Significance
Dengue is the most prevalent mosquito-borne viral disease, endemic in more than a hundred tropical and subtropical countries. NS5, the largest viral protein, is a key replication enzyme with both methyltransferase and RNA polymerase activities. We present to our knowledge the first crystal structure of the full-length NS5 protein from dengue virus bound to the authentic 5′-end viral RNA fragment. This structure captures the viral enzyme in the act of transferring a methyl group to the 2′-O-ribose of the first nucleotide of the viral genome, providing an atomic-level understanding of specific 2′-O-methylation and cap formation by the flavivirus methyltransferase. The structure also suggests an evolutionary origin for the methyltransferase domain of NS5 and strategies for designing novel antiviral inhibitors.
Keywords: dengue virus, nonstructural protein 5 methyltransferase-polymerase, 2′-O-ribose methyltransferase, cap-0 RNA, innate immunity evasion
Abstract
Dengue virus (DENV) causes several hundred million human infections and more than 20,000 deaths annually. Neither an efficacious vaccine conferring immunity against all four circulating serotypes nor specific drugs are currently available to treat this emerging global disease. Capping of the DENV RNA genome is an essential structural modification that protects the RNA from degradation by 5′ exoribonucleases, ensures efficient expression of viral proteins, and allows escape from the host innate immune response. The large flavivirus nonstructural protein 5 (NS5) (105 kDa) has RNA methyltransferase activities at its N-terminal region, which is responsible for capping the virus RNA genome. The methyl transfer reactions are thought to occur sequentially using the strictly conserved flavivirus 5′ RNA sequence as substrate (GpppAG-RNA), leading to the formation of the 5′ RNA cap: G0pppAG-RNA→m7G0pppAG-RNA (“cap-0”)→m7G0pppAm2′-O-G-RNA (“cap-1”). To elucidate how viral RNA is specifically recognized and methylated, we determined the crystal structure of a ternary complex between the full-length NS5 protein from dengue virus, an octameric cap-0 viral RNA substrate bearing the authentic DENV genomic sequence (5′-m7G0pppA1G2U3U4G5U6U7-3′), and S-adenosyl-l-homocysteine (SAH), the by-product of the methylation reaction. The structure provides for the first time, to our knowledge, a molecular basis for specific adenosine 2′-O-methylation, rationalizes mutagenesis studies targeting the K61-D146-K180-E216 enzymatic tetrad as well as residues lining the RNA binding groove, and offers previously unidentified mechanistic and evolutionary insights into cap-1 formation by NS5, which underlies innate immunity evasion by flaviviruses.
Several members of the Flavivirus genus from the Flaviviridae family are major human pathogens, such as the four serotypes of dengue virus (DENV1–4), West Nile virus (WNV), Japanese encephalitis virus (JEV), and yellow fever virus (YFV). Recent large-scale DENV vaccine trials using a tetravalent formulation and three dose injections have shown only limited protection against the four DENV serotypes, and no specific antiviral drug has reached the market so far (1–3). The flavivirus genome consists of a (+)-sense single-stranded RNA of ∼11 kb with a type 1 cap structure, followed by the strictly conserved dinucleotide sequence “AG”: 5′-m7GpppAm2′-O-G-3′ (4, 5). Addition of the cap moiety to the 5′ end of the viral genome is crucial for viral replication, because this structure ensures efficient production of viral polyproteins by the host translation machinery and protection against degradation by 5′-3′ exoribonucleases, and also because it conceals the triphosphorylated (or diphosphorylated) end from recognition by host cell innate immune sensors (6–9). Following (+)-strand RNA synthesis by the C-terminal RNA-dependent RNA polymerase (RdRp) domain of nonstructural protein 5 (NS5), cap formation in flaviviruses results from several sequential enzymatic reactions carried out by (i) the RNA triphosphatase activity of the NS3 protease-helicase that hydrolyzes the γ-phosphate group of the viral 5′ untranslated region (UTR), yielding a diphosphate RNA, (ii) a guanylyl-transferase activity proposed to reside in the methyltransferase (MTase) domain of NS5, which transfers a GMP molecule to the 5′-diphosphate RNA, and (iii) NS5-mediated sequential N-7- and 2′-O-methylations according to the following scheme: G0pppAG-RNA→m7G0pppAG-RNA (“cap-0”)→m7G0pppAm2′-O-G-RNA (“cap-1”) (5, 10–12).
During flavivirus RNA replication, 5′-guanosine N-7-methylation is shown to be essential for translation of the viral polyprotein (13), whereas 2′-O-methylation on the penultimate A nucleotide conceals the viral genome from host immune sensors, notably RIG-I (14), MDA5 (15), and IFIT1 (16–18). Specifically, WNV carrying the E218A mutation in NS5 (E216A in DENV3 NS5) devoid of 2′-O (but not N-7) MTase activity was attenuated in wild-type but not Ifit1−/− cells (16). Furthermore, the translation of JEV viral proteins was inhibited by IFIT1 through direct binding to the 5′-capped 2′-O-unmethylated mRNA (17). More recently, 2′-O-methylation at internal adenosines (but not at G, C, or U positions) by the flavivirus NS5 protein was demonstrated. The functional consequence of methylation at internal adenosines was an attenuation of viral RNA translation and replication (12). In vitro, the MTase domain of NS5 catalyzes these two enzymatic reactions with distinct requirements of RNA substrates and buffers: 5′-guanosine N-7 methyl transfer is optimal on a 211-nt segment of the 5′UTR at pH 6 and is inhibited by MgCl2, whereas adenosine ribose 2′-O-methylation only requires a short RNA with “AG” as the first two RNA nucleotides and is maximal at pH 9–10 in the presence of Mg2+ ions. Thus, NS5 plays a crucial role both in virus replication and evasion of the host innate immune response, and therefore constitutes an attractive therapeutic target for antiviral drug and vaccine development (2, 19).
