Significance
Mounting epidemiological and genetic evidence suggests that the disruption of circadian rhythms is at the origin of pathologies. It is known that people who are engaged in shift work and exhibit a shifted feeding schedule often develop a cohort of metabolic pathologies including diabetes, obesity, and metabolic syndrome. However, the molecular mechanisms that are at the origin of these pathologies are poorly understood. Using mice, we now revealed at the molecular level how metabolic alterations generated on shifting the eating schedule from the normal active phase to the rest phase creates a misalignment’ between the central and peripheral circadian clocks. Importantly, we demonstrate that this misalignment progressively induces a metabolic pathological syndrome similar to that observed in shift workers.
Keywords: circadian clocks misalignment, shift work, diabetes, metabolic syndrome, mouse
Abstract
The light-entrained master central circadian clock (CC) located in the suprachiasmatic nucleus (SCN) not only controls the diurnal alternance of the active phase (the light period of the human light-dark cycle, but the mouse dark period) and the rest phase (the human dark period, but the mouse light period), but also synchronizes the ubiquitous peripheral CCs (PCCs) with these phases to maintain homeostasis. We recently elucidated in mice the molecular signals through which metabolic alterations induced on an unusual feeding schedule, taking place during the rest phase [i.e., restricted feeding (RF)], creates a 12-h PCC shift. Importantly, a previous study showed that the SCN CC is unaltered during RF, which creates a misalignment between the RF-shifted PCCs and the SCN CC-controlled phases of activity and rest. However, the molecular basis of SCN CC insensitivity to RF and its possible pathological consequences are mostly unknown. Here we deciphered, at the molecular level, how RF creates this misalignment. We demonstrate that the PPARα and glucagon receptors, the two instrumental transducers in the RF-induced shift of PCCs, are not expressed in the SCN, thereby preventing on RF a shift of the master SCN CC and creating the misalignment. Most importantly, this RF-induced misalignment leads to a misexpression (with respect to their normal physiological phase of expression) of numerous CC-controlled homeostatic genes, which in the long term generates in RF mice a number of metabolic pathologies including diabetes, obesity, and metabolic syndrome, which have been reported in humans engaged in shift work schedules.
Under physiological conditions, the light-entrained central master circadian clock (CC), which is located in the suprachiasmatic nucleus (SCN), synchronizes the ubiquitous peripheral CCs (PCCs) and generates a diurnal alternance of phases of activity and rest, both of which are at the origin of rhythmic variations of gene expression, which are essential to maintain metabolic and behavioral homeostasis (1–3). It is well established that shifting the feeding time in the mouse from the “active” to the “rest” phase [so-called restricted feeding (RF)] leads to a 12-h shift in the expression of PCC components (4). As the SCN CC is not affected during RF (4), this situation leads to a misalignment between the diurnal active and rest phases and the expression of PCC components. We recently unveiled in mice the origin and the identity of the molecular signals through which RF leads to this 12-h shift in the expression of PCC components (5). However, the molecular mechanisms that confer to the SCN CC an insensitivity to RF, as well as the consequences of the misalignment between the PCCs and the master SCN CC on homeostasis, are still largely unexplored (3, 6). In the present study, we elucidated, at the molecular level, how the SCN CC is protected against the RF-induced shift of PCCs, which is induced by metabolic alterations (5), and how the misalignment between the master SCN CC and the PCCs generates a metabolic syndrome-like pathology, similar to that exhibited by shift workers (3, 6–9).
Results and Discussion
The Glycogen Synthase Kinase 3β Plays a Crucial Role Both in the Long-Term Maintenance of the RF-Induced CC Shift and Its Reversal on Return to Normal Feeding.
We recently elucidated (5) how the RF-induced decrease in insulin (INS) blood level during the active phase triggers an aberrant activation of nuclear receptor subfamily 1, group D, member 1 (Nr1d1/RevErbα) through phosphorylation by active glycogen synthase kinase 3β (GSK3β; Fig. 1F) (5). This RevErbα phosphorylation prevents its proteasome degradation (10) and is crucial for the repression of RORα/RevErbα response element (RORE)-DNA binding sequence (DBS)-containing genes (e.g., Bmal1, Cry1), which is a critical event in the initiation of the RF-induced shift of PCCs (5).
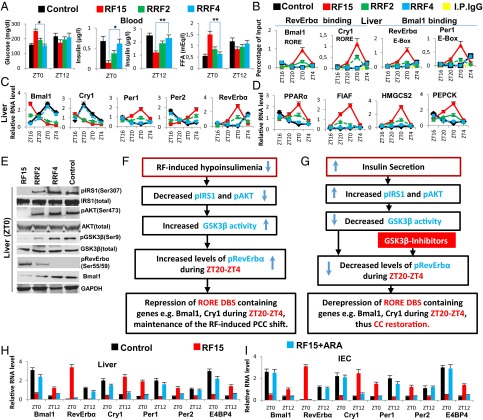
Fig. 1.
GSK3β-dependent RevErbα phosphorylation is critical for maintaining the RF-induced CC shift. (A) Levels of blood components in control, RF15 mice, and in mice after 2 and 4 d of reversal of RF (RRF2 and RRF4). (B) ChIP-qPCR assays in liver to analyze the RevErbα and the Bmal1 recruitment to their respective DBSs in the genes as indicated. (C) RNA transcript levels of CC components in liver of control, RF15, RRF2, and RRF4 mice. (D) RNA transcript levels of genes, as indicated, in the liver of control, RF15, RRF2, and RRF4 mice. (E) Immunoblot analyses, at ZT0, of control, RF15, RRF2, and RRF4 livers with indicated antibodies. (F) A schematic representation of how RF-induced PCCs shift is maintained. (G) A schematic representation of how restoration of insulin signaling leads to the reversal of RF-induced PCCs shift. (H) RNA transcript levels of CC components in liver of control, RF15, and RF15+ARA mice. (I) RNA transcript levels of CC components in IEC of control, RF15, and RF15+ARA mice. All values are mean ± SEM. *P < 0.05, **P < 0.01.
As this PCC shift is maintained in various tissues throughout the RF regime (Fig. 1C and Fig. S1 C and D and In RF Mice, the Expression of PCC-Controlled Output Genes Is Shifted by 12 h with Respect to the Diurnal Active and Rest Phases Controlled by the Central SCN CC), we explored the molecular basis of its maintenance and found that the active phase RF hypoinsulinemia is a recurring event during long-term RF regime (Fig. 1A and In RF Mice, the Expression of PCC-Controlled Output Genes Is Shifted by 12 h with Respect to the Diurnal Active and Rest Phases Controlled by the Central SCN CC), which, as expected (5, 11), leads to an increase in GSK3β-mediated phosphorylated RevErbα (pRevErbα) level at “Zeitgeber” (ZT) 0 (Fig. 1 E and F and Fig. S1K). Accordingly, in long-term RF mice, the level of pRevErbα bound to the RORE DBS of aryl hydrocarbon receptor nuclear translocator-like protein 1 (ARNTL1/Bmal1) and cryptochrome 1 (Cry1) is permanently increased during the ZT20–ZT0 period (Fig. 1B), thereby leading to a permanent PCCs shift (Fig. 1C and Fig. S1 C and D).
Fig. S1.
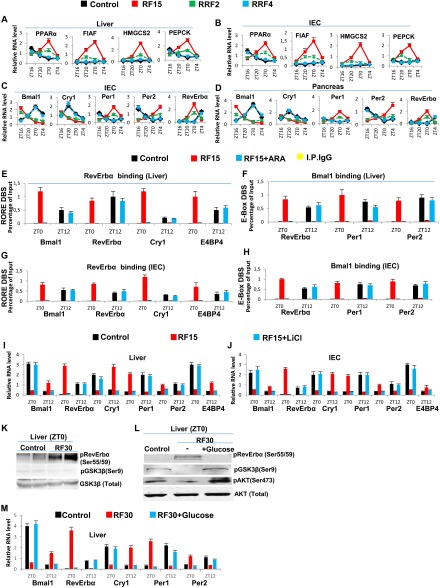
RF-induced increase in GSK3β activity plays a critical role in maintaining the peripheral CCs shift. (A and B) RNA transcript levels of genes, as indicated, in liver (A) and IECs (B) of control, RF15, and reversal of RF (RRF; RRF2 and RRF4) mice. (C and D) RNA transcript levels of CC components genes in IECs (C) and pancreas (D) of control, RF15, RRF2, and RRF4 mice. (E) ChIP-qPCR analysis of RevErbα recruitment to RORE DBSs present in genes, as indicated, in liver of RF mice, with or without i.p. administration of the GSK3β inhibitor ARA. (F) ChIP-qPCR analysis of Bmal1 recruitment to E-box DBS present in genes, as indicated, in liver of RF mice, with or without i.p. administration of the GSK3β inhibitor ARA. (G) ChIP-qPCR analysis of RevErbα recruitment to RORE DBSs present in genes, as indicated, in IECs of RF mice, with or without i.p. administration of the GSK3β inhibitor ARA. (H) ChIP-qPCR analysis of Bmal1 recruitment to E-box DBS present in genes, as indicated, in IECs of RF mice, with or without i.p. administration of the GSK3β inhibitor ARA. (I and J) RNA transcript levels of CC components in liver (I) and IECs (J) of RF mice with or without i.p. administration of LiCl. (K) Immunoblot analyses, at ZT0, of control and RF30 livers, with indicated antibodies. (L) Immunoblot analyses, at ZT0, of RF30 livers treated with or without i.p. administration of glucose, with indicated antibodies. (M) RNA transcript levels of CC components in the liver of control, RF30, and RF30+Glucose mice. All values are mean ± SEM.
