Significance
Oil spills are a significant source of hydrocarbon inputs into the ocean. In response to oil spills, chemical dispersants are applied to the oil-contaminated seawater to disperse surface slicks into smaller droplets that are presumed to be more bioavailable to microorganisms. We provide evidence that chemical dispersants applied to either deep water or surface water from the Gulf of Mexico did not stimulate oil biodegradation. Direct measurement of alkane and aromatic hydrocarbon oxidation rates revealed either suppression or no stimulation of oil biodegradation in the presence of dispersants. However, dispersants affected microbial community composition and enriched bacterial populations with the ability to use dispersant-derived compounds as growth substrates, while oil-alone amendments enriched for natural hydrocarbon degraders.
Keywords: oceanography, microbial dynamics, hydrocarbon cycling, chemical dispersants, oil spills
Abstract
During the Deepwater Horizon oil well blowout in the Gulf of Mexico, the application of 7 million liters of chemical dispersants aimed to stimulate microbial crude oil degradation by increasing the bioavailability of oil compounds. However, the effects of dispersants on oil biodegradation rates are debated. In laboratory experiments, we simulated environmental conditions comparable to the hydrocarbon-rich, 1,100 m deep plume that formed during the Deepwater Horizon discharge. The presence of dispersant significantly altered the microbial community composition through selection for potential dispersant-degrading Colwellia, which also bloomed in situ in Gulf deep waters during the discharge. In contrast, oil addition to deepwater samples in the absence of dispersant stimulated growth of natural hydrocarbon-degrading Marinobacter. In these deepwater microcosm experiments, dispersants did not enhance heterotrophic microbial activity or hydrocarbon oxidation rates. An experiment with surface seawater from an anthropogenically derived oil slick corroborated the deepwater microcosm results as inhibition of hydrocarbon turnover was observed in the presence of dispersants, suggesting that the microcosm findings are broadly applicable across marine habitats. Extrapolating this comprehensive dataset to real world scenarios questions whether dispersants stimulate microbial oil degradation in deep ocean waters and instead highlights that dispersants can exert a negative effect on microbial hydrocarbon degradation rates.
Crude oil enters marine environments through geophysical processes at natural hydrocarbon seeps (1) at a global rate of ∼700 million liters per year (2). In areas of natural hydrocarbon seepage, such as the Gulf of Mexico (hereafter, the Gulf), exposure of indigenous microbial communities to oil and gas fluxes can select for microbial populations that use petroleum-derived hydrocarbons as carbon and energy sources (3, 4). The uncontrolled deep-water oil well blowout that followed the explosion and sinking of the Deepwater Horizon (DWH) drilling rig in 2010 released about 750 million liters of oil into the Gulf. Seven million liters of chemical dispersants were applied (5) with the goal of dispersing hydrocarbons and stimulating oil biodegradation. A deep-water (1,000–1,300 m) plume, enriched in hydrocarbons (6–11) and dioctyl sodium sulfosuccinate (DOSS) (12, 13), a major component of chemical dispersants (14), formed early in the discharge (7). The chemistry of the hydrocarbon plume significantly altered the microbial community (11, 15–17), driving rapid enrichment of low-abundance bacterial taxa such as Oceanospirillum, Cycloclasticus, and Colwellia (18). The natural hydrocarbon degraders in Gulf waters were either in low abundance or absent in DWH deep-water plume samples (18).
Chemical dispersants emulsify surface oil slicks, reduce oil delivery to shorelines (19), and increase dissolved oil concentrations, which should make oil more bioavailable (20) and stimulate biodegradation (21). The efficacy of dispersants in stimulating oil biodegradation is debated (22) and negative environmental effects have been documented (23). Dispersant application often requires ecological tradeoffs (24). Surprisingly little is known about the impacts of dispersants on the activity and abundance of hydrocarbon-degrading microorganisms (25). This work addressed three key questions: (i) Do dispersants influence microbial community composition? (ii) Is the indigenous microbial community as effective at oil biodegradation as microbial populations following dispersant/dispersed oil exposure? (iii) Does chemically dispersed oil stimulate hydrocarbon biodegradation rates?
Laboratory experiments were used to unravel the effects of oil-only (supplied as a water-accommodated fraction, “WAF”), Corexit 9500 (“dispersant-only”), oil–Corexit 9500 mixture (chemically enhanced water-accommodated fraction, CEWAF) or a CEWAF with nutrients (CEWAF + nutrients) (SI Appendix) on Gulf deep-water microbial populations (SI Appendix, SI Text and Figs. S1 and S2). Experimental conditions (SI Appendix, Table S1) mimicked those prevailing in the DWH deep-water hydrocarbon plume (6–13, 18), the chemistry of which varied substantially over space and time (18). Amending samples with WAFs and CEWAFs assured that observed differences in microbial community composition and activity would be driven by compositional differences (e.g., the presence or absence of dispersants) in the dissolved organic carbon (DOC) pool rather than by differences in the bulk DOC concentration (26, 27). We developed an improved radiotracer method to directly quantify hydrocarbon oxidation rates. The microbial community composition was monitored over time using 16S rRNA amplicon sequencing. Dispersant application selected for specific microbial taxa and oligotypes with 16S rRNA gene sequences similar to those recovered in situ during the DWH discharge. Surprisingly, CEWAF (± nutrients) addition did not enhance microbial activity or microbial oil-degradation rates.
Results and Discussion
Dispersant Significantly Altered Microbial Community Composition.
