Significance
Protein interactions mediated by modular domains, such as PDZ and SH2 domains, play critical roles in biology. The modules typically recognize a linear motif in their ligands, with a few residues in the motif determining the specificity. We report a crystal structure of the complex between the cytoplasmic region of PlexinB2 and the PDZ domain of PDZ–RhoGEF. The structure shows that, in addition to the PDZ/motif interaction, a secondary interface is formed between the three-dimensional domains of the two proteins. We further show that the secondary interface enhances the affinity between plexin and PDZ–RhoGEF and is important for plexin signaling. Our analyses suggest that secondary interface-mediated interactions may be a broadly used mechanism for modular domains to achieve high specificity.
Keywords: PDZ, plexin, signaling, protein interaction module, specificity
Abstract
PDZ domains are abundant protein interaction modules and typically recognize a short motif at the C terminus of their ligands, with a few residues in the motif endowing the binding specificity. The sequence-based rules, however, cannot fully account for the specificity between the vast number of PDZ domains and ligands in the cell. Plexins are transmembrane receptors that regulate processes such as axon guidance and angiogenesis. Two related guanine nucleotide exchange factors (GEFs), PDZ–RhoGEF and leukemia-associated RhoGEF (LARG), use their PDZ domains to bind class B plexins and play critical roles in signaling. Here, we present the crystal structure of the full-length cytoplasmic region of PlexinB2 in complex with the PDZ domain of PDZ–RhoGEF. The structure reveals that, in addition to the canonical C-terminal motif/PDZ interaction, the 3D domain of PlexinB2 forms a secondary interface with the PDZ domain. Our biophysical and cell-based assays show that the secondary interface contributes to the specific interaction between plexin and PDZ–RhoGEF and to signaling by plexin in the cell. Formation of secondary interfaces may be a general mechanism for increasing affinity and specificity of modular domain-mediated interactions.
Plexins are cell surface receptors for semaphorins, extracellular cues that control essential processes such as neuronal axon guidance and vasculature development (1). Binding of semaphorin to the extracellular region of plexin induces formation of the active dimer of the cytoplasmic region, which transduces signal to downstream pathways (2–7). The plexin cytoplasmic region contains a juxtamembrane segment (JM-segment), a RhoGTPase binding domain (RBD), and a GTPase activating protein (GAP) domain (8–10). The GAP domain, activated by the dimerization, transduces signal through converting its substrate GTPase Rap from the GTP-bound active to the GDP-bound inactive state (2, 3). The RBD regulates plexin activity in response to binding of Rho family GTPases, such as Rac1 (reviewed in ref. 11).
In addition to the common signaling pathways through the domains shared by all plexins, class B plexins (B1, B2, and B3) mediate a pathway through their unique C terminus. The conserved “VTDL” motif at the C terminus of these plexins binds to the N-terminal PDZ (PSD-95/Discs-large/ZO-1) domains of two related guanine nucleotide exchange factors (GEFs), PDZ–RhoGEF, and leukemia-associated RhoGEF (LARG) (12–17). This interaction recruits PDZ–RhoGEF and LARG to the plasma membrane, where they promote the exchange of GDP for GTP on RhoA. GTP-bound RhoA binds its downstream effectors and contributes to plexin signaling (13–15, 18). A recent study has shown that deletion of the C terminus of PlexinB2 causes defects in the development of the liver vasculature in mice, highlighting the critical role of the PDZ–RhoGEF/LARG–RhoA pathway in plexin function in vivo (19).
More than 250 PDZ domains exist in the human proteome, constituting one of the most abundant protein interaction modules (20, 21). Correspondingly, there are ∼600 ligands for the PDZ domains (22). A high degree of mutual specificity is expected between PDZ domains and their respective ligands to ensure fidelity of signaling in the cell. The fold of PDZ domains is composed of a six-stranded β-barrel and two α-helices. The canonical interaction mode between the PDZ domains and their ligands involves the binding of the C terminus of the ligand in an extended β-strand conformation to the groove between βB and αB in the PDZ domain. The C-terminal carboxyl group of the ligand forms two hydrogen bonds with the backbone of the conserved “GΦGF” motif (Φ: hydrophobic residue) in the βA–βB loop of the PDZ domain, which underlies the strong preference of PDZ domains for C termini. Previous studies have established the general rules of ligand specificity for PDZ domains (23). Class I PDZ domains recognize C-terminal motifs with the “T/S-X-Φ” (X: any residue) sequence, whereas class II PDZ domains prefer the “Φ-X-Φ” sequence. The structural basis for this selectivity is relatively well understood (20). However, many PDZ domains are promiscuous toward short peptidic ligands and defy these simple rules of specificity. Particularly, the PDZ domains in PDZ–RhoGEF and LARG, categorized as class I PDZ domains, bind with similar affinities to many peptides of both classes I and II (24).
Binding of PDZ–RhoGEF and LARG to full-length class B plexins has been detected by various in vitro and cell-based experiments, whereas many other PDZ domains showed no binding under similar conditions (12–16). In contrast, isolated C-terminal peptides from class B plexins and the PDZ domains of PDZ–RhoGEF/LARG only exhibit modest affinity, with the dissociation constant (Kd) in the range of 10–40 μM (24–26). This discrepancy suggests that the specificity between class B plexins and PDZ–RhoGEF/LARG is not fully recapitulated by the peptide/PDZ interaction. More broadly, the sequence-based rules described above are likely an oversimplification of the mechanisms underlying specificity between PDZ domains and their ligands. Most previous structural and binding analyses focused on isolated peptidic binding motifs derived from PDZ ligands, leaving open the question of whether other regions in the ligands are involved in the interaction.
To address these questions, we determined the crystal structure of the complex between the full-length cytoplasmic region of PlexinB2 and the PDZ domain of PDZ–RhoGEF. The structure reveals a secondary interface between PlexinB2 and the PDZ domain, in addition to the canonical interaction mediated by the C-terminal “VTDL” motif of PlexinB2. Our structure-based mutational analyses show that the secondary interface plays an important role in the specific interaction between class B plexins and PDZ–RhoGEF.
Results
Crystal Structure of the PlexinB2/PDZ Complex Reveals a Secondary Binding Interface.