Several crystal structures of flavivirus MTases have been reported either as free enzymes or bound to GTP (20), to the broad antiviral nucleoside analog ribavirin (21), to short cap analogs (22, 23), and to a capped-RNA octamer (24). Collectively, these structures uncovered a GTP binding site, the S-adenosyl-methionine (SAM) methyl-donor binding pocket, and a basic cleft at the protein surface that was proposed to accommodate the incoming RNA substrate. However, in the absence of a viral RNA in a catalytically meaningful position, the mechanism accounting for specific viral RNA methylation, including the structural basis for specific adenosine 2′-O-methylation, remains elusive. Moreover, the size of the RNA substrate that can be accommodated by the putative RNA binding cleft is also unknown, as well as any requirement for a specific RNA conformation. Determination of the structure of NS5 bound to a viral RNA would give valuable information to guide the design of specific NS5 inhibitors.
To elucidate how the flavivirus RNA is recognized and methylated, we determined the crystal structure of a ternary complex between the full-length NS5 protein (DENV serotype 3), an authentic cap-0 viral RNA substrate (5′-m7G0pppA1G2U3U4G5U6U7-3′), and S-adenosyl-l-homocysteine (SAH), the by-product of the methylation reaction. Together with mutagenesis data informed by the present structure, this work reveals a unique and specific interaction between the protein and viral RNA and provides the molecular basis for the methyl transfer reaction. Furthermore, despite a low sequence identity between the two proteins, the RNA recognition mode is reminiscent of how the human 2′-O-ribose methyltransferase CMTr1 binds mRNA for cap formation, suggesting that the viral methyltransferase might derive from its eukaryotic homolog.
Results and Discussion
Overall Structure of the Ternary Complex of DENV3 NS5, Cap-0 Viral RNA, and SAH.
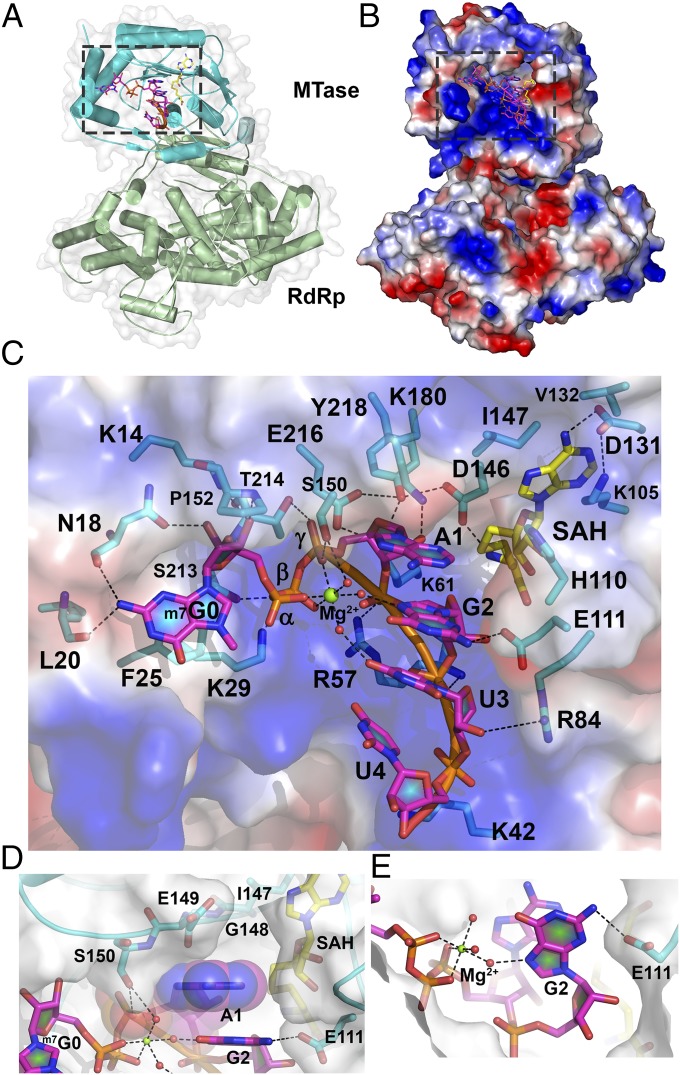
An overall view of bound RNA in the MTase domain of NS5 is given in Fig. 1A. The electrostatic surface representation in Fig. 1B reveals that the RNA moiety occupies a significant portion of a large basic patch of the NS5 protein. The ordered part of the single-strand RNA substrate consists of the 5′-m7G0pppA1G2U3U4-3′ sequence (Fig. 1C, SI Appendix, Fig. S1, and Movie S1). The crystal structure of the ternary complex between NS5, cap-0 RNA, and SAH (Fig. 1) was refined to Rwork 0.1827, Rfree 0.2436 at a resolution of 2.6 Å with good stereochemical parameters (Table 1). Bound RNA extends between F25, which is stacked with m7G0 at the 5′ end, and the methyl donor pocket, spanning an overall distance of ∼17 Å across the RNA binding groove. The m7G0 ring makes hydrogen bonds through its N2 atom with the carbonyl oxygen atom of L20 and N18 as well as via its ribose 2′-OH with the N18 side chain (Fig. 1C) (13, 20). The N-7-methyl of the m7G0 enhances stacking interactions with F25. The negative charges of the triphosphate group that links m7G0 and A1 are neutralized by a hexacoordinated Mg2+ ion (via three phosphate and three water molecule oxygen atoms, respectively) and hydrogen-bond with residues S213 and S150 that project from the protein surface (Fig. 1C and SI Appendix, Table S1A). The remainder of the RNA (G2U3U4) adopts an α-helical conformation along an axis that crisscrosses with the m7G0ppp moiety, giving an overall “L” shape to the bound cap-0 RNA (Fig. 1C). The observation of four stacked bases, A1G2U3U4, in addition to m7G0ppp (Fig. 1C) is consistent with previous RNase footprinting assays showing that a total of 4 nt becomes protected by the flavivirus MTase during 2′-O-methylation (13, 25). Only weak electron density is visible for G5U6U7 that extends out of the MTase binding groove. In the present conformation, no constraint on the length of the RNA substrate that can be accommodated by the MTase domain seems imposed by the NS5 RdRp domain (Fig. 1 and SI Appendix, Fig. S1). To assess possible conformational changes upon substrate binding, we superimposed the present NS5-capped RNA complex with the free NS5 enzyme [Protein Data Bank (PDB) ID code 4V0Q]. This returned an rms value of 0.281 Å for 758 Cα atoms, suggesting an essentially preformed RNA binding groove with no domain reorientation between the MTase and RdRp domains upon binding capped RNA (26). Remarkably, the RNA substrate is positioned such that the 2′-O atom of residue A1 lies next to the sulfur atom of SAH and adjacent to the K180 side chain from the “K61-D146-K180-E216” enzymatic motif, poised to accept a methyl group from a SAM methyl donor (Figs. 1C and 2A).