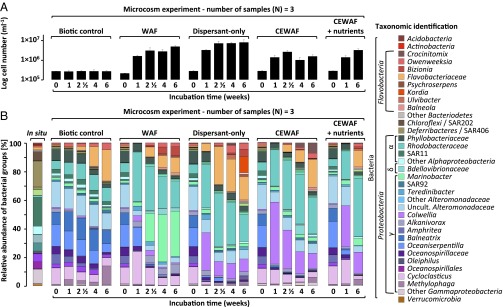
We hypothesized that dispersants would alter microbial community composition in the deepwater samples and that selection of one population over another would drive differences in hydrocarbon-degradation rates, altering the oil-degradation efficiency. We explored patterns in microbial abundance (Fig. 1A) using microscopy and community composition by Illumina paired-end sequencing of bacterial 16S rRNA gene amplicons (Fig. 1B). We resolved closely related bacterial taxa using oligotyping analysis (28) (Fig. 2 and SI Appendix, Fig. S3). We elucidated the ecological preference of specific taxa using statistical correspondence analysis (SI Appendix, Figs. S4–S8).
Fig. 1.
Dispersants affect the evolution of oil-degrading microbial populations. (A) Average and standard deviation (SD) of cell numbers from sample triplicates (log scale) monitored for 6 wk in microcosms. (B) Relative abundance of bacterial groups in Gulf of Mexico deep water in situ samples and in the microcosms (average of triplicate samples). Reads of the V4V5 regions of the 16S rRNA gene were clustered into operational taxonomic units and taxonomy was assigned with Global Alignment for Sequence Taxonomy (GAST).
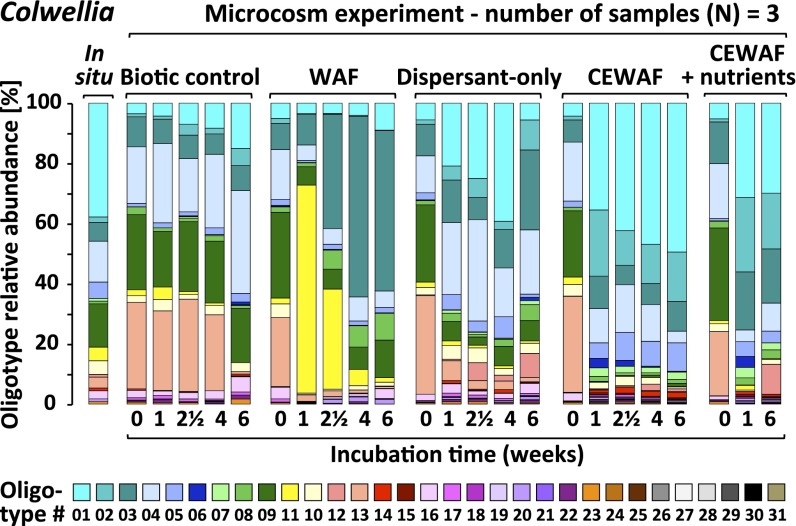
Fig. 2.
Different microbial oligotypes respond to dispersants or oil (WAF). Oligotyping enabled the interpretation of 16S rRNA gene sequence diversity at the level of specific oligotypes. Relative abundance (averaged across biological triplicates) of Colwellia oligotypes in microcosms, simulating DWH spill-like plumes.
All dispersant-amended treatments showed ingrowth of Colwellia (SI Appendix, Fig. S4), a group containing both hydrocarbon and dispersant degraders (29). After 1 wk, the relative abundance of Colwellia increased from 1% to 26–43% in dispersant-only and CEWAF (± nutrients) treatments (Fig. 1B). In contrast, Colwellia was a minority (1–4%) in WAF treatments. Selective enrichment of Colwellia in dispersant-only treatments indicates that dispersant components served as growth substrates (29). The relative abundance of Colwellia oligotypes 01, 02, and 05 increased in dispersant treatments (Fig. 2 and SI Appendix, Fig. S5), whereas oligotypes 03 and 10 increased in treatments receiving oil only, underscoring the role of dispersants in driving variation in Colwellia taxa. Phylogenetic analysis of the 16S rRNA gene amplicons confirmed that these oligotypes were closely related to species detected in DWH plume samples in situ (9, 16, 18) (SI Appendix, Fig. S9), verifying the environmental relevance of these organisms during the DWH discharge.
The dominant microbial responder to WAF addition was Marinobacter, whose relative abundance increased from 2% to 42% after 4 wk (Fig. 1B). In contrast, in dispersant-only and CEWAF (± nutrients) treatments, Marinobacter comprised only 1–5% of all sequences. The correspondence analysis emphasized the dominance of Marinobacter in WAF samples (SI Appendix, Fig. S6) and the same Marinobacter oligotypes occurred across all treatments, illustrating that dispersants did not select for specific Marinobacter taxa, as was the case for Colwellia (SI Appendix, Fig. S3A). Marinobacter (SI Appendix, Fig. S10) degrade a wide variety of hydrocarbons, including pristane, hexadecane, octane, toluene, benzynes, and phenanthrene (30–32) and are likely dominant hydrocarbon degraders under natural conditions. However, their abundance clearly declined in the presence of dispersants. Whether Colwellia outcompetes Marinobacter or whether Marinobacter is inhibited by some component of Corexit 9500 or the CEWAF remains to be resolved (SI Appendix).
Like Marinobacter, the abundance of Cycloclasticus increased primarily in the WAF treatments, where their relative abundance increased from 12% to 23% after 1 wk and an oligotype (type 03) closely related to Cycloclasticus pugetii (SI Appendix, Figs. S3B and S11), which degrades naphthalene, phenanthrene, anthracene, and toluene as sole carbon sources (33), increased substantially. Cycloclasticus also increased slightly in relative abundance in the CEWAF + nutrients treatment (Fig. 1B), but less so than in the WAF treatment.
Oceaniserpentilla (also known as DWH Oceanospirillum) (34) abundance decreased consistently across treatment, regardless of the presence or absence of WAF, dispersant, or CEWAF (± nutrients) (Fig. 1B and SI Appendix, Figs. S3C and S8). The observed oligotypes closely resembled those observed in situ during the DWH incident (18) (SI Appendix, Fig. S12). The DWH Oceanospirillum oxidize n-alkanes and cycloalkanes (17); cycloalkanes are absent in surrogate Macondo oil, possibly explaining the low abundance of Oceanospirillum in the microcosms.