Reciprocally, we found that, on a 4-day return to a normal active phase feeding after RF15 (RRF4 following RF15), the levels of several metabolic parameters (including INS) were normalized (Fig. 1 A and D and Fig. S1 A and B). Importantly, this treatment also normalized the expression of all CC components in liver, intestinal epithelial cells (IECs), and pancreas (Fig. 1C and Fig. S1 C and D). As expected (11), the RRF restoration of INS signaling during the active phase led to an increased level of pAKT, which through phosphorylation of GSK3β inhibited its activity, thereby reducing drastically the level of pRevErbα at ZT0 (Fig. 1 E and G). This RRF-induced decrease in pRevErbα level, which is correlated with its reduced binding to the RORE DBS of Bmal1 and Cry1 (Fig. 1B), then restored through derepression the normal (ZT20–ZT4) expression of these genes (5) and consequently the return to the original normal PCCs (Fig. 1C and Fig. S1 C and D). Most interestingly, we also found that increasing the INS blood level through administration of glucose for 4 consecutive d to RF30 mice at ZT18 (after removal of food at ZT12), activated pAKT (due to the INS increase) and led to a reduction in pRevErbα level at ZT0 (Fig. S1L), thereby normalizing the PCC (Fig. S1 L and M). Taken altogether, these experiments demonstrate that the restoration of INS signaling in RF mice during the active phase triggers the PCC normalization (Fig. 1 A, E, and G).
Even though, on RF cessation, RF mice do return to apparent normality in a few days (see above), it is noteworthy that such a return can also be achieved under continuous RF regime by administration of GSK3β inhibitors, which prevents RevErbα phosphorylation (Fig. 1G). Indeed, we found that pharmacological inhibition of GSK3β activity, through a 5-day administration to RF mice (at ZT18) of either one of the two inhibitors of GSK3β activity, AR-A014418 (ARA) (12) and LiCl (10), is sufficient to prevent the RF-induced ZT0 binding of RevErbα to the RORE DBSs present in PCC components Fig. S1 E and G), thereby restoring normal PCCs under RF conditions (Fig. 1 H and I and Fig. S1 F and H–J). Most notably, as pharmacological inhibitors of GSK3β (e.g., LiCl and valproic acid, among others) are used in clinics to treat anxiety and mood disorders, it is tempting to speculate that, under shift work conditions, their use may prove to be beneficial to normalize the misalignment of circadian clocks, thereby preventing the progression of metabolic pathologies toward insulin resistance.
The Lack of PPARα and Glucagon Receptors Immunizes the SCN Central Clock Against RF-Induced Metabolic Perturbations.
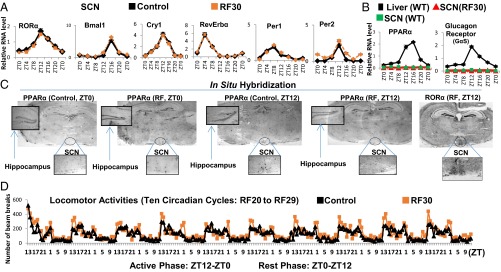
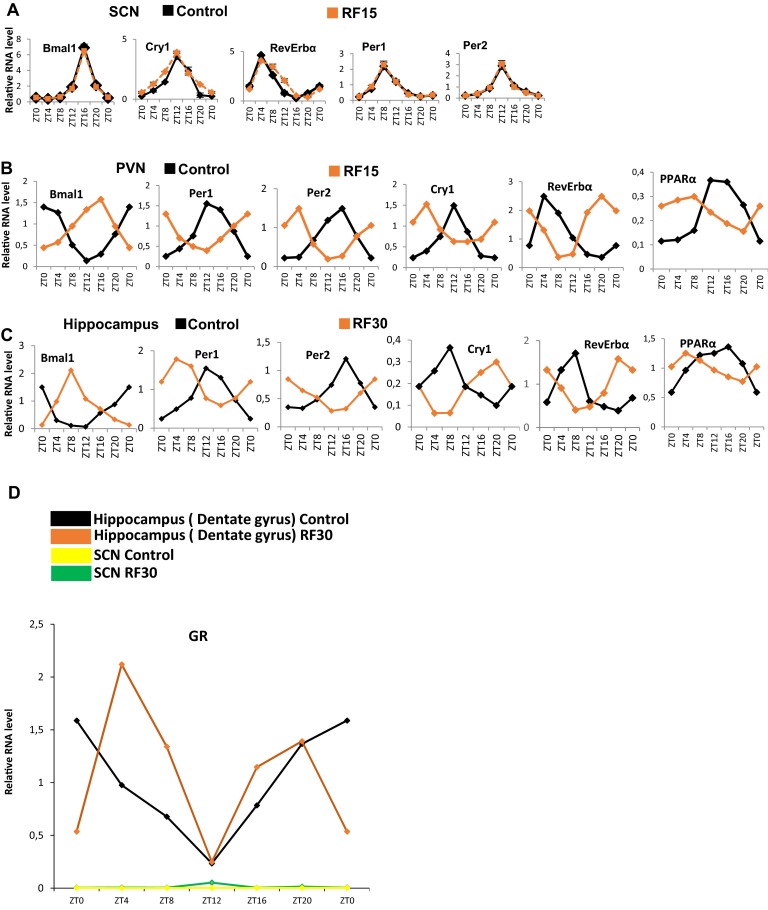
The SCN expression of Period 1 and 2 (Per1 and Per2) was reported to be unaltered after 9 d of RF (RF9) (4). Of note, transcript analyses on RF15 and RF30 failed to detect any variation in the expression of not only Per1 and Per2, but also of the other circadian clock components RORα, Bmal1, Cry1, and RevErbα (Fig. 2A and Fig. S2A). This raised the possibility that the SCN CC may either lack or be unresponsive (due to the absence of their transducers) to the RF metabolic signals triggering the shift of PCCs (5). Most notably, on quantitative RT-PCR (qRT-PCR) of microdissected SCN, we could not detect any transcripts for the peroxisome proliferator-activated receptor alpha (PPARα) and glucagon (subunit GαS) receptors, which orchestrate the CC shift in peripheral tissues (Fig. 2B) (5). Accordingly, in situ hybridization of SCN sections of both control and RF mice failed to reveal PPARα transcripts (Fig. 2C). In marked contrast, PPARα expression was readily detected in the hippocampus and paraventricular nucleus (PVN), in which a peripheral 12-h CC shift was also observed (Fig. 2B and Fig. S2 B and C). These CC shifts in PVN and hippocampus are noteworthy, as (i) the PVN, which is connected with the SCN via both afferent and efferent projections, has been proposed to function as a “relaying center” for SCN-generated signals (13), and (ii) the proper functioning of peripheral circadian clocks in the hippocampus is known to facilitate memory formation (14), which suggests that RF could interfere with memory processes (15).
Fig. 2.
The SCN CC is insensitive to the RF-induced metabolic alterations. (A) RNA transcript levels of CC components in the SCN of control and RF30 mice. (B) RNA transcript levels of PPARα and glucagon receptors in the SCN of control and RF30 mice. Expression of these genes in liver was evaluated as a control. (C) In situ hybridization of brain sections of control and RF mice with PPARα riboprobes. RORα expression was used as a marker for localizing the SCN. (D) Actimetric analyses of the circadian locomotor activity in control and RF30 mice. All values are mean ± SEM.
