Significance
Productive viral replication requires overcoming many barriers posed by the host innate immune system. Human sterile alpha motif domain-containing 9 (SAMD9) is a newly identified antiviral factor that is specifically targeted by poxvirus proteins belonging to the C7 family of host-range factors. Here we provide the first, to our knowledge, atomic view of two functionally divergent proteins from the C7 family and determine the molecular basis that dictates whether they can target SAMD9 effectively. Our studies of distantly related C7 family members suggest that SAMD9 is an evolutionally conserved immune barrier that has been overcome successfully by diverse mammalian poxviruses.
Keywords: host-range factors, C7L, poxvirus, crystal structure, SAMD9
Abstract
Human sterile alpha motif domain-containing 9 (SAMD9) protein is a host restriction factor for poxviruses, but it can be overcome by some poxvirus host-range proteins that share homology with vaccinia virus C7 protein. To understand the mechanism of action for this important family of host-range factors, we determined the crystal structures of C7 and myxoma virus M64, a C7 family member that is unable to antagonize SAMD9. Despite their different functions and only 23% sequence identity, the two proteins have very similar overall structures, displaying a previously unidentified fold comprised of a compact 12-stranded antiparallel β-sandwich wrapped in two short α helices. Extensive structure-guided mutagenesis of C7 identified three loops clustered on one edge of the β sandwich as critical for viral replication and binding with SAMD9. The loops are characterized with functionally important negatively charged, positively charged, and hydrophobic residues, respectively, together forming a unique “three-fingered molecular claw.” The key residues of the claw are not conserved in two C7 family members that do not antagonize SAMD9 but are conserved in distantly related C7 family members from four poxvirus genera that infect diverse mammalian species. Indeed, we found that all in the latter group of proteins bind SAMD9. Taken together, our data indicate that diverse mammalian poxviruses use a conserved molecular claw in a C7-like protein to target SAMD9 and overcome host restriction.
Viral infection of host cells is associated with the production of pathogen-associated molecular patterns, which are detected by cellular pattern recognition receptors, resulting in the secretion of interferons and inflammatory cytokines. IFNs induce the expression of hundreds of IFN-stimulated genes (ISGs) and play a critical role in antiviral responses. To replicate in the cells successfully, viruses must overcome the formidable barriers posed by the cellular innate immune system, often by deploying viral proteins to antagonize critical host antiviral factors, many of which are products of ISGs. The HIV-1 Vpu protein, for example, specifically targets Tetherin, which is an ISG product that inhibits particle release of many enveloped viruses (1). Many similar examples of the “arms race” between hosts and pathogens can be found in poxviruses, a family of large, complex DNA viruses. The poxvirus family includes members that infect different mammalian species and cause highly significant human or animal diseases (2). Smallpox was caused by variola virus, which was eradicated through global vaccination with the related vaccinia virus (VACV). VACV is the best-studied poxvirus and has been used as a vaccine vector for other infectious diseases and cancer. VACV encodes a large number of proteins that are dedicated to evading host immune responses, including secreted cytokine-binding proteins and viral host-range factors (3).
One of the most critical host-range factors for mammalian poxviruses belongs to a family of proteins that share sequence homology with VACV C7. In VACV, the function of C7 can be complemented by a nonhomologous host-range factor K1 (4), but the deletion of both C7L and K1L (the genes that encode C7 and K1) from VACV results in abortive replication of the mutant (VACV-K1L−C7L−) in many mammalian cells and severe attenuation in animal models (5–8). The replication defect can be rescued by restoring either K1L or C7L to VACV-K1L−C7L− (7). In permissive human cell lines, the replication of VACV-K1L−C7L−, but not of viruses with either K1L or C7L, can be blocked effectively by treatment with type I IFN or overexpression of IFN regulatory factor 1 (9, 10), indicating that the host restriction factor for the mutant is an ISG product. Although K1L is present only in VACV and a few closely related poxviruses, homologs of C7L are present in almost all poxviruses that infect mammalian species (8), including myxoma virus (MYXV, infecting rabbit), Yaba-like diseases virus (YLDV, infecting monkey), swinepox virus (SWPV), and sheeppox virus (SPPV). MYXV, in particular, has three C7 homologs. Functionally, the C7 family can be divided into two subgroups based on their roles in viral replication. One subgroup includes the majority of the C7 family members, which could function interchangeably with VACV-C7 in sustaining the replication of VACV in human cells (referred to as “C7-equivalent homologs” hereafter). The C7 homologs from YLDV (YLDV-67), SPPV (SPPV-63), SWPV (SWPV-64), and one of the C7 homologs from MYXV (MYXV-M62) can rescue the replication defect of VACV-K1L−C7L− and render the virus resistant to type I IFNs (8, 10). Similar to VACV-K1L−C7L−, M62R-deletion MYXV replicates abortively in most mammalian cells (11). The other subgroup of the C7 family includes two C7 homologs from MYXV, MYXV-M63, and MYXV-M64, which fail to rescue the replication of VACV-K1L−C7L− in human cells (8). Unlike VACV-K1L−C7L−, M64R-deletion MYXV has no replication defect in cell culture (12), whereas M63R-deletion MYXV has a replication defect only in rabbit cells (13).
An IFN-stimulated antiviral factor was discovered recently through studies of the C7 family of poxvirus host-range factors (11, 14). Human alpha sterile motif domain-containing 9 (SAMD9) protein was identified as a protein that binds directly to MYXV-M62, and knocking down SAMD9 in human cells rescued the replication of M62R-deletion MYXV (11). Although other C7 family members initially were reported not to bind SAMD9 (11), a recent study showed that both C7 and K1 bind SAMD9 (15), and knocking down SAMD9 rescued the replication of VACV-K1L−C7L− (14, 15). The exact function of SAMD9 is poorly understood, but it has antiviral activities against Japanese encephalitis virus (16), and its expression is induced by type I IFNs (17). SAMD9 also is a tumor suppressor, and mutations in the human SAMD9 gene are responsible for a rare life-threatening human disease, normophosphatemic familiar tumoral calcinosis (18).
The C7 family of host-range factors has no recognizable sequence motifs and no homologs outside the poxvirus family. To gain insight into their mechanism of action, we determined the crystal structures of two functionally divergent C7 family members, VACV-C7 and MYXV-M64. To our knowledge, the structures provide the first atomic view of the C7 family of proteins. Our structure-guided functional studies revealed the molecular basis for antagonizing SAMD9 by diverse mammalian poxviruses.
Results
The Structures of VACV-C7 and MYXV-M64.
