Significance
Gene silencing by Polycomb group (PcG) proteins is required to maintain tissue and stage-specific patterns of gene expression. PcG complexes assemble on special elements, Polycomb response elements (PREs), and function to silence gene activity in the surrounding domain. The activity of PREs is subject to regulation and can be epigenetically switched from “on” (silencing) to “off” (permissive for transcription) as development proceeds. Previous studies suggested that a burst of transcription through a PRE displaces PcG complexes, switching the PRE off in a heritable fashion. Here, we directly tested the transcription read-through hypothesis. We show that transcription through a PRE is not sufficient for induce a heritable switch. In fact, continuous transcription through a PRE fails to displace PcG complexes or eliminate repressive chromatin marks.
Keywords: Polycomb, chromatin silencing, bithorax, intergenic transcription, Trithorax
Abstract
In Drosophila, Polycomb (PcG) and Trithorax (TrxG) group proteins are assembled on Polycomb response elements (PREs) to maintain tissue and stage-specific patterns of gene expression. Critical to coordinating gene expression with the process of differentiation, the activity of PREs can be switched “on” and “off.” When on, the PRE imposes a silenced state on the genes in the same domain that is stably inherited through multiple rounds of cell division. When the PRE is switched off, the domain is in a state permissive for gene expression that can be stably inherited. Previous studies have suggested that a burst of transcription through a PRE sequence displaces PcG proteins and provides a universal mechanism for inducing a heritable switch in PRE activity from on to off; however, the evidence favoring this model is indirect. Here, we have directly tested the transcriptional read-through mechanism. Contrary to previous suggestions, we show that transcription through the PRE is not sufficient for inducing an epigenetic switch in PRE activity. In fact, even high levels of continuous transcription through a PRE fails to dislodge the PcG proteins, nor does it remove repressive histone marks. Our results indicate that other mechanisms involving adjacent DNA regulatory elements must be implicated in heritable switch of PRE activity.
During development of multicellular organisms, cell-type specific patterns of gene expression must be established and then stably transmitted. Stable transmission of repressed or active states depends upon Polycomb (PcG) and Trithorax (TrxG) group proteins. PcG proteins maintain repression, whereas TrxG proteins promote gene activity (1–4). Misregulation of PcG and TrxG genes leads to abnormalities in development and cancer (5–8).
In Drosophila, PcG and TrxG proteins are recruited to special sequences called Polycomb response elements (PREs). PREs harbor sites for a collection of DNA-binding proteins that provide a scaffold for the binding of PcG complexes. The DNA-binding proteins include Pho, Pho-like, GAF, Pipsqueak, Zeste, Spps, and Grainyhead (9). The PcG proteins are preassembled into three main complexes (2, 3). The PRC1 complex contains PC, PH, dRing, and Psc core subunits (10–12), whereas the PRC2 complex contains E(z), Esc, Su(z)12, and Caf1 core subunits (13–16). The SET domain of the PRC2 complex E(z) protein has methyltransferase activity and is responsible for generating the repressive H3K27me3 mark on nucleosomes (13–16). This mark is, in turn, recognized by the chromodomain of the PRC1 complex protein PC (17, 18). The third complex, PhoRC contains dSfmbt protein and the DNA-binding factor Pho (19).
In its endogenous setting, the activity of the PREs can be switched “on” and “off.” When on, they function to establish and maintain the silenced state. When off, they are permissive for the activity of genes encompassed in the same regulatory domain (4). This on–off regulation has been recapitulated in transgene experiments, either by neighboring enhancers or upon stimulation by an inducible GAL4 activator (4). In the GAL4 system, sites for the GAL4 protein are included in a transgene carrying a reporter and a test PRE (20–22). In the absence of GAL4, the PRE is on and functions to silence reporter expression. Silencing is relieved when GAL4 is expressed. However, when GAL4 expression is shutoff, silencing activity is reestablished.
Although PRE-silencing activity is typically reestablished after GAL4 disappears, in a number of cases, it has been possible to switch a PRE from on (silencing) to off (permissive) in a manner that could be epigenetically transmitted (20–22). Epigenetic switching requires the coupling of the PRE to other sequences. Rank et al. (22) found that transient GAL4 induction could epigenetically switch the activity state of a Bithorax complex (BX-C) PRE that is contained in a 3.6-kb fragment from Fab-7 region of the complex. Although several mechanisms could potentially explain how GAL4 is able to inheritably switch this PRE off, the mechanism that Rank et al. favored is transcriptional read through by RNA polymerase II (RNAPII). In this model, transcription displaces the PcG complexes from the PRE, turning off the silencing activity of the PRE. RNAPII could additionally reset the epigenetic state by recruiting chromatin modifying and remodeling complexes that imprint marks for active chromatin. Consistent with a transcriptional mechanism, strand-specific noncoding (nc)RNAs spanning the Fab-7 fragment were detected during GAL4 induction. The Fab-7 fragment used in these studies contained not only the major nuclease hypersensitive site associated with the iab-7PRE but also the Fab-7 boundary and ∼1.8 kb of flanking DNA from the adjacent iab-6 cis-regulatory domain. The iab-6 sequences are essential both for a heritable switch and for the expression of the ncRNA. When these sequences were deleted, GAL4 induced expression of the ncRNAs is lost, as is the epigenetic switch. Instead, the PRE regains full active after a transient GAL4 pulse and silences genes linked in the same transgene. Rank et al. found that heritable GAL4 induced switching of two other BX-C PREs, bxd and Mcp, is also accompanied by the expression of ncRNAs. Like the Fab-7 element, the bxd and Mcp fragments contained additional sequences that provide a promoter for the ncRNA transcripts. A similar correlation between transcriptional read-through, in this case, strand-specific, has been described for the switching of a “vestigial” PRE (23).