Fig. S2.
The central SCN CC is unaffected by the RF. (A) RNA transcript levels of CC components in the SCN of control and RF15 mice. (B) RNA transcript levels of CC components in the PVN of control and RF15 mice. (C) RNA transcript levels of CC components genes in the hippocampus (dentate gyrus) of control and RF30 mice. (D) RNA transcript level of GR in the hippocampus (dentate gyrus) and SCN of control and RF30 mice. All values are mean ± SEM.
Importantly and in keeping with the established role of the SCN in controlling the diurnal alternance of active (ZT12–ZT0) and rest (ZT0–ZT12) phases (1–3), analyses of the locomotor activity of RF mice revealed that, on RF, this alternance was unaffected (Fig. 2D). Thus, by excluding the expression of two transducers (PPARα and glucagon receptor) necessary to induce the PCC shift (5), the neurons of the SCN ensure the constancy of the master SCN CC during RF, as well as the proper control of the diurnal alternance of active and rest phases (1–3).
From the teleological perspective, this insensitivity of the SCN CC during a RF starvation-like state is an expected necessity, as the SCN-controlled active phase has to occur during the mouse dark cycle (the human light cycle), even though, due to the misalignment of the PCCs, this active period corresponds to the rest phase in peripheral tissues. This evolutionary design, which maintains the original active phase (wakefulness) in the SCN CC, as well as the rest phase (sleep) during RF-like conditions, is indeed advantageous as it provides the opportunity for a starving organism to efficiently look for food during the normal active phase, thereby increasing the chance to put an end to starvation and to readily realign the PCCs with the unchanged SCN master clock, as soon as feeding is restored.
It was previously suggested that the RF insensitivity of the SCN CC is related to the lack of the glucocorticoid (GC) receptor (GR) in SCN (16), as such a lack (that we confirmed; Fig. S2D) would prevent a SCN CC shift through an alteration of Per1/Per2 expression on RF extra-corticosterone production (16, 17). Our present results do not exclude this possibility. However, we found that the lacks of both PPARα and glucagon receptors in SCN are sufficient on their own to prevent a shift of the whole SCN CC on RF, as taken together they prevent the RF-induced CC shifts of RevErbα through PPARα increase and of Per1/Per2 through glucagon/CREB activation (5).
In RF Mice, the Expression of PCC-Controlled Output Genes Is Shifted by 12 h with Respect to the Diurnal Active and Rest Phases Controlled by the Central SCN CC.
Under physiological conditions, the level of expression of nearly 10–15% of the genes expressed in mice is controlled by PCCs, such that specific sets of genes are selectively expressed during the active and rest phases, their expression being directly controlled by CC components or indirectly by their output genes (1–3). Notably, it has been demonstrated that the normal expression of RORE DBS-bearing genes in different tissues is at their zenith during the circadian active phase (while at nadir during the rest phase), whereas E- and D-Box DBS-bearing genes are at their zenith during the circadian rest phase (18–20).
As the RF CC shifts occur in peripheral tissues, but not in SCN, we investigated whether the misalignment between the unchanged SCN CC-controlled genes expressed during the active and rest phases will lead to an “out of phase” expression of PCC-controlled output genes, i.e., whether the PCC active phase genes would be expressed during the SCN rest phase, whereas the PCC rest phase genes would be expressed during the SCN active phase. A bioinformatic search in the mouse genome for genes harboring D-Box DBS in their promoter-enhancer regions revealed numerous candidates, of which ∼2,000 have human orthologs (Dataset S1 and SI Methods). Among these, we chose 40 D-Box–containing genes having known functions. We also chose 40 RORE DBS-containing genes all having known homeostatic functions (19). When analyzed at RF15 in liver and IECs the circadian expression of such RORE DBS- and D-Box–containing genes, which in WT mice also displayed a circadian variation (Tables S1–S4), confirmed a 12-h shift (misalignment) in the expression, such that the expression of the normally active phase-restricted RORE genes was shifted to the rest phase (Tables S1 and S2), whereas the initially rest phase-restricted D-Box genes were expressed during the active phase (Tables S3 and S4). Importantly, this misaligned expression of numerous CC-controlled output genes (Tables S1–S4), including those of endocrine factors (INS, IGF1, and FGF21), key transcription factors (c-Jun, IRF3, ATF5, and FXRβ), critical enzymes (NAMPT, Hsd3b5, FAS, and HMGCR), receptors, and transporters (INSR, IRS2, Glut2, and TLRs), all controlling essential physiological homeostatic processes, sets the stage for the development of RF-associated pathologies (see below).
Table S1.
Circadian expression of mouse liver genes containing RORE DBS after RF15
| Genes | Function | Circadian expression | |
| Ad libitum feeding (ZT20–4 > ZT12–16) | RF15 (ZT12–16 > ZT20–4) | ||
| ATF5 | Transactivator | + | + |
| BAX | Apoptosis | + | + |
| c-Jun | Transactivator | + | + |
| CDC25c | Cell cycle regulator | + | + |
| CDKN1A | Cell cycle regulator | + | + |
| CDK8 | Transcriptional coactivator | + | + |
| CHEK1 | Regulator of DNA damage signaling | + | + |
| CREB1 | Transactivator | = | = |
| CSNK2A1 | Casein kinase2, α1 subunit | = | = |
| CSNK1D | Casein kinase1, δ subunit | = | = |
| CUL5 | Ubiquitin ligase | = | = |
| CYFIP1 | FMR1 interacting protein | + | + |
| CYP8B1 | Bile acid synthesis | + | + |
| DAG1 | Structural maintenance | + | + |
| DR5 | Apoptosis | + | + |
| DLAT | Member of pyruvate dehydrogenase complex | = | = |
| ELOVL3 | Fatty acid synthesis | + | + |
| FGF18 | Osteocyte differentiation | = | = |
| GCK | Glucose metabolism | + | + |
| GCKR | Glucose metabolism | + | + |
| HSD3B5 | Cholesterol metabolism | + | + |
| HIC53 | Phosphatidic acid phosphatase | = | = |
| IGFBP4 | IGF1 signaling | + | + |
| INSIG2A | Regulator of lipogenesis | + | + |
| IRF1 | Transactivator | + | + |
| KLF15 | Transactivator | + | + |
| LIPC | Lipoprotein metabolism | + | + |
| LRIG1 | Regulator of ERBB signaling | = | = |
| MGA | Transactivator | = | = |
| NOD2 | Receptor for inflammosome activation | + | + |
| PDK4 | Glucose metabolism | + | + |
| PEX3 | Peroxisomal biogenesis | + | + |
| PEX16 | Peroxisomal biogenesis | + | + |
| POFUT1 | Fucosyl transferase | + | + |
| RAF1 | Regulator of MAPK signaling | = | = |
| RAB25 | Endocytosis | = | = |
| ROCK2 | Rho-associated kinase | = | = |
| U1 | Spliceosome assembly | = | = |
| UBE2QL1 | Ubiquitin conjugating enzyme | = | = |
| WDR5B | WD-40 repeat protein, subunit 5B | = | = |
+, genes with circadian variation in the transcript levels; =, represent genes with no circadian variation in the transcript levels.
Table S4.