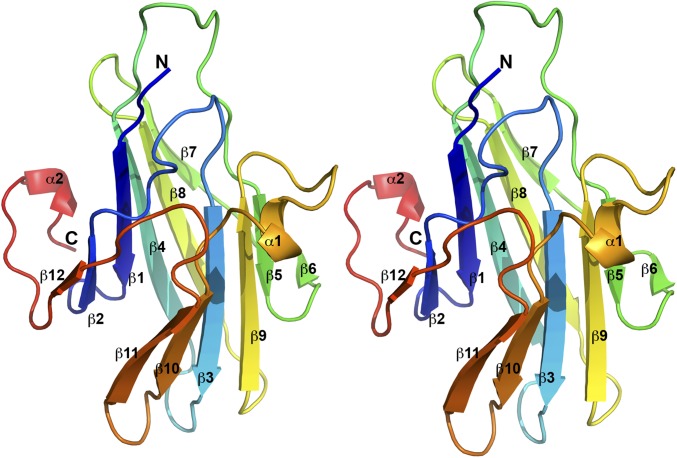
VACV-C7 and MYXV-M64 proteins were expressed in Escherichia coli and purified to homogeneity. Crystals of selenomethionine (SeMet)-substituted C7 protein were obtained and used to determine the C7 structure with the single-wavelength anomalous dispersion method. The structure of C7 includes all but the first two and last residues. It adopts a compact β-sandwich fold with an overall dimension of ∼45 × 35 × 30 Å (Fig. 1). It consists mainly of two curved layers, each comprising a six-stranded antiparallel β sheet. One layer comprises strands β1, β2, β4, β7, β8, and β12. The other layer comprises strands β3, β5, β6, β9, β10, and β11. The strands in the center of the sheets (β3, β4, β8, and β9) are significantly longer than the ones at the ends. Loops of varying length connect the β strands, and two short α helices (α1 and α2) on the opposite side of the β sandwich embrace the β sheets. Helix α1 links strands β9 and β10. Helix α2 is located at the C-terminal end.
Fig. 1.
The structure of VACV C7 displays a previously unidentified fold. A stereo view of the C7 structure is depicted. The secondary structures are labeled and shown in rainbow color with the N terminus in blue and the C terminus in red. The structure of C7 consists mainly of two curved layers, each comprised of a six-stranded antiparallel β sheet. Structure homology search did not find any match, suggesting that C7 adopts a novel fold.
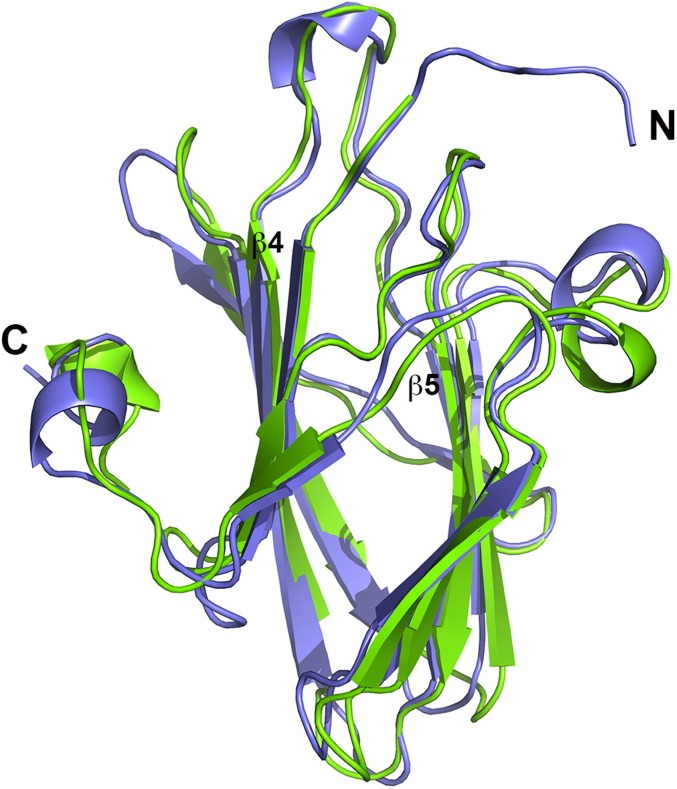
The structure of MYXV-M64 was solved by molecular replacement using the C7 structure as the template (Fig. S1). It includes residues 1–150 but lacks the 53 C-terminal residues, which are not conserved in the C7 family (Fig. S2). The two structures resemble each other closely with an rmsd of 0.74 Å over 136 aligned Cα atoms (Fig. S1), although the proteins share only 23% sequence identity (Fig. S2). Notable differences between the two structures include the presence of an α helix between strands β4 and β5 of M64 instead of a loop in C7 and a longer N terminus in M64 (Fig. S1). A structural similarity search using the secondary structure-matching (SSM) server (19) and DALI server (20) did not yield any significant match to the structures of VACV-C7 and MYXV-M64.
Fig. S1.
Comparison of the structures of VACV-C7 and MYXV-M64. The structures of VACV-C7 and MYXV-M64 are shown in green and blue, respectively. The two structures adopt the same β-sandwich fold and superimpose well with a 0.74-Å rmsd over 136 aligned Cα atoms. The N terminus and C terminus are labeled.
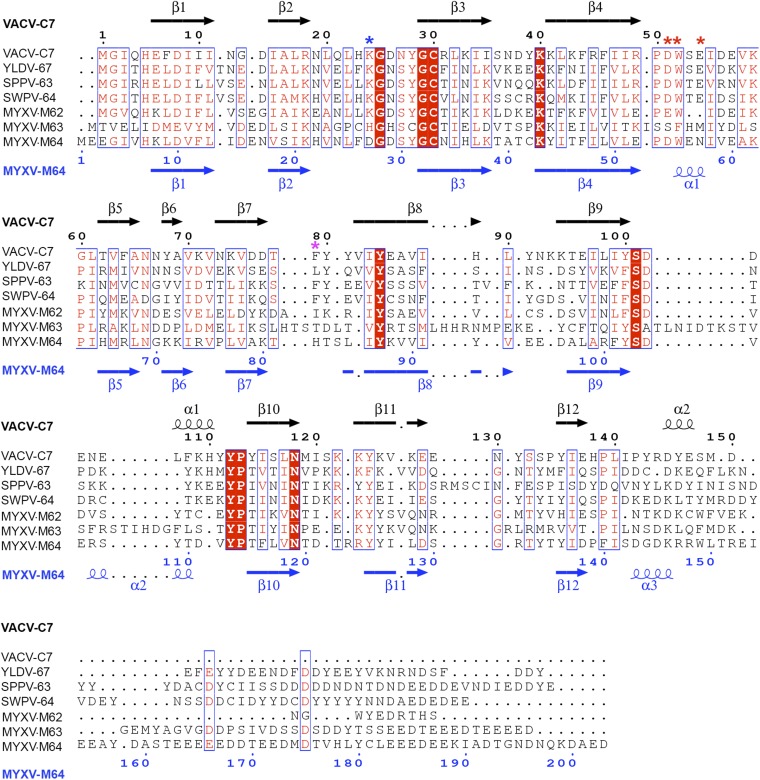
Fig. S2.
Structure-based sequence alignment of the C7 family of Poxvirus host-range factors. The alignment was performed with the SSM server (19) by using the crystal structures of VACV-C7 and MYXV-M64 as the templates, and the figure was created with ESPript (38). VACV-C7 and MYXV-M64 secondary structures and residue numbers are denoted above and below the alignment, respectively. Colored asterisks indicate key residues on three distinct loops that were found to be critical for the function of VACV-C7 (red: acidic loop; blue: basic loop; magenta: hydrophobic loop).
A Unique Three-Fingered Molecular Claw Is Critical for the Host-Range Function of C7.
