 10.1186/1746-4811-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tardif M, et al. PredAlgo: A new subcellular localization prediction tool dedicated to green algae. Mol Biol Evol. 2012;29(12):3625–3639. doi: 10.1093/molbev/mss178. [DOI] [PubMed] [Google Scholar]
- 27.Terashima M, Specht M, Naumann B, Hippler M. Characterizing the anaerobic response of Chlamydomonas reinhardtii by quantitative proteomics. Mol Cell Proteomics. 2010;9(7):1514–1532. doi: 10.1074/mcp.M900421-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grefen C, Obrdlik P, Harter K. The determination of protein-protein interactions by the mating-based split-ubiquitin system (mbSUS) Methods Mol Biol. 2009;479:217–233. doi: 10.1007/978-1-59745-289-2_14. [DOI] [PubMed] [Google Scholar]
- 29.Li X, et al. A galactoglycerolipid lipase is required for triacylglycerol accumulation and survival following nitrogen deprivation in Chlamydomonas reinhardtii. Plant Cell. 2012;24(11):4670–4686. doi: 10.1105/tpc.112.105106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hugly S, Kunst L, Browse J, Somerville C. Enhanced thermal tolerance of photosynthesis and altered chloroplast ultrastructure in a mutant of Arabidopsis deficient in lipid desaturation. Plant Physiol. 1989;90(3):1134–1142. doi: 10.1104/pp.90.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen HM, et al. The green microalga Chlamydomonas reinhardtii has a single ω-3 fatty acid desaturase that localizes to the chloroplast and impacts both plastidic and extraplastidic membrane lipids. Plant Physiol. 2013;163(2):914–928. doi: 10.1104/pp.113.223941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dörmann P, Balbo I, Benning C. Arabidopsis galactolipid biosynthesis and lipid trafficking mediated by DGD1. Science. 1999;284(5423):2181–2184. doi: 10.1126/science.284.5423.2181. [DOI] [PubMed] [Google Scholar]
- 33.Yang W, Catalanotti C, Wittkopp TM, Posewitz MC, Grossman AR. Algae after dark: Mechanisms to cope with anoxic/hypoxic conditions. Plant J. 2015;82(3):481–503. doi: 10.1111/tpj.12823. [DOI] [PubMed] [Google Scholar]
- 34.Pollock SV, Colombo SL, Prout DL, Jr, Godfrey AC, Moroney JV. Rubisco activase is required for optimal photosynthesis in the green alga Chlamydomonas reinhardtii in a low-CO(2) atmosphere. Plant Physiol. 2003;133(4):1854–1861. doi: 10.1104/pp.103.032078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang W, et al. Alternative acetate production pathways in Chlamydomonas reinhardtii during dark anoxia and the dominant role of chloroplasts in fermentative acetate production. Plant Cell. 2014;26(11):4499–4518. doi: 10.1105/tpc.114.129965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heinnickel ML, et al. Novel thylakoid membrane GreenCut protein CPLD38 impacts accumulation of the cytochrome b6f complex and associated regulatory processes. J Biol Chem. 2013;288(10):7024–7036. doi: 10.1074/jbc.M112.427476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aksoy M, Pootakham W, Grossman AR. Critical function of a Chlamydomonas reinhardtii putative polyphosphate polymerase subunit during nutrient deprivation. Plant Cell. 2014;26(10):4214–4229. doi: 10.1105/tpc.114.129270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reynolds ES. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 40.Benning C, Somerville CR. Isolation and genetic complementation of a sulfolipid-deficient mutant of Rhodobacter sphaeroides. J Bacteriol. 1992;174(7):2352–2360. doi: 10.1128/jb.174.7.2352-2360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.