Cell Growth and Exopolymer Formation.
Initially, cell abundance was similar across treatments (3 × 105 cells⋅mL−1; Fig. 1A). At the experiment’s termination, microbial abundance in WAF treatments had increased by a factor of 60, which was significantly higher (T4: P < 0.0001) than microbial abundance in CEWAF (± nutrients) treatments. Microbial abundance in dispersant-only treatments increased by a factor of 29, less than in WAF treatments but showing clear stimulation of growth by dispersant alone.
Marine oil snow, here defined as particles >0.5 mm in diameter, formed in WAF, dispersant-only, and CEWAF (± nutrients) microcosms, but differed in appearance, size, and abundance across treatments (SI Appendix). Microbial exopolymeric substances, including transparent exopolymer particles (TEP), are a matrix for marine snow formation (35). Oil-degrading bacteria produce TEP as biosurfactants (36). TEP production increased in the WAF microcosms relative to controls, underscoring the metabolic activities of oil-degrading bacteria (SI Appendix, Table S1). The abundance of TEP could not be quantified in dispersant treatments (SI Appendix) but extensive formation of oil snow was observed in the CEWAF + nutrients treatments (SI Appendix), inferring that TEP levels were likely elevated. The macroscopic particles observed in these experiments resembled marine oil snow observed in situ during the DWH oil spill (SI Appendix, Fig. S13 F and G). Catalyzed reporter deposition in combination with fluorescence in situ hybridization (CARD–FISH) revealed that Gammaproteobacteria and Alteromonadales, which includes the Colwellia, dominated microaggregate populations in CEWAF + nutrients treatments (SI Appendix, Fig. S13 P–R and SI Text). These findings suggest that Colwellia plays an important role in marine oil snow formation in the presence of dispersants.
Microbial Activity and Oil and Dispersant Degradation.
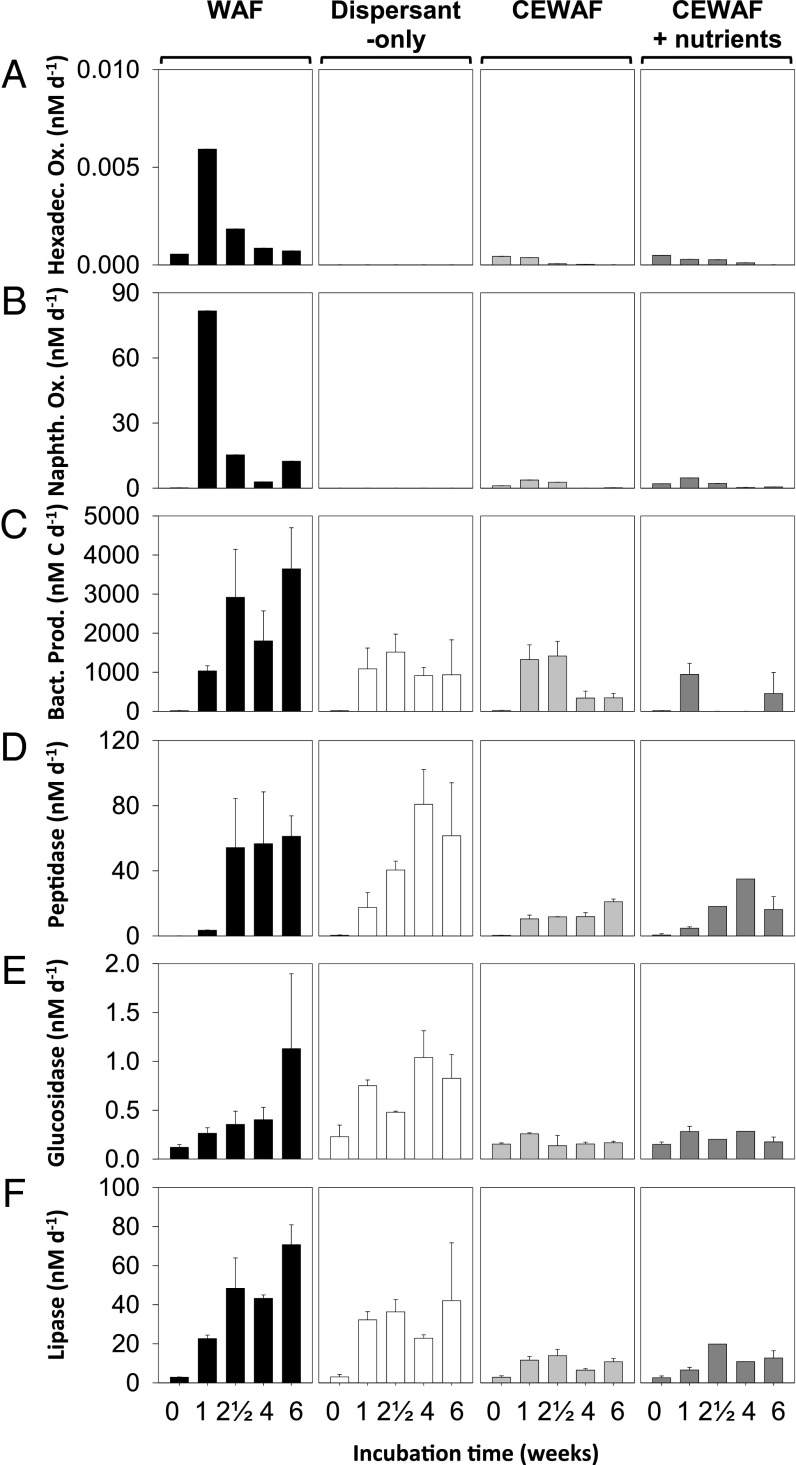
Dispersant addition did not enhance bacterial oil degradation or microbial activity in general, as reflected in rates of hydrocarbon oxidation, bacterial protein production, and exoenzyme activities. Radiotracer assays allowed direct quantification of alkane ([1-14C]-hexadecane) and polycyclic aromatic hydrocarbon (PAH) ([1-14C]-naphthalene) oxidation rates across treatments (SI Appendix) (Fig. 3 A and B and SI Appendix). Hexadecane oxidation rates were significantly reduced (T3 and T4: P = 0.004) in dispersant-only and CEWAF (± nutrients) treatments (Fig. 3A), implying that dispersants suppressed hexadecane degradation. Similarly, naphthalene oxidation rates in the WAF treatments were higher than those in dispersant-only and CEWAF (± nutrients) treatments (T3 and T4: P < 0.0001), inferring that dispersants did not stimulate microbial naphthalene degradation (Fig. 3B). When substrate turnover constants instead of concentration-dependent rates were considered, inhibition of hexadecane turnover remained apparent, whereas naphthalene turnover was comparable between WAF and CEWAF treatments (SI Appendix, Fig. S14). Together, these data show a clear concentration-independent inhibition of hexadecane oxidation by dispersants and further show that dispersants did not stimulate naphthalene biodegradation rates.