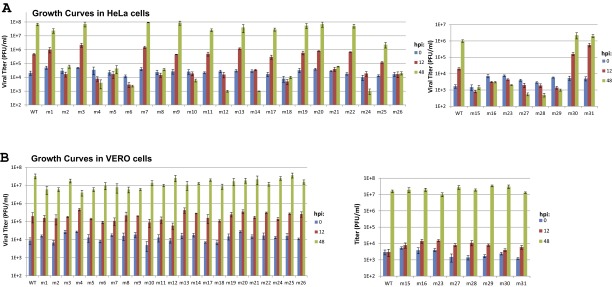
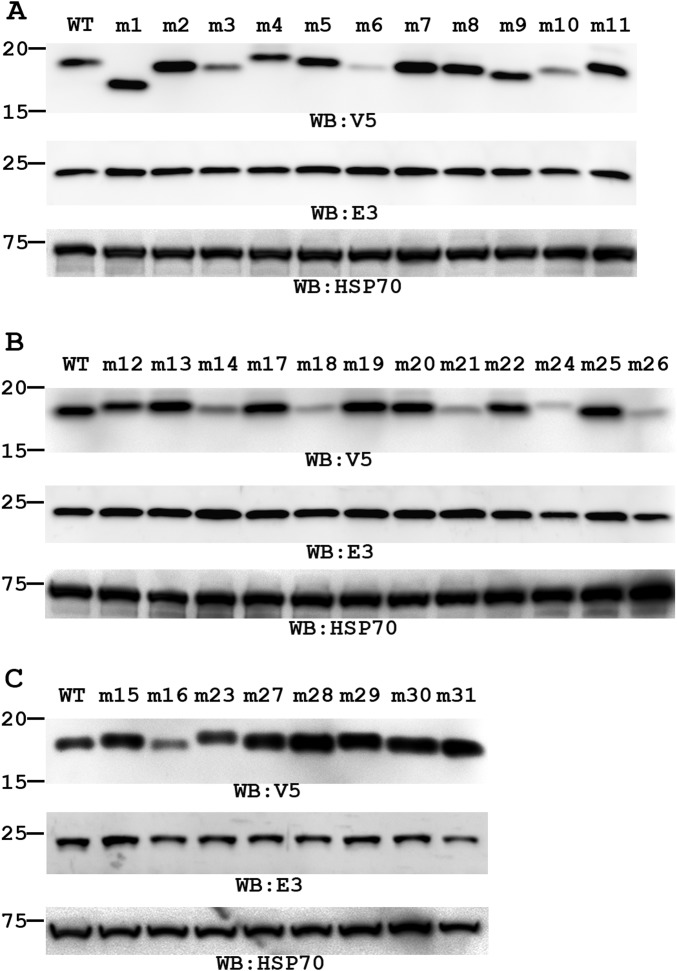
Based on the structure, we performed a systematic mutagenesis study of C7 to identify residues that are critical for the host-range function. VACV mutants with various mutations in C7L and a deletion in K1L were constructed on Vero cells, which permit the replication of VACV without K1 and C7. As expected, on VERO cells all mutants replicated similarly to VACV expressing wild-type C7 (referred to as “WT VACV”) (Fig. S3). To assess whether the mutations had any significant impact on C7 protein stability, the level of C7 protein in infected cells was determined by Western blot (Fig. S4). To determine the effect of the mutations on the host-range function, viral growth was determined on a representative human cell line, HeLa cells (Fig. S3). The results are summarized in Table 1.
Fig. S3.
Growth curves of C7 mutant VACVs. HeLa (A) and VERO (B) cells in 12-well plates were infected with the indicated viruses at an MOI of 0.05 pfu per cell. Viral titers at 0, 12, and 48 hpi were determined by plaque assay on VERO cells. The mutants were studied in groups in two separate experiments, each with a WT VACV control. The experiment was repeated one time with similar results.
Fig. S4.
Western blot of viral proteins expressed by C7 mutant VACVs. VERO cells were infected with the indicated virus at an MOI of 5 pfu per cell. C7 and E3 protein levels at 8 hpi were determined by Western blot with antibodies against V5 and E3, respectively. E3 is a viral protein unrelated to C7, serving as a control for infectivity and gel loading. The levels of a host protein (Hsp70) were determined also as an additional loading control. The size of the molecular weight marker in kilodaltons is shown on the left. The mutants were studied in groups in three separate experiments (A, B, and C), each with a WT control. The figure shows a representative result of two similar experiments.
Table 1.
Summary of the C7 mutagenesis study
| Name | Mutations | Protein level* | Viral growth† | SAMD9 precipitation‡ |
| WT | None | +++ | +++ | +++ |
| m1 | Δ(141-150) | +++ | +++ | + |
| m2 | K24E | +++ | − | − |
| m3 | K40E, K41E, K43E | ++ | +++ | +++ |
| m4 | D51K, E54R, D56K, E57K | +++ | − | − |
| m5 | W52A | +++ | − | − |
| m6 | K59E | + | − | + |
| m7 | F64A | +++ | +++ | +++ |
| m8 | F79S, Y80S, Y81S | +++ | − | − |
| m9 | K93E, K94E, E96K | +++ | +++ | + |
| m10 | D102K, D103K, E104K, E106K | ++ | − | + |
| m11 | K123E, K125E, K127D | +++ | +++ | + |
| m12 | D51K, E54R | +++ | − | − |
| m13 | D56K, E57K | +++ | +++ | +++ |
| m14 | E6K, D8K | + | − | − |
| m15 | D26K, N27A | +++ | − | − |
| m16 | R31E, K33E | ++ | − | − |
| m17 | D38K, Y39A | +++ | +++ | +++ |
| m18 | R45E, R49E | + | − | − |
| m19 | Y68A | +++ | +++ | ++ |
| m20 | K71E, K74E, D76K, D77K | +++ | +++ | +++ |
| m21 | E85K, H89A, Y91A | + | − | + |
| m22 | Y99A | +++ | +++ | +++ |
| m23 | F108A, K109E, H110A | +++ | − | − |
| m24 | Y111A, Y112A, Y114A | + | − | − |
| m25 | E128K, E129K, Y131A | +++ | ++ | + |
| m26 | Y135A, E137K | + | − | − |
| m27 | D51K | +++ | − | − |
| m28 | E54R | +++ | − | − |
| m29 | W52I | +++ | − | − |
| m30 | D102K, D103K | +++ | +++ | +++ |
| m31 | E104K, E106K | +++ | +++ | +++ |
Relative C7 protein level in VERO cells as shown in Fig. S4 and quantified by band intensity, indicating the effect of the mutations on protein stability. +++, >50% of the WT level; ++, ∼50% of the WT level, +, <20% of the WT level.
Viral growth in HeLa cells as shown in Fig. S3. +++, WT level of viral titers at 12 and 48 hpi; ++, ∼1/100 of the WT titer at 48 hpi; −, no growth.
The C termini of the C7 family members vary in length and sequence (Fig. S2). In both C7 and M64 structures, the C terminus after residue 140 is not part of the compact β sandwich structure. The truncation of the C terminus of C7 after residue 140 did not affect C7 stability, because the VACV mutant (m1) encoding the truncated C7 [Δ(141–150)] expressed a level of C7 protein similar to that of WT VACV (Fig. S4). In HeLa cells, the mutant replicated to a level similar to that of WT VACV (Fig. S3), indicating that the C terminus is not essential for C7 function. This finding suggests that the functional core for the C7 family is the conserved β sandwich.
