We have the capability of completing the first draft of the description of life on Earth within the lifetime of many of the youngest scientists among us. This description is an enormous challenge and will be one of the great accomplishments of modern science. It will yield fundamental paradigm shifts in biology and revolutionary breakthroughs in medicine, agriculture, and biotechnology. Like life, a meaningful description of life will be enormously complex. Although it will certainly comprise grand generalizations and natural laws, it will also include sufficient detail to capture the nuances of life and unite molecular biology with natural history. The PNAS paper by Rubin et al. (1) is a major step toward this goal by providing a detailed examination of gene function in a cyanobacterium, one of the dominant groups of oxygenic photosynthetic organisms on Earth.
This modern description of life will have four components. The first will be a complete census of the major eukaryotic and prokaryotic lineages on Earth. Depending upon how major is defined, the census of macrobiota might be considered largely complete (2). Even for microorganisms, rapid progress is being made. For instance, it is likely that most of the diversity of the 16S rRNA genes representing the major lineages of prokaryotes will have been discovered by the end of this decade (3). The second component will be sampling of genome sequences of representatives of each of the major lineages. Although this task remains daunting, especially for the single-celled eukaryotes, rapid progress is being made among the prokaryotes. Noteworthy are the efforts to sequence genomes of all of the named prokaryotes (4) as well representatives of the very diverse and uncultured lineages often called “microbial dark matter” (5–7). The third component will be developing an understanding of the evolutionary processes that have and will continue to occur within living organisms, especially the prokaryotes and single-celled eukaryotes (8, 9). In addition to revealing the history of life on Earth, this understanding will allow us to infer the properties of organisms based upon those of their relatives (10). The availability of genome sequences will be key to understanding these processes. The fourth and most challenging task will be linking the wealth of phylogenetic and genomic information to the properties of the living organisms. The paper by Rubin et al. (1) contributes to this important goal for the oxygenic phototroph Synechococcus elongatus PCC7942.
In this work, Rubin et al. use a combination of high-density transposon mutagenesis and next-generation sequencing called TnSeq to evaluate the contribution of each gene to fitness during growth under laboratory conditions (1). These experiments provide a wealth of information about gene function, but they are especially challenging because they require very high rates of genetic transformation. In fact, of the 34 experiments of this type performed on prokaryotes listed in a recent database, only 6 were performed on environmentally important organisms (11). This fact reflects the difficulty of manipulating many of these largely understudied and mysterious organisms. Despite their abundance and major roles in biogeochemical cycles, only a minute fraction have yielded to laboratory investigation. As a consequence, little is known about their most basic biological properties.
To my knowledge, Synechococcus elongatus PCC7942 is the first cyanobacterium and first phototroph interrogated by this technique. Because it is an obligate photolithotroph, these experiments yield insights into photosynthesis and autotrophic CO2 assimilation that have been previously unavailable. Of particular value was the ability to test the essentiality of all of the components of the photosystems and associated electron transport chains, providing a rigorous test of the state of our knowledge about these complex, multiprotein systems. Similarly, the pathways of central metabolism that convey carbon from CO2 through the reductive pentose phosphate pathway into amino acids and other biosynthetic precursors are of special interest in these autotrophs. Here, the experiments of Rubin et al. reveal an unknown flexibility and ability to dispense with a major segment of the tricarboxylic acid cycle. As important, these experiments open new areas of investigation into regulation by noncoding RNAs. The dataset generated is extensive, and more than 246,000 mutations were mapped onto the 2.80-Mbp genome. Thus, it will be the starting point for many additional studies of gene function and a valuable resource for many laboratories with different interests in cyanobacteria.
Synechococcus elongatus PCC7942 was a good choice for these experiments. A unicellular cyanobacterium, it possesses many features typical of this group, including the complex intracytoplasmic membranes called thylakoids that are the site of photosynthesis (Fig. 1) and carboxysomes, an inclusion body formed largely of a paracrystalline array of the key enzyme of the reductive pentose phosphate pathway RuBisCO. Moreover, they share features with many heterotrophic bacteria, such as a complex cell wall structure and cellular appendages composed of the protein pilin. Species of the genus Synechococcus are common in fresh as well as marine surface waters. Although not the most abundant cyanobacterial phototrophs in most marine waters, they still represent one of the most abundant microorganisms on Earth (12). Although S. elongatus PCC7942 is a freshwater strain and not closely related to the marine strains, it shares enough properties with them to serve as a good model for their basic biology (13). Moreover, its carbon and nitrogen metabolism, responses to iron and other nutrient stresses, adaptations to temperature and light intensity, and circadian clock have also been extensively studied. With methods to conveniently transfer in DNA being readily available, it is also a platform for metabolic engineering production of commodity chemicals (14) and host for a commercially available protein expression kit.
Fig. 1.