Fig. 1.
Structure of the ternary complex between DENV3 NS5, capped RNA, and SAH. (A) Crystal structure of NS56–896 (MTase domain, cyan; RdRp domain, green) bound to RNA (5′-m7G0pppA1G2U3U4GUU-3′; magenta sticks) and SAH (yellow sticks). The RNA/SAH binding site is boxed. (B) Electrostatic surface representation of NS5 (positive charges are in blue, and negative charges are in red). Capped RNA (magenta) and SAH (yellow) are shown as sticks. (C) Close-up view of the RNA and SAH binding sites as boxed in B. Protein residues binding RNA or SAH are represented as sticks (cyan) and labeled. Capped RNA nucleotides are labeled. Dashed lines indicate polar interactions. Mg2+ is displayed as a green sphere, and water molecules are shown as red spheres. (D) Interactions established between adenine A1 from the bound RNA and NS5, illustrating the tight shape complementarity with adenine only. (E) Hydrogen bonds formed between E111, the capped RNA second nucleotide G2, and the bound Mg2+ ion.
Table 1.
Data collection and refinement statistics
| Parameters | NS5: Cap-0 RNA: SAH |
| Data collection statistics | |
| Resolution range, Å | 47.33–2.60 (2.72–2.60)* |
| Space group | P 21212 |
| Unit cell parameters a, b, c, Å | 94.66, 151.39, 69.43 |
| Measured reflections | 183,254 (22,200) |
| Unique reflections | 31,327 (3,722) |
| Rmerge† | 0.175 (1.041) |
| CC1/2 | 0.992 (0.641) |
| Multiplicity | 5.8 (6.0) |
| Completeness, % | 99.9 (99.1) |
| Mean I/sigma(I) | 9.9 (1.8) |
| Refinement statistics | |
| Resolution range, Å | 47.33–2.60 (2.69–2.60)* |
| Rwork‡ | 0.1827 (0.2857) |
| Rfree§ | 0.2436 (0.3382) |
| No. of nonhydrogen atoms | 7,228 |
| Macromolecules | 6,950 |
| Ligands | 148 |
| Water | 241 |
| Protein residues | 852 |
| Rms, bonds, Å | 0.003 |
| Rms, angles, ° | 0.779 |
| Ramachandran plot, % | |
| Favored | 95.04 |
| Allowed | 4.6 |
| Outliers | 0.35 |
| Clash score | 6.53 |
| Average B factor | 45.8 |
| Macromolecules | 46.0 |
| Ligands | 66.6 |
| Solvent | 36.1 |
The numbers in parentheses refer to the last (highest) resolution shell.
Rmerge = ∑|Ij − < I >|/∑Ij, where Ij is the intensity of an individual reflection and < I > is the average intensity of that reflection.
Rwork = ∑||Fo| − |Fc||/∑|Fc|, where Fo denotes the observed structure factor amplitude and Fc denotes the structure factor amplitude calculated from the model.
Rfree is as for Rwork but calculated with 5% (3,044) of randomly chosen reflections omitted from the refinement.
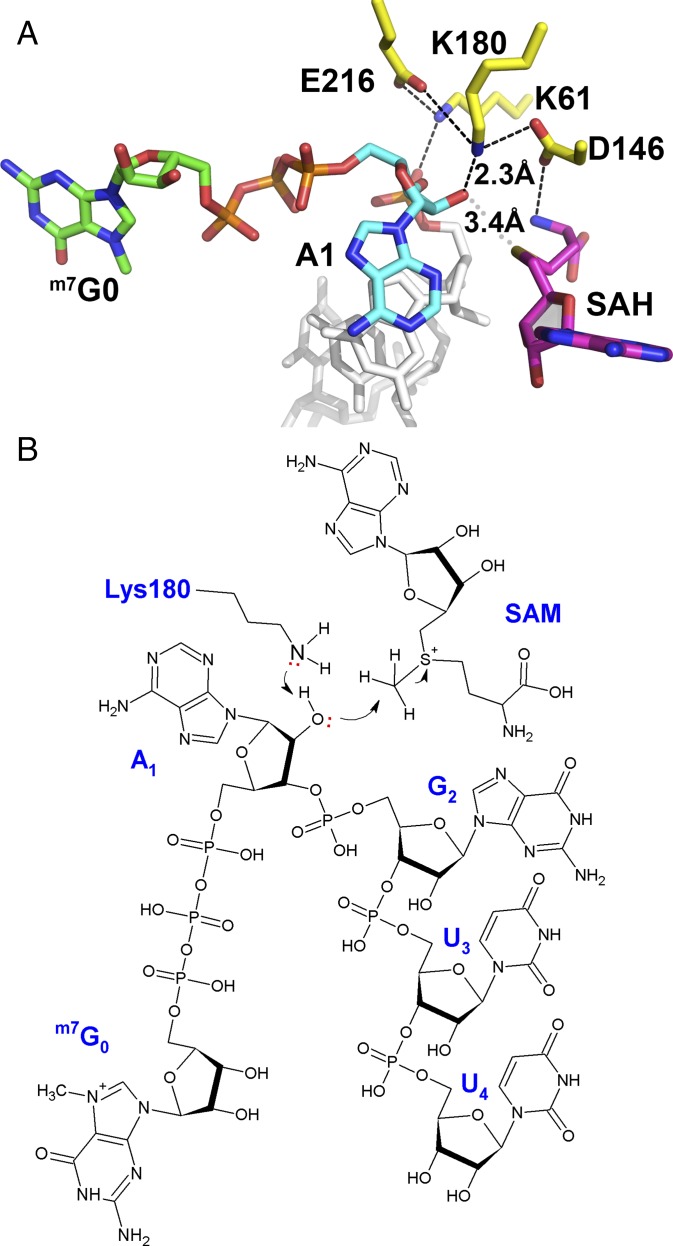
Fig. 2.