Although these experiments are consistent with the idea that a burst of transcription through a PRE is itself sufficient to induce a heritable switch in PRE activity, the evidence is indirect and alternative explanations are equally plausible. Moreover, PcG proteins were shown to bind at intronic PREs in Ubx and abd-A in cells where these genes are transcribed, suggesting that transcription does not obligatory leads to dislodgement of PcG proteins (24–26). For this reason, we have directly tested the transcriptional read-through hypothesis. Using a GAL4-inducible promoter and only PRE sequences, we show that a transient burst of transcription through the PRE is not sufficient to induce a heritable epigenetic switch in the activity state of the PRE. When GAL4 protein expression is turned off, the silencing activity of the PRE is reestablished. This finding is not surprising because even continuous GAL4 induction fails to displace PcG complexes, nor does it completely remove silencing histone modifications from PRE-linked transgenes. Finally, because PRE silencing can be relieved by the GAL4 activator independent of the orientation of the GAL4-inducible promoter, the activator must be able to counteract the effects of the PRE by a mechanism that does not require RNAPII movement through the PRE sequences.
Results and Discussion
Experimental Design.
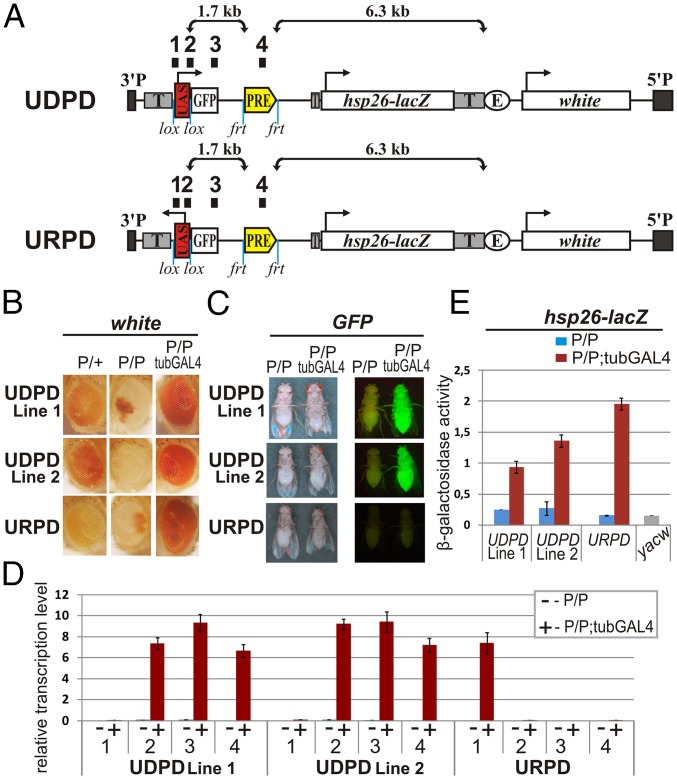
The studies of Rank et al. indicated that epigenetic switching of the iab-7PRE is not an intrinsic feature of this PRE but instead required sequences from the iab-6 cis-regulatory domain (22). Although read-through from a cryptic promoter element in the iab-6 sequence could potentially be responsible for the epigenetic inheritance, it is not known what other regulatory elements and activities might be included in the iab-6 sequences. For this reason, we decided to combine a minimal PRE with a GAL4-inducible promoter. For this purpose, we selected the bxdPRE because the sequences needed for its silencing functions in transgenes have been extensively characterized (22, 27, 28). In their experiments, Rank et al. used a 2.3-kb fragment from the bxd/pbx region of BX-C that contained not only the same bxdPRE but also an additional ∼1,900 bp of flanking bxd/pbx regulatory DNA. We used a smaller 660-bp bxdPRE fragment that is sufficient to assemble fully functional PcG complexes and set repression. The sequence organization of the two transgenes used to test the effects of transcriptional read-through on bxdPRE activity is shown in Fig. 1A. The transgenes contain an upstream activating sequence (UAS) element linked to the hsp70 promoter. In the UDPD transgene, GFP-coding sequences and the bxdPRE are located downstream of the hsp70 promoter. In the control URPD transgene, the hsp70 promoter is flipped so that GAL4-induced transcription is in the opposite direction toward the sequences flanking the transgene insert. The transgenes also have two reporters: hsp26-lacZ and white, the white enhancer and transcription termination elements to block possible genome transcription.
Fig. 1.
Experimental design. (A) Maps of transgenes. The UAS-hsp70 promoter drives GAL4-inducible transcription through GFP and PRE sequences in the UDPD transgene. In the URPD transgene, transcription is directed toward genomic sequences at the site of insertion. Labels: “T,” terminators of transcription; hsp26-lacZ and white, reporters; “E,” enhancer of the white gene. Numbers on top of the construct (1, 2, 3, and 4) indicate positions of primers used for RT–quantitative PCR (RT-qPCR) in D. (B and C) Analysis of the white (B) and GFP (C) in UDPD (line 1 and line 2) and URPD flies in adult animals. P/+, hemizygous; P/P, homozygous; P/P tubGAL4, homozygous with GAL4 activator. (D) RT-qPCR. Individual transcript levels were normalized relative to Ras64B. (E) β-Galactosidase assay. yacw, background line without transgene insertion. (D and E) Error bars indicate SDs.