Fig. 3.
Microbial activity, hydrocarbon oxidation and enzymatic activities are not enhanced by dispersed oil (CEWAF ± nutrients). (A and B) Oxidation rates of 14C-hexadecane and 14C-naphthalene as model compounds for alkanes and PAHs degradation, respectively (SI Appendix, Table S1). (C) Rates of bacterial production increased up to three orders of magnitude in the 2 wk between the first and second sampling point (SI Appendix, Table S1). (D–F) Potential activities of peptidase, glucosidase, and lipase measured using fluorogenic substrate analogs were up to one order of magnitude higher in the WAF and dispersant-only compared with the CEWAF ± nutrients treatments. All data are illustrated as average of biological triplicates and error bars show SD of the mean (note that a lack of error bars indicates SDs were too small to be shown on the plot scale).
To validate the patterns of rates in these deepwater samples in another Gulf habitat, we determined hydrocarbon turnover of hexadecane and naphthalene in highly oil-contaminated (SI Appendix) surface seawater samples with and without dispersant addition (dispersant to seawater dilution was 1:100,000 vol/vol). Application of the radiotracer assay demonstrated that hexadecane turnover was inhibited significantly by dispersant amendments and that naphthalene turnover was not stimulated (SI Appendix, Fig. S15). These findings mirror those observed in the deepwater microcosms and underscore their broad relevance.
Further, in the deepwater experiments, not only were rates of hydrocarbon oxidation highest in the WAF treatments, rates of bacterial protein synthesis and exoenzyme activities indicative of potential bacterial degradation rates of carbohydrate- and protein-rich exopolysaccharides (EPSs) were also maximal in WAF treatments (Fig. 3C and SI Appendix, Table S1). All enzyme assays exhibited up to one order of magnitude higher activities in the WAF and dispersant-only treatments compared with the CEWAF (± nutrients) treatments (Fig. 3 D–F and SI Appendix, Table S1), underscoring that dispersant-only and CEWAF (± nutrients) did not stimulate bacterial production (T3 and T4: P < 0.001) relative to the WAF treatments.
Results from gas chromatography-mass spectrometry (GC-MS) and excitation/emission matrix spectra (EEMS) in deepwater samples further confirmed the patterns of hydrocarbon degradation across deepwater treatments. Concentrations of n-alkanes and hexadecane decreased more significantly in WAF treatments (SI Appendix, Fig. S16). In the WAF treatment, microorganisms preferentially degraded low molecular weight n-alkanes (<C20) relative to high molecular weight (≥C21) compounds and the isoprenoids, pristane and phytane. In the dispersant treatments, this pattern was not observed (SI Appendix, Fig. S17). The temporal changes in n-alkane concentration (SI Appendix, Fig. S16) supported the rate data (SI Appendix, Table S1) and emphasized the fact that oil degradation was highest in WAF treatments and that addition of CEWAF, even in the presence of additional nutrients, did not generate higher overall hydrocarbon degradation rates.
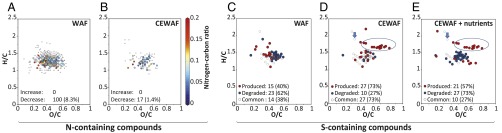
Biodegradation of anionic surfactant DOSS to α/β-ethyhexylsulfosuccinate (EHSS) occurs under aerobic conditions (37). In the dispersant-only treatment, a significant (P < 0.05) decrease (8%) of DOSS and an increase of EHSS (15%) was observed at T3 (SI Appendix, Fig. S18 A and B). The nonionic surfactants were consumed within 1 wk driving concentrations below detection (20 µg L−1; SI Appendix, Fig. S18 C and D). In the CEWAF (± nutrients) treatments, DOSS decreased significantly (P < 0.05) after 6 wk (SI Appendix, Fig. S18A). No significant change in EHSS concentrations was observed in CEWAF (± nutrients) treatments (SI Appendix, Fig. S18B), indicating that DOSS was converted to other products, an observation supported by formation of sulfur-containing compounds detected by ultrahigh-resolution Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR-MS) (38) (Fig. 4 D and E).
Fig. 4.
Dispersants impact microbial turnover of dissolved organic matter. Analysis of molecular-level patterns in Van Krevelen diagrams (hydrogen-to-carbon, H/C, and oxygen-to-carbon, O/C ratios; each circle represents a molecular formula). (A and B) Van Krevelen diagrams showing nitrogen-containing formulae (color scale depicts N/C ratios; open circles, formula contained no nitrogen). (C–E) Van Krevelen diagrams presenting changes in the presence or absence of sulfur-containing compounds (red circles, produced compounds, i.e., absent at T0 but present at T4; blue circles, degraded compounds, i.e., absent at T4 but present at T0; open circles, common compounds present at T0 and T4). DOSS (molecular formula C20H38O7S, marked by arrow) was present at T0 and T4. Several sulfur-containing compounds were exclusively produced in the dispersant-amended treatments (molecular formulae marked by an ellipse).