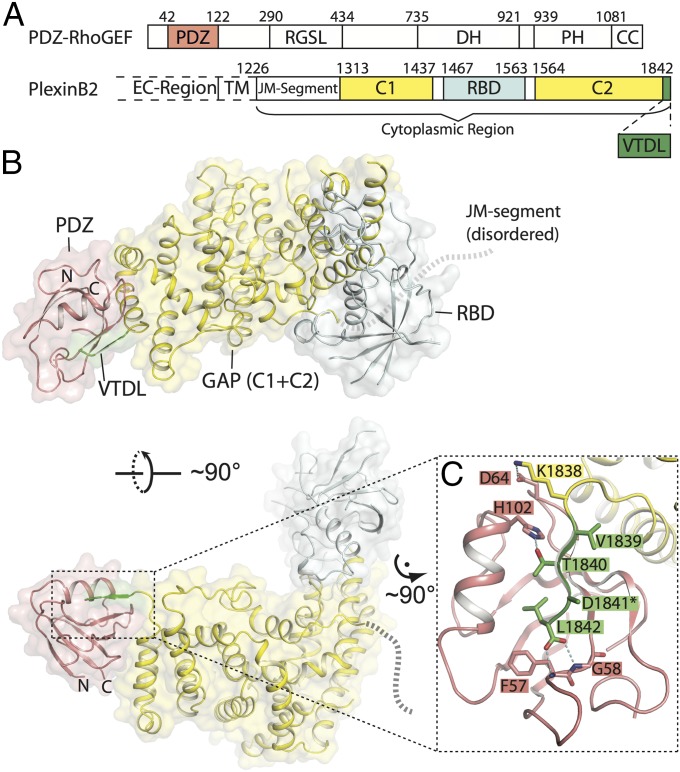
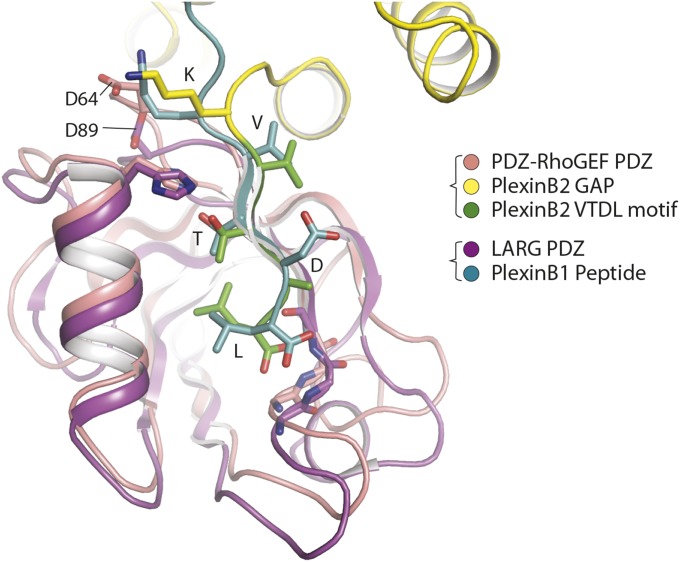
We determined the crystal structure of the full-length cytoplasmic region of mouse PlexinB2 (PlexinB2cyto) in complex with the PDZ domain from human PDZ–RhoGEF (Fig. 1A). The diffraction data are anisotropic, extending to 3.2-Å resolution in the a* and b* directions and ∼5 Å resolution in the c* direction (Table S1). The asymmetric unit of the crystal contains one PlexinB2/PDZ complex (Fig. S1). The structure of PlexinB2cyto, except for the disordered JM-segment, is similar to reported structures of other plexins (2, 3, 8–10). The PDZ domain here is similar to the NMR structure of the PDZ domain of LARG (Fig. S2). PlexinB2cyto and the PDZ domain form a 1:1 complex, with the PlexinB2 “VTDL” motif and the PDZ domain interacting in the typical PDZ/type I ligand binding mode (Fig. 1 B and C). These interactions are similar to those seen in the NMR structure of the complex between the octameric tail peptide from PlexinB1 and the PDZ domain from LARG (Fig. 1 B and C and Fig. S2) (25). In addition, Lys-1838 in PlexinB2, immediately upstream of the VTDL motif, makes an electrostatic contact with Asp-64 in the PDZ domain (Fig. 1C). The equivalent interaction, between Lys-2131 in the human PlexinB1 peptide and Asp-89 in the LARG PDZ domain, is also present in the NMR structure (Fig. S2) (25). Similar interactions have been extensively characterized for many PDZ domains and their ligands (20).
Fig. 1.
Crystal structure of the complex between PlexinB2cyto and the PDZ domain from PDZ–RhoGEF. (A) Domain structure of PlexinB2 and PDZ–RhoGEF. Residue numbers are based on human PDZ–RhoGEF and mouse PlexinB2, respectively. CC, coiled-coil; DH, Dbl-homology domain; EC-region, extracellular region; PH, pleckstrin-homology domain; RGSL, regulator of G protein signaling-like domain; TM, transmembrane region. The C1 and C2 segments in plexin together form the GAP domain. (B) Structure of the PlexinB2cyto/PDZ complex. The color scheme is the same as in A. N and C indicate the N and C termini of the PDZ domain, respectively. The dotted lines indicate the approximate location of the disordered JM-segment. (C) Interface between the PDZ domain and the VTDL motif in PlexinB2. *The side chain of D1841 is not built because of poor density.
Table S1.
Data collection and structure refinement statistics
| Data collection | |
| Wavelength, Å | 0.979237 |
| Space group | P6222 |
| Cell dimensions | |
| a, b, c, Å | 126.2, 126.2, 211.5 |
| α, β, γ, ° | 90, 90, 120 |
| Resolution, Å | 50.0–3.20 (3.26–3.20)* |
| Rsym, % | 9.9 (>100) |
| Rpim, % | 2.9 (84.6) |
| I/σ | 56.0 (1.3) |
| Completeness, % | 99.9 (100) |
| Redundancy | 30.1 (27.4) |
| CC1/2 of the highest-resolution shell | 0.74 |
| Refinement | |
| Resolution, Å | 48.5–3.2 (3.46–3.22) |
| No. of reflections | 14538 (2394) |
| Rwork/Rfree, % | 25.4 (39.9)/29.7 (45.8) |
| No. of atoms | 4478 |
| Average B factor | 137.9 |
| r.m.s. deviations: | |
| Bond lengths, Å | 0.004 |
| Bond angles, ° | 0.754 |
| Ramachandran plot | |
| Favored, % | 92.1 |
| Allowed, % | 7.2 |
| Disallowed, % | 0.7 |
Values in parentheses are for the highest-resolution shell.
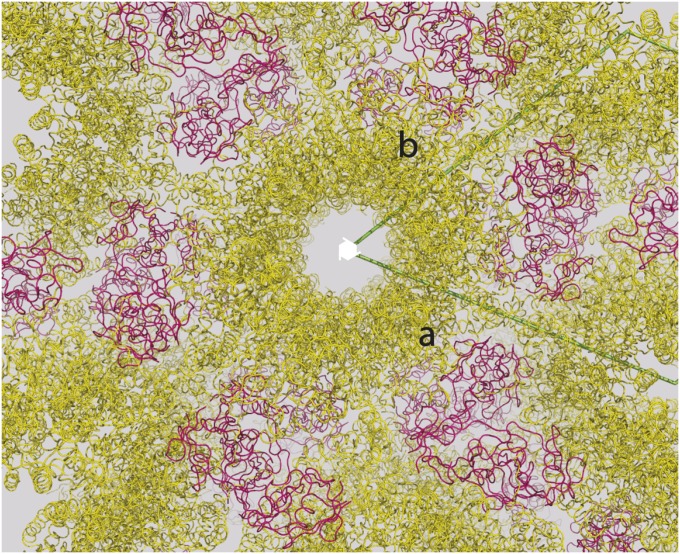
Fig. S1.
Crystal packing of the PlexinB2cyto/PDZ complex. PlexinB2 and the PDZ domain are colored yellow and red, respectively. PlexinB2 molecules related by the crystallographic 62-fold screw axis form “helical columns.” These columns do not contact one another, and PlexinB2 does not form any significant dimeric interactions in the crystal. One PDZ domain binds each PlexinB2 and also mediates crystal contacts between different PlexinB2 columns.
Fig. S2.
The PDZ domains from LARG and PDZ–RhoGEF bind the C-terminal motif in class B plexins in the same mode. The NMR structure of the LARG PDZ domain in complex with the C-terminal motif from PlexinB1 (PDB ID code 2OS6) is superimposed on the PlexinB2/PDZ structure reported here.