Multiple lines for each transgene were generated and characterized (SI Appendix, Tables S1 and S2). Fig. 1B shows two representative lines for the UDPD transgene and one line for the URPD transgene selected for detailed analysis. These and most other lines exhibited enhanced (pairing-sensitive) silencing when homozygous. Moreover, silencing was relieved by GAL4 expression under the control of the tubulin promoter. We confirmed that silencing activity depends in each case on the bxdPRE by deleting it (SI Appendix, Fig. S1 and Tables S1 and S2).
GAL4 Activates the hsp70 Promoter and Generates Transcripts That Read-Through the PRE.
We next determined whether GAL4 induces the expression of GFP. Fig. 1C shows that GAL4 activates robust GFP expression in adult flies in both of the UDPD lines, whereas, as expected, no GFP is detected in the URPD line, where the promoter is directed away from the GFP-coding sequences. GFP expression is also induced by GAL4 in the UDPD lines at other stages of development (SI Appendix, Fig. S2).
In the UDPD lines, the transcripts generated by the hsp70 promoter in response to GAL4 are expected to include not only GFP sequences but also sequences spanning the bxdPRE. We used RT-PCR with four sets of primer pairs to examine the RNAs produced by the different transgenes with or without GAL4 induction. These experiments show that in the presence of GAL4, RNAPII transcribes through GFP and bxdPRE sequences in both UDPD lines but not in the URPD line (Fig. 1D).
GAL4 Induces lacZ Expression and Disrupts white Silencing by the bxdPRE.
In addition to turning on the hsp70 promoter, constitutively expressed GAL4 activates hsp26-lacZ. Fig. 1E shows that there is low level of β-galactosidase in the absence of GAL4. When GAL4 is present, it up-regulates β-galactosidase expression between ∼5- and 20-fold in the transgenic lines. Although the fold induction of β-galactosidase in the two UDPD lines is less than in the URPD line, we suspect that this difference is attributable to chromosomal position effects rather than to the orientation of hsp70 promoter.
Constitutive GAL4 expression not only activates the hsp70 and hsp26 promoters but also counteracts the silencing activity of the bxdPRE. Fig. 1B shows that the pairing-sensitive silencing of white in both of the UDPD transgenic lines is suppressed by constitutive GAL4 expression. Moreover, the GAL4 activator can overcome the silencing activity of the bxdPRE without inducing transcription through the PRE sequences (see URPD, Fig. 1B). Thus, when there is constitutive GAL4 activation, relief of silencing activity does not require transcription through the PRE sequences. We used RT-PCR to confirm that GAL4 relieves the silencing of white in all three transgenic lines (SI Appendix, Fig. S3).
RNAPII Transcription Through the bxdPRE Is Not Sufficient to Induce a Heritable Epigenetic Switch.
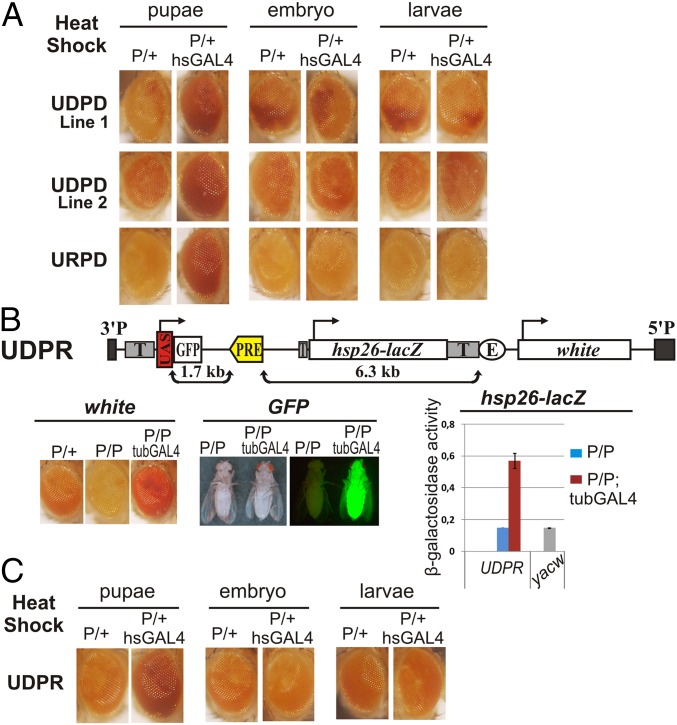
We next determined whether a pulse of transcription through the bxdPRE is sufficient to induce a heritable switch in the silencing activity of this PRE from on to off. To generate a pulse of transcription, we introduced an hsp70:GAL4 transgene into the bxdPRE lines and then heat-shocked at different stages of development. The white gene must be expressed in the eye discs during the pupal stage for eye pigmentation. As expected, heat shock during the pupal stage interfered with bxdPRE silencing and is sufficient to give adult flies that have uniformly pigmented eyes (Fig. 2A). As with continuous GAL4, this disruption of silencing activity does not require transcriptional read-through and was observed in both the UDPD and URPD transgenic lines.
Fig. 2.
Transcription through PRE is not sufficient for epigenetic switch. (A) UDPD and URPD flies were out crossed to wild-type (P/+) or to hsp70-GAL47-1 (P/+ hsGAL4) and heat-shocked at different stages of development (pupa, embryo, and larva). (B) Map of control UDPR transgene containing PRE in reverse orientation. Diagram shows the result of β-galactosidase assay. (C) Effect of hsGAL4 pulse on the white gene expression in the UDPR flies.