Molecular Characterization of Dissolved Organic Matter.
High-resolution FT-ICR-MS analysis provides a much more robust way to assess the molecular diversity of hydrocarbons in oil than does conventional GC-MS analyses (39, 40). The FT-ICR-MS results further suggest that significantly more oil-derived dissolved organic molecules were degraded in the WAF compared with CEWAF (± nutrients) treatments, again leading to the conclusion that more extensive biodegradation occurred in the absence of dispersant (Fig. 4 and SI Appendix, Fig. S19). Between 50% and 74% of the degraded compounds were highly unsaturated molecular formulae containing only the elements C, H, and O (SI Appendix, Fig. S19 A and B), which include the common aromatic hydrocarbons abundant in Macondo crude oil (39).
Oil-derived nitrogen-containing dissolved organic matter (DOM) compounds also decreased during the incubations (between 26% and 43% of the decreasing formulae, Fig. 4 A and B), agreeing with previous studies reporting that crude oil (40), including Macondo oil (39), contains numerous biodegradable polar and water-soluble organic nitrogen compounds. The WAF treatments exhibited the highest rates of degradation of oil-derived nitrogen-containing compounds (∼8% vs. ∼1% in the CEWAF treatment, Fig. 4 A and D) (38). In the WAF treatments, protein synthesis rates significantly exceeded those in the dispersant-amended treatments (T4: P = 0.0002), and a 31% decrease of seawater- and oil-derived dissolved organic nitrogen (DON) concentrations showed that the generation of microbial biomass required significant rates of nitrogen assimilation (SI Appendix, Table S1). The enhanced uptake of oil-derived organic nitrogen illustrates that oil can serve as an important nitrogen source when oil-degrading microbial communities are nitrogen limited (41).
Organic sulfur compounds are abundant in Macondo oil (39). The FT-ICR-MS results imply complex processing of sulfur-containing oil-derived and dispersant-derived DOM, including degradation of oil-derived sulfur compounds and formation of new organic sulfur compounds (Fig. 4 C–E). The FT-ICR-MS detected DOSS (molecular formula C20H38O7S; see arrow in Fig. 4 D and E) in all dispersant-amended treatments after 6 wk of incubation. The formation of new organic sulfur-compounds was particularly pronounced in the CEWAF (± nutrients) samples (circled area in Fig. 4 D and E), signaling that their formation was stimulated by dispersant addition. Elevated relative abundances of Colwellia in post-DWH discharge seawater along with enhanced expression of genes involved in the degradation of sulfur-containing organic matter (e.g., alkanesulfonate monooxygenase) (42) infer a role for Colwellia in organic sulfur cycling in situ during the DWH incident. The genome of C. psychrerythraea strain 34H has a remarkable potential for sulfur metabolism (43). Thus, we hypothesize that Colwellia played an important role in the observed turnover of DOSS-derived sulfur compounds as a result of their capability to metabolize the organic sulfur compounds in dispersants; they may have exhibited similar metabolic abilities in situ during the DWH incident.
Factors Regulating Microbial Activity.
To further unravel factors that regulate activity of key bacterial taxa, we determined statistically significant relationships between experimental conditions (geochemistry, cell counts, and microbial activity) and oligotype abundances. Distinct trends were apparent for Colwellia, Marinobacter, Oceaniserpentilla, and Cycloclasticus, as were correlations for specific oligotypes (SI Appendix, Table S2). Of the 24 detected Colwellia oligotypes, many correlated positively with concentrations of DOC (88%), ammonium (50%), cell counts (46%), and bacterial production (79%) as well as peptidase, glucosidase, and lipase (38–79%) activities. The majority of Colwellia oligotypes correlated negatively with concentration of total n-alkanes, hexadecane, naphthalene, and phenanthrene (71–79%), supporting the hypothesis that oligotypes of this taxon are predominantly responsible for dispersant breakdown. A considerable number of the 24 Marinobacter oligotypes correlated positively with cell counts (79%), bacterial production (79%), as well as peptidase and lipase (67–71%) activities. In contrast to Colwellia, Marinobacter oligotypes correlated positively to total petroleum concentrations (83%) and hexadecane oxidation (71%), highlighting a key role for these microorganisms in hexadecane degradation in the absence of dispersants. Oceaniserpentilla and Cycloclasticus oligotypes (30 and 31 types, respectively) correlated positively with nitrate and total n-alkanes, hexadecane, naphthalene, and phenanthrene (71–80%) concentrations. In addition, Cycloclasticus abundance positively correlated with naphthalene oxidation (61%), supporting their involvement in PAH degradation.
Evaluating the Utility of Dispersants.
Dispersants are used regularly as a response action after oil spills to disperse oil slicks, enhance the relative oil surface area in water, and to stimulate microbial hydrocarbon degradation. During the DWH incident, the deep-sea application of dispersants was unprecedented. Prior studies about microbial dispersant impacts generated confounding results (for review see ref. 25) most likely because nonspecific metrics were used, e.g., microbial cell counts or the production of CO2. Though changes in these two metrics reflect changes in microbial growth or activity, they do not specifically signify changes in hydrocarbon degradation rates. Further, it is quite possible that microorganisms stimulated by dispersant addition may outcompete natural hydrocarbon degraders. Thus, a direct quantification of hydrocarbon oxidation, accomplished here by direct determination of hydrocarbon oxidation using radiotracer assays in tandem with hydrocarbon quantification by GC-MS, is necessary to elucidate the impacts of dispersants on microbial populations and activities. The data obtained do not support dispersant stimulation of oil biodegradation, questioning the utility of dispersant application to pelagic ocean ecosystems.