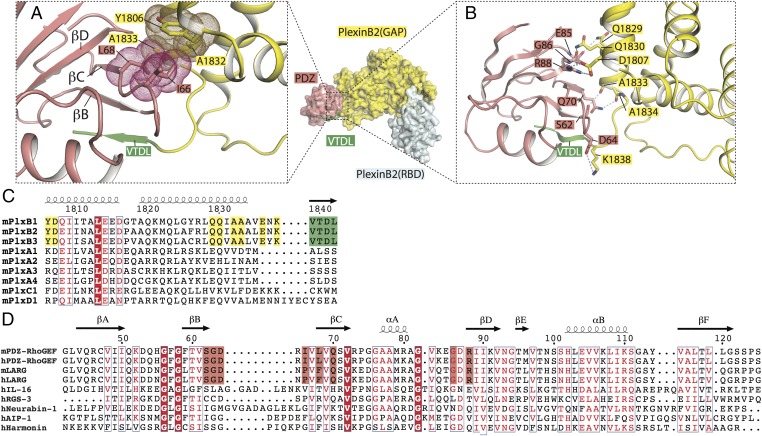
In addition to the canonical interface, our structure shows a secondary interface between the last two helices in the GAP domain of PlexinB2 and one face of the PDZ domain composed of β strands B–D (Fig. 2A). The two interfaces together bury ∼1,500 Å2 of solvent-accessible surface area, with the canonical and the secondary interfaces contributing ∼600 and ∼900 Å2, respectively. The center of the secondary interface is mediated by hydrophobic interactions between Leu-68 from the PDZ domain and two alanine residues (Ala-1832 and -1833) from PlexinB2 (Fig. 2A). Ile-66 of the PDZ and Tyr-1806 of PlexinB2 contribute additional hydrophobic contacts at the periphery of the interface. The interface is further stabilized by polar interactions. Arg-88 and Asp-64 from the PDZ domain interact with Asp-1807 and Lys-1838 from PlexinB2cyto, respectively (Fig. 2B). Tandem glutamine residues from PlexinB2 (Gln-1829 and -1830) interact with the backbone carbonyls of Gly-86 and Asp-87 from the PDZ domain. Ser-62 and Gln-70 from the PDZ domain make polar interactions with the C terminus of the last helix in the PlexinB2 GAP domain (Fig. 2B). Some of these interactions may be mediated by hydrogen bonds, but are not assigned because of the moderate resolution of the structure.
Fig. 2.
Secondary interface between PlexinB2 and the PDZ domain. (A) Central hydrophobic interactions in the secondary interface. van der Waals surfaces of residues are shown as dots. (B) Peripheral polar interactions in the secondary interface. (C) Sequence alignment of the PDZ-binding region of mouse PlexinB2 with other plexin family members. Residues involved in the secondary interface are highlighted in yellow. Residue numbers of mouse PlexinB2 are shown at the top. (D) Sequence alignment of PDZ domains. The sequences of the PDZ domains from PDZ–RhoGEF and LARG are aligned with several diverse PDZ domains. Residues involved in the secondary interface in PDZ–RhoGEF/LARG are highlighted in pink. Residue numbers at the top are for human PDZ–RhoGEF. In both C and D, conserved residues are in red, and identical residues are highlighted by red background. h, human; m, mouse.
The residues in PlexinB2 mediating the secondary interface are conserved across class B plexins, but not among class A, C, and D plexins (Fig. 2C). Ile-66 and Leu-68 in the PDZ domain of PDZ–RhoGEF are replaced by a proline and phenylalanine residue, respectively, in LARG, maintaining the hydrophobicity (Fig. 2D). Other residues in the secondary interface are identical between PDZ–RhoGEF and LARG from both human and mouse (Fig. 2D). These residues, however, are not conserved in PDZ domains from other proteins (Fig. 2D). These patterns of sequence conservation suggest that class B plexins and PDZ–RhoGEF/LARG have coevolved the secondary interface to enhance their mutual specificity, and the secondary interface is functionally important.
Tight Binding Between PlexinB2cyto and the PDZ Domain from PDZ–RhoGEF.
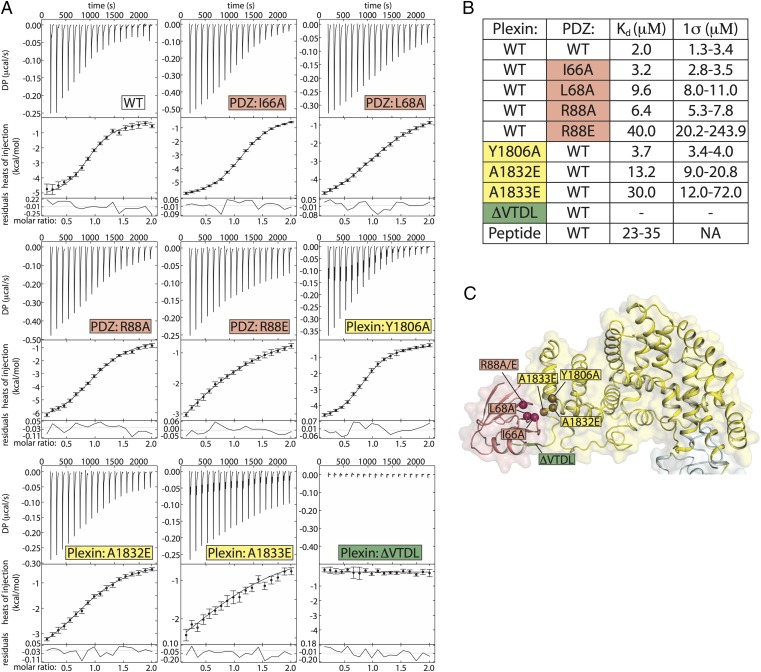
By using fluorescence-based methods or isothermal titration calorimetry (ITC), several studies have determined the Kd value between isolated C-terminal peptides from PlexinB1 and the PDZ domain of PDZ–RhoGEF in the range of 30–36 μM under various buffer conditions (24, 26). The PDZ domain of LARG shows similar affinity to the tail peptide from PlexinB1 (24–26). The peptides used in the studies of PDZ–RhoGEF included no more than the C-terminal six residues of human PlexinB1, which are identical to those in mouse PlexinB2. We analyzed the binding between PlexinB2cyto and the PDZ domain of PDZ–RhoGEF using ITC. PlexinB2cyto with the C-terminal four residues truncated (ΔVTDL) showed no detectable binding to the PDZ domain, consistent with the notion that the VTDL motif is critical for the interaction (Fig. 3). PlexinB2cyto binds the PDZ domain with a Kd value of 2 μM (Fig. 3), >10-fold tighter than that of the isolated C-terminal motif. These results suggest that the secondary interface as observed in our crystal structure substantially enhances the plexin/PDZ interaction.
Fig. 3.
Mutational analysis of the secondary interface by ITC. (A) Representative baseline-subtracted ITC thermograms (Top), integrated titration heats (circles) with fits shown (lines; Middle), and residuals plots (Bottom). (B) Values of Kd and 1σ confidence intervals from data in A. The Kd values and confidence intervals are derived from a global analysis of triplicate datasets for each binding pair. Kd values for the tail peptide are from previous reports. (C) Locations of the mutations in the crystal structure.
Mutational Analyses of the Secondary Interface.