To test whether read-through transcription can induce a heritable epigenetic switch in the activity of the bxdPRE, we next heat-shocked embryos and larvae. Previous studies showed that a pulse of the GAL4 activator during the embryonic stage could switch off the silencing activity of large fragments containing PREs in a manner that is epigenetically inherited (20–22). For the Fab-7 fragment, this effect was shown to be stage-specific, because a GAL4 pulse later in development only alleviated silencing transiently, and once GAL4 was removed, silencing was reestablished (20, 21). Fig. 2A shows that a GAL4 pulse during the embryonic stage is not sufficient to switch the bxdPRE off in a heritable fashion in any of the transgenic lines. In fact, although GAL4 induces transcription through the bxdPRE in both the UDPD transgenes, the effects on silencing activity are no different from those observed for the control transgene in which transcription is directed away from PRE (Fig. 2A). Similarly, a pulse of GAL4 during the larval stages also does not induce a heritable switch in the activity of the bxdPRE (Fig. 2).
Changing the 5′→3′ Orientation of the bxdPRE Does Not Induce an Epigenetic Switch.
One plausible explanation for our failure to induce an epigenetic switch with a pulse of GAL4 is that the transcriptional effects on PRE activity are stand-specific. In this case, RNAPII must read through the PRE in the opposite direction from that in our UDPD transgene. To test this possibility, we generated a third transgene, UDPR, in which the orientation of the bxdPRE is the opposite of that in the UDPD transgene (Fig. 2B). As observed for the other transgenes, the bxdPRE in this transgene silences white, and silencing is enhanced in homozygous UDPR flies (Fig. 2B and SI Appendix, Table S3). Like the UDPD transgene, GFP expression is induced at all stages when combined with the tublin-GAL4 driver and transcription reads through the bxdPRE sequence (SI Appendix, Fig. S4). The tubulin-GAL4 driver also relieves silencing of white. Fig. 2C shows that transcription through the reversed bxdPRE in either embryos or larvae is not sufficient to switch silencing activity off in heritable fashion; the bxdPRE-induced silencing of white is relieved only when GAL4 is expressed during the pupal stage.
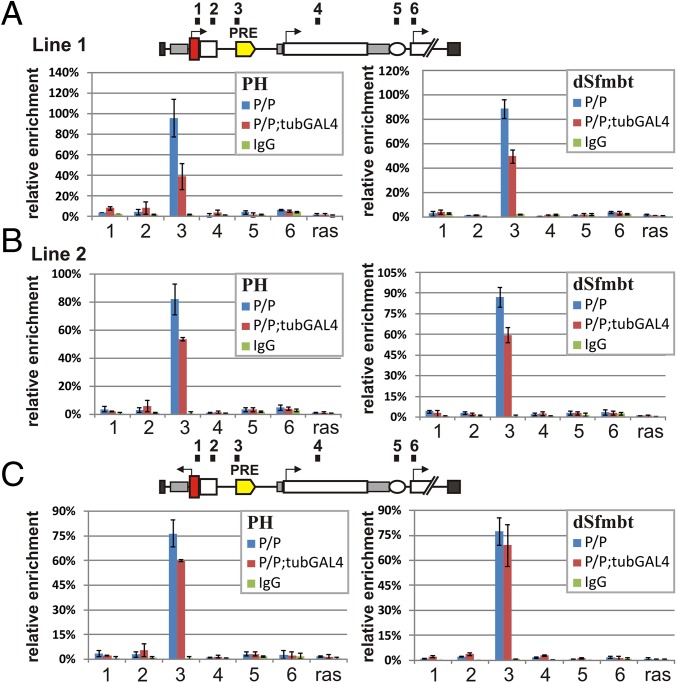
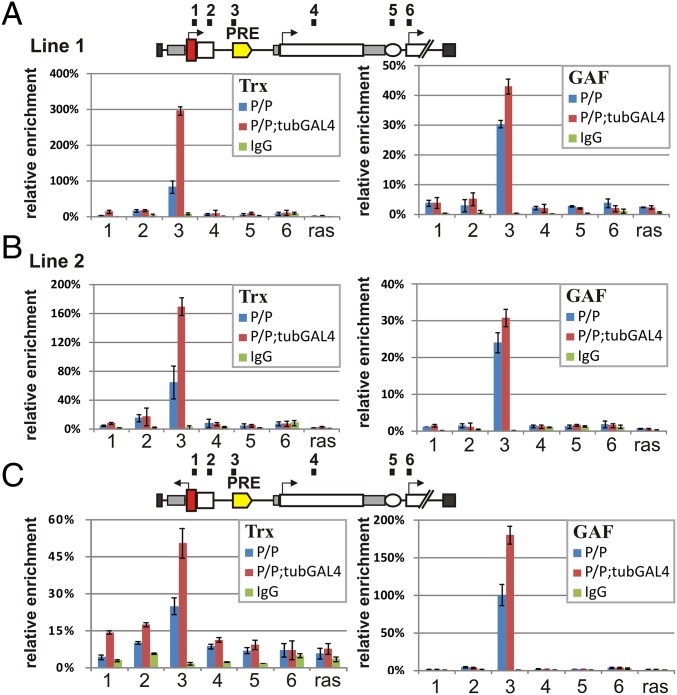
Transcriptional Read-Through Reduces but Does Not Eliminate in Vivo Binding of dSfmbt and PH to the bxdPRE.