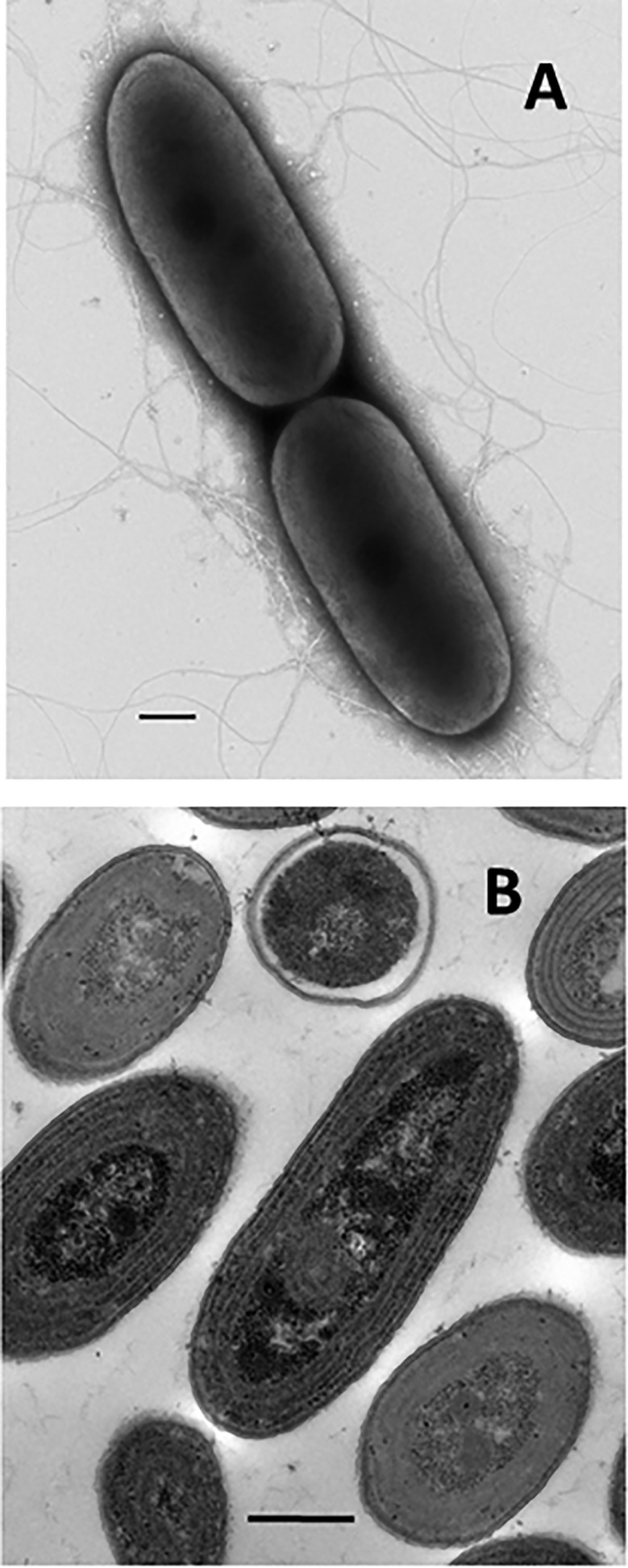
Morphology of Synechococcus elongatus PCC7942. (A) Cells are short rods about 1.5 μm in diameter that divide by binary fission. Although nonmotile, they possess numerous pili of unknown function. (Scale bar: 0.5 μm.) Cells and pili were visualized in this transmission electron micrograph of intact cells by negative staining with uranyl acetate by Timo Meerloo (Electron Microscopy Facility, University of California at San Diego, La Jolla, CA). (B) The cell interior is packed with concentric thylakoid membranes that are the site of photosynthesis. (Scale bar: 0.5 μm.) Cells were embedded in resin and ultrathin sectioned by Ying Jones (Electron Microscopy Facility, University of California at San Diego, La Jolla, CA). Samples and images courtesy of Ryan Simkovsky (University of California at San Diego, La Jolla, CA).
Footnotes
The author declares no conflict of interest.
See companion article on page E6634.
References
- 1.Rubin BE, et al. The essential gene set of a photosynthetic organism. Proc Natl Acad Sci USA. 2015;112:E6634–E6643. doi: 10.1073/pnas.1519220112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mora C, Tittensor DP, Adl S, Simpson AG, Worm B. How many species are there on Earth and in the ocean? PLoS Biol. 2011;9(8):e1001127. doi: 10.1371/journal.pbio.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yarza P, et al. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol. 2014;12(9):635–645. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- 4.Kyrpides NC, et al. Genomic encyclopedia of bacteria and archaea: Sequencing a myriad of type strains. PLoS Biol. 2014;12(8):e1001920. doi: 10.1371/journal.pbio.1001920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rinke C, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499(7459):431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 6.Spang A, et al. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. 2015;521(7551):173–179. doi: 10.1038/nature14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown CT, et al. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature. 2015;523(7559):208–211. doi: 10.1038/nature14486. [DOI] [PubMed] [Google Scholar]
- 8.Doolittle WF. Rethinking the tree of life. Microbe. 2015;10(8):319–323. [Google Scholar]
- 9.Hinchliff CE, et al. Synthesis of phylogeny and taxonomy into a comprehensive tree of life. Proc Natl Acad Sci USA. 2015;112(41):12764–12769. doi: 10.1073/pnas.1423041112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langille MGI, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31(9):814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo H, Lin Y, Gao F, Zhang CT, Zhang R. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res. 2014;42(Database issue):D574–D580. doi: 10.1093/nar/gkt1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scanlan DJ, West NJ. Molecular ecology of the marine cyanobacterial genera Prochlorococcus and Synechococcus. FEMS Microbiol Ecol. 2002;40(1):1–12. doi: 10.1111/j.1574-6941.2002.tb00930.x. [DOI] [PubMed] [Google Scholar]
- 13.Herdman M, et al. Form-genus XIII. Synechococcus. In: Boone DR, Castenholz RW, editors. Bergey’s Manual of Systematic Bacteriology. Vol 1. Springer; New York: 2001. pp. 508–512. [Google Scholar]
- 14.Angermayr SA, Gorchs Rovira A, Hellingwerf KJ. Metabolic engineering of cyanobacteria for the synthesis of commodity products. Trends Biotechnol. 2015;33(6):352–361. doi: 10.1016/j.tibtech.2015.03.009. [DOI] [PubMed] [Google Scholar]