To assess the contribution of the secondary interface to the plexin/PDZ interaction, we examined the effects of a series of mutations at this interface by ITC. Three mutations—Y1806A, A1832E, and A1833E—were introduced individually into PlexinB2cyto. The Y1806A mutant displayed approximately twofold reduction in binding (Kd = 3.7 μM) compared with the wild-type (WT) PlexinB2cyto (Fig. 3). The A1832E and A1833E mutants showed much weaker binding, with Kd values of 13 and 30 μM, respectively. The A1833E mutation reduces the affinity to the reported level of the isolated C-terminal peptide of PlexinB1, effectively eliminating the contribution of the secondary interface. Tyr-1806 is located at the periphery of the interface, and the alanine mutation removes a portion of the hydrophobic interaction, but does not sterically occlude binding. Introducing a charged and larger residue to the hydrophobic core of the interface, as in the case of the A1832E and A1833E mutations, however, disrupts the secondary interface.
The PDZ mutations also attenuated binding to various degrees, consistent with the features of the secondary interface. The PDZ R88A mutant increased Kd by approximately threefold to 6.4 μM (Fig. 3). Loss of the peripheral interaction between Arg-88 in the PDZ domain and Asp-1807 in PlexinB2cyto is not expected to actively disrupt the interface core or the ability of other contacts to form. The R88E mutation, which introduces charge/charge repulsion with Asp-1807 in PlexinB2cyto, reduced the affinity by 20-fold (Kd = 40 μM). An alanine mutation of Leu-68 in the PDZ domain, which docks into the hydrophobic patch created by Ala-1832 and -1833 in PlexinB2cyto, reduces the affinity by approximately fivefold. Mutating a peripheral hydrophobic residue in the PDZ domain, I66A, attenuated binding by less than twofold. Together, these data demonstrate that the secondary interface is responsible for the >10-fold tighter binding to the PDZ domain by PlexinB2cyto compared with the isolated PDZ-binding motif.
Both Binding Interfaces Contribute to Recruitment of PDZ–RhoGEF by Plexin.
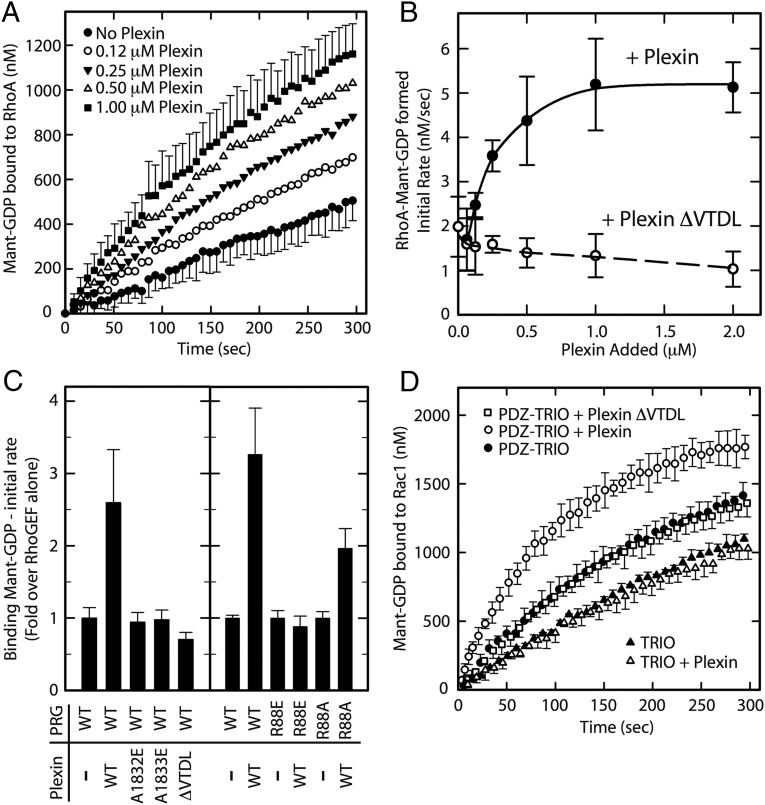
We have shown previously that the GEF activity of PDZ–RhoGEF and its homologs can be facilitated by membrane recruitment to membrane-delimited substrates such as RhoA (27) and that this functional recruitment can mediate hormone signaling in cells (28). In the reconstituted system, GEFs were recruited to the surface of lipid vesicles that contained regulatory partners and substrates tethered to the membrane surface through interaction of polyhistidine tags with Ni–NTA-conjugated lipids. We used this system to examine the interaction of full-length PDZ–RhoGEF with PlexinB2cyto (Fig. 4 A–C). PDZ–RhoGEF in solution displayed modest nucleotide exchange activity toward RhoA tethered to lipid vesicles. Addition of His6–PlexinB2cyto produced a concentration-dependent increase in the exchange activity (Fig. 4 A and B), suggesting functional recruitment of PDZ–RhoGEF by His6–PlexinB2cyto to membrane-localized RhoA. Deletion of the C-terminal motif from PlexinB2 (ΔVTDL) eliminated the GEF activity increase (Fig. 4 B and C), consistent with an anticipated reduction in the recruitment. We further tested mutations in the secondary interface, two in the PDZ domain (R88E and R88A) and two in PlexinB2cyto (A1832E and A1833E). All four mutants attenuated the stimulatory effect of PlexinB2 on the GEF activity of PDZ–RhoGEF (Fig. 4C). The partial reduction by the R88A mutation is consistent with its more moderate effect on binding affinity (Fig. 3). These results implicate both the primary and secondary interfaces in the recruitment of PDZ–RhoGEF by plexin. The sufficiency of this interaction is demonstrated by the recruitment of a chimeric protein containing the PDZ domain of PDZ–RhoGEF linked to an unrelated RacGEF, TRIO (triple functional domain protein) (Fig. 4D). His6–PlexinB2cyto stimulates nucleotide exchange of membrane delimited Rac1 by the chimeric PDZ–TRIO, but not TRIO alone.
Fig. 4.
Recruitment of PDZ–RhoGEF by His6–PlexinB2cyto to membrane-localized substrate. Nucleotide exchange assays with lipid vesicles containing immobilized RhoA– or Rac1–His6 are based on the change in fluorescence between bound and free N-methylanthraniloyl–GDP (mant-GDP). (A) Time course of mant–GDP association to RhoA in the presence of 10 nM PDZ–RhoGEF (PRG) and increasing concentrations of His6-PlexinB2cyto. In two curves, the SDs among four measurements are shown as error bars. Errors of other curves are similar but omitted for clarity. (B) Initial rates of exchange in the presence of increasing concentrations of the WT or ΔVTDL mutant of His6–PlexinB2. Results are the average of four titrations as in A; error bars are SD. (C) Initial rates of RhoA exchange stimulated by 10 nM WT or mutants of PRG in the presence of 0.5 μM WT or mutants of His6–PlexinB2cyto. Results are averages of nine measurements; errors are SD. All mutants showed no significant stimulation of PRG activity with one exception; the activity of PRG–R88A was stimulated by WT PlexinB2cyto, but less so than PRG–WT (P < 0.0001 for both comparisons; Student’s t test). (D) Stimulation of chimeric PDZ–TRIO (150 nM) by 0.5 μM WT or ΔVTDL mutant of His6–PlexinB2cyto. Results represent the average and SD of triplicate measurements. Stimulation of PDZ–TRIO by twofold to fourfold was significant (P < 0.05) in two such experiments; no significant stimulation was observed with TRIO or with PlexinB2ΔVTDL.
Secondary Interface Contributes to Plexin-Mediated Activation of PDZ–RhoGEF in Cells.