It has been suggested that transcriptional read-through permanently switches PRE activity off by displacing PcG complexes and inducing a chromatin state that is permissive for gene activity. Because our experiments argue that this hypothesis is likely incorrect, we sought to understand what effects read-through transcription has on components of the PcG silencing system. We used chromatin immunoprecipitation (ChIP) in embryos and adults to examine the association of the PRC1 complex protein PH and the PhoRC complex protein dSfmbt with sequences in six different regions of the transgene: (i) the hsp70 promoter; (ii) the GFP ORF; (iii) the bxdPRE; (iv) the lacZ ORF; (v) the white enhancer; and (vi) the white promoter. To standardize the amount of PH and dSfmbt associated with each region of the transgene in different samples, we calculated the extent enrichment relative to a positive internal control—a sequence adjacent to the 660 bxdPRE in BX-C. For a negative control, we used the Ras64B coding region.
Fig. 3 shows that under conditions in which the PRE is fully functional and represses the white gene, the association of PH and dSfmbt in adults is restricted to the bxdPRE. Similar results were obtained in embryos (SI Appendix, Fig. S5). These findings are consistent with genome-wide studies showing that both proteins preferentially localize to PREs but are not found at high levels elsewhere in silenced domains (19, 26, 29, 30). We next examined the association of PH and dSfmbt under conditions in which constitutive expression of GAL4 from the tubulin promoter disrupts the silencing activity of the bxdPRE. Several findings are of interest. First, although tethering GAL4 to the transgene UAS clearly disrupts bxdPRE silencing, neither PH nor dSfmbt is dislodged from the PRE either in adults or in embryos (Fig. 3 A and B and SI Appendix, Fig. S5). Instead, there is only a modest reduction in the level of the two proteins bound to the bxdPRE. Second, when comparing UDPD (Fig. 3 A and B) and URPD (Fig. 3C), it appears that transcription through the bxdPRE contributes, at least in part, to the modest reduction in the binding of dSfmbt in the presence of GAL4. In the case of PH, the differences between the UDPD and URPD transgenes with and without GAL4 are too small to draw any conclusions.
Fig. 3.
PH (PRC1) and dSfmbt (PhoRC) proteins remain bound to PRE during transcriptional read-through. (A) UDPD construct line 1. (B) UDPD construct line 2. (C) URPD construct. The ChIP experiments were performed with chromatin isolated from adult flies. Numbers on top of the construct (1, 2, 3, 4, 5, and 6) indicate regions amplified by qPCR. The results of ChIP are presented as a percentage of input DNA normalized relative to the endogenous positive control region adjacent to bxdPRE in the genome. The coding part of the Ras64B gene was used as negative control. The blue bars indicate relative cross-linked ChIP (X-ChIP) signal levels in homozygote lines (P/P), red bars indicate relative X-ChIP signal levels in homozygote lines with GAL4 activator (P/P;+GAL4), and green bars indicate signal levels with nonspecific antibodies. Vertical lines indicate SDs.
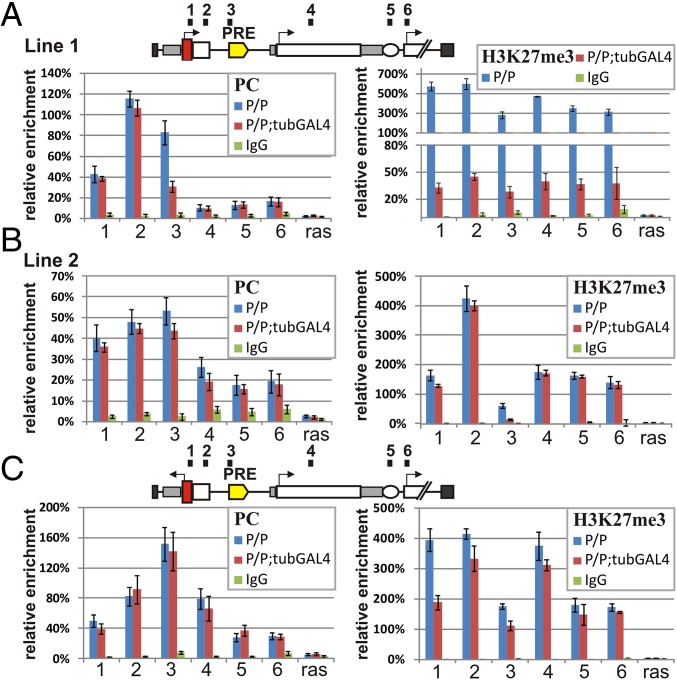
PC (PRC1) and H3K27me3 Remain Associated with bxdPRE in the Presence of Read-Through Transcription.
The PRC1 complex PC protein specifically interacts with histone 3 trimethylated at K27 (H3K27me3), the nucleosome mark stably associated with PcG-repressed chromatin (30, 31). Unlike other PRC core components, the distribution of PC and H3K27me3 in PcG silenced domains is not limited to the PRE but instead extends outward from the PRE (26, 30, 31). We obtained similar results: ChIP experiments indicate that PC and H3K27me3 are associated with transgene sequences to either side of the PRE including white gene (Fig. 4 and SI Appendix, Fig. S6). The effects of GAL4 induction on PC association are minimal (with one exception: UDPD sequence 3) (Fig. 4). A more complicated position-dependent pattern is observed for the H3K27me3 mark. In one of the UDPD lines (UDPD line 1), this mark is present throughout the transgene and is significantly depressed in the presence of GAL4 (Fig. 4). A lower level of the H3K27me mark is detected in UDPD line 2 and in the URPD line in the absence of GAL4, whereas only minimal changes are observed at most sites in the presence of GAL4.
Fig. 4.
Persistence of PC (PRC1) and H3K27me3 during transcription. (A) UDPD construct line 1. (B) UDPD construct line 2. (C) URPD construct. For other designations, see the legend of Fig. 3.
Recruitment of TrxG Proteins to bxdPRE in the Presence of Read-Through Transcription.