Circadian expression of mouse IEC genes containing D-Box DBS after RF15
| Genes | Function | Circadian expression | |
| Ad libitum feeding (ZT20–4 > ZT12–16) | RF15 (ZT12–16 > ZT20–4) | ||
| AIF | Apoptosis | + | + |
| ATG14 | Autophagy | + | + |
| CHERP | Calcium homeostasis | = | = |
| CGGBP1 | CGG-binding protein | + | + |
| CLPTM1 | Development of palate | = | = |
| DUSP12 | Negative regulator of MAPK signaling | + | + |
| DUSP22 | Negative regulator of MAPK signaling | + | + |
| ELP4 | Regulator of transcriptional elongation | = | = |
| ELVOL7 | Fatty acid synthesis | = | = |
| ERCC3 | DNA-repair | = | = |
| FASL | Apoptosis | + | + |
| GCH1 | GTP-hydrolysis | = | = |
| GSK3β | Signal transduction | = | = |
| HSF2 | Transactivator | + | = |
| KDM3A | Lysine demethylase | = | = |
| LARP7 | La ribonucleoprotein 7 | = | = |
| MRPL1 | Ribosome biogenesis | = | = |
| MRPL3 | Ribosome biogenesis | = | = |
| MRC1 | Mannose receptor, C-type | = | = |
| MTND3 | Mitochondrial NAD dehydrogenase | + | + |
| MUT | Methylmalonyl CoA mutase | + | + |
| NDUFB9 | NADH dehydrogenase subunit | + | + |
| NOL7 | Nucleolar organizer | = | = |
| OSBP | Oxysterol binding | + | + |
| PAIP1 | Regulator of transcription termination | + | + |
| PDK1 | Glucose metabolism | + | + |
| PER3 | Regulator of circadian clock | + | + |
| PHF17 | Transcriptional regulation | + | + |
| PTEN | Negative regulator of PI3K signaling | + | + |
| RAP1GDS1 | Rap1 GTP-GDP exchange stimulator | + | + |
| RPS27A | Ribosomal protein | + | + |
| SMAD5 | Transactivator | = | = |
| SPT16 | Suppressor of Ty16 | = | = |
| SUMO2 | Posttranslational modification | = | = |
| TEF | Transactivator | + | + |
| TGM1 | Glutamine metabolism | = | = |
| TGFBR1 | TGFβ receptor | = | = |
| TSG101 | Tumor suppressor | = | = |
| VDBP | Vitamin (D3) metabolism | = | = |
| WEE1 | Cell cycle regulator | + | + |
+, genes with circadian variation in the transcript levels; =, represent genes with no circadian variation in the transcript levels.
Table S2.
Circadian expression of mouse IEC genes containing RORE DBS after RF15
| Genes | Function | Circadian expression | |
| Ad libitum feeding (ZT20–4 > ZT12–16) | RF15 (ZT12–16 > ZT20–4) | ||
| ATF5 | Transactivator | = | = |
| BAX | Apoptosis | + | + |
| c-Jun | Transactivator | + | + |
| CDC25c | Cell cycle regulator | + | + |
| CDKN1A | Cell cycle regulator | + | + |
| CDK8 | Transcriptional coactivator | + | + |
| CHEK1 | Regulator of DNA damage signaling | + | + |
| CREB1 | Transactivator | = | = |
| CSNK2A1 | Casein kinase 2, α1 subunit | = | = |
| CSNK1D | Casein kinase 1, δ subunit | = | = |
| CUL5 | Ubiquitin ligase | = | = |
| CYFIP1 | FMR1 interacting protein | + | + |
| CYP8B1 | Bile acid synthesis | + | + |
| DAG1 | Structural maintenance | = | = |
| DR5 | Apoptosis | + | + |
| DLAT | Member of pyruvate dehydrogenase complex | = | = |
| ELOVL3 | Fatty acid synthesis | + | + |
| FGF18 | Osteocyte differentiation | = | = |
| GCK | Glucose metabolism | + | + |
| GCKR | Glucose metabolism | + | + |
| HSD3B5 | Cholesterol metabolism | + | + |
| HIC53 | Phosphatidic acid phosphatase | = | = |
| IGFBP4 | IGF1 signaling | = | = |
| INSIG2A | Regulator of lipogenesis | + | + |
| IRF1 | Transactivator | + | + |
| KLF15 | Transactivator | + | + |
| LIPC | Lipoprotein metabolism | + | + |
| LRIG1 | Regulator of ERBB signaling | + | + |
| MGA | Transactivator | = | = |
| NOD2 | Receptor for inflammosome activation | + | + |
| PDK4 | Glucose metabolism | + | + |
| PEX3 | Peroxisomal biogenesis | + | + |
| PEX16 | Peroxisomal biogenesis | + | + |
| POFUT1 | Fucosyl transferase | = | + |
| RAF1 | Regulator of MAPK signaling | = | = |
| RAB25 | Endocytosis | = | = |
| ROCK2 | Rho-associated kinase | = | = |
| U1 | Spliceosome assembly | = | = |
| UBE2QL1 | Ubiquitin conjugating enzyme | = | = |
| WDR5B | WD-40 repeat protein, subunit 5B | = | = |
+, genes with circadian variation in the transcript levels; =, represent genes with no circadian variation in the transcript levels.
Table S3.
Circadian expression of mouse liver genes containing D-Box DBS after RF15
| Genes | Function | Circadian expression | |
| Ad libitum feeding (ZT20–4 > ZT12–16) | RF15 (ZT12–16 > ZT20–4) | ||
| AIF | Apoptosis | + | + |
| ATG14 | Autophagy | + | + |
| CHERP | Calcium homeostasis | = | = |
| CGGBP1 | CGG-binding protein | = | = |
| CLPTM1 | Development of palate | = | = |
| DUSP12 | Negative regulator of MAPK signaling | + | + |
| DUSP22 | Negative regulator of MAPK signaling | + | + |
| ELP4 | Regulator of transcriptional elongation | + | + |
| ELVOL7 | Fatty acid synthesis | + | + |
| ERCC3 | DNA-repair | + | + |
| FASL | Apoptosis | + | + |
| GCH1 | GTP-hydrolysis | + | + |
| GSK3β | Signal transduction | = | = |
| HSF2 | Transactivator | = | = |
| KDM3A | Lysine demethylase | = | = |
| LARP7 | La ribonucleoprotein 7 | + | + |
| MRPL1 | Ribosome biogenesis | = | = |
| MRPL3 | Ribosome biogenesis | = | = |
| MRC1 | Mannose receptor, C-type | = | = |
| MTND3 | Mitochondrial NAD dehydrogenase | + | + |
| MUT | Methylmalonyl CoA mutase | + | + |
| NDUFB9 | NADH dehydrogenase subunit | + | + |
| NOL7 | Nucleolar organizer | = | = |
| OSBP | Oxysterol binding | + | + |
| PAIP1 | Regulator of transcription termination | + | + |
| PDK1 | Glucose metabolism | + | + |
| PER3 | Regulator of circadian clock | + | + |
| PHF17 | Transcriptional regulation | + | + |
| PTEN | Negative regulator of PI3K signaling | = | = |
| RAP1GDS1 | Rap1 GTP-GDP exchange stimulator | + | = |
| RPS27A | Ribosomal protein | + | + |
| SMAD5 | Transactivator | = | = |
| SPT16 | Suppressor of Ty16 | + | + |
| SUMO2 | Posttranslational modification | = | = |
| TEF | Transactivator | + | + |
| TGM1 | Glutamine metabolism | = | = |
| TGFBR1 | TGFβ receptor | = | = |
| TSG101 | Tumor suppressor | + | + |
| VDBP | Vitamin (D3) metabolism | = | = |
| WEE1 | Cell cycle regulator | + | + |
+, genes with circadian variation in the transcript levels; =, represent genes with no circadian variation in the transcript levels.
Mice Selectively Fed During the Rest Phase Display a Metabolic Syndrome-Like Pathology due to the Misalignment of the Peripheral Clocks with the Central SCN CC.
The physiological alignment of PCCs with the master central SCN CC has emerged as an important factor for the homeostatic maintenance of an organism, as misalignments of PCCs with the SCN CC, such as those associated with shift work and RF, both of which correspond to activities performed during the SCN CC-controlled rest phase while being physiologically controlled and exerted during the SCN CC-controlled active phase, have been associated with increased risk of developing diabetes (hypoinsulinemia, hyperglycemia, reduced glucose tolerance), high free fatty acid (FFA) level, hypertriglyceridemia, obesity, and metabolic syndrome (6–9, 21–23). It is, however, largely unknown how these pathologies are mechanistically related to the PCC/SCN CC misalignments (3, 6–9). Most interestingly, our present study reveals, at the molecular level, that all of these metabolic pathologies eventually develop in RF mice, as a consequence of the RF-induced 12-h misalignment between the PCCs and rest and active phases of activity of the SCN CC.
A Misalignment-Induced Decrease in INS Production Leads to Diabetes in RF Mice.