Dispersant impacts on pelagic environments that are not impacted by natural oil seepage remain largely unknown. However, it seems unlikely that dispersants would stimulate hydrocarbon degradation in a system that lacks a substantial population of hydrocarbon degraders when they had no stimulatory effect in samples from a system that was primed for oil degradation (e.g., oil degraders account for 7–10% of the natural microbial population at site GC600) (18). In fact, the presence of dispersant selected against the most effective hydrocarbon degrading microorganisms (i.e., Marinobacter). This multidisciplinary data set strongly suggests that dispersants did not stimulate microbial hydrocarbon-degradation rates, as maximal oil-degradation rates were observed in the WAF treatments. Though we quantified degradation rates of only two hydrocarbons, hexadecane and naphthalene, biodegradation of other n-alkanes and PAHs could be similarly affected by dispersants. Quantification of the total crude oil also showed that the highest levels of oil biodegradation occurred in treatments without dispersants.
Whereas microbial activities in CEWAF (± nutrients) microcosms were comparable for 1 wk, rates were stimulated by nutrients in the later time points (e.g., CEWAF + nutrient hydrocarbon oxidation rates after 4 and 6 wk), suggesting progressive nutrient limitation. Clearly, the Gulf’s deepwater microbial community is able to degrade oil efficiently in the absence of dispersant. Therefore, caution is advised when considering dispersant applications as a primary response for future oil spills in deepwater environments similar to the Gulf. A full understanding of dispersant impacts on microbial populations requires immediate and careful evaluation of dispersant impacts across a variety of habitats.
Materials and Methods
Microcosm Setup and Sampling.
Seawater (160 L) was sampled from 1,178 m at an active natural hydrocarbon seep in the northern Gulf on March 7, 2013 (site GC600, latitude 27.3614, longitude −90.6018; SI Appendix, Fig. S1). After sampling, seawater was transferred to 20 L carboys and stored at 4 °C onboard the ship for 3 d. The carboys were transported at 4 °C to the laboratory at University of Georgia where the experiment and sampling were conducted in an 8 °C cold room. Setup and sampling of microcosms are described in detail in SI Appendix, SI Materials and Methods. In brief, 72 2-L glass bottles (1.8-L sample per bottle) were incubated on a roller table (SI Appendix, Fig. S2). Treatments (WAF, dispersant-only, and CEWAF ± nutrients) and controls (abiotic and biotic) were run in triplicate for each time point. Sampling (except for the CEWAF + nutrients treatment) was performed after 0 d (T0), 1 wk (T1), 2.5 wk (16 d; T2), 4 wk (T3), and 6 wk (T4); CEWAF + nutrients treatments were sampled at T0, T1, and T4. CEWAFs were prepared by mixing pasteurized seawater with oil and/or dispersants for 48 h at room temperature and subsequently subsampling CEWAFs, excluding contamination by oil or dispersants phases (SI Appendix). In addition, hydrocarbon turnover was determined in oil-contaminated surface seawater samples obtained along a transect from the Taylor Energy oil platform to the Mississippi River plume. Oil-contaminated surface seawater samples were used directly (untreated samples) or amended with dispersants (SI Appendix). Hydrocarbon turnover was analyzed using the newly adapted radiotracer assays (SI Appendix).
Molecular, Microbiological, and Geochemical Analyses.
Nutrients (nitrate, nitrite, phosphate, and ammonium), dissolved inorganic carbon, and oxygen as well as hydrocarbons (44) and dispersant concentrations were monitored during the course of the experiment (SI Appendix). Microbial community evolution and cell numbers were investigated for each sample using 16S rRNA amplicon Illumina sequencing (Bioproject accession PRJNA253405), computational oligotyping analysis (28), and total cell counts (SI Appendix). Activity measurements were performed using enzyme assays (peptidase, glucosidase, lipase) (45), 3H-leucine incorporation analysis (46), as well as the newly developed method for the analysis of 14C-hexadecane and 14C-naphthalene oxidation (SI Appendix). TEP analyses were carried out for controls and oil-only treatments (47) and CARD–FISH analysis (48) were performed in particular for microbial-aggregate formations in nutrient treatments (SI Appendix). Oil-derived hydrocarbons were extracted from water samples using a mixture of hexane:dichloromethane (1:1, vol/vol). After concentration, hydrocarbon compounds were identified and quantified by GC/MSD using conditions described previously (49) (SI Appendix). Analysis of the surfactant components of the dispersant Corexit was performed by LC-MS/MS as described elsewhere (13), with minor modification (SI Appendix). FT-ICR-MS was carried out to analyze DOM (50) (SI Appendix). Statistical analyses were used to unravel factors that drive microbial community evolution and microbial activities (SI Appendix).
Supplementary Material
Acknowledgments
We thank the captain and shipboard party of R/V Pelican cruise PE 529, especially Laura Lapham, for collecting the seawater used in the experiments; Julie Huber and Wade Jeffrey for sharing protocols for DNA extraction and WAF preparation, respectively; Kim Hunter for conducting nutrient and DOC analyses; Vladimir Samarkin for assistance during radiotracer assay development; and the Microbial Diversity Course (coordinated by Steven Zinder and Daniel H. Buckley) at the Marine Biological Laboratory, for providing supplies for CARD–FISH and access to the laser-scanning fluorescence microscope. This research was supported by a grant from British Petroleum/the Gulf of Mexico Research Initiative to support the “Ecosystem Impacts of Oil and Gas Inputs to the Gulf (ECOGIG)” consortium. P.M.M. also acknowledges funding from the National Science Foundation (OCE-1057683). This is ECOGIG contribution no. 347 and the data are archived at Gulf of Mexico Research Initiative Information and Data Cooperative data set number R1.x132.135:0012.
Footnotes
The authors declare no conflict of interest.
Data deposition: 16S rRNA amplicon Illumina sequencing data were deposited in the GenBank database (BioProject accession no. PRJNA253405).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1507380112/-/DCSupplemental.
References
- 1.Kvenvolden KA, Cooper CK. Natural seepage of crude oil into the marine environment. Geo-Mar Lett. 2003;23(3):140–146. [Google Scholar]
- 2.National Research Council . Committee on Oil in the Sea III: Inputs, Fates, and Effects. National Academies Press; Washington, DC: 2003. p. 280. [PubMed] [Google Scholar]
- 3.Widdel F, Knittel K, Galushko A. 2010. Anaerobic hydrocarbon-degrading microorganisms: An overview. Handbook of Hydrocarbon and Lipid Microbiology, eds Timmis KN, McGenity T, van der Meer JR, de Lorenzo V (Springer, Berlin), Vol 3, pp 1997–2021.