PcG repression is associated with a competition with TrxG proteins (4). For example, it was shown that Trx could be recruited to PREs in both activating and in repressing states (26, 29). Another factor—GAF protein—was originally identified as a TrxG member (32); however, further studies indicated involvement of GAF in PRE-silencing function (33–37).
To gain further insights into what molecular changes in PRE architecture might be induced by GAL4, we analyzed the recruitment of Trx and GAF in the absence and presence of GAL4 (Fig. 5 and SI Appendix, Fig. S7). ChIP experiments show that both are associated with the bxdPRE in the absence of GAL4. In the presence of GAL4, the level of these two protein changes to a greater or lesser extent depending upon the insertion site. However, instead of decreasing, as observed for PH and dSfmbt, the level of Trx and GAF enrichment increased (Fig. 5).
Fig. 5.
Trx and GAF protein association increases during transcription. (A) UDPD construct line 1. (B) UDPD construct line 2. (C) URPD construct. For other designations, see the legend of Fig. 3.
Conclusions
PcG-dependent silencing in Drosophila depends upon special cis-acting elements called PREs. These elements recruit PCR1, PCR2, and PhoRC and, in doing so, impose PcG-dependent silencing on the domain in which the PRE resides. In transgene assays, PREs exhibit several characteristic properties. Although silencing activity is subject to position effects, PREs in transgenes are mostly active (on). PREs silence linked reporter genes throughout development without any intrinsic stage or tissue specificity (4, 38). Their silencing activity is also often enhanced by pairing of PREs in trans (38).
Although PREs in transgenes are constitutively active silencers, in their endogenous context, PREs’ activity state can be switched from on to off (and vice versa). For example, the bxdPRE is located in the BX-C bxd/pbx cis-regulatory domain (27, 28). The bxd/pbx domain is responsible for directing Ultrabithorax (Ubx) expression in parasegment PS6 (39). In parasegments anterior to PS6, the bxd/pbx domain is silenced by a mechanism that depends upon the ability of the bxdPRE (and other PRE-like elements in the regulatory domain) to recruit PcG complexes (40). The on activity state of the bxdPRE in anterior parasegments is established in the early embryo and is then maintain during the remainder of development (4, 39). By contrast, in PS6 and more posterior parasegments, the bxdPRE is switch off in the early embryo and is then maintained in the off state during the rest of development. As a consequence, the bxd/pbx cis-regulatory domain is stably activated and can direct Ubx expression. The differences between the on and off state of the bxdPRE (and other PRE-like elements in bxd/pbx) is evident from the H3K27me3 ChIP experiments of earlier studies (24, 26) showing that nucleosomes of the bxd/pbx cis-regulatory domain have high levels of H3K27me3 when the target gene is silent and only low levels if it is active.
In the BX-C, the activity state of the PREs (on or off) and of the cis-regulatory domain in which the PREs reside is determined by special elements called parasegment-specific initiators (39). Initiators function during early embryogenesis and their parasegment specificity is determined by maternal and zygotic segmentation genes. Although initiators must be able to switch the PREs in the cis-regulatory domain off when they activate the domain, it is not understood how the initiators communicate with the PREs. One model for switching PREs from on to off is that the initiators activate transcription through the PREs. In this model, RNAPII transcription read-through displaces PcG complexes from the PRE, allowing chromatin-modifying and -remodeling complexes to establish a chromatin structure that prevents the reassociation of PcG complexes. This model has been supported by studies showing that ncRNAs complementary to the PREs in different BX-C cis-regulatory domains are expressed in parasegment-specific patterns that mimic the parasegment-specific activation of the regulatory domain (41). It has also been supported by transgene studies showing that the epigenetic state of a PRE can be switched when the PRE is linked to sequences from the surrounding region of BX-C that have cryptic, GAL4-inducible promoters (22).
In the studies reported here, we have used a minimal bxdPRE and a GAL4-inducible promoter to test the transcriptional read through model. As has been reported for other PREs (20–22), we find that the silencing activity of the 660-bp bxdPRE can be relieved by GAL4. When binding to the UAS in the transgene, GAL4 turns on the expression of the linked reporter genes and alleviates PRE-dependent silencing of the white gene. The effect of GAL4 on PRE-dependent silencing of white (and the other reporters in the transgene) does not require the transcription of RNAPII through the PRE. It is observed whether the GAL4-inducible promoter is directed toward or away from the PRE. Furthermore, we find that a pulse of RNAPII transcription through the bxdPRE in response to GAL4 is not sufficient to induce a heritable switch in the activity of the PRE from on to off. Instead, when expression of GAL4 activator is turned off, silencing by the PRE is reestablished.
Our ChIP experiments provide a plausible explanation for why transcriptional read-through is unable to permanently disrupt PRE function. These experiments show that even in the presence of a steady source of the GAL4 activator, and thus continuous and high level transcription, the PcG proteins PH, dSfmbt, and PC remain associated with the bxdPRE. Transcriptional read-through has essentially no effect on PC association, whereas for PH and dSfmbt, there is at most only about a twofold reduction in their binding. As for the PcG-specific histone mark, H3K27me3, the effectiveness of the GAL4 activator depends upon chromosomal position. For one of the UDPD lines, there is a marked reduction in H3K27me3 at all of the transgene sequences tested. For the other UDPD line, the GAL4 activator has little effect on the level the H3K27me3 modification. The same is true for URPD transgene, where GAL4-inducible transcription is directed away from the PRE.