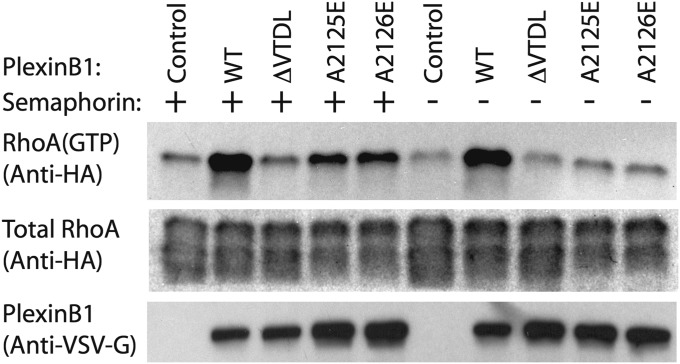
Activation of class B plexins by semaphorin activates PDZ–RhoGEF and LARG, leading to increased levels of GTP-bound RhoA in cells (12–17). To examine whether this plexin/PDZ–RhoGEF-mediated activation of RhoA is dependent on the secondary interface, we used a pull-down assay for quantifying RhoA(GTP) levels in the cell (29). Full-length human PlexinB1, which is nearly identical to mouse PlexinB2 in both the C-terminal motif and the secondary interface, was used in these experiments. Consistent with previous studies, the results show that overexpression of PlexinB1 increased the levels of RhoA(GTP) in HEK293T cells, which express PDZ–RhoGEF endogenously (Fig. 5) (13, 18). RhoA activation was further enhanced by semaphorin treatment. Deletion of the VTDL motif in PlexinB1 (ΔVTDL) abolished RhoA activation in the presence or absence of semaphorin treatment. These results are also consistent with previous studies (13, 14). We then tested the effects on RhoA activation of the A2125E and A2126E mutations of PlexinB1, equivalent to the secondary interface mutations A1832E and A1833E of PlexinB2, respectively. These mutations both attenuated RhoA activation, although the reduction was not as severe as that caused by ΔVTDL (Fig. 5). Together, our data strongly support the hypothesis that the C-terminal motif and secondary interface in class B plexins together contribute to optimal recruitment of PDZ–RhoGEF and activation of RhoA for signal transduction in the cell.
Fig. 5.
Contribution of the secondary interface between Plexin and PDZ–RhoGEF to RhoA activation in cells. Total expression levels of VSV-G–tagged PlexinB1 and HA-tagged RhoA were probed by anti–VSV-G and anti-HA antibodies, respectively. GTP-bound RhoA was pulled down by GST–Rotekin and probed by the same anti-HA antibody. Results shown are from one of the three independent repeats.
Discussion
Protein interactions between modular domains, such as PDZ, SH2, and SH3 domains, and linear binding motifs are a common theme in biology (30). An established paradigm is that sequence variations of one or a few residues in the binding motifs determine the specificity. However, modules of the same type often show similar sequence preferences for the binding motifs, and similar or identical motifs often exist in different proteins. This paradigm is therefore inadequate for rationalizing the diverse yet specific interactions mediated by modular domains. In the case of PDZ domains, binding motifs of different classes do not always show large differences in affinity to cross-typed PDZ domains. Extensive investigations, including large-scale peptide library screening and bioinformatics analyses, have shown that residues beyond positions 0 and −2 in PDZ binding motifs are involved in fine-tuning the specificity (20, 21). Based on these studies, as many as 16 classes have been defined to account for the specificity profiles of PDZ domains (31). A recent study has shown that the PDZ domain in the protein PALS1 forms a structural supramodule with the SH3 and guanylate kinase domains, with all of the three domains collectively contributing to specific binding to the C terminus of its ligand crumbs (32).
The above analyses are largely limited to linear binding motifs. Our finding of the secondary interface mediated by the folded domains of PlexinB2 and PDZ–RhoGEF provides an additional dimension to the mechanism by which PDZ domains achieve specificity. Interestingly, a recent study has shown that the C-terminal SH2 domain in phospholipase Cγ forms a secondary interface with the kinase domain of fibroblast growth factor receptor, in addition to the SH2/phosphoryl–peptide interaction (33). Similar secondary interactions have also been seen for a SH3 domain and a small GTPase (34, 35). The 3D domain-mediated secondary interfaces may be broadly used by modular domains to increase affinity and specificity, but are underappreciated because they involve residues not adjacent in sequence to the binding motifs and cannot be easily identified by sequence analyses. Structural studies of larger constructs beyond the modules and their binding motifs are required to reveal such interactions.
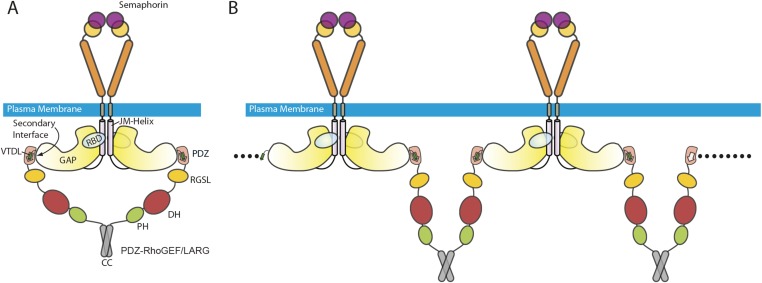
It is unclear how the interaction between class B plexins and PDZ–RhoGEF/LARG is regulated by semaphorin binding to plexin. Before activation, plexin exists as an inactive monomer or inhibitory dimer (3, 9, 36). Semaphorin binding induces the formation of the active dimer of plexin (2, 4–7). PDZ–RhoGEF and LARG also form dimers or oligomers through their C-terminal coiled-coil region (37). The plexin active dimer and the PDZ–RhoGEF/LARG dimer may form a 2:2 complex (Fig. S3A). Alternatively, dimeric PDZ–RhoGEF/LARG may bridge two copies of the plexin dimer, leading to their clustering on the cell surface (Fig. S3B). The dimerization/oligomerization may increase the avidity between plexin and PDZ–RhoGEF/LARG. PDZ–RhoGEF and LARG both have other domains that help target them to the plasma membrane, such as the Pleckstrin-homology domain that interacts with GTP-bound RhoA (38). These factors together may facilitate the interaction between plexin and PDZ–RhoGEF/LARG and thereby enhance their ability to discriminate against nonspecific interactions.
Fig. S3.
Model for the recruitment of PDZ–RhoGEF/LARG to the plasma membrane by class B plexins in the active state. The PDZ/C-terminal motif interaction and the secondary interface contribute to the interaction together. (A) The interaction may be further strengthened by bivalent engagement between dimeric PDZ–RhoGEF/LARG and the semaphorin-induced active dimer of plexin as shown. (B) An oligomeric complex may also form as shown. Potential additional interactions with the membrane mediated by the PH and RGSL domains of PDZ–RhoGEF/LARG are not shown.
Materials and Methods
Full-length PDZ–RhoGEF was expressed in Spodoptera frugiperda cells. Other proteins were expressed in Escherichia coli. Proteins were purified by affinity chromatography in combination with ion exchange and gel filtration chromatography. Detailed experimental procedures are included in SI Materials and Methods.
SI Materials and Methods
Protein Expression and Purification.