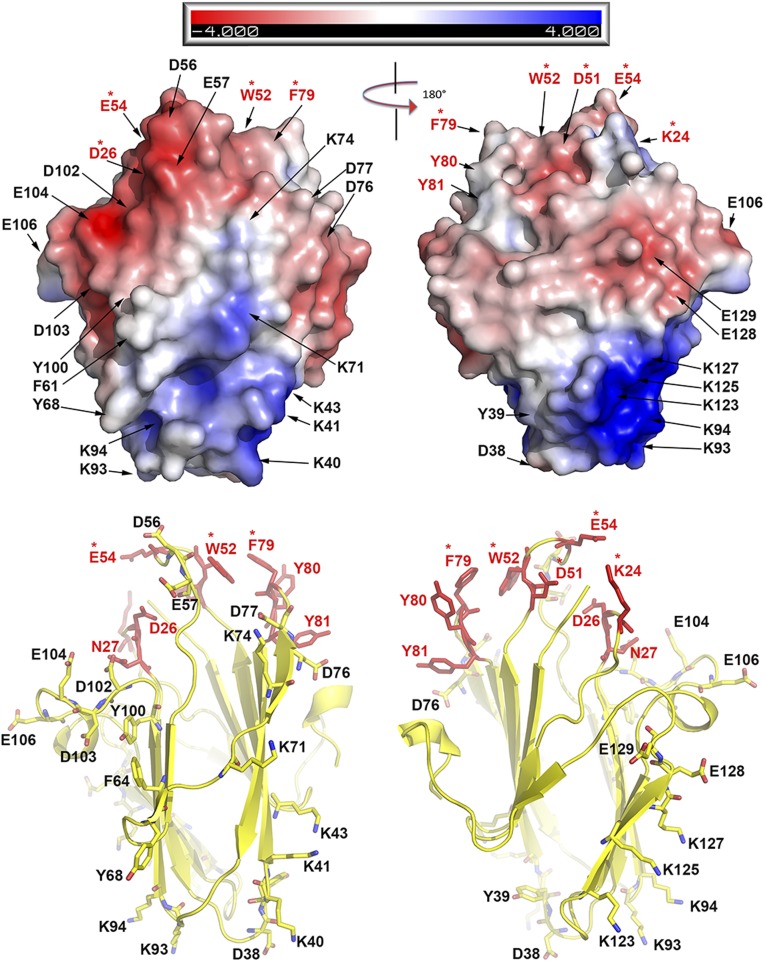
Next, the entire surface of the C7 protein was divided into different patches, which were mutated individually by charge-reversal of the polar residues and alanine or serine substitution of the nonpolar residues (Fig. 2 and Table 1). Many mutations on the β strands and the loops at the bottom of the β sandwich had no significant effect on the host-range function of C7 (Fig. 2). These include mutations of D38/Y39 (mutant m17) on the loop connecting strands β3 and β4 (loop ß3–β4), of K40/K41/K43 (m3) at the tip of strand β4, of F64 (m7) on β5, of Y68 (m19) at the tip of β6, of K71/K74/D76/D77 (m20) on loop ß6–β7 and β7, of Y99 (m22) at the end of β9, and of K123/K125/K127 (m11) on β11. Mutations of some residues at the top of the β sandwich, including D56/E57 (m13) on loop ß4–β5, D102/D103 (m30), and E104/E106 (m31) on loop ß9–α1, also did not affect C7 function. However, mutations of nine residues on three loops at the top of the β sandwich completely disrupted C7 function (Fig. 2 and Fig. S3). They are mutations of K24 (m2), D26/N27 (m15) on loop ß2–β3, D51 (m27), W52 (m5 and m29), E54 (m28) on loop ß4–β5, and F79/Y80/Y81 (m8) on loop ß7–β8. In particular, single amino acid substitutions of K24, D51, W52, and E54 completely abolished C7 function. Notably, these mutations had no negative effect on C7 stability, because a normal level of C7 protein was detected in VERO cells, suggesting that the three loops (β2–β3, β4–β5, and β7–β8) are directly involved in C7 function. Additional mutations abolished the host-range function, but the majority of these mutations (m6, m10, m14, m16, m18, m21, m24, and m26 in Table 1) appeared to affect protein folding, because the C7 protein level in VERO cells was significantly reduced. The F108A/K109E/H110A triple mutation (m23) also is likely to affect the protein conformation, because F108 is involved in hydrophobic interactions with aliphatic side chains of R31 (β3), L98, Y100 (β9), and Y112 (β9–β10). Furthermore, F108/K109/H110 are not conserved in other C7-equivalent homologs (Fig. S2). Therefore, these mutations may disrupt C7 function indirectly by affecting C7 structure. Overall, our data indicate that the functional site for C7 centers on three loops, β2–β3, β4–β5, and β7–β8, which together form a unique “three-fingered molecular claw.”
Fig. 2.
The functional site for C7 centers at three spatially clustered loops. (Upper) Electropotential surface of C7 shown in two views with 180° rotation. The residues that were mutated in this study are indicated by arrows. The residues whose mutations had no effect on viral replication are labeled in black. The residues whose mutations abolished viral replication are indicated in red; those that are conserved in C7-equivalent homologs are marked with red asterisks. (Lower) C7 is shown in a cartoon presentation with the secondary structures. The coloring scheme is the same as above. Notice that the key functional residues are clustered on the top of the β sandwich. The three functional loops form a unique molecular claw.
The Molecular Claw Is Conserved in C7-Equivalent Homologs from Diverse Mammalian Poxviruses and Is Essential for SAMD9 Binding.
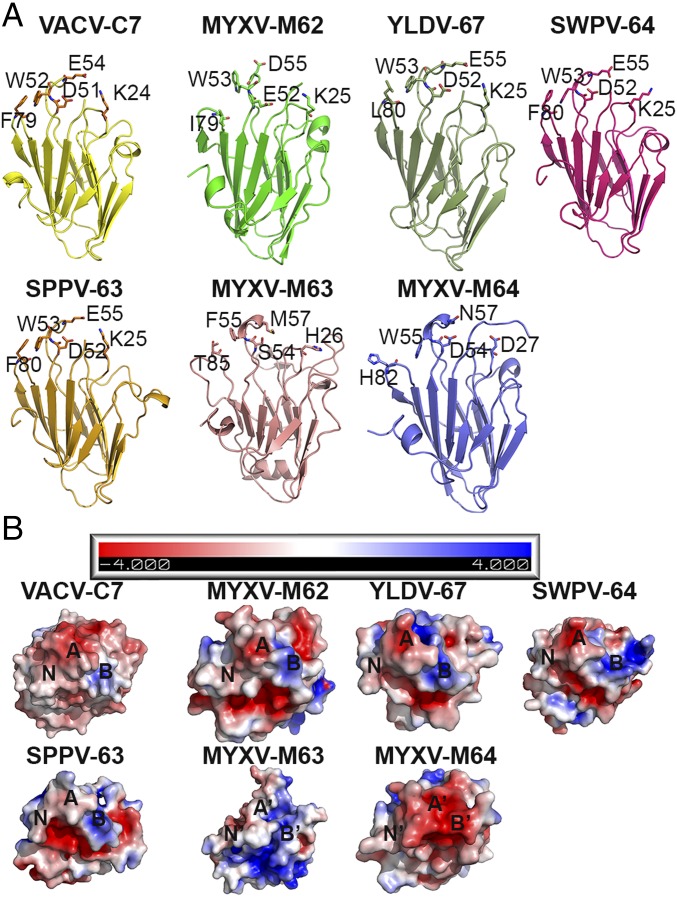
Although VACV-C7 and MYXV-M64 share limited sequence identity and perform different functions, their overall structures are almost identical, suggesting that all C7 family members adopt the same protein fold. We therefore carried out homology modeling of other C7 family members, including YLDV-67, SWPV-64, SPPV-63, MYXV-M62, and MYXV-M63 (Fig. 3). Among the nine C7 residues we identified as being critical for its function, five residues (K24, D51, W52, E54, and F79) are highly conserved among C7-equivalent homologs (MYXV-M62, YLDV-67, SWPV-64, and SPPV-63). Specifically, at the corresponding positions of the C7-equivalent homologs, K24 and W52 have identical residues, D51 has an acidic residue (Asp or Glu), and F79 has a hydrophobic residue (Phe, Leu, or Ile). E54 has an identical residue at the corresponding position of all homologs except MYXV-M62, which lacks two residues at the corresponding loop (Fig. S2). However, in MYXV-M62 an acidic residue is located two residues downstream, which could still result in an acidic residue at the position in MYXV-M62 that corresponds to E54. In contrast, only one or two of these five residues are conserved in MYXV-M63 and MYXV-M64 (Fig. S2), which are known to function differently than VACV-C7.