Proposed enzymatic mechanism for 2′-O-methylation by the flavivirus NS5 MTase. (A) Close-up view of the MTase active site with the K61-D146-K180-E216 catalytic tetrad shown as yellow sticks and SAH in magenta sticks. Color code for the bound RNA: G0 carbon atoms, green; A1, cyan; G2U3U4, gray. The close contacts between the 2′-oxygen atom of adenine A1, the amino group of K180, and the sulfur group of SAH are indicated by dashed lines, and the corresponding distances are given. (B) Schematic view of an active ternary complex comprising cap-0 RNA and the methyl donor SAM, based on the present structure. The stereochemistry of the reactants (distances and angles) conforms to what is expected from an inline SN2 reaction.
Specificity of Viral RNA Recognition by NS5 MTase.
The crystal structure explains the specific recognition of the flavivirus RNA 5′ cap by NS5 and in particular the strict requirement for an adenosine at position 1 and the preference for a guanosine at position 2 (Fig. 1 D and E and SI Appendix, Fig. S2). The first nucleotide of the flavivirus genome, always an adenosine (A1), resides ∼8 Å away from and nearly perpendicular to m7G0, to which it is connected via the triphosphate 5′–5′ linkage (Fig. 1 C and D). Remarkably, the A1 base fits snugly in a pocket shaped by residues I147-G148-E149-S150 from NS5 and SAH (Fig. 1D). To understand the origin of specific adenosine 2′-O-methylation by flaviviruses, we modeled X1 = G, U, or C (m7G0pppX1-RNA) in place of A1 and found that the N2 amine group from G1 would sterically collide with SAH (nearest distance ∼1 Å) (SI Appendix, Fig. S2A). Conversely, pyrimidine bases would leave a large empty space in the pocket, leading to energetically less favorable van der Waals interactions with the protein (SI Appendix, Fig. S2A). Consistently, specific 2′-O-methylations have been identified in viral genomic RNA internal adenosines and polyA but not on polyG, polyC, or polyU substrates (12).
G2, the second nucleotide of the viral genome, stacks with A1 and makes a hydrogen bond with the carboxylic side chain of E111 via its N2 atom and with a water molecule coordinating the Mg2+ ion (Fig. 1E and SI Appendix, Fig. S2B). We explore below the importance of this polar contact in the context of DENV replication. Modeling X2 = A, U, or C (7meG0pppAX2-RNA) in place of G2 showed that bases other than G disrupt favorable contacts and, in the case of a pyrimidine base, introduces unfavorable charge–charge repulsion between the carboxylic group of E111 and the O2 atom of X2 = U or C (SI Appendix, Fig. S2B). Thus, specific recognition of the 5′ end of the flavivirus genome is seen as the consequence of optimum shape complementarity and specific hydrogen bonding between the NS5 protein, the SAM methyl donor, and the 5′ terminus of viral genomic RNA. This observation is consistent with the strict conservation of the 5′ G0pppA1G2 sequence during flavivirus genome evolution (SI Appendix, Fig. S2C) (27). Likewise, residues from the NS5 MTase domain that form the tip of the RNA binding pocket and establish specific polar interactions with the A1G2 dinucleotide are also conserved.
Enzymatic Mechanism for Viral RNA 2′-O-Methylation.
RNA is bound in the DENV MTase catalytic site, such that the 2′-O-ribose of residue A1 is placed next to the SAH moiety. This observation suggests that the present complex represents a snapshot along the 2′-O-methylation reaction pathway, possibly the reaction product (Fig. 2 and SI Appendix, Fig. S3A): (i) The side chain of K180 activates the 2′-OH of the adenosine ribose of the RNA methyl acceptor (28), and (ii) the activated ribose 2′-O makes a nucleophilic attack on the methyl group of the positively charged sulfur center via an inline SN2 reaction. At pH 8.5, a value close to the pKa of the lysine ε-amino-group side chain, where the 2′-O-MTase reaction is maximal (25, 29), the side chain of K180 can be easily deprotonated. Moreover, because K180 is sandwiched between the acidic side chains of D146 and E216, its side chain could alternate between a charged and uncharged species at various stages of the 2′-O-methylation cycle. This scheme is consistent with the available mutagenesis data: The D146A single mutant is completely devoid of 2′-O-methylation activity (SI Appendix, Table S1A), and its side-chain carboxylic group is at the right distance to stabilize the electrophilic sulfur group of SAM as well as K180, following activation of the 2′-OH adenosine ribose (Fig. 2A and SI Appendix, Fig. S3) (30). Mutagenesis experiments also indicate that residues F25, R57, and K61 play a major role in the reaction: Alanine mutations at these positions essentially abolish the 2′-O-MTase activity (SI Appendix, Table S1A). Moreover, the pattern of RNA-enhanced thermostabilization for the DENV MTase is consistent with the observation that amino acids F25, R57, and R61 all make key contacts with RNA in the present complex (29). One attractive hypothesis suggested by the present structure is that the SAM cofactor could act as a “sensor” by probing for the base identity and for the presence of a methyl group(s) in the RNA substrate. Depending on its methylation state, the RNA substrate could either be in a favorable position in the binding groove conducive to methyl transfer or sterically collide with the methyl donor, favoring its dissociation from the MTase domain.