The construct for PlexinB2cyto (residues 1,226–1,842) was obtained from Open Biosystems and subcloned into a modified pET28 vector (Novagen), which encodes an N-terminal His6 tag and a cleavage site for the human rhinovirus C3 protease (3). PlexinB2cyto proteins were expressed by using the bacteria strain ArcticExpress (Stratagene) and purified as described (2, 3, 9). For the lipid vesicle assays, the N-terminal His6 tag was left intact for tethering PlexinB2cyto to the Ni–NTA-conjugated lipids. In all other cases, the His6 tag was removed by treatment with the human rhinovirus C3 proteases. Mutant constructs of PlexinB2 were generated by using PCR-based mutagenesis.
The PDZ domain from human PDZ–RhoGEF (residues 42–125) was subcloned into the same modified pET28 vector as PlexinB2. The PDZ domain tends to form an interchain disulfide linkage by Cys-47 (26). To avoid this issue, a C47S mutant was made for the ITC experiments. All interface mutations of the PDZ domain were introduced in the C47S background. Proteins were expressed by using the bacteria strain BL21(DE3) and affinity-purified by using a 1-mL nickel conjugated column (HisTrap FF; GE Healthcare). The N-terminal His6 tag was removed by the human rhinovirus C3 protease. After protease digestion, proteins were further purified by using a size-exclusion column (Superdex 75 10/300 GL; GE Healthcare), and the monomeric species was collected.
The full-length human cDNA of PDZ–RhoGEF (PRG) was cloned into pFastbac1 (Invitrogen) modified with an N-terminal EE-tag (EYMPME), expressed in Spodoptera frugiperda (SF9) cells with the Bac-to-Bac system (Invitrogen), and purified by affinity chromatography with anti-EE coupled Sepharose (BAbCO) as described (39). Mutations in the PDZ domain were introduced by site-directed mutagenesis. Preparation of full-length RhoA with a C-terminal His-tag replacing the last four amino acids (RhoA–His6) was performed as described (27); Rac1 with a His6-tag on its C terminus (Rac1–His6) was inserted into the same expression vector and purified similarly. The PDZ domain of PRG (residues 2–127) was cloned with a linker sequence (GSGTGSGIDGTGTGTG) onto the N terminus of the first Dbl-homology (DH)-Pleckstrin homology (PH) domains of human Trio (residues 1,226–1,535) to create PDZ–TRIO, inserted into pGEX–KG–TEV, expressed in E. coli strain BL21 (DE3), and purified as described (38, 40).
The RBD construct of mouse Rhotekin (residues 7–89) was cloned into the pGEX–2T vector (GE Healthcare). The Rhotekin RBD was expressed by using the bacteria strain BL21 (DE3) and affinity-purified by using a 1-mL GST column (GSTrap FF; GE Healthcare), followed by a size-exclusion column (HiLoad 16/60 Superdex 200; GE Healthcare).
Crystallization and Structure Determination.
Various constructs of human PlexinsB1, mouse PlexinB2 and B3, and the PDZ domain from human PDZ–RhoGEF were subjected to cocrystallization trials. The PDZ domains from human and mouse PDZ–RhoGEF only have one nonidentical residue, located outside of the plexin-binding interface (Fig. 2D). Therefore, the species mismatch is not expected to affect the plexin/PDZ interaction. Initial crystal screens were conducted with PlexinB2 and the PDZ domains mixed at a 1:1 molar ratio by using sitting-drop 96-well plates. The PlexinB2cyto/PDZ complex crystallized at 20 °C against a well solution containing 0.2 M K/Na tartrate, 0.1 M Na citrate (pH 5.6), and 2.0 M ammonium sulfate, with a mixture of PlexinB2 at a concentration of 6 mg/mL and the PDZ domain at a concentration of 0.78 mg/mL. Larger crystals were grown by hanging-drop vapor diffusion at 20 °C against a well solution containing 0.2 M K/Na tartrate, 0.1 M Na citrate (pH 5.3), and 1.4 M ammonium sulfate. Crystals were cryoprotected by using the crystallization solution supplemented with 25% (vol/vol) glycerol and flash-cooled in liquid nitrogen. Diffraction data were collected at 100 K at the Advanced Photon Source (Argonne National Laboratory) on beamline 19ID. HKL2000 was used to index and scale the data (41). Diffraction of crystals was consistent with the P6222 space group. Data were highly anisotropic, with diffraction in the c* direction being much weaker (extending to ∼5 Å) than in the a* and b* directions (extending to 3.2 Å). Data were therefore processed with the “auto corrections” option in HKL2000, which truncates and scales anisotropic data. The Phaser module of the Phenix software package was used for molecular replacement phasing (42, 43). The structures of the cytoplasmic region of mouse PlexinA3 [Protein Data Bank (PDB) ID code 3IG3] and the PDZ domain of human LARG (PDB ID code 2OS6) were used as the search models.
Model building, refinement, and validation were performed by using the Coot, Phenix, and MolProbity programs, respectively (42, 44, 45). The average B factor of the refined model is high. Accordingly, detailed interactions such as hydrogen bonds were not assigned to avoid overinterpretation. Data collection and refinement statistics are listed in Table S1. All structure figures were generated by using the PyMOL program (PyMOL Molecular Graphics System; Schrodinger). Buried surface area was calculated by using the Get_Area function in PyMOL.
ITC Experiments.
All ITC experiments were conducted in triplicate by using a MicroCal ITC200 instrument (GE Healthcare). Proteins of the PDZ domain from PDZ–RhoGEF used in these experiments contained the C47S mutation to avoid the interchain disulfide formation as mentioned above (26). The mutation site is distal to the binding interface and not expected to affect binding. Purified and concentrated proteins (PlexinB2cyto and PDZ) were dialyzed overnight at 4 °C against 1 L of buffer containing 10 mM Tris (pH 8.0), 150 mM NaCl, 10% (vol/vol) glycerol, and 1 mM Tris(2-carboxyethyl)phosphine. All proteins were thoroughly degassed before titration. PDZ proteins (300 or 500 μM) were placed in the syringe, and PlexinB2cyto proteins (30 or 50 μM) were placed in the cell. Titrations of WT PDZ into WT PlexinB2cyto were carried out by using 300 and 30 μM protein solutions, respectively. Titration of WT PDZ into PlexinB2cyto with the C-terminal four-residue truncation was also performed at concentrations of 300 and 30 μM, respectively. All other mutant titrations were carried out by using 50 μM PlexinB2cyto and 500 μM PDZ solutions. The PDZ was titrated into the cell over the course of 18 injections of 2 μL. All titrations were conducted at 20 °C with continuous stirring at 1,000 rpm and 2-min intervals between injections. Data were integrated by using NITPIC (46) and passed to SEDPHAT (47) for fitting and global analysis. The 1:1 Hetero-Association model was used for all analysis with Kd, change in enthalpy (ΔH), and fraction of binding incompetent protein being fit. Global fit values for Kd and 1σ confidence intervals are reported. Confidence intervals were calculated in SEDPHAT by using the “automatic confidence interval search with projection method” option. Representative isotherms and thermograms from each set of titrations are shown. All ITC data figures were generated with the GUSSI program (48).
Vesicle-Based GEF Activity Assays.