- 4.Head IM, Jones DM, Röling WFM. Marine microorganisms make a meal of oil. Nat Rev Microbiol. 2006;4(3):173–182. doi: 10.1038/nrmicro1348. [DOI] [PubMed] [Google Scholar]
- 5.National Commission on the BP Deepwater Horizon Oil Spill and Offshore Drilling 2011. The use of surface and subsea dispersants during the BP Deepwater Horizon oil spill. Available at 1.usa.gov/1qtH0YS. Accessed October 19, 2015.
- 6.Diercks A-R, et al. Characterization of subsurface polycyclic aromatic hydrocarbons at the Deepwater Horizon site. Geophys Res Lett. 2010;37(20):L20602. [Google Scholar]
- 7.Camilli R, et al. Tracking hydrocarbon plume transport and biodegradation at Deepwater Horizon. Science. 2010;330(6001):201–204. doi: 10.1126/science.1195223. [DOI] [PubMed] [Google Scholar]
- 8.Kessler JD, et al. A persistent oxygen anomaly reveals the fate of spilled methane in the deep Gulf of Mexico. Science. 2011;331(6015):312–315. doi: 10.1126/science.1199697. [DOI] [PubMed] [Google Scholar]
- 9.Valentine DL, et al. Propane respiration jump-starts microbial response to a deep oil spill. Science. 2010;330(6001):208–211. doi: 10.1126/science.1196830. [DOI] [PubMed] [Google Scholar]
- 10.Joye SB, MacDonald IR, Leifer I, Asper V. Magnitude and oxidation potential of hydrocarbon gases released from the BP oil well blowout. Nat Geosci. 2011;4(3):160–164. [Google Scholar]
- 11.Reddy CM, et al. Composition and fate of gas and oil released to the water column during the Deepwater Horizon oil spill. Proc Natl Acad Sci USA. 2012;109(50):20229–20234. doi: 10.1073/pnas.1101242108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kujawinski EB, et al. Fate of dispersants associated with the deepwater horizon oil spill. Environ Sci Technol. 2011;45(4):1298–1306. doi: 10.1021/es103838p. [DOI] [PubMed] [Google Scholar]
- 13.Place BJ, et al. Trace analysis of surfactants in Corexit oil dispersant formulations and seawater. Deep-Sea Res Pt II, in press. [DOI] [PMC free article] [PubMed]
- 14.US Environmental Protection Agency 2014. Questions and Answers on Dispersants. Available at 1.usa.gov/1MzgGnc. Accessed October 19, 2015. [PubMed]
- 15.Valentine DL, et al. Dynamic autoinoculation and the microbial ecology of a deep water hydrocarbon irruption. Proc Natl Acad Sci USA. July 31, 2012;109(50):20286–20291. doi: 10.1073/pnas.1108820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Redmond MC, Valentine DL. Natural gas and temperature structured a microbial community response to the Deepwater Horizon oil spill. Proc Natl Acad Sci USA. 2012;109(50):20292–20297. doi: 10.1073/pnas.1108756108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mason OU, et al. Metagenome, metatranscriptome and single-cell sequencing reveal microbial response to Deepwater Horizon oil spill. ISME J. 2012;6(9):1715–1727. doi: 10.1038/ismej.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleindienst S, et al. Diverse, rare microbial taxa responded to the Deepwater Horizon deep-sea hydrocarbon plume. ISME J. July 31, 2015 doi: 10.1038/ismej.2015.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lunel T, et al. 1996. Shoreline clean up during the Sea Empress incident: the role of surf washing (clay-oil flocculation), dispersants and bioremediation. Proceedings of the Nineteenth Arctic and Marine Oilspill Program (AMOP) Technical Seminar, (Environment Canada, Ottawa, Ontario), p 2v.
- 20.Mu J, Jin F, Ma X, Lin Z, Wang J. Comparative effects of biological and chemical dispersants on the bioavailability and toxicity of crude oil to early life stages of marine medaka (Oryzias melastigma) Environ Toxicol Chem. 2014;33(11):2576–2583. doi: 10.1002/etc.2721. [DOI] [PubMed] [Google Scholar]
- 21.Prince RC, Butler JD. A protocol for assessing the effectiveness of oil spill dispersants in stimulating the biodegradation of oil. Environ Sci Pollut Res Int. 2014;21(16):9506–9510. doi: 10.1007/s11356-013-2053-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.National Research Council 2005. Committee on Understanding Oil Spill Dispersants: Efficacy and Effects. Oil Spill Dispersants: Efficacy and Effects (National Academies Press, Washington, DC)
- 23.Smith JE. 1968. ‘Torrey Canyon’ Pollution and Marine Life. A Report by the Plymouth Laboratory of the Marine Biological Association of the United Kingdom (Cambridge Univ Press, Cambridge)
- 24.Peterson CH, et al. A tale of two spills. Biosci Biotechnol Biochem. 2012;62:461–469. [Google Scholar]
- 25.Kleindienst S, Paul JH, Joye SB. Using dispersants after oil spills: Impacts on the composition and activity of microbial communities. Nat Rev Microbiol. 2015;13(6):388–396. doi: 10.1038/nrmicro3452. [DOI] [PubMed] [Google Scholar]