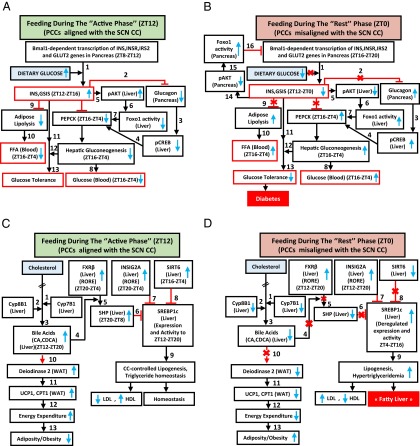
Under ad libitum feeding during the active phase (the ZT8–ZT12 period; Fig. 4A), the Bmal1 activity of the pancreatic CC transcribes INS, insulin receptor (INSR), insulin receptor substrate 2 (IRS2), and glucose transporter 2 (GLUT2) genes, thus aligning INS synthesis and signaling with the ZT12 feeding time (Fig. 3I) (24) and enabling maximal postprandial (ZT12–ZT16) glucose-stimulated INS secretion (GSIS) (Fig. 3A). This GSIS (step 1 in Fig. 4A), at the beginning of the active phase, is critical to prevent lipolysis within the adipose tissue (11), thereby decreasing the FFA blood level (Fig. 3A and steps 9–10 in Fig. 4A). Moreover, the INS blood increase (i) counteracts glucagon secretion to prevent pCREB activity in liver (steps 2–4 in Fig. 4A) and (ii) activates pAKT (Fig. 1E and Fig. S1L) to inactivate the FOXO1 protein in liver (11) (steps 5–7 in Fig. 4A). This combined decrease in pCREB and FOXO1 reduces the PEPCK expression in liver, thereby preventing hepatic gluconeogenesis (ZT16–ZT4) and maintaining the physiological glucose blood level (step 8 in Fig. 4A), which ensures glucose tolerance (steps 11–13 in Fig. 4A).
Fig. 4.
A misalignment of PCCs with the SCN CC under RF regime progressively induces a metabolic syndrome. (A) Schematic representation of how PCCs alignment with the SCN CC maintains INS, glucose, and FFA homeostasis. (B) A schematic representation of how PCCs misalignment with the SCN CC in RF mice leads to hypoinsulinemia, hyperglycemia, and increased FFA level. (C) A schematic representation of how PCCs alignment with the SCN CC maintains lipogenesis and TG level and prevents adiposity. (D) A schematic representation of how PCCs misalignment with the SCN CC in RF mice leads to an increase in lipogenesis, hypertriglyceridemia, and adiposity.
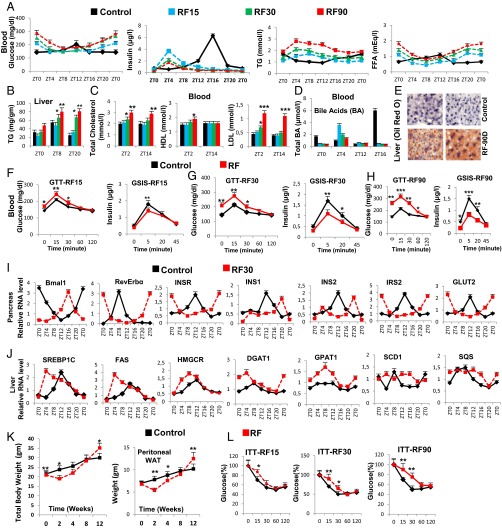
Fig. 3.
Prolonged feeding during the rest phase leads to the development of diabetes and fatty liver. (A) Levels of blood components, as indicated, after RF15, RF30, and RF90. (B) As in A, but measuring TG levels in liver. (C) As in A, but measuring blood levels of total, HDL, and LDL cholesterol. (D) As in A, but measuring total bile acids. (E) Oil red O staining to detect TG deposition in control and RF90 liver. (F–H) Glucose tolerance tests (GTTs) and GSIS after RF15 (F), RF30 (G), and RF90 (H). (I) RNA transcript levels of genes, as indicated, in pancreas of control and RF30 mice. (J) RNA transcript levels of genes as indicated in liver of control and RF30 mice. (K) Total body weight and weight of the peritoneal adipose tissue in control and RF mice. (L) Insulin tolerance tests (ITT) after RF15, RF30, and RF90 days. All values are mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
We found by RF7, that the CC shift was complete in pancreas, which resulted in a permanent misalignment in the expression of genes critically involved in insulin synthesis and signaling (see below) and led to a recurrent hypoinsulinemia (in RF mice) during the active phase, thereby accounting for all of the pathological metabolic features typically associated with diabetes (Fig. 4B). Due to this RF CC shift, Bmal1-dependent transcriptions in pancreas were shifted to the ZT16–ZT20 period (Fig. 3I), at a time where dietary glucose is unavailable due to the rest phase feeding at ZT0 (Fig. 4B).The ensuing RF-hypoinsulinemia (ZT12–ZT0; Fig. 3A) induced the FOXO1 activity in pancreas, as a consequence of a reduction in pAKT level (11, 25, 26), thus leading to a repression (25) of the transcription of the INS gene at ZT0 (steps 14–16 in Fig. 4B and Fig. 3I) and to a decreased postprandial GSIS in RF mice (Fig. 3A). Moreover, as a result of the RF hypoinsulinemia, there was an increase in PEPCK expression due to enhanced pCREB and FOXO1 activity (Fig. S3D and steps 2–7 in Fig. 4B), which resulted in RF hyperglycemia (Fig. 3A and step 8 in Fig. 4B). Taken together, this RF hypoinsulinemia and hyperglycemia account for the reduced glucose tolerance in RF mice (Fig. 3 F–H and steps 11–13 in Fig. 4B). However, even at RF90, an INS-resistant state was not achieved, although there was a progressive deterioration from RF15 to RF90 (Fig. 3L). Importantly, the reduction in INS level in RF mice also accounted for the inability of these mice to suppress lipolysis in adipose tissue and the resulting increase in FFA blood level (Fig. 3A and steps 9–10 in Fig. 4B).
Fig. S3.
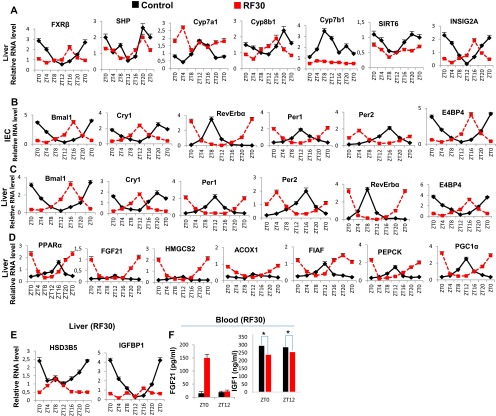
Metabolic features of RF30 mice. (A) RNA transcript levels of genes, as indicated, in liver of control and RF30 mice. (B) RNA transcript levels of CC components in IECs of control and RF30 mice. (C) RNA transcript levels of CC components liver of control and RF30 mice. (D) RNA transcript levels of genes, as indicated, in liver of control and RF30 mice. (E) RNA transcript levels of HSD3B5 and IGFBP1 in liver of control and RF30 mice. (F) FGF21 and IGF1 levels in blood of control and RF30 mice. All values are mean ± SEM. *P < 0.05.
In summary, taken together, our present results establish how an RF-induced misalignment of PCCs and SCN CC triggers the development of diabetes (hypoinsulinemia, hyperglycemia, reduced glucose tolerance) and increased FFA level through misexpression during the active phase of PCC genes normally expressed during the rest phase.
A Misalignment-Induced Increase in SREBP1c Leads to Hypertriglyceridemia and Hypercholesterolemia in RF Mice.
Because, in addition to diabetes, shift workers also develop hypertriglyceridemia and hypercholesterolemia (6, 7, 22, 23), we investigated whether these metabolic alterations could also originate from RF-induced clock misalignment. Under ad libitum feeding during the active phase (ZT12–ZT20; Fig. 4C), the liver CC components are involved in the control of expression and activity of SREBP1c, which is well known to act as the master transcriptional regulator of triglyceride (TG) synthesis and lipogenesis (2, 3, 27). Multiple mechanisms are involved in this regulation. (i) In liver, cholesterol is converted into bile acids (BAs), cholic acid (CA), and cheno deoxy-cholic acid (CDCA) through the action of two enzymes Cyp8B1 and Cyp7B1, respectively (steps 1–2 in Fig. 4C); this blood BA postprandial increase (Fig. 3D and step 3 in Fig. 4C) enables the RORE-controlled farnesoid X receptor (FXR) receptor (28) (Fig. S3A) to activate transcription of the small heterodimer partner (SHP) repressor during the ZT20–ZT8 period (Fig. S3A and steps 4–5 in Fig. 4C), which in turn inhibits the expression of SREBP1c (29) (step 6 in Fig. 4C). (ii) The nuclear import of SREBP1c from endoplasmic reticulum and Golgi vesicles is inhibited by INSIG2A (30), the expression of which is also controlled (30) by a RORE DBS (ZT20–ZT4, Fig. S3A and step 7 in Fig. 4C). (iii) The chromatin recruitment of SREBP1c to DBSs present in multiple genes is inhibited by SIRT6 (31) (step 8 in Fig. 4C). Taken together, these mechanisms ensure a circadian pattern for SREBP1c expression and activity, which maintains homeostatic TG levels in blood and tissues (steps 6–9 in Fig. 4C).