Fig. 3.
The C7 family is related by a common protein fold but is distinguished by sequences at the molecular claws. (A) The crystal structures of VACV-C7, MYXV-M64, and the homology modeled structures of MYXV-M62, YLDV-67, SWPV-64, and SPPV-63 are shown in ribbon configuration. The homology models were generated by SWISS-Model interface (swissmodel.expasy.org) (37) using the crystal structure of C7 as the template. The conserved key residues of VACV-C7 and their corresponding residues in other homologs are shown as sticks. Notice that all C7 family members adopt a conserved β-sandwich fold but with substantial variations in the three functional loops (the molecular claw) between the C7 equivalents (VACV-C7, MYXV-M62, YLDV-67, SWPV-64, and SPPV-63) and the nonequivalent homologs (MYXV-M63 and MYXV-M64). (B) Electropotential surfaces of VACV-C7 and its homologs. The three loops are indicated as A (acidic), B (basic), and N (nonpolar). Notice that MYXV-M63 and MYXV-M64 display surface features that are drastically different from those of VACV-C7 and other functional equivalents.
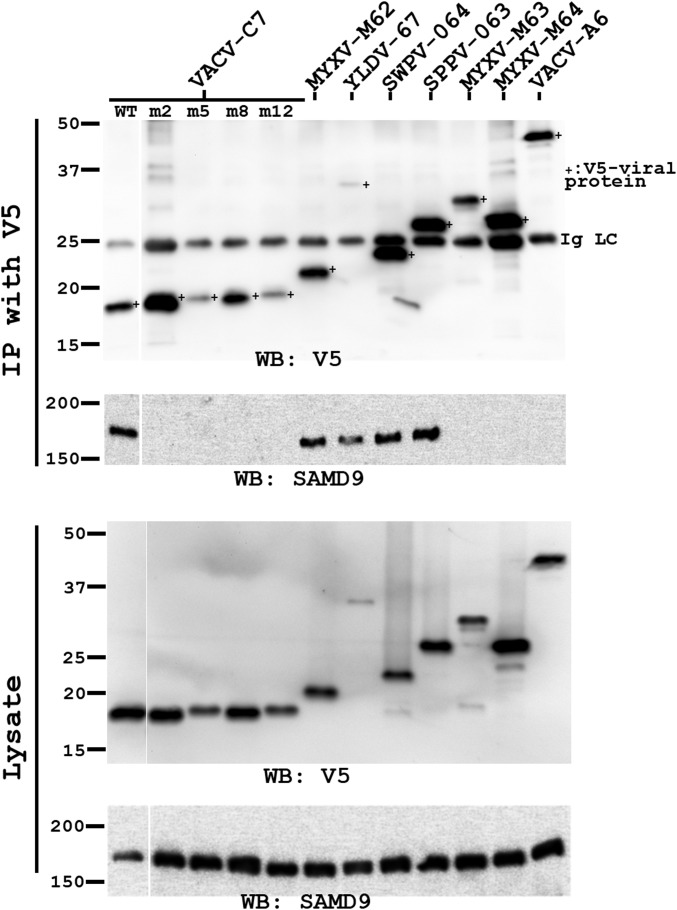
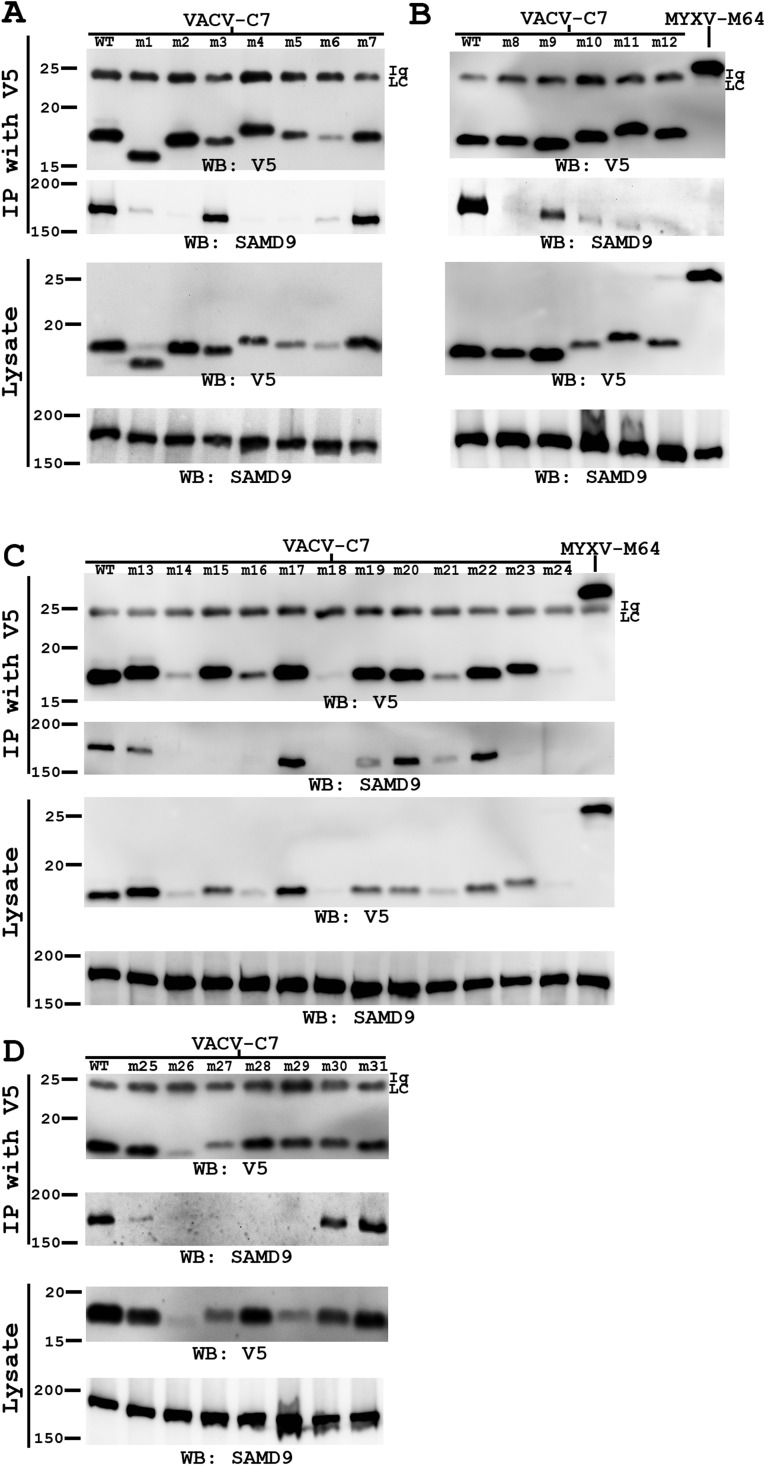
MYXV-M62 and VACV-C7 have been shown to bind SAMD9 (11, 15), but it is unclear whether other C7 family members do so and whether the binding is essential for their host-range function. We therefore examined the abilities of C7 family members and C7 mutants to bind SAMD9. We infected HeLa cells with VACV expressing epitope-tagged C7 homologs or mutated C7, immunoprecipitated the tagged proteins from the cells, and examined the precipitates for the presence of SAMD9 by Western blot (Fig. 4 and Fig. S5). As reported previously (11, 15), MYXV-M62 and VACV-C7, but not MYXV-M63, coimmunoprecipitated SAMD9. MYXV-M64 and an unrelated VACV protein A6 (21) failed to precipitate SAMD9, but YLDV-67, SWPV-64, and SPPV-63 were able to precipitate SAMD9 protein from HeLa cells. Furthermore, mutations at the C7 molecular claw, including the single amino acid substitutions of K24, D51, W32, and E54 (m2, m5, m27, m28, and m29, respectively), abolished the ability of C7 to precipitate SAMD9 (Fig. 4 and Table 1). In contrast, mutations that have no effect on the host-range function did not disrupt the ability to precipitate SAMD9 [although some of these mutations (m1 and m11) appeared to have a reduced ability to precipitate SAMD9]. Overall, the ability of C7 family members to overcome host restriction correlates with their ability to precipitate SAMD9.