The catalytic mechanism proposed for NS5 is shared with the human mRNA cap-specific 2′-O-ribose methyltransferase CMTr1 and the vaccinia virus VP39 proteins (whose structures with capped RNA are known) and other RNA 2′-O-MTases across all animal kingdoms and viruses whose free enzyme structures have been reported (SI Appendix, Fig. S4 and Table S2) (31, 32). However, in contrast to the human CMTr1 or vaccinia virus VP39 that methylates cap-0 RNA substrates in a sequence-independent manner, the flavivirus NS5 MTase has a demonstrated RNA substrate sequence specificity, which is evident in our structure analysis and functional studies (Figs. 1, 3, and 4). A requirement for Mg2+ ions was reported for the COMT MTase as well as for FtsJ 2′-O-methylation activities (33, 34), and Mg2+ has stimulatory effects on DENV 2′-O-MTase activity (35). The structure of COMT suggested that Mg2+ ions play an important role for RNA stabilization but not for catalysis. Given the close similarity between the active sites of VP39 and FtsJ, Mg2+ ions are thought to be dispensable for 2′-O-methylation (34). Thus, activity enhancement in the presence of Mg2+ in DENV NS5 appears to be due to structural and electrostatic stabilization of the protein–RNA complex.
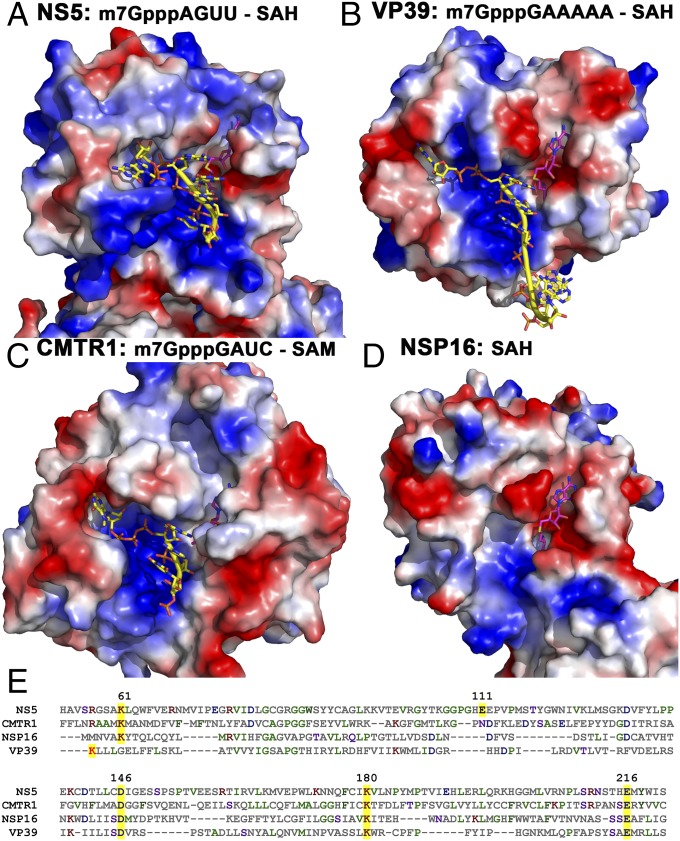
Fig. 3.
Comparison of NS5 MTase with other human and viral 2′-O-MTases. (A) The DENV3 NS5 MTase (this work). (B) Vaccinia virus MTase VP39. (C) Human CMTr1. (D) Severe acute respiratory syndrome (SARS) coronavirus MTase nonstructural protein 16 (NSP16). The comparison highlights the variety of shape complementarity and surface charge distributions between the enzymes, cofactor, and RNA substrate. The VP39 G0 pocket is deeper and more distant from the RNA binding groove compared with NS5 and CMTr1. As a result, G0 is deeply buried, with a protruding ribose. In contrast, no apparent G0 binding pocket is found in NSP16. Triphosphates display various conformations: extended in VP39 and CMTr1, and bent in NS5. In all three complexes, four RNA bases occupy the positively charged groove. The RNA binding groove in nsp16 is shallow and less positively charged. The SAM binding pocket is the most conserved feature, except at the adenine binding site: In NS5, a positively charged pocket is located next to N-7, whereas in CMTr1, the corresponding pocket is deeper and larger; in VP39, it is open and negatively charged. In nsp16, no apparent pocket is visible. Positive protein surfaces are shown in blue, and negative surfaces are in red; RNA moieties (yellow carbons) and SAH/SAM (magenta carbons) are shown as sticks. (E) Multiple sequence alignment. The K-D-K-E catalytic tetrad is highlighted in yellow. E111 from DENV NS5 is also highlighted.
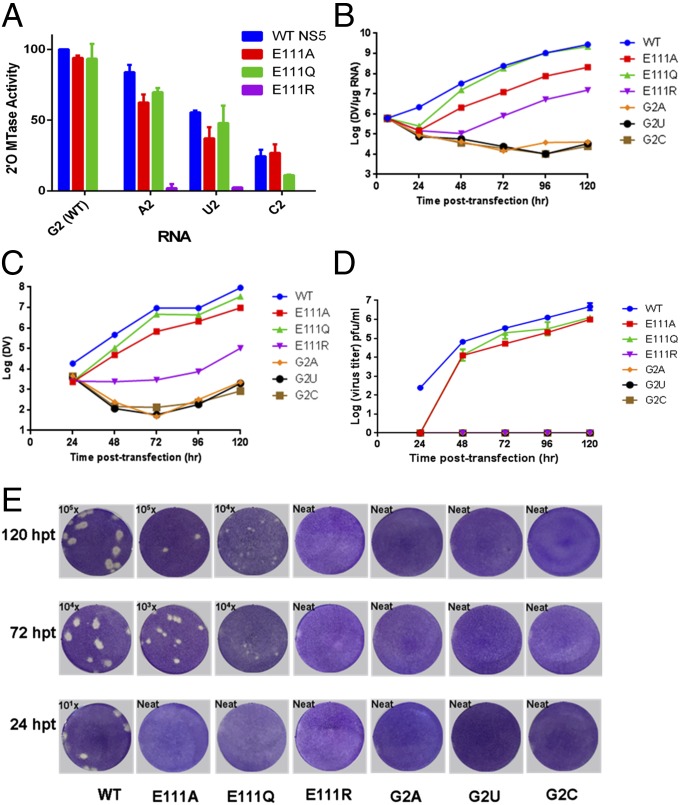
Fig. 4.