Assays with unilamellar phospholipid vesicles followed described procedures (27). Briefly, vesicles of ∼100-nm diameter were prepared by extrusion through an Mini-Extruder by using a 100-nm polycarbonate membrane (Avanti Polar Lipids). The 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, and 1,2-dioleoyl-sn-glycero-3-{[N-(5-amino-1-carboxypentyl)iminodiacetic acid]succinyl} [nickel salt; 18:1 DGS–NTA(Ni)] were incorporated in a mole ratio of 4.75:5:0.25, respectively. Vesicles (1.5 mM phospholipid) and mant-GDP (5 μM) were mixed with 2 μM RhoA– or Rac1–His6 as indicated. After incubation at 25 °C for 1 min, exchange reactions were initiated by addition of the indicated GEF and monitored by a Fluorolog-3 spectrofluorometer. When included, His-tagged plexins were added to vesicles at the same time as His-tagged GTPases. In experiments using the PDZ–TRIO chimera, assay solutions were supplemented with 20 mM imidazole to reduce nonspecific binding of the protein to the Ni–NTA lipids; this low concentration of imidazole did not affect functional association of polyhistidine-tagged proteins. An increase in fluorescence was measured as mant–GDP exchanged onto the GTPases in place of GDP. Total fluorescence change was measured with saturating concentrations of GEF and used to calculate molar binding of mant–GDP, assuming a linear change for bound mant–GDP and saturation of the GTPase present in the assay. Initial rates of exchange were calculated with linear analysis or curve fitting, depending on the shape of the response.
Cell-Based RhoA Activation Assay.
The full-length human PlexinB1 construct with an N-terminal VSV-G tag in the pcDNA3.1(+) vector (Invitrogen) was obtained from K. L. Guan (University of California at San Diego, La Jolla). Empty pcDNA3.1(+) served as a vector control. The full-length human RhoA construct with an N-terminal HA-tag in the pCMV5 vector (ATCC) was obtained from M. Cobb (University of Texas Southwestern Medical Center). The construct for the PlexinB1 ligand, Semaphorin4D, was obtained from R. Giger (University of Michigan, Ann Arbor) and used to express the ligand as described (9). PlexinB1 mutants were generated by using PCR-based mutagenesis.
Levels of RhoA activation in cells cotransfected with RhoA and WT or mutant PlexinB1 were quantified by pulling down RhoA(GTP) using a GST–Rhotekin RBD as described (29). Briefly, HEK293T cells were seeded onto 60-mm dishes and grown to 70–80% confluence in Dubecco’s Modified Eagle Medium (DMEM) medium supplemented with 10% (vol/vol) FBS. Cells were transiently transfected with 0.5 μg of the RhoA plasmid and 4.5 μg of the PlexinB1 plasmid or the control vector by using Lipofectamine2000 (Life Technologies). At 36 h after transfection, cells were serum-starved for 4 h in DMEM supplemented with 0.2% FBS. Cells were treated with 10 nM Semaphorin4D or control medium for 15 min and then washed in cold PBS and lysed in 300 μL of lysis buffer (50 mM Tris, pH 7.5, 30 mM MgCl2, 5 mM EDTA, 150 mM NaCl, 1% Nonidet P-40, 2 mM DTT, and Pierce Protease Inhibitor Mini Tablets no. 88666). Lysates were cleared by centrifugation for 10 min at 18,000 × g at 4 °C. RhoA(GTP) was captured from lysates by incubation with 20 μg of GST–Rhotekin and 25 μL of Glutathione Sepharose 4B bead slurry (GE Healthcare) for 1 h at 4 °C. Beads were washed three times with lysis buffer, and the supernatant was removed. Bound proteins were eluted by the addition of 30 μL of SDS loading buffer [50 mM Tris, pH 6.8, 5% (vol/vol) β-mercaptoethanol, 1% SDS, 10% (vol/vol) glycerol, and 0.02% Bromophenol Blue] before SDS/PAGE and Western blot analysis. GTP-loaded and total HA-tagged RhoA populations were detected by immunoblotting with a monoclonal anti-HA antibody (Cell Signaling; no. 3724S). VSV-G–PlexinB1 expression was probed by immunoblotting cell lysates with a monoclonal anti–VSV-G antibody (Clone P5D4; Sigma).
Acknowledgments
We thank T. Scheuermann for assistance in isothermal titration calorimetry (ITC); I. White for discussions; and R. Hibbs, the staff of the Structural Biology Laboratory at the University of Texas Southwestern Medical Center (UTSW), and the staff of beamline 19ID at Advanced Photon Source (APS) for assistance in X-ray data collection. X.Z. is a Virginia Murchison Linthicum Scholar in Medical Research at UTSW. The work is supported in part by National Institutes of Health (NIH) Grants GM088197 (to X.Z.) and GM031954 (to P.C.S.); Welch Foundation Grant I-1702 (to X.Z.); and the Alfred and Mabel Gilman Chair in Molecular Pharmacology (to P.C.S). H.G.P. is supported in part by NIH Training Grant GM008203. Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at APS. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5E6P).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1508931112/-/DCSupplemental.
References
- 1.Tran TS, Kolodkin AL, Bharadwaj R. Semaphorin regulation of cellular morphology. Annu Rev Cell Dev Biol. 2007;23:263–292. doi: 10.1146/annurev.cellbio.22.010605.093554. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Pascoe HG, Brautigam CA, He H, Zhang X. Structural basis for activation and non-canonical catalysis of the Rap GTPase activating protein domain of plexin. eLife. 2013;2:e01279. doi: 10.7554/eLife.01279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, et al. Plexins are GTPase-activating proteins for Rap and are activated by induced dimerization. Sci Signal. 2012;5(207):ra6. doi: 10.1126/scisignal.2002636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nogi T, et al. Structural basis for semaphorin signalling through the plexin receptor. Nature. 2010;467(7319):1123–1127. doi: 10.1038/nature09473. [DOI] [PubMed] [Google Scholar]
- 5.Liu H, et al. Structural basis of semaphorin-plexin recognition and viral mimicry from Sema7A and A39R complexes with PlexinC1. Cell. 2010;142(5):749–761. doi: 10.1016/j.cell.2010.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janssen BJ, et al. Structural basis of semaphorin-plexin signalling. Nature. 2010;467(7319):1118–1122. doi: 10.1038/nature09468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janssen BJ, et al. Neuropilins lock secreted semaphorins onto plexins in a ternary signaling complex. Nat Struct Mol Biol. 2012;19(12):1293–1299. doi: 10.1038/nsmb.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell CH, Aricescu AR, Jones EY, Siebold C. A dual binding mode for RhoGTPases in plexin signalling. PLoS Biol. 2011;9(8):e1001134. doi: 10.1371/journal.pbio.1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He H, Yang T, Terman JR, Zhang X. Crystal structure of the plexin A3 intracellular region reveals an autoinhibited conformation through active site sequestration. Proc Natl Acad Sci USA. 2009;106(37):15610–15615. doi: 10.1073/pnas.0906923106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tong Y, et al. Structure and function of the intracellular region of the plexin-b1 transmembrane receptor. J Biol Chem. 2009;284(51):35962–35972. doi: 10.1074/jbc.M109.056275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pascoe HG, Wang Y, Zhang X. Structural mechanisms of plexin signaling. Prog Biophys Mol Biol. 2015;118(3):161–168. doi: 10.1016/j.pbiomolbio.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aurandt J, Vikis HG, Gutkind JS, Ahn N, Guan KL. The semaphorin receptor plexin-B1 signals through a direct interaction with the Rho-specific nucleotide exchange factor, LARG. Proc Natl Acad Sci USA. 2002;99(19):12085–12090. doi: 10.1073/pnas.142433199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perrot V, Vazquez-Prado J, Gutkind JS. Plexin B regulates Rho through the guanine nucleotide exchange factors leukemia-associated Rho GEF (LARG) and PDZ-RhoGEF. J Biol Chem. 2002;277(45):43115–43120. doi: 10.1074/jbc.M206005200. [DOI] [PubMed] [Google Scholar]