- 26.Li D, et al. Dissolved organic carbon influences microbial community composition and diversity in managed aquifer recharge systems. Appl Environ Microbiol. 2012;78(19):6819–6828. doi: 10.1128/AEM.01223-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eiler A, Langenheder S, Bertilsson S, Tranvik LJ. Heterotrophic bacterial growth efficiency and community structure at different natural organic carbon concentrations. Appl Environ Microbiol. 2003;69(7):3701–3709. doi: 10.1128/AEM.69.7.3701-3709.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eren AM, et al. Oligotyping: Differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol. 2013;4(12):1111–1119. doi: 10.1111/2041-210X.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chakraborty R, Borglin SE, Dubinsky EA, Andersen GL, Hazen TC. Microbial response to the MC-252 oil and corexit 9500 in the Gulf of Mexico. Front Microbiol. 2012;3:357. doi: 10.3389/fmicb.2012.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singer E, et al. Genomic potential of Marinobacter aquaeolei, a biogeochemical “opportunitroph”. Appl Environ Microbiol. 2011;77(8):2763–2771. doi: 10.1128/AEM.01866-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huu NB, Denner EB, Ha DT, Wanner G, Stan-Lotter H. Marinobacter aquaeolei sp. nov., a halophilic bacterium isolated from a Vietnamese oil-producing well. Int J Syst Bacteriol. 1999;49(Pt 2):367–375. doi: 10.1099/00207713-49-2-367. [DOI] [PubMed] [Google Scholar]
- 32.Gauthier MJ, et al. Marinobacter hydrocarbonoclasticus gen. nov., sp. nov., a new, extremely halotolerant, hydrocarbon-degrading marine bacterium. Int J Syst Bacteriol. 1992;42(4):568–576. doi: 10.1099/00207713-42-4-568. [DOI] [PubMed] [Google Scholar]
- 33.Dyksterhouse SE, Gray JP, Herwig RP, Lara JC, Staley JT. Cycloclasticus pugetii gen. nov., sp. nov., an aromatic hydrocarbon-degrading bacterium from marine sediments. Int J Syst Bacteriol. 1995;45(1):116–123. doi: 10.1099/00207713-45-1-116. [DOI] [PubMed] [Google Scholar]
- 34.Hazen TC, et al. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science. 2010;330(6001):204–208. doi: 10.1126/science.1195979. [DOI] [PubMed] [Google Scholar]
- 35.Passow U. Formation of rapidly-sinking, oil-associated marine snow. Deep Sea Res Part II Top Stud Oceanogr. November 6, 2014 doi: 10.1016/j.dsr1012.2014.1010.1001. [DOI] [Google Scholar]
- 36.Gutierrez T, et al. Role of bacterial exopolysaccharides (EPS) in the fate of the oil released during the Deepwater Horizon oil spill. PLoS One. 2013;8(6):e67717. doi: 10.1371/journal.pone.0067717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campo P, Venosa AD, Suidan MT. Biodegradability of Corexit 9500 and dispersed South Louisiana crude oil at 5 and 25 °C. Environ Sci Technol. 2013;47(4):1960–1967. doi: 10.1021/es303881h. [DOI] [PubMed] [Google Scholar]
- 38.Seidel M, Kleindienst S, Dittmar T, Joye SB, Medeiros PM. Biodegradation of crude oil and dispersants in deep seawater from the Gulf of Mexico: Insights from ultra-high resolution mass spectrometry. Deep-Sea Res Pt II. May 30, 2015 doi: 10.1016/j.dsr2.2015.05.012. [DOI] [Google Scholar]
- 39.McKenna AM, et al. Expansion of the analytical window for oil spill characterization by ultrahigh resolution mass spectrometry: Beyond gas chromatography. Environ Sci Technol. 2013;47(13):7530–7539. doi: 10.1021/es305284t. [DOI] [PubMed] [Google Scholar]
- 40.Corilo YE, et al. Oil spill source identification by principal component analysis of electrospray ionization Fourier transform ion cyclotron resonance mass spectra. Anal Chem. 2013;85(19):9064–9069. doi: 10.1021/ac401604u. [DOI] [PubMed] [Google Scholar]
- 41.Lu Z, et al. Microbial gene functions enriched in the Deepwater Horizon deepsea oil plume. ISME J. 2012;6(2):451–460. doi: 10.1038/ismej.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rivers AR, et al. Transcriptional response of bathypelagic marine bacterioplankton to the Deepwater Horizon oil spill. ISME J. 2013;7(12):2315–2329. doi: 10.1038/ismej.2013.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Methé BA, et al. The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc Natl Acad Sci USA. 2005;102(31):10913–10918. doi: 10.1073/pnas.0504766102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joye SB, Bowles MW, Samarkin VA, Hunter KS, Niemann H. Biogeochemical signatures and microbial activity of different cold-seep habitats along the Gulf of Mexico deep slope. Deep Sea Res Part II Top Stud Oceanogr. 2010;57(21-23):1990–2001. [Google Scholar]
- 45.Hoppe HG. Significance of exoenzymatic activities in the ecology of brackish water: Measurements by means of methylumbelliferyl-substrates. Mar Ecol Prog Ser. 1983;11:299–308. [Google Scholar]
- 46.Smith DC, Azam F. A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar Microb Food Webs. 1992;6(2):107–111. [Google Scholar]
- 47.Passow U, Alldredge AL. A dye-binding assay for the spectrophotometric measurement of transparent exopolymer particles (TEP) Limnol Oceanogr. 1995;40(7):1326–1335. [Google Scholar]
- 48.Kleindienst S, Ramette A, Amann R, Knittel K. Distribution and in situ abundance of sulfate-reducing bacteria in diverse marine hydrocarbon seep sediments. Environ Microbiol. 2012;14(10):2689–2710. doi: 10.1111/j.1462-2920.2012.02832.x. [DOI] [PubMed] [Google Scholar]
- 49.Medeiros PM, Bícego MC. Investigation of natural and anthropogenic hydrocarbon inputs in sediments using geochemical markers. I. Santos, SP--Brazil. Mar Pollut Bull. 2004;49(9-10):761–769. doi: 10.1016/j.marpolbul.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 50.Seidel M, et al. Biogeochemistry of dissolved organic matter in an anoxic intertidal creek bank. Geochim Cosmochim Acta. 2014;140(0):418–434. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.