Specific recognition of the capped viral RNA is essential for virus replication. (A) In vitro 2′-O-MTase activities of WT NS5 and E111 mutants using WT and mutant RNA templates. Data are normalized to the activity of the WT NS5 MTase on WT viral 5′UTR RNA, which is set to 100% (corresponding to ∼4 nM 3H-methyl incorporated into the RNA; signal-to-background count ratios are approximately four- to fivefold). Each data point is the average for three replicates, and error bars show the standard deviations. (B) Intracellular viral RNA levels detected by quantitative (q)RT-PCR. (C) Extracellular viral RNA levels in the supernatants detected by qRT-PCR. (D) Virus titers based on the plaque assay shown in E. (E) Plaque assay for WT and mutants at 24, 72, and 120 h posttransfection (hpt). The limits of detection for DENV genomic RNA by qRT-PCR and plaque assays are 100 copies and 1 infectious virus particle, respectively.
Flavivirus NS5 MTase Is Closest to the Human mRNA Cap-Specific 2′-O-Ribose CMTr1.
To gain insights into the evolution leading to the present NS5 MTase domain, we performed a homology search against the various available MTase structures (36). The flavivirus NS5 protein belongs to a family of SAM-dependent RNA MTases that methylate diverse RNA species including mRNA, rRNA, tRNA, and siRNA, either at the bases or at 2′-hydroxyl groups of specific nucleosides. Within the Flavivirus genus, both the primary sequence and the 3D structures of the MTase domain are highly conserved, with sequence identities ranging from 51% to 100% (Z scores of 32.2–43.8). Surprisingly, despite a low sequence identity of 14% between the two proteins, the closest NS5 structural homolog with an rmsd of 2.9 Å for 222 of the 406 CMTr1 α-carbon atoms and a Z score of 15.8 is the human mRNA cap-specific 2′-O-ribose MTase CMTr1 (PDB ID code 4N48). Moreover, the seven next-closest homologs are also of nonviral origin (Fig. 3 and SI Appendix, Fig. S4 and Table S2). These structures share the general αβα-Rossmann-fold core domain that shapes the SAM binding pocket and the highly conserved K-D-K-E catalysis tetrad, which is consistent with a common catalytic mechanism for methyl transfer. Structural divergence beyond the core domain likely reflects the present variety of RNA substrate preferences (e.g., 2′-O-ribose of capped RNA vs. ribosomal RNA methylation) (37, 38). Thus, the flavivirus NS5 MTase domain might derive from its eukaryotic homolog. Interestingly, the Flavivirus genus is the only clade in the Flaviviridae family that encodes an MTase at the N terminus of NS5. Considering that the RdRp domain is highly conserved in all viruses from the Flaviviridae family but is absent from the host cells, the present flavivirus NS5 protein might have originated from a domain fusion event through the acquisition of an RNA 2′-O-MTase domain from the host following the separation from the Hepatitis, Pestivirus, and Pegivirus of the Flaviviridae family (26).
Residue E111 Is Key for Virus Replication.
To investigate the functional implications of the present ternary complex for virus replication, we performed a mutagenesis study targeting the highly conserved residue E111 of NS5, which establishes the sole direct specific polar contact with the second base (G2) of the flavivirus RNA (Figs. 1C and 4 and SI Appendix, Fig. S2B). We evaluated the effects of E111 mutants both in vitro, using an NS5 MTase enzymatic assay (Fig. 4A and SI Appendix, Table S1B), and also in cell culture by examining the growth kinetics of the corresponding virus mutants (Fig. 4 B–E). Reversing the charge through the E111R mutation abolishes 2′-O-MTase activity (Fig. 4A). In contrast, mutants E111A and E111Q retain >93% activity, which is consistent with a limited impact of these mutations on protein–RNA interaction (Fig. 4A and SI Appendix, Table S1B). Modeling the E111A and E111Q mutations in the NS5–cap-0 RNA suggests that these two mutations have a smaller impact on the ability of the protein to bind cap-0 viral RNA compared with the E111R mutation, which is likely to introduce steric hindrance with the RNA moiety (SI Appendix, Fig. S2D). These modeling results are in line with experimental thermostability measurements of the corresponding NS5 wild-type and mutant proteins in their free states and in the presence of cap-0 RNA: The E111R mutant has the most decreased thermostability both in the presence and absence of the RNA ligand (SI Appendix, Table S1C).
Substituting X2 = U2 or C2 in place of G2 (7meG0pppAX2-RNA) introduces unfavorable electrostatic contacts between their O2 atom and the carboxylic group of E111 and, for X2 = A2, between N3 and E111 (SI Appendix, Fig. S2B). This is consistent with the observation that the consensus sequence G0pppA1G2X found in flaviviruses (SI Appendix, Fig. S2C) binds more tightly than G0pppA1C2X to the DENV MTase (23). Moreover, mutations of G2 to U, C, or A reduce the 2′-O-methylation activity of NS5 down to 33%, 19%, and 100%, respectively, in WNV (31) and to 55%, 24%, and 84% in DENV4 (SI Appendix, Table S1B). The corresponding DENV mutants G2A, G2U, and G2C completely fail to grow (Fig. 4 B–E), showing that the virus does not tolerate changes at position G2. Because these substitutions preserve 2′-O-MTase activity appreciably, their impact on virus growth is likely to derive at least in part from a different process, possibly the de novo initiation of polymerization by NS5, where a strict requirement for pppA1G2 was demonstrated (39).
We also found that E111 plays a crucial role in viral replication (Fig. 4 B–E). The E111Q and E111A mutations attenuated virus replication and produced three to four times less infectious virus, whereas the E111R mutation severely impaired viral replication and yielded no virus (Fig. 4E). Mutants such as WNV-E218A abolishing 2′-O-MTase activity yet growing to relatively wild-type levels in BHK or Vero cells have been described (16). Therefore, the origin of the strong attenuation observed for the E111R mutant virus might be multifactorial: E111R is less thermostable than WT protein (SI Appendix, Table S1B) and has lower elongation polymerase activity, probably due to its poorer ability to bind RNA (SI Appendix, Table S1C). Together, these effects are likely to cause the mutant virus to be highly attenuated.