- 14.Swiercz JM, Kuner R, Behrens J, Offermanns S. Plexin-B1 directly interacts with PDZ-RhoGEF/LARG to regulate RhoA and growth cone morphology. Neuron. 2002;35(1):51–63. doi: 10.1016/s0896-6273(02)00750-x. [DOI] [PubMed] [Google Scholar]
- 15.Driessens MH, Olivo C, Nagata K, Inagaki M, Collard JG. B plexins activate Rho through PDZ-RhoGEF. FEBS Lett. 2002;529(2-3):168–172. doi: 10.1016/s0014-5793(02)03323-9. [DOI] [PubMed] [Google Scholar]
- 16.Oinuma I, Katoh H, Harada A, Negishi M. Direct interaction of Rnd1 with Plexin-B1 regulates PDZ-RhoGEF-mediated Rho activation by Plexin-B1 and induces cell contraction in COS-7 cells. J Biol Chem. 2003;278(28):25671–25677. doi: 10.1074/jbc.M303047200. [DOI] [PubMed] [Google Scholar]
- 17.Hirotani M, et al. Interaction of plexin-B1 with PDZ domain-containing Rho guanine nucleotide exchange factors. Biochem Biophys Res Commun. 2002;297(1):32–37. doi: 10.1016/s0006-291x(02)02122-8. [DOI] [PubMed] [Google Scholar]
- 18.Basile JR, Barac A, Zhu T, Guan KL, Gutkind JS. Class IV semaphorins promote angiogenesis by stimulating Rho-initiated pathways through plexin-B. Cancer Res. 2004;64(15):5212–5224. doi: 10.1158/0008-5472.CAN-04-0126. [DOI] [PubMed] [Google Scholar]
- 19.Worzfeld T, et al. Genetic dissection of plexin signaling in vivo. Proc Natl Acad Sci USA. 2014;111(6):2194–2199. doi: 10.1073/pnas.1308418111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye F, Zhang M. Structures and target recognition modes of PDZ domains: Recurring themes and emerging pictures. Biochem J. 2013;455(1):1–14. doi: 10.1042/BJ20130783. [DOI] [PubMed] [Google Scholar]
- 21.Luck K, Charbonnier S, Travé G. The emerging contribution of sequence context to the specificity of protein interactions mediated by PDZ domains. FEBS Lett. 2012;586(17):2648–2661. doi: 10.1016/j.febslet.2012.03.056. [DOI] [PubMed] [Google Scholar]
- 22.Kim J, et al. Rewiring of PDZ domain-ligand interaction network contributed to eukaryotic evolution. PLoS Genet. 2012;8(2):e1002510. doi: 10.1371/journal.pgen.1002510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Songyang Z, et al. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275(5296):73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 24.Smietana K, et al. Degenerate specificity of PDZ domains from RhoA-specific nucleotide exchange factors PDZRhoGEF and LARG. Acta Biochim Pol. 2008;55(2):269–280. [PubMed] [Google Scholar]
- 25.Liu J, et al. Conformational change upon ligand binding and dynamics of the PDZ domain from leukemia-associated Rho guanine nucleotide exchange factor. Protein Sci. 2008;17(6):1003–1014. doi: 10.1110/ps.073416508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paduch M, et al. Bivalent peptides as models for multimeric targets of PDZ domains. ChemBioChem. 2007;8(4):443–452. doi: 10.1002/cbic.200600389. [DOI] [PubMed] [Google Scholar]
- 27.Medina F, et al. Activated RhoA is a positive feedback regulator of the Lbc family of Rho guanine nucleotide exchange factor proteins. J Biol Chem. 2013;288(16):11325–11333. doi: 10.1074/jbc.M113.450056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carter AM, Gutowski S, Sternweis PC. Regulated localization is sufficient for hormonal control of regulator of G protein signaling homology Rho guanine nucleotide exchange factors (RH-RhoGEFs) J Biol Chem. 2014;289(28):19737–19746. doi: 10.1074/jbc.M114.564930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren XD, Schwartz MA. Determination of GTP loading on Rho. Methods Enzymol. 2000;325:264–272. doi: 10.1016/s0076-6879(00)25448-7. [DOI] [PubMed] [Google Scholar]
- 30.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300(5618):445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 31.Tonikian R, et al. A specificity map for the PDZ domain family. PLoS Biol. 2008;6(9):e239. doi: 10.1371/journal.pbio.0060239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, et al. Structure of Crumbs tail in complex with the PALS1 PDZ-SH3-GK tandem reveals a highly specific assembly mechanism for the apical Crumbs complex. Proc Natl Acad Sci USA. 2014;111(49):17444–17449. doi: 10.1073/pnas.1416515111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bae JH, et al. The selectivity of receptor tyrosine kinase signaling is controlled by a secondary SH2 domain binding site. Cell. 2009;138(3):514–524. doi: 10.1016/j.cell.2009.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee CH, Saksela K, Mirza UA, Chait BT, Kuriyan J. Crystal structure of the conserved core of HIV-1 Nef complexed with a Src family SH3 domain. Cell. 1996;85(6):931–942. doi: 10.1016/s0092-8674(00)81276-3. [DOI] [PubMed] [Google Scholar]
- 35.Ostermeier C, Brunger AT. Structural basis of Rab effector specificity: Crystal structure of the small G protein Rab3A complexed with the effector domain of rabphilin-3A. Cell. 1999;96(3):363–374. doi: 10.1016/s0092-8674(00)80549-8. [DOI] [PubMed] [Google Scholar]
- 36.Marita M, et al. Class A plexins are organized as preformed inactive dimers on the cell surface. Biophys J. 2015;109(9):1937–1945. doi: 10.1016/j.bpj.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chikumi H, et al. Homo- and hetero-oligomerization of PDZ-RhoGEF, LARG and p115RhoGEF by their C-terminal region regulates their in vivo Rho GEF activity and transforming potential. Oncogene. 2004;23(1):233–240. doi: 10.1038/sj.onc.1207012. [DOI] [PubMed] [Google Scholar]
- 38.Chen Z, et al. Activated RhoA binds to the pleckstrin homology (PH) domain of PDZ-RhoGEF, a potential site for autoregulation. J Biol Chem. 2010;285(27):21070–21081. doi: 10.1074/jbc.M110.122549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wells CD, et al. Mechanisms for reversible regulation between G13 and Rho exchange factors. J Biol Chem. 2002;277(2):1174–1181. doi: 10.1074/jbc.M105274200. [DOI] [PubMed] [Google Scholar]
- 40.Chen Z, Guo L, Sprang SR, Sternweis PC. Modulation of a GEF switch: Autoinhibition of the intrinsic guanine nucleotide exchange activity of p115-RhoGEF. Protein Sci. 2011;20(1):107–117. doi: 10.1002/pro.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 42.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 43.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Keller S, et al. High-precision isothermal titration calorimetry with automated peak-shape analysis. Anal Chem. 2012;84(11):5066–5073. doi: 10.1021/ac3007522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Houtman JC, et al. Studying multisite binary and ternary protein Interactions by global analysis of isothermal titration calorimetry data in SEDPHAT: application to adaptor protein complexes in cell signaling. Protein Sci. 2007;16(1):30–42. doi: 10.1110/ps.062558507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brautigam CA. Calculations and publication-quality illustrations for analytical ultracentrifugation data. Methods Enzymol. 2015;562:109–133. doi: 10.1016/bs.mie.2015.05.001. [DOI] [PubMed] [Google Scholar]