In contrast, we found that in RF liver there was a decrease in Cyp7B1 and Cyp8B1 expression (Fig. S3A; see above), which was correlated with a decrease in postprandial BA secretion (Fig. 3D and steps 1–3 in Fig. 4D). Moreover, as a consequence of the RF CC shift in liver (5), the RORE-dependent FXRβ expression was shifted to the ZT12–ZT20 period (Fig. S3A), which in conjunction with the reduced BA levels in RF mice decreased the FXR-dependent expression of SHP (Fig. S3A and steps 3–5 in Fig. 4D). Furthermore, the SIRT6 expression was reduced in RF mice (Fig. S3A and step 8 in Fig. 4D), whereas the RORE-dependent expression of INSIG2A (Fig. S3A and step 7 in Fig. 4D) was shifted due to the permanent shift of RevErbα expression in RF liver (5). Thus, alterations of all of these SREBP1c negative regulators (2, 3, 32, 33) increased SREBP1c expression and activity in RF liver (Fig. 3J and steps 6–9 in Fig. 4D), which resulted in an induction of the genes critically involved in the lipogenesis (27) (i.e., FAS, HMGCR, DGAT1, GPAT1, SCD1, and SQS; Fig. 3J), thereby leading to an increase in hepatic de novo lipogenesis and finally to hypertriglyceridemia and hypercholesterolemia (Fig. 3 B and C).
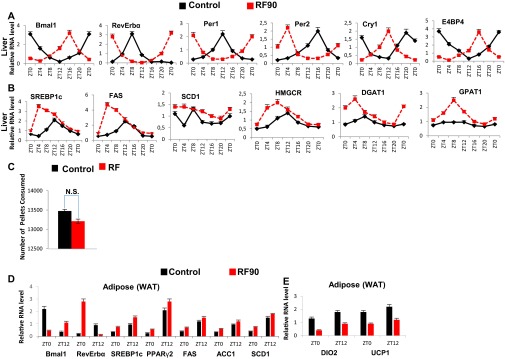
By RF90, the expression pattern of CC genes in various tissues remained unchanged (compare Fig. S3D with Fig. S4A) (5). However, at this time, RF mice displayed a fatty liver phenotype, as revealed by an increase in hepatic TG levels and Oil red O staining (Fig. 3 B and E). This was accompanied in RF liver by a further up-regulation of SREBP1c-driven expression of lipogenic genes (Fig. S4B), which disrupted the normal circadian variation (2, 3) in liver and blood TG levels (2, 3) (Fig. 3 A and B; compare the progressive deteriorations from RF15 to RF90). Importantly, this RF increase in lipogenesis enhanced blood levels of both total and LDL cholesterol (Fig. 3C), whereas the HDL-LDL ratio was reduced (Fig. 4D), which is a hallmark of atherosclerosis (27) and is also associated with shift work (6, 23).
Fig. S4.
Progressive deterioration of metabolism on prolonged RF. (A) RNA transcript levels of CC components in liver of control and RF90 mice. (B) RNA transcript levels of genes, as indicated, in liver of control and RF90 mice. (C) The total number of pellets consumed by control and RF90 mice. (D and E) RNA transcript levels of genes, as indicated, in WAT of control and RF90 mice. All values are mean ± SEM.
Of note, RF mice after an initial loss of weight, slowly started gaining weight, despite equal food intake, and by 12 wk of RF, weighed more than control mice, with a selective increase in the weight of visceral white adipose tissue (WAT; Fig. 3K and Fig. S4C). In keeping with this weight gain, transcript analyses of WAT revealed (i) an increase of the SREBP1c level, which is known to stimulate (34) the expression of adipogenic genes (PPARγ2, FAS, SCD1, ACC1, and DGAT1; Fig. S4D), and (ii) a decrease in BA-controlled expression of iodothyronine deiodinase 2 (DIO2) (29) and of the uncoupling protein 1 (UCP1) (Fig. S4E and steps 10–13 in Fig. 4D), which are known to increase energy expenditure (29). Remarkably, we also found that even after RF90, both the RF-induced CC shift and all of the above metabolic perturbations (Fig. 4 A–D) were still reversible within 15 d (RRF15) on return to the normal active phase feeding (Fig. 5 A–D).
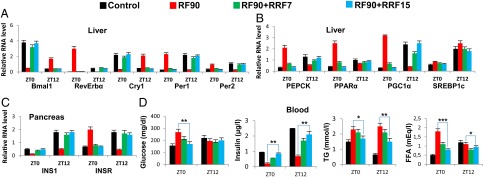
Fig. 5.
Prolonged RF-induced metabolic alterations are reversible on returning to normal feeding regime. (A) RNA transcript levels of CC components in liver of control and RF90 mice, and in mice after reversal of RF (RRF7 and RRF15). (B) RNA transcript levels of genes, as indicated, in liver of control and RF90 mice, and in mice after reversal of RF (RRF7 and RRF15). (C) RNA transcript levels of genes, as indicated, in pancreas of control and RF90 mice, and in mice after reversal of RF (RRF7 and RRF15). (D) Levels of blood components, as indicated, in control and RF90 mice, and in mice after reversal of RF (RRF7 and RRF15). All values are mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
In conclusion, it is striking that the pathological metabolic perturbations that are frequently observed in shift workers are similar to those occurring in mice on a prolonged RF-induced misalignment between the feeding time and the diurnal rest and active phases controlled by the master SCN clock, which validates the use of RF mice as a model for further studies on the consequences of shift work.
SI Methods
Mice.
All experiments were performed under light-dark (L/D) conditions, with ZT0 being the start of the light period (6:00 AM) and ZT12 the start of the dark period (6:00 PM). Mice were killed at 4-h intervals starting from ZT12 or ZT0. Food was removed from the RF cages at 6:00 PM and reintroduced at 6:00 AM. Food intake was measured (in 12 control and 20 RF mice) by continuously measuring the weight of the food supplied for individual animals (individual pellets weighing 20 mg). Overall food intake was measured by determining the weight of consumed food in bins of 1 wk. All mice were fed the normal laboratory chow diet.
Isolation and Laser Capture Microdissection of SCN and PVN.
The brains were isolated and frozen in iso-pentane (Sigma Aldrich) kept on dry ice and immediately stored at −70 °C. The frozen brains were fixed by Cryomatrix (Thermo Scientific), and 12-µm-thick sections were cut using a cryotome. The sections were mounted on PET-Membrane frame slides (Leica no. 11505151), which were first fixed in 70% (vol/vol) ethanol for 1 min and then stained in cresyl violet (Sigma Aldrich) for 3 min, followed by rinsing in 70% and 100% ethanol for 30 s each. Finally, the slides were air dried and used for laser capture microdissection. The locations of the SCN and PVN was confirmed using the brain atlas. The laser-cut SCN and PVN samples were collected in the caps of RNase-free 200-µL PCR tubes containing RLT buffer (a guanidine-thiocyanate–containing lysis buffer) (Qiagen). RNA was extracted using the RNeasy micro kit (Qiagen) following the manufacturer’s instruction manual. A standard reverse transcription protocol was used to convert the isolated RNA into cDNA, and purity of SCN samples was verified by measuring the transcripts for arginine vasopressin (AVP) and vasoactive intestinal polypeptide (VIP), expressed relative to GAPDH.
In Situ Hybridization.