Concluding Remarks
The structure presented here suggests possible routes for the design of inhibitors targeting the NS5 MTase catalytic site. One possible strategy would consist of extending SAM analogs toward the RNA binding groove, for instance by linking the A1 moiety to nucleoside analogs (e.g., ribavirin) that bind at the GTP binding site, next to residue F25. This strategy may lead to larger but more specific and potent flavivirus MTase inhibitors (40). Also, the effect of MTase mutant on polymerase activities suggests the interdependence of two domains in viral replication (41). Our study also indicates that attenuated viruses could be designed by targeting the E111 residue of NS5.
Materials and Methods
Methods used in this study are briefly summarized below. Full descriptions are given in SI Appendix, Materials and Methods.
Protein Expression and Purification.
The protocol for NS5 protein expression and purification was as previously described (26).
Crystallization and Data Collection.
Crystallization was set up at 20 °C using the hanging-drop vapor-diffusion method and published conditions (26). Native crystals obtained over 2–5 d by mixing a volume of 1 µL NS5 (residues 6–895) at 4–6 mg/mL with 1 µL precipitation solution [0.2 M magnesium acetate, 0.1 M sodium cacodylate, pH 6.4, 10–20% (wt/vol) PEG 8000] were soaked with 1 mM 5′-7meG0pppA1G2U3U4G5U6U7-3′ overnight.
Structure Solution and Refinement.
The crystal had a Matthews coefficient (Vm) of 2.47 (42). Refinement was initiated with the structure PDB ID code 4V0Q using REFMAC5 (43) and PHENIX (44) and interspersed with manual model-rebuilding sessions using Coot (45). The refined coordinates were deposited in the PDB under ID code 5DTO.
Construction of Mutant m7Gppp-DENV4 5′UTR Nt-110 RNAs for 2′-O-MTase Assay.
DENV4 5′UTR nt-110 DNA with substitutions at the position of the 5′ nucleotide G2 was engineered by PCR using Phusion DNA Polymerase with standard procedures.
DENV4 2′-O-MTase Assay.
DENV4 2′-O-MTase assay was performed as described previously (11, 35).
Construction of Mutant DENV2 Full-Length Infectious Clone.
DENV2 full-length cDNA clones with NS5 E111A, E111Q, and E111R mutations were constructed with a pACYC-NGC FL and a TA-NGC (shuttle E) vector as previously described (46).
Supplementary Material
Acknowledgments
We thank Drs. Liew Chong Wai and Chen Ming Wei and scientists from Swiss Light Source beamline PXIII and Taiwan Republic of China National Synchrotron Radiation Research Centre beamline ID13 for their help with diffraction data collection. We thank Dr. George Chandy for critically reading our manuscript and Dr. Gérard Bricogne (Global Phasing) for advice on refinement with Buster. This work was supported by (i) a start-up grant to the D.L. laboratory from Lee Kong Chian School of Medicine, Nanyang Technological University, (ii) National Medical Research Council Grant CBRG14May051 (to J.L.), (iii) an MOE Tier 1 Complexity grant (to the J.L. laboratory), and (iv) National Medical Research Council Grant NMRC/1315/2011 (to the S.G.V. laboratory). Y.Z. is supported by a National University of Singapore Graduate School for Integrative Sciences and Engineering (NGS) scholarship, and T.S.S. is supported by an Industrial Postgraduate Programme Scholarship from the Economic Development Board, Singapore.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5DTO).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1514978112/-/DCSupplemental.
References
- 1.Capeding MR, et al. CYD14 Study Group Clinical efficacy and safety of a novel tetravalent dengue vaccine in healthy children in Asia: A phase 3, randomised, observer-masked, placebo-controlled trial. Lancet. 2014;384(9951):1358–1365. doi: 10.1016/S0140-6736(14)61060-6. [DOI] [PubMed] [Google Scholar]
- 2.Lim SP, et al. Ten years of dengue drug discovery: Progress and prospects. Antiviral Res. 2013;100(2):500–519. doi: 10.1016/j.antiviral.2013.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Low JG, et al. Efficacy and safety of celgosivir in patients with dengue fever (CELADEN): A phase 1b, randomised, double-blind, placebo-controlled, proof-of-concept trial. Lancet Infect Dis. 2014;14(8):706–715. doi: 10.1016/S1473-3099(14)70730-3. [DOI] [PubMed] [Google Scholar]
- 4.Lindenbach BD, Thiel HJ, Rice CM. Flaviviridae: The Viruses and Their Replication. Lippincott-Raven; Philadelphia: 2007. [Google Scholar]
- 5.Decroly E, Ferron F, Lescar J, Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol. 2012;10(1):51–65. doi: 10.1038/nrmicro2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314(5801):997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 7.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314(5801):994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 8.Rehwinkel J, et al. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 2010;140(3):397–408. doi: 10.1016/j.cell.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 9.Goubau D, et al. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5′-diphosphates. Nature. 2014;514(7522):372–375. doi: 10.1038/nature13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ray D, et al. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J Virol. 2006;80(17):8362–8370. doi: 10.1128/JVI.00814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung KY, et al. Higher catalytic efficiency of N-7-methylation is responsible for processive N-7 and 2′-O methyltransferase activity in dengue virus. Virology. 2010;402(1):52–60. doi: 10.1016/j.virol.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 12.Dong H, et al. 2′-O methylation of internal adenosine by flavivirus NS5 methyltransferase. PLoS Pathog. 2012;8(4):e1002642. doi: 10.1371/journal.ppat.1002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong H, et al. Flavivirus RNA methylation. J Gen Virol. 2014;95(Pt 4):763–778. doi: 10.1099/vir.0.062208-0. [DOI] [PubMed] [Google Scholar]