Our findings clearly demonstrate that transcriptional read-through per se does not disrupt or block the association of PcG complexes with the bxdPRE, nor does transcriptional read-through prevent the accumulation of the PcG-specific, histone modification H3K27me3 across the transgene insert. There are other reasons to think that transcriptional read-through cannot be the (sole) mechanism for inactivating a PRE such as the bxdPRE in its natural setting. The small effects on PcG protein binding and H3K27me3 that we see are dependent on chromosomal position and likely reflect the fact that the environment near the transgene insert is generally permissive for transcription. This is not the case in silenced BX-C cis-regulatory domains. In fact, Mihaly et al. (42) have shown that a deletion of the iab-7PRE is not sufficient to disrupt silencing of the iab-7 cis-regulatory domain. The reason for this finding is that there are other PRE-like elements in the iab-7 domain that can compensate for the loss of the primary PRE.
Although our experiments argue that other mechanisms must be in place to induce an epigenetic switch from on (silencing) to off (permissive), what these mechanisms are is unclear. The experiments of Rank et al. (22) indicate that sequences outside of the PRE in the fragments containing BX-C PREs bxd, Mcp, and iab-7 are required for the epigenetic switch. One possibility is that these sequences contain enhancer-like elements that not only counteract the repressive effects of the PRE but can also do so in a heritable fashion. Another possibility is that transcripts produced from the cryptic promoters in these fragments contain RNA motifs (distinct from the PRE sequences) that, for example, are able to recruit TrxG complexes to the PRE and block PcG activity. In either case, it should be noted that PcG proteins can still be detected at PREs that are located within BX-C regulatory domains that are active and lack the H3K27me3 mark (24, 26). This finding would argue that regulating PRE activity might not involve a mechanism requiring displacement of PcG proteins from PREs. Clearly, the next step will be to discover this mechanism.
Materials and Methods
Drosophila strains, germ line transformation, genetic crosses, and plasmid construction are described in SI Appendix, Supplemental Methods.
β-Galactosidase Activity Assay.
Twenty adult flies were homogenized in buffer I (1 mM MgCl2; 50 mM KH2PO4, pH 7.6; 1 mM PMSF). Homogenates were incubated with chlorophenol Red-β-D-galactopyranoside (CPRG) for 20 min, and then optical density was determined at 595 nm. The results obtained were normalized to the total amount of protein calculated using the BCA Protein Assay kit (Thermo Scientific) according to the manufacturer’s instructions.
RT-PCR.
RNA was isolated from ∼20 adult flies homozygote for the construct. Experimental procedures were described previously (43), and the details of experimental procedures are given in SI Appendix, Supplemental Methods. Primers are listed in SI Appendix, Table S4.
Cross-Linked ChIP–Quantitative PCR.
For each experiment, 150–200 mg of the initial material (0- to 16-h embryos or from 2- to 5-d-old adult flies homozygote for the construct) was collected. Experimental procedures were described previously (43), and the details of experimental procedures are given in SI Appendix, Supplemental Methods. Primers are listed in SI Appendix, Table S5.
Antibodies.
Antibodies used are listed in SI Appendix, Table S6.
Supplementary Material
Acknowledgments
We acknowledge support from Russian Foundation for Basic Research Grant 15-34-20581-mol-а-ved (to D.C.), the Ministry of Education and Science of the Russian Federation (Grant 14.B25.31.0022), and National Institutes of Health Grant R01GM043432.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.E.K. is a guest editor invited by the Editorial Board.
See Commentary on page 14755.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1515276112/-/DCSupplemental.
References
- 1.Beisel C, Paro R. Silencing chromatin: Comparing modes and mechanisms. Nat Rev Genet. 2011;12(2):123–135. doi: 10.1038/nrg2932. [DOI] [PubMed] [Google Scholar]
- 2.Müller J, Verrijzer P. Biochemical mechanisms of gene regulation by polycomb group protein complexes. Curr Opin Genet Dev. 2009;19(2):150–158. doi: 10.1016/j.gde.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Schwartz YB, Pirrotta V. A new world of Polycombs: Unexpected partnerships and emerging functions. Nat Rev Genet. 2013;14(12):853–864. doi: 10.1038/nrg3603. [DOI] [PubMed] [Google Scholar]
- 4.Steffen PA, Ringrose L. What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat Rev Mol Cell Biol. 2014;15(5):340–356. doi: 10.1038/nrm3789. [DOI] [PubMed] [Google Scholar]
- 5.Aloia L, Di Stefano B, Di Croce L. Polycomb complexes in stem cells and embryonic development. Development. 2013;140(12):2525–2534. doi: 10.1242/dev.091553. [DOI] [PubMed] [Google Scholar]
- 6.Brockdorff N. Chromosome silencing mechanisms in X-chromosome inactivation: Unknown unknowns. Development. 2011;138(23):5057–5065. doi: 10.1242/dev.065276. [DOI] [PubMed] [Google Scholar]
- 7.Grossniklaus U, Paro R. Transcriptional silencing by polycomb-group proteins. Cold Spring Harb Perspect Biol. 2014;6(11):a019331. doi: 10.1101/cshperspect.a019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weaver JR, Bartolomei MS. Chromatin regulators of genomic imprinting. Biochim Biophys Acta. 2014;1839(3):169–177. doi: 10.1016/j.bbagrm.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McElroy KA, Kang H, Kuroda MI. Are we there yet? Initial targeting of the Male-Specific Lethal and Polycomb group chromatin complexes in Drosophila. Open Biol. 2014;4:140006. doi: 10.1098/rsob.140006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Francis NJ, Saurin AJ, Shao Z, Kingston RE. Reconstitution of a functional core polycomb repressive complex. Mol Cell. 2001;8(3):545–556. doi: 10.1016/s1097-2765(01)00316-1. [DOI] [PubMed] [Google Scholar]
- 11.Saurin AJ, Shao Z, Erdjument-Bromage H, Tempst P, Kingston RE. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature. 2001;412(6847):655–660. doi: 10.1038/35088096. [DOI] [PubMed] [Google Scholar]