In situ hybridization was performed using digioxigenin (DIG)-labeled riboprobes as previously described (35). PPARα and RORα cDNA was PCR amplified from mouse liver and subsequently cloned into the pGEM-T easy vector system (Promega) and sequenced to verify the gene identify. DIG-labeled sense and antisense riboprobes were synthesized using the in vitro transcription kit (Promega). Brain sections (14 µm thick) were cut and mounted on superfrost plus (Thermo Scientific) slides. The sections were stored at −80 °C. For in situ hybridization, the slides were first defrosted and then air dried at room temperature for 30 min. The sections were postfixed in ethanol gradation of 70%, 90%, and 100% for 5 min each and finally rinsed in PBS. The sections were hybridized overnight using 0.1 µg/mL RORα riboprobe and 1 μg/mL PPARα riboprobe in hybridization solution and incubated at 65 °C in a humidified chamber. The slides were washed in 5× SSC for 15 min followed by 0.2× SSC for 2 h at 72 °C. The blocking was performed for 2 h in blocking solution [maleic acid buffer (MAB), pH 7.5, Tween-20 0.1%, blocking reagent 2% (Roche), and 20% heat inactivated normal goat serum]. Next, alkaline phosphatase-conjugated anti-DIG Fab fragment (Roche) was added on the glass sides at a dilution of 1:2,000 in blocking solution and incubated overnight at 4 °C under glass coverslip. The slides were then washed five times in MAB with 10% Tween-20 for 5 min each and finally rinsed in alkaline phosphatase buffer. Nitro blue tetrazolium (NBT)/5-Bromo 4-chloro-3-indolyl phosphate (BCIP) (Roche) was used as staining solution for developing slides to detect alkaline phosphatase activity. The sections were incubated in dark for color development for 1 d (RORα) and 3 d (PPARα) at room temperature. After color development, the slides were rinsed in PBS, air dried, and mounted in PERTEX. Photographs were taken using Leica M420 macroscope and DMLB/DM4000B microscopes equipped with Photometrics digital cameras and CoolSnap imaging software (Roger Scientific). Primer sequences for the preparation of riboprobes are available on request.
RNA Transcript Determination.
Freshly isolated IECs, liver, pancreas, and hippocampus samples were used for RNA isolation using TRI reagent (Molecular Research Center). Subsequent to the verification of RNA quality (gel electrophoresis), 1 μg total RNA (determined spectrophotometrically) was reverse transcribed using random hexamers and SuperScript II reagents (Invitrogen) per the manufacturer’s instructions. The synthesized cDNA was used for qRT-PCR with SYBRgreen (QIAGEN) and expressed relative to the hypoxanthine phosphoribosyltransferase (HPRT) levels, as previously described (19). Primer sequences are available on request. For transcript determination, four control and four RF mice were killed at each time point, and each experiment was replicated three times.
Protein Immunoblots.
Immunoblots from isolated liver were performed following standard SDS/PAGE procedures. Proteins were visualized following ECL (Pierce), and images were captured using CCD. The primary antibodies for IRS1 (2390), pIRS1 Ser307 (2381), pAkt Ser473 (4060), Akt (4685), pGSK3β Ser9 (5558), GSK3β (12456), and pRevErbα Ser55/59 (2129) were obtained from Cell Signaling Technology. RevErbα (ab115552) and Bmal1 (ab93806) antibodies were obtained from abcam. GAPDH antibody was from Millipore.
FGF21 Measurement.
FGF21 was assayed from the plasma samples obtained from the blood of control and RF mice (eight animals were used per group), using the FGF21 quantikine ELISA kit (R&D; MF2100) per the manufacturer’s instructions.
ChIP Assays.
ChIP was performed as reported (19) with some minor modifications. Briefly, isolated IEC suspension in PBS was cross-linked with 1% formaldehyde for 15 min at room temperature; cross-linking was stopped by addition of 2 M glycine (0.125 M final concentration) at room temperature for 5 min. Cells were pelleted, and 500 μL lysis buffer was added in presence of protease inhibitors (Roche) on ice. For liver samples, identical lobes of liver from different groups of mice were disrupted using a dounce homogenizer; samples were then cross-linked and processed as described above. For adrenal glands, 12 adrenal glands were pulled and homogenized for each antibody per time point. Following cell lysis, the samples were sonicated (Bioruptor; Diagenode) to generate fragments of average length of 200–500 bp. Cellular debris were removed by centrifugation at 4 °C for 10 min (10,000 × g), and supernatant was precleared with Protein A/G-Sepharose (Roche) beads and preblocked with salmon DNA and BSA for 60 min at 4 °C. Beads were pelleted and discarded; 10% of the lysate was stored from each sample as the source of Input, and the remaining lysate was diluted eight times (five times for adrenal glands) in dilution buffer [16.7 mM Tris⋅HCl, pH 8.1, 0.01% (wt/vol) SDS, 1.1% (vol/vol) Triton X-100, 1.2 mM EDTA, 16.7 mM NaCl, protease inhibitor mixture] in the presence of different primary antibodies for 14 h at 4 °C, on a flip-flop rocker. Ninety microliters Protein A/G-Sepharose beads (preblocked with salmon DNA and BSA) was then added for 90 min at 4 °C. Immunecomplexes were recovered by centrifugation at 500 × g for 1 min. Beads were washed extensively at 4 °C in low salt buffer [20 mM Tris⋅HCl, pH 8.1, 0.1% (wt/vol) SDS, 1% (vol/vol) Triton X-100, 2 mM EDTA, 150 mM NaCl], in high salt buffer [20 mM Tris⋅HCl, pH 8.1, 0.1% (wt/vol) SDS, 1% (vol/vol) Triton X-100, 2 mM EDTA, 500 mM NaCl], in LiCl buffer [10 mM Tris⋅HCl, pH 8.1, 250 mM LiCl, 1% (vol/vol) Nonidet P-40, 1% (wt/vol) sodium deoxycholate, 1 mM EDTA], and finally in 1 mL TE buffer (10 mM Tris⋅HCl, pH 8.0, 1 mM EDTA). The bound chromatin was released from the beads by intermittent vortexing at room temperature in 200 μL elution buffer [1 (wt/vol) SDS and 100 mM NaHCO3]. One milliliter of 10 mg/mL RnaseA and 5 M NaCl (200 mM final concentration) was added to the eluate and incubated overnight at 65 °C and then treated with Proteinase K for 1 h at 55 °C; DNA was purified using the QIAGEN PCR purification kit in a final volume of 50 μL. The qPCR was done using this eluted DNA and SYBRgreen reagent (QIAGEN). PCR cycles were verified to be within the linear range of amplification. Primer sequences are available on request.
Plasma Metabolic Analysis.
Blood glucose, INS, TG, FFA, cholesterol (total, LDL, HDL), bile acids, and IGF1 levels were measured from control and RF mice (eight animals per group per time point) at indicated ZTs. Blood glucose levels were determined on blood collected from the tail vein using a handheld Accu-check active glucometer (Roche). For all other measurements, blood were collected by retro-orbital puncture in EDTA-coated vials, plasma was separated, and measurements were done in the metabolomics unit of the IGBMC/ICS.
GTTs and ITTs.
GTTs and ITTs were performed after 15, 30, and 90 d of RF. Blood glucose was measured at the indicated times after i.p. administration of glucose (1 g/kg) of body weight. GSIS was also similarly studied after i.p. administration of glucose. ITTs were carried out after i.p. administration of 0.75 U INS/kg of body weight (Humulin R; EliLilly). INS level at the beginning of the experiment was considered as 100%. Eight to 10 control and RF mice were used for GTT and ITT experiments. GTT and GSIS were performed from the same animals. For glucose administration-dependent reversal of RF PCC shift, glucose (2 g/kg) was i.p. administered for 4 consecutive d at ZT18.
Pharmacological Inhibition of GSK3β.
GSK3β activity was inhibited by i.p. administration of either LiCl (Merck) or AR-A014418 (ARA; Sigma Aldrich; 3230) at ZT18 to RF mice. LiCl (25 mEq/kg) was prepared in PBS, whereas ARA (8 mg/kg) was dissolved in DMSO (Sigma Aldrich) and diluted in PBS before injection. Both inhibitors were administered for 5 consecutive RF days (injected every day at ZT18). One group of RF mice were mock i.p. administered with DMSO + PBS.
Oil Red O Staining.
Following death, liver samples were frozen in Cryomatrix and kept at −20 °C. Eight-micrometer sections were prepared and stained in 0.5% Oil red O stain (prepared in glycerol) for lipid (TG) and then in hematoxylin for 5 s, and images were captured.
Quantification of TG Levels in Liver.
Hepatic TG levels were analyzed as per a previous report (33). Briefly liver extracts were prepared by homogenization in 0.25% sucrose with 1 mmol/L EDTA, and lipids were extracted using chloroform/methanol [2:1 (vol/vol)] and suspended with 5% fatty acid-free BSA. TG level was measured using triglyceride assay reagents (Sigma). The assay was performed in the metabolomics unit of ICS.
Circadian Locomotor Activity and Ingestive Behaviors.