Fig. 4.
C7-equivalent homologs from diverse mammalian poxviruses bind SAMD9, and the binding is disrupted by mutations at the molecular claw. HeLa cells in six-well plate were infected with VACV expressing the indicated viral proteins at a multiplicity of infection (MOI) of 5 pfu per cell. All viral proteins had a C-terminal V5 epitope tag. VACV-A6, a viral protein involved in viral assembly, was used as a negative control. At 8 hpi, the cells were lysed. V5-tagged viral proteins were immunoprecipitated from the cell lysates with agarose conjugated with a monoclonal V5 antibody (IP with V5). The levels of SAMD9 and V5-tagged viral proteins in the cell lysates and precipitates were determined by Western blot with antibody against SAMD9 and V5, respectively. The experiment was performed twice with similar results, and a representative result is shown. Ig LC, the light chain of the anti-V5 antibody.
Fig. S5.
Coimmunoprecipitation studies of additional C7 mutants. HeLa cells in a six-well plate were infected with VACV expressing the indicated C7 mutants at an MOI of 5 pfu per cell. At 8 hpi, the cells were lysed. V5-tagged C7 proteins were immunoprecipitated from the cell lysates with agarose conjugated with a monoclonal V5 antibody. The levels of SAMD9 and V5-tagged viral proteins in the cell lysates and precipitates were determined by Western blot with antibody against SAMD9 and V5, respectively. The mutants were studied in groups in four separate experiments (A, B, C, and D), each with a WT control. The experiment was performed twice with similar results, and a representative result is shown. Ig LC, the light chain of the anti-V5 antibody.
Discussion
The C7 family of poxvirus host-range factors is grouped together based on shared sequence homology, but the family members can be clearly divided into two subgroups based on their different functions in viral replication. The majority of the C7 family members could function interchangeably with VACV-C7 in sustaining the replication of VACV in human cells, but other C7 family members, including MYXV-M63 and MYXV-M64, do not (8, 10). The former subgroup of proteins is essential for productive poxvirus replication in many mammalian cells, but the latter group either is not essential for viral replication or is essential only in specific cells. Our structural and functional studies of C7 family members from the two different subgroups illustrate that all C7 family members are related by a common protein scaffold but perform distinct functions, possibly through sequence variations at a molecular claw that bind different proteins.
As is consistent with the C7 family having no homologs outside the poxvirus family, the structures of VACV-C7 and MYXV-M64 are dissimilar to any existing structures in the Protein Data Bank (PDB) database. Although VACV-C7 and MYXV-M64 share only 23% sequence identity, their overall structures are almost identical. Because all the other C7 family members have more than 23% sequence identity to VACV-C7 or MYXV-M64, all C7 family members are predicted to adopt the same protein fold, and their structures based on homology modeling could be quite accurate. To our knowledge, these structures provide the first atomic view of this important family of viral host-range proteins and will be valuable in guiding functional studies.
Although the two subgroups of C7 proteins have no major differences in their overall structures, we found that they differ in their sequences at the three-fingered molecular claw and in their abilities to bind the host restriction factor SAMD9. When this article was submitted, MYXV-M62 was the only C7 family member found to bind SAMD9, and VACV-C7 was reported to be unable to bind SAMD9 (11). More recently, VACV-C7 and VACV-K1 also have been found to bind SAMD9 (15), thus explaining why these two structurally different proteins could complement each other functionally in viral replication. In this study, we confirmed the binding between VACV-C7 and SAMD9. Furthermore, we found that YLDV-67, SWPV-64, and SPPV-63 also bind SAMD9. In contrast, MYXV-M64 and MYXV-M63 could not bind SAMD9. The natural hosts for MYXV, YLDV, SWPV, and SPPV are very different mammalian species, but the C7 homologs from all these viruses can bind human SAMD9 and overcome host restriction in human cells. This finding indicates that SAMD9 orthologs from different mammalian species are sufficiently similar so that the C7 homologs from these viruses antagonize SAMD9 not only from their natural hosts but also human SAMD9. SAMD9 appears to have originated after the mammalian radiation, and all mammals with a completely sequenced genome encode SAMD9 and/or its paralog SAMD9L (22). Interestingly, a C7-equivalent homolog is present in almost every poxvirus that infects mammals but not in poxviruses that infect birds or insects (8, 23). Therefore, the C7 family of host-range factors may have been acquired by the poxviruses specifically to overcome the SAMD9-mediated host restriction in mammals. To confirm the significance of SAMD9/SAMD9L in restricting poxvirus infection, it will be interesting to test whether VACV-K1L−C7L−, which is highly attenuated in mice, has increased virulence in SAMD9L-knockout mice.
Through this structure-guided mutagenesis study we have determined the molecular mechanism by which C7 targets SAMD9 and have shown that SAMD9 binding is essential for the host-range function. We show that the main body of the β sandwich serves as a protein scaffold and that three spatially clustered loops with distinct features (basic, acidic, and hydrophobic) at one side of the β sandwich structure are the functional site. We identified nine residues (K24, D26/N27, D51, W52, E54, and F79/Y80/Y81) on these loops as being critical for viral replication in human cells and for binding with SAMD9. Five of these residues (K24, D51, W52, E54, and F79) are conserved in all C7-equivalent homologs. D26 and N27 are not conserved, but they are located near the conserved K24 residue. The D26K/N27A mutation may disrupt C7 function indirectly by perturbing K24 orientation. Y82 is not conserved, and Y81 is not conserved in MYXV-M62 , which has a Lys instead. Triple mutations of F79, Y80, and Y81 may disrupt C7 function mainly by affecting the conserved F79. Based on both the C7 mutagenesis result and sequence comparison between C7 family members, we conclude that five residues (K24, D51, W52, E54, and F79) probably are involved directly in binding SAMD9. Most of these five residues are not conserved in MYXV-M63 and MYXV-M64, perhaps explaining their inability to bind SAMD9. At the basic loop, the equivalent position of K24 is H26 in MYXV-M63 and D27 in MYXV-M64. Notably, the single K24E mutation of C7 abolished viral replication and SAMD9 binding. At the acidic loop, the equivalent position of E54 is a neutral residue, M57 in MYXV-M63 and N57 in MYXV-M64. The single E54R mutation of C7 abolishes SAMD9 binding and consequently host-range function. D51 is present in MYXV-M64 (D54) but is replaced by S54 in MYXV-M63. The single D51K mutation of C7 also abolishes SAMD9 binding and the host-range function. At the nonpolar loop, the equivalent position of F79 is a polar residue, T85 in MYXV-M63 and H82 in MYXV-M64. To convert MYXV-M64 into a SAMD9-binding molecule, a triple mutation of D27K, N57E, and H82F would be required, and additional surface remodeling might be necessary to remove possible clashes with the SAMD9 surface. Although MYXV-M63 and MYXV-M64 do not target SAMD9, they play distinct roles in MYXV pathogenesis and host tropism (12, 13). It is tempting to speculate that they have acquired different functions by evolving their molecular claws to bind different host proteins.