- 12.Shao Z, et al. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98(1):37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 13.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298(5595):1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 14.Czermin B, et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111(2):185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 15.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16(22):2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Müller J, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111(2):197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 17.Fischle W, et al. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17(15):1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17(15):1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klymenko T, et al. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 2006;20(9):1110–1122. doi: 10.1101/gad.377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cavalli G, Paro R. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell. 1998;93(4):505–518. doi: 10.1016/s0092-8674(00)81181-2. [DOI] [PubMed] [Google Scholar]
- 21.Cavalli G, Paro R. Epigenetic inheritance of active chromatin after removal of the main transactivator. Science. 1999;286(5441):955–958. doi: 10.1126/science.286.5441.955. [DOI] [PubMed] [Google Scholar]
- 22.Rank G, Prestel M, Paro R. Transcription through intergenic chromosomal memory elements of the Drosophila bithorax complex correlates with an epigenetic switch. Mol Cell Biol. 2002;22(22):8026–8034. doi: 10.1128/MCB.22.22.8026-8034.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herzog VA, et al. A strand-specific switch in noncoding transcription switches the function of a Polycomb/Trithorax response element. Nat Genet. 2014;46(9):973–981. doi: 10.1038/ng.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bowman SK, et al. H3K27 modifications define segmental regulatory domains in the Drosophila bithorax complex. eLife. 2014;3:e02833. doi: 10.7554/eLife.02833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwong C, et al. Stability and dynamics of polycomb target sites in Drosophila development. PLoS Genet. 2008;4(9):e1000178. doi: 10.1371/journal.pgen.1000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papp B, Müller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 2006;20(15):2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan CS, Rastelli L, Pirrotta V. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 1994;13(11):2553–2564. doi: 10.1002/j.1460-2075.1994.tb06545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Müller J, Bienz M. Long range repression conferring boundaries of Ultrabithorax expression in the Drosophila embryo. EMBO J. 1991;10(11):3147–3155. doi: 10.1002/j.1460-2075.1991.tb04876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beisel C, et al. Comparing active and repressed expression states of genes controlled by the Polycomb/Trithorax group proteins. Proc Natl Acad Sci USA. 2007;104(42):16615–16620. doi: 10.1073/pnas.0701538104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuettengruber B, et al. Functional anatomy of polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol. 2009;7(1):e13. doi: 10.1371/journal.pbio.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwartz YB, et al. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38(6):700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- 32.Farkas G, et al. The Trithorax-like gene encodes the Drosophila GAGA factor. Nature. 1994;371(6500):806–808. doi: 10.1038/371806a0. [DOI] [PubMed] [Google Scholar]
- 33.Busturia A, et al. The MCP silencer of the Drosophila Abd-B gene requires both Pleiohomeotic and GAGA factor for the maintenance of repression. Development. 2001;128(11):2163–2173. doi: 10.1242/dev.128.11.2163. [DOI] [PubMed] [Google Scholar]
- 34.Hagstrom K, Muller M, Schedl P. A Polycomb and GAGA dependent silencer adjoins the Fab-7 boundary in the Drosophila bithorax complex. Genetics. 1997;146(4):1365–1380. doi: 10.1093/genetics/146.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hodgson JW, Argiropoulos B, Brock HW. Site-specific recognition of a 70-base-pair element containing d(GA)(n) repeats mediates bithoraxoid polycomb group response element-dependent silencing. Mol Cell Biol. 2001;21(14):4528–4543. doi: 10.1128/MCB.21.14.4528-4543.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horard B, Tatout C, Poux S, Pirrotta V. Structure of a polycomb response element and in vitro binding of polycomb group complexes containing GAGA factor. Mol Cell Biol. 2000;20(9):3187–3197. doi: 10.1128/mcb.20.9.3187-3197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strutt H, Cavalli G, Paro R. Co-localization of Polycomb protein and GAGA factor on regulatory elements responsible for the maintenance of homeotic gene expression. EMBO J. 1997;16(12):3621–3632. doi: 10.1093/emboj/16.12.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kassis JA. Pairing-sensitive silencing, polycomb group response elements, and transposon homing in Drosophila. Adv Genet. 2002;46:421–438. doi: 10.1016/s0065-2660(02)46015-4. [DOI] [PubMed] [Google Scholar]
- 39.Maeda RK, Karch F. The open for business model of the bithorax complex in Drosophila. Chromosoma. 2015;124(3):293–307. doi: 10.1007/s00412-015-0522-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kassis JA, Brown JL. Polycomb group response elements in Drosophila and vertebrates. Adv Genet. 2013;81:83–118. doi: 10.1016/B978-0-12-407677-8.00003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bae E, Calhoun VC, Levine M, Lewis EB, Drewell RA. Characterization of the intergenic RNA profile at abdominal-A and Abdominal-B in the Drosophila bithorax complex. Proc Natl Acad Sci USA. 2002;99(26):16847–16852. doi: 10.1073/pnas.222671299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mihaly J, Hogga I, Gausz J, Gyurkovics H, Karch F. In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development. 1997;124(9):1809–1820. doi: 10.1242/dev.124.9.1809. [DOI] [PubMed] [Google Scholar]
- 43.Erokhin M, et al. Transcription through enhancers suppresses their activity in Drosophila. Epigenetics Chromatin. 2013;6(1):31. doi: 10.1186/1756-8935-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.