Spontaneous locomotor activity of control and RF mice in the front and rear portions of the cages were simultaneously recorded from 24 individual boxes equipped with infrared captors, connected to computers. The quantity of water and food consumed was measured during the test period using an automated pellet feeder and lickometer (Imetronic, Pessac, France). For RF cages, the automated pellet feeder was disconnected manually at 6:00 PM (ZT12), whereas it was connected back to the cages at 6:00 AM (ZT0). Mice were tested for 32 h to measure habituation to the apparatus, as well as nocturnal and diurnal activities, following which the locomotor activity data are recorded for 10 consecutive light-dark periods. Results are expressed per 1-h periods.
Bioinformatics Analysis.
For identification of D-Box DNA binding sequences present across the human and mouse genome, human (hg19, Ensembl version: 67), downloaded from Ensembl and mouse (mm9, Ensembl version: 67; ftp://ftp.ensembl.org/pub/current_fasta/mus_musculus/dna/) repeat masked genome assembly was used. The database had 37,991 mouse genes and 57,945 human genes. The database was searched for D-Box–containing genes with the consensus sequence RT(G/T)AYGTAAY [where R is a purine and Y is a pyrimidine residue (36)] and no mismatch in both the DNA strands, within −20 kb upstream and +5 kb downstream of the transcriptional start sites for each genes using a JAVA-based homemade program to reveal gene annotation: Ensembl Gene ID, Associated Gene Name, Chromosome Name, Gene Start, Gene End, chromosome strand, and location. The analysis revealed 12,933 mouse genes and 18,311 human genes containing at least one D-Box sequence. The D-Box containing 2,110 ortholog genes between mice and humans was found using the table of gene orthology (Ensembl) with the help of biomart (www.biomart.org). Gene functional annotation was done using the DAVID program (37).
Methods
Mice and Treatments.
Eight- to 12-wk-old C57BL6/J male WT mice (Charles River Laboratories) were used. Control mice were provided food and water ad libitum, under 12-h light (6:00 AM–6:00 PM) and 12-h dark (6:00 PM–6:00 AM) conditions. RF mice were provided food during the entire light period (4). Breeding, maintenance, and experimental manipulations were approved by the Animal care and Use Committee of Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC)/Institut Clinique de la souris (ICS).
Statistics.
Data are represented as mean ± SEM of at least three independent experiments and were analyzed by SyStat and Microsoft Excel statistics software using the Student t test (RNA transcripts) and one-way ANOVA (blood metabolic analysis). P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank the staffs of animal house facilities in Institut de Génétique et de Biologie Moléculaire et Cellulaire (IGBMC)/Institut Clinique de la souris (ICS) for help. This work was supported by the Centre national de la recherche scientifique (CNRS), Institut national de la santé et de la recherche médicale (INSERM), University of Strasbourg Institute for Advanced Studies, and the Association pour la Recherche a l’IGBMC (ARI). A.M., A.K., M.D., and N.M. were supported by fellowships from ARI.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1519807112/-/DCSupplemental.
References
- 1.Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13(2):125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 2.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330(6009):1349–1354. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asher G, Sassone-Corsi P. Time for food: The intimate interplay between nutrition, metabolism, and the circadian clock. Cell. 2015;161(1):84–92. doi: 10.1016/j.cell.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 4.Damiola F, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14(23):2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mukherji A, Kobiita A, Chambon P. Shifting the feeding of mice to the rest phase creates metabolic alterations, which, on their own, shift the peripheral circadian clocks by 12 hours. Proc Natl Acad Sci USA. 2015;112:E6683–E6690. doi: 10.1073/pnas.1519735112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paschos GK. Circadian clocks, feeding time, and metabolic homeostasis. Front Pharmacol. 2015;6(112):112. doi: 10.3389/fphar.2015.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leproult R, Holmbäck U, Van Cauter E. Circadian misalignment augments markers of insulin resistance and inflammation, independently of sleep loss. Diabetes. 2014;63(6):1860–1869. doi: 10.2337/db13-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esquirol Y, Bongard V, Ferrieres J, Verdier H, Perret B. Shiftwork and higher pancreatic secretion: Early detection of an intermediate state of insulin resistance? Chronobiol Int. 2012;29(9):1258–1266. doi: 10.3109/07420528.2012.719959. [DOI] [PubMed] [Google Scholar]
- 9.McHill AW, et al. Impact of circadian misalignment on energy metabolism during simulated nightshift work. Proc Natl Acad Sci USA. 2014;111(48):17302–17307. doi: 10.1073/pnas.1412021111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin L, Wang J, Klein PS, Lazar MA. Nuclear receptor Rev-erbalpha is a critical lithium-sensitive component of the circadian clock. Science. 2006;311(5763):1002–1005. doi: 10.1126/science.1121613. [DOI] [PubMed] [Google Scholar]
- 11.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414(6865):799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 12.Ly PT, et al. Inhibition of GSK3β-mediated BACE1 expression reduces Alzheimer-associated phenotypes. J Clin Invest. 2013;123(1):224–235. doi: 10.1172/JCI64516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moga MM, Weis RP, Moore RY. Efferent projections of the paraventricular thalamic nucleus in the rat. J Comp Neurol. 1995;359(2):221–238. doi: 10.1002/cne.903590204. [DOI] [PubMed] [Google Scholar]
- 14.Gerstner JR, Yin JC. Circadian rhythms and memory formation. Nat Rev Neurosci. 2010;11(8):577–588. doi: 10.1038/nrn2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Talani G, et al. Enhanced glutamatergic synaptic plasticity in the hippocampal CA1 field of food-restricted rats: Involvement of CB1 receptors. Neuropsychopharmacology. September 10, 2015 doi: 10.1038/npp.2015.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balsalobre A, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289(5488):2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 17.Le Minh N, Damiola F, Tronche F, Schütz G, Schibler U. Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. EMBO J. 2001;20(24):7128–7136. doi: 10.1093/emboj/20.24.7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koike N, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338(6105):349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153(4):812–827. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 20.Fang B, et al. Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell. 2014;159(5):1140–1152. doi: 10.1016/j.cell.2014.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris CJ, et al. Endogenous circadian system and circadian misalignment impact glucose tolerance via separate mechanisms in humans. Proc Natl Acad Sci USA. 2015;112(17):E2225–E2234. doi: 10.1073/pnas.1418955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suwazono Y, et al. A longitudinal study on the effect of shift work on weight gain in male Japanese workers. Obesity (Silver Spring) 2008;16(8):1887–1893. doi: 10.1038/oby.2008.298. [DOI] [PubMed] [Google Scholar]
- 23.Sookoian S, et al. Effects of rotating shift work on biomarkers of metabolic syndrome and inflammation. J Intern Med. 2007;261(3):285–292. doi: 10.1111/j.1365-2796.2007.01766.x. [DOI] [PubMed] [Google Scholar]
- 24.Marcheva B, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466(7306):627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitamura T, et al. The forkhead transcription factor Foxo1 links insulin signaling to Pdx1 regulation of pancreatic β cell growth. J Clin Invest. 2002;110(12):1839–1847. doi: 10.1172/JCI200216857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inagaki T, et al. Inhibition of growth hormone signaling by the fasting-induced hormone FGF21. Cell Metab. 2008;8(1):77–83. doi: 10.1016/j.cmet.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horton JD, Goldstein JL, Brown MS. SREBPs: Activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109(9):1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126(4):801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 29.Houten SM, Watanabe M, Auwerx J. Endocrine functions of bile acids. EMBO J. 2006;25(7):1419–1425. doi: 10.1038/sj.emboj.7601049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Martelot G, et al. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7(9):e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masri S, et al. Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell. 2014;158(3):659–672. doi: 10.1016/j.cell.2014.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu S, et al. Polysome profiling in liver identifies dynamic regulation of endoplasmic reticulum translatome by obesity and fasting. PLoS Genet. 2012;8(8):e1002902. doi: 10.1371/journal.pgen.1002902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim HS, et al. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 2010;12(3):224–236. doi: 10.1016/j.cmet.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim JB, Wright HM, Wright M, Spiegelman BM. ADD1/SREBP1 activates PPARgamma through the production of endogenous ligand. Proc Natl Acad Sci USA. 1998;95(8):4333–4337. doi: 10.1073/pnas.95.8.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chotteau-Lelièvre A, Dollé P, Gofflot F. Expression analysis of murine genes using in situ hybridization with radioactive and nonradioactively labeled RNA probes. Methods Mol Biol. 2006;326:61–87. doi: 10.1385/1-59745-007-3:61. [DOI] [PubMed] [Google Scholar]
- 36.Mitsui S, Yamaguchi S, Matsuo T, Ishida Y, Okamura H. Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev. 2001;15(8):995–1006. doi: 10.1101/gad.873501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.