Materials and Methods
Protein Purification and Crystallization.
VACV C7L was cloned in a modified pET vector with an N-terminal 6xHis tag followed by a tobacco etch virus (TEV) protease cleavage site. The recombinant C7 and the SeMet-substituted protein were expressed in E. coli and purified by Ni-NTA as previously described (24). Briefly, C7 protein was first purified from soluble cell lysate using an Ni-NTA affinity column. The eluted protein subsequently was subjected to TEV protease cleavage and was collected as flow through of a second subtracting Ni-NTA column. The purified protein was concentrated to 3 mg/mL, flash frozen, and stored at −80 °C until use (25). C7 protein crystals were obtained at room temperature in a solution containing 0.1 M Tris (pH 8.5) and 3.5 M NaCl. Glycerol [25% (vol/vol)] was added to the mother liquid as cryoprotectant. MYXV-M64 was cloned, expressed, and purified the same way as C7. A single M64 protein crystal was obtained after 5 mo at room temperature in a solution containing 28% (vol/vol) PEG2000, monomethyl ether, and 0.1 M Bis-Tris (pH 6.5). Glycerol [20% (vol/vol)] was added to the mother liquid as cryoprotectant.
Structure Determination.
A set of data was collected from a SeMet-substituted C7 protein crystal at the beamline 19-ID at the Advanced Photon Source (APS), Argonne National Laboratory. The structure was solved by the single-wavelength anomalous dispersion method using program HKL3000 (26). A nearly complete model was constructed from the experimental phases obtained from the SeMet crystal data. This model was used to solve the native C7 structure by molecular replacement method using the Phaser program (27). A set of data was collected on a single M64 crystal at beamline 19-ID at APS. The structure was solved by the molecular replacement method using the Phaser program (27) with the structure of C7 as the model template. The structure lacks the 53 C-terminal residues, presumably because these residues were disordered in the crystal packing or degraded during the extended crystallization period. For both structures, the PHENIX program (28) was used for the refinement, and Coot (29) was used for the iterative manual model building. Translation, libration, and screw-rotation displacement (TLS) groups used in the refinement were defined by the TLMSD server (30). The current models are of good geometry and refinement statistics (Table S1). Electrostatic surface potentials were determined by using PDB2PQR (31, 32) and APBS (33) plugin tools from PyMOL (34) and were visualized over a range of −4 kT/e to +4 kT/e. All molecular graphic figures were generated with PyMOL.
Table S1.
Crystallographic data and statistics
| Data collection | VACV-C7 Se peak | VACV-C7 native | MYXV-M64 |
| Beamline | 19-ID, APS | 19-ID, APS | 19-ID, APS |
| Wavelength, Å | 0.97948 | 0.97948 | 0.97921 |
| Space group | P213 | P213 | P212121 |
| Cell parameters a, b, c, Å | 100.5, 100.5, 100.5 | 100.7, 100.7, 100.7 | 51.8, 70.9, 115.4 |
| Resolution, Å | 50.0–2.5 (2.59–2.50) | 50.0–2.0 (2.07–2.0) | 50.0–2.5 (2.54–2.50) |
| Total reflections | 257,049 | 458,131 | 141,699 |
| Unique reflections | 11,974 (1,122) | 22,967 (1,987) | 15,289 (739) |
| Redundancy | 21.5 (5.4) | 19.9 (3.4) | 9.3 (8.2) |
| Completeness, % | 99.4 (93.7) | 98.7 (86.6) | 100 (99.9) |
| I/σ | 62.8 (1.25) | 50.8 (1.8) | 22.6 (2.5) |
| Rsym, % | 9.3 (78.0) | 6.8 (54.5) | 12.7 (82.0) |
| Refinement statistics | |||
| Resolution range used, Å | 45.0–2.00 (2.09–2.00) | 44.75–2.50 (2.65–2.50) | |
| No. reflections used | 22,941 (2,437) | 15,168 (2,314) | |
| Rwork/Rfree | 15.3/18.2 | 20.7/23.2 | |
| Rmsd bond lengths, Å | 0.020 | 0.003 | |
| Rmsd bond angles, ° | 1.656 | 0.699 | |
| Number of atoms per average B, Å2 | |||
| Protein | 1,267 (37.1) | 2,479 (57.6) | |
| Ligand | 12 (74.7) | 8 (82.0) | |
| Water | 160 (47.6) | 52 (50.8) | |
| Ramachandran values | |||
| Preferred regions, % | 97.3 | 97.6 | |
| Allowed regions, % | 2.7 | 2.4 | |
Values in parentheses are for the highest-resolution shell. Rsym = Σ |Iobs − Iavg|/Σ Iavg; Rwork = Σ|| Fobs | − |Fcalc||/Σ Fobs. Rfree was calculated using 5% of data.
Cells and Viruses.
VERO and HeLa cells were cultured in DMEM (Invitrogen) supplemented with 10% FBS. All recombinant viruses were propagated on VERO cells.
Plasmid Construction.
All plasmids for making C7L mutants were derived from the transfer plasmid described previously (8). The transfer plasmid contains (i) 300 bp of the downstream flanking region of C7L; (ii) GFP under the control of VACV late promoter P11; (iii) C7 with a C-terminal V5 tag; and (iv) 300 bp of the upstream flanking region of C7L including the C7L promoter. Specific mutations of C7 were introduced into the plasmid through recombinant PCR as described previously (35). All constructs were confirmed by DNA sequencing.
Recombinant Virus Construction.
Various C7 mutant viruses were constructed by homologous recombination of vK1L−C7L− with the specific transfer plasmid described above. Briefly, the transfer plasmids containing the C7 mutation were transfected into VERO cells that were infected with vK1L−C7L−. Recombinant viruses encoding GFP and V5-tagged C7 were picked under the fluorescence microscope and purified by four rounds of plaque purification on VERO cells. vK1L−C7L−/GFP+, vK1L−C7L−, vVACV-C7L (labeled as WT VACV here), vYLDV-67R, vMYXV-M62R, vMYXV-M63R, vMYXV-M64R, vSPPV-063, and vSWPV-064 were described previously (8, 9, 35).
Growth Curve Analysis of C7 Mutants.
VERO or HeLa cells in 12-well plates were incubated with 0.05 pfu per cell of different C7 mutant viruses for 2 h at room temperature. Following adsorption, the cells were washed twice with PBS and were moved to an incubator at 37 °C to initiate viral entry and replication. The cells were harvest at 0, 12, and 48 h postinfection (hpi). The viral titers in the cell lysates were determined by plaque assays on VERO cells.