- 14.Schuberth-Wagner C, et al. A conserved histidine in the RNA sensor RIG-I controls immune tolerance to N1-2′O-methylated self RNA. Immunity. 2015;43(1):41–51. doi: 10.1016/j.immuni.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Züst R, et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol. 2011;12(2):137–143. doi: 10.1038/ni.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daffis S, et al. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468(7322):452–456. doi: 10.1038/nature09489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura T, et al. Ifit1 inhibits Japanese encephalitis virus replication through binding to 5′ capped 2′-O unmethylated RNA. J Virol. 2013;87(18):9997–10003. doi: 10.1128/JVI.00883-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Habjan M, et al. Sequestration by IFIT1 impairs translation of 2′O-unmethylated capped RNA. PLoS Pathog. 2013;9(10):e1003663. doi: 10.1371/journal.ppat.1003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li SH, et al. Rational design of a flavivirus vaccine by abolishing viral RNA 2′-O methylation. J Virol. 2013;87(10):5812–5819. doi: 10.1128/JVI.02806-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egloff MP, Benarroch D, Selisko B, Romette JL, Canard B. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: Crystal structure and functional characterization. EMBO J. 2002;21(11):2757–2768. doi: 10.1093/emboj/21.11.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benarroch D, et al. A structural basis for the inhibition of the NS5 dengue virus mRNA 2′-O-methyltransferase domain by ribavirin 5′-triphosphate. J Biol Chem. 2004;279(34):35638–35643. doi: 10.1074/jbc.M400460200. [DOI] [PubMed] [Google Scholar]
- 22.Bollati M, et al. Recognition of RNA cap in the Wesselsbron virus NS5 methyltransferase domain: Implications for RNA-capping mechanisms in Flavivirus. J Mol Biol. 2009;385(1):140–152. doi: 10.1016/j.jmb.2008.10.028. [DOI] [PubMed] [Google Scholar]
- 23.Egloff MP, et al. Structural and functional analysis of methylation and 5′-RNA sequence requirements of short capped RNAs by the methyltransferase domain of dengue virus NS5. J Mol Biol. 2007;372(3):723–736. doi: 10.1016/j.jmb.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 24.Yap LJ, et al. Crystal structure of the dengue virus methyltransferase bound to a 5′-capped octameric RNA. PLoS One. 2010;5(9):e12836. doi: 10.1371/journal.pone.0012836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Y, et al. Structure and function of flavivirus NS5 methyltransferase. J Virol. 2007;81(8):3891–3903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Y, et al. A crystal structure of the dengue virus NS5 protein reveals a novel inter-domain interface essential for protein flexibility and virus replication. PLoS Pathog. 2015;11(3):e1004682. doi: 10.1371/journal.ppat.1004682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paranjape SM, Harris E. 2009. in Control of Dengue Virus Translation and Replication, Current Topics in Microbiology and Immunology, Vol 338, ed Rothman AL (Springer, Berlin), pp 15–34.
- 28.Li C, Xia Y, Gao X, Gershon PD. Mechanism of RNA 2′-O-methylation: Evidence that the catalytic lysine acts to steer rather than deprotonate the target nucleophile. Biochemistry. 2004;43(19):5680–5687. doi: 10.1021/bi0359980. [DOI] [PubMed] [Google Scholar]
- 29.Dong H, et al. Biochemical and genetic characterization of dengue virus methyltransferase. Virology. 2010;405(2):568–578. doi: 10.1016/j.virol.2010.06.039. [DOI] [PubMed] [Google Scholar]
- 30.Dong H, et al. West Nile virus methyltransferase catalyzes two methylations of the viral RNA cap through a substrate-repositioning mechanism. J Virol. 2008;82(9):4295–4307. doi: 10.1128/JVI.02202-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smietanski M, et al. Structural analysis of human 2′-O-ribose methyltransferases involved in mRNA cap structure formation. Nat Commun. 2014;5:3004. doi: 10.1038/ncomms4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hodel AE, Gershon PD, Quiocho FA. Structural basis for sequence-nonspecific recognition of 5′-capped mRNA by a cap-modifying enzyme. Mol Cell. 1998;1(3):443–447. doi: 10.1016/s1097-2765(00)80044-1. [DOI] [PubMed] [Google Scholar]
- 33.Vidgren J, Svensson LA, Liljas A. Crystal structure of catechol O-methyltransferase. Nature. 1994;368(6469):354–358. doi: 10.1038/368354a0. [DOI] [PubMed] [Google Scholar]
- 34.Hager J, Staker BL, Bügl H, Jakob U. Active site in RrmJ, a heat shock-induced methyltransferase. J Biol Chem. 2002;277(44):41978–41986. doi: 10.1074/jbc.M205423200. [DOI] [PubMed] [Google Scholar]
- 35.Lim SP, et al. A scintillation proximity assay for dengue virus NS5 2′-O-methyltransferase—Kinetic and inhibition analyses. Antiviral Res. 2008;80(3):360–369. doi: 10.1016/j.antiviral.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 36.Holm L, Rosenstrom P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Web Server issue):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghosh A, Lima CD. Enzymology of RNA cap synthesis. Wiley Interdiscip Rev RNA. 2010;1(1):152–172. doi: 10.1002/wrna.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang SP, Deng L, Ho CK, Shuman S. Phylogeny of mRNA capping enzymes. Proc Natl Acad Sci USA. 1997;94(18):9573–9578. doi: 10.1073/pnas.94.18.9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Selisko B, et al. Molecular basis for nucleotide conservation at the ends of the dengue virus genome. PLoS Pathog. 2012;8(9):e1002912. doi: 10.1371/journal.ppat.1002912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim SP, et al. Small molecule inhibitors that selectively block dengue virus methyltransferase. J Biol Chem. 2011;286(8):6233–6240. doi: 10.1074/jbc.M110.179184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao Y, et al. Flexibility of NS5 methyltransferase-polymerase linker region is essential for dengue virus replication. J Virol. 2015;89(20):10717–10721. doi: 10.1128/JVI.01239-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33(2):491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 43.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 44.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Xie X, Gayen S, Kang C, Yuan Z, Shi PY. Membrane topology and function of dengue virus NS2A protein. J Virol. 2013;87(8):4609–4622. doi: 10.1128/JVI.02424-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.