Immunoprecipitation and Western Blot Analysis.
Infected cells were harvested 8 hpi and were lysed on ice with a lysis buffer [0.1% (wt/vol) Nonidet P-40, 50 mM Tris (pH 7.4), 150 mM NaCl] supplemented with protease inhibitor mixture tablets (Roche Molecular Biochemicals). The cleared cell lysates were mixed with 50 µL of 50% (vol/vol) V5-agarose beads (Sigma-Aldrich) for 30 min at 4 °C. After washing with lysis buffer, the beads were resuspended in SDS sample buffer, and the eluted proteins were resolved by SDS/PAGE and detected with Western blot as described previously (35). The detection antibodies were mouse monoclonal antibodies against V5 (clone V5-10; Sigma-Aldrich) and VACV E3 (36) and rabbit anti-SAMD9 polyclonal antibody (HPA-21319; Sigma-Aldrich).
Acknowledgments
We thank the staff of Beamline 19ID at the Advanced Photon Source for their support. This work was supported by NIH Grant AI079217 (to Y.X.). The crystallography study in the J.D. laboratory was supported by NIH Grant AI113539 (to J.D.) and NIH Institutional Development Award Grant P20GM103640.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors reported in this paper have been deposited in the Protein Data Bank database (accession nos. 5CYW and 5CZ3).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515354112/-/DCSupplemental.
References
- 1.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451(7177):425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 2.Moss B. Poxviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Vol 2. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 2905–2946. [Google Scholar]
- 3.Johnston JB, McFadden G. Poxvirus immunomodulatory strategies: Current perspectives. J Virol. 2003;77(11):6093–6100. doi: 10.1128/JVI.77.11.6093-6100.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y, Meng X, Xiang Y, Deng J. Structure function studies of vaccinia virus host range protein k1 reveal a novel functional surface for ankyrin repeat proteins. J Virol. 2010;84(7):3331–3338. doi: 10.1128/JVI.02332-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drillien R, Koehren F, Kirn A. Host range deletion mutant of vaccinia virus defective in human cells. Virology. 1981;111(2):488–499. doi: 10.1016/0042-6822(81)90351-2. [DOI] [PubMed] [Google Scholar]
- 6.Gillard S, Spehner D, Drillien R, Kirn A. Localization and sequence of a vaccinia virus gene required for multiplication in human cells. Proc Natl Acad Sci USA. 1986;83(15):5573–5577. doi: 10.1073/pnas.83.15.5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perkus ME, et al. Vaccinia virus host range genes. Virology. 1990;179(1):276–286. doi: 10.1016/0042-6822(90)90296-4. [DOI] [PubMed] [Google Scholar]
- 8.Meng X, Chao J, Xiang Y. Identification from diverse mammalian poxviruses of host-range regulatory genes functioning equivalently to vaccinia virus C7L. Virology. 2008;372(2):372–383. doi: 10.1016/j.virol.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meng X, et al. Vaccinia virus K1L and C7L inhibit antiviral activities induced by type I interferons. J Virol. 2009;83(20):10627–10636. doi: 10.1128/JVI.01260-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng X, et al. C7L family of poxvirus host range genes inhibits antiviral activities induced by type I interferons and interferon regulatory factor 1. J Virol. 2012;86(8):4538–4547. doi: 10.1128/JVI.06140-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Wennier S, Zhang L, McFadden G. M062 is a host range factor essential for myxoma virus pathogenesis and functions as an antagonist of host SAMD9 in human cells. J Virol. 2011;85(7):3270–3282. doi: 10.1128/JVI.02243-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, et al. Myxoma virus M064 is a novel member of the poxvirus C7L superfamily of host range factors that controls the kinetics of myxomatosis in European rabbits. J Virol. 2012;86(9):5371–5375. doi: 10.1128/JVI.06933-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrett JW, et al. Myxoma virus M063R is a host range gene essential for virus replication in rabbit cells. Virology. 2007;361(1):123–132. doi: 10.1016/j.virol.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, McFadden G. SAMD9 is an innate antiviral host factor with stress response properties that can be antagonized by poxviruses. J Virol. 2015;89(3):1925–1931. doi: 10.1128/JVI.02262-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sivan G, Ormanoglu P, Buehler EC, Martin SE, Moss B. Identification of Restriction Factors by Human Genome-Wide RNA Interference Screening of Viral Host Range Mutants Exemplified by Discovery of SAMD9 and WDR6 as Inhibitors of the Vaccinia Virus K1L-C7L- Mutant. MBio. 2015;6(4):e01122. doi: 10.1128/mBio.01122-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang LK, Chai F, Li HY, Xiao G, Guo L. Identification of host proteins involved in Japanese encephalitis virus infection by quantitative proteomics analysis. J Proteome Res. 2013;12(6):2666–2678. doi: 10.1021/pr400011k. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka M, Shimbo T, Kikuchi Y, Matsuda M, Kaneda Y. Sterile alpha motif containing domain 9 is involved in death signaling of malignant glioma treated with inactivated Sendai virus particle (HVJ-E) or type I interferon. Int J Cancer. 2010;126(8):1982–1991. doi: 10.1002/ijc.24965. [DOI] [PubMed] [Google Scholar]
- 18.Topaz O, et al. A deleterious mutation in SAMD9 causes normophosphatemic familial tumoral calcinosis. Am J Hum Genet. 2006;79(4):759–764. doi: 10.1086/508069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 20.Holm L, Rosenstrom P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meng X, Embry A, Sochia D, Xiang Y. Vaccinia virus A6L encodes a virion core protein required for formation of mature virion. J Virol. 2007;81(3):1433–1443. doi: 10.1128/JVI.02206-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemos de Matos A, Liu J, McFadden G, Esteves PJ. Evolution and divergence of the mammalian SAMD9/SAMD9L gene family. BMC Evol Biol. 2013;13:121–136. doi: 10.1186/1471-2148-13-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Rothenburg S, McFadden G. The poxvirus C7L host range factor superfamily. Curr Opin Virol. 2012;2(6):764–772. doi: 10.1016/j.coviro.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krumm B, Meng X, Li Y, Xiang Y, Deng J. Structural basis for antagonism of human interleukin 18 by poxvirus interleukin 18-binding protein. Proc Natl Acad Sci USA. 2008;105(52):20711–20715. doi: 10.1073/pnas.0809086106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng J, et al. An improved protocol for rapid freezing of protein samples for long-term storage. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 1):203–204. doi: 10.1107/s0907444903024491. [DOI] [PubMed] [Google Scholar]
- 26.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: The integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 8):859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 27.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr. 2007;63(Pt 1):32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 30.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 4):439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 31.Dolinsky TJ, et al. PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35(Web Server issue):W522–525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32(Web Server issue):W665–667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeLano WL. 2002 The PyMOL molecular graphics system. Available at www.pymol.org. Accessed September 22, 2015.
- 35.Meng X, Xiang Y. Vaccinia virus K1L protein supports viral replication in human and rabbit cells through a cell-type-specific set of its ankyrin repeat residues that are distinct from its binding site for ACAP2. Virology. 2006;353(1):220–233. doi: 10.1016/j.virol.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 36.Kolli S, et al. Structure-function analysis of vaccinia virus H7 protein reveals a novel phosphoinositide binding fold essential for poxvirus replication. J Virol. 2015;89(4):2209–2219. doi: 10.1128/JVI.03073-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biasini M, et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42(Web Server issue):W252–258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31(13):3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]