Significance
Despite decades of research, we still have a very incomplete understanding of what is special about the human brain compared with the brains of our closest fossil and living relatives. Parsing the genetic versus environmental factors that govern the structure of the cerebral cortex in humans and chimpanzees may shed light on the evolution of behavioral flexibility in the human lineage. We show that the morphology of the human cerebral cortex is substantially less genetically heritable than in chimpanzees and therefore is more responsive to molding by environmental influences. This anatomical property of increased plasticity, which is likely related to the human pattern of development, may underlie our species’ capacity for cultural evolution.
Keywords: brain evolution, plasticity, hominins, neocortex, altriciality
Abstract
The study of hominin brain evolution has focused largely on the neocortical expansion and reorganization undergone by humans as inferred from the endocranial fossil record. Comparisons of modern human brains with those of chimpanzees provide an additional line of evidence to define key neural traits that have emerged in human evolution and that underlie our unique behavioral specializations. In an attempt to identify fundamental developmental differences, we have estimated the genetic bases of brain size and cortical organization in chimpanzees and humans by studying phenotypic similarities between individuals with known kinship relationships. We show that, although heritability for brain size and cortical organization is high in chimpanzees, cerebral cortical anatomy is substantially less genetically heritable than brain size in humans, indicating greater plasticity and increased environmental influence on neurodevelopment in our species. This relaxed genetic control on cortical organization is especially marked in association areas and likely is related to underlying microstructural changes in neural circuitry. A major result of increased plasticity is that the development of neural circuits that underlie behavior is shaped by the environmental, social, and cultural context more intensively in humans than in other primate species, thus providing an anatomical basis for behavioral and cognitive evolution.
Compared with nonhuman primates, human brains are significantly enlarged, reorganized, and have a disproportionately expanded neocortex (1–3). The fossil evidence demonstrates that these changes occurred in the hominin lineage over the last ∼6–8 My (4–9) in parallel with modifications to neurodevelopmental rates (10–13). Although some of these changes have been linked to certain genetic variants in the human lineage [either shared with other late hominin species or exclusive to modern humans (14, 15)], exploring brain evolution in hominins is challenging because of the limitations of the endocranial fossil record (4, 5). Comparisons of chimpanzee and human brains therefore are essential to reveal the neural traits that differ between the two species, that underlie their behavioral specializations, and that must have evolved after they split from their last common ancestor.
Human behavioral and cognitive development is highly dependent on cultural influences and social learning (16, 17). Notably, modern human behavioral adaptations for living in diverse habitats depend on skills and information learned from others (18). Similarly, it has been demonstrated that enculturated great apes perform better in different tasks related to physical and, especially, social cognition (19), underscoring the importance of environmental influences in shaping behavior. These observations are congruent with experimental studies in mouse models showing that variation in sensory experience early in postnatal life causes reorganization of neural circuits that underlie behavior (20). However, the clear differences in behavioral and cognitive development between enculturated apes and humans point to particular neural specializations that make the human brain—but not the brain of great apes—extremely responsive to exogenous influences. In this light, several comparative studies have shown molecular and microstructural specializations in the human brain indicating an increased level of synaptic plasticity (21, 22), which might be linked to increased learning abilities.
The potential role that changes in life history and developmental patterns may have had in human brain evolution has been highlighted in paleoanthropology and primatology (10, 13). It is generally assumed that the extended period of growth and delayed maturation of humans in the context of a complex social environment is related to our species’ cognitive specializations (13). It remains to be clarified, however, if the human brain is indeed more extensively modeled by environmental factors than the brain of our closest living and fossil relatives. In the current study we evaluated heritability of brain size and cortical organization in chimpanzees and humans to assess the relative contribution of genes and environment to neural development. Heritability is defined as the proportion of total phenotypic variance in a population that has a genetic basis. The heritability of traits can be calculated from phenotypic similarities between individuals with different degrees of genetic similarity.
The sample included MRI scans of 206 chimpanzees and 218 humans. A well-documented pedigree is available for the chimpanzees, and the human sample includes monozygotic twins, nonmonozygotic twins, and nontwin siblings. MRI scans were used to measure brain volume and to reconstruct 3D models of the cortical surface. Cortical organization was characterized through a set of anatomically homologous landmarks (SI Text, Fig. S1, and Table S1), which were analyzed using linear distances (Fig. S2 and Table S2), and a geometric morphometric approach (Datasets S1–S3). Linear metrics and principal components of shape were obtained after each individual brain was scaled to a common size through Procrustes superimposition, removing the effects of differences in overall brain size. Consequently, these metrics do not reflect absolute size but instead represent relative lobe proportions and sulcal measurements. This homology-based method allows comparability across species despite differences in cortical anatomy and variation in scanning procedures. Also, the geometric morphometric approach captures important information about the position and orientation of different cortical regions that is overlooked when focusing on the volumes or surface areas of these regions. Additionally, this landmark-based approach avoids intensive automatic processing of anatomical data, which has been demonstrated to have a significant effect on neuroanatomical studies (23). Because of the importance of differential cortical expansion and reorganization in both evolution and development (24), we selected variables related to the morphology of sulci across the cerebral cortex. Sulcal variation shows a close correspondence with primary sensory and motor cytoarchitectonic areas (25) but a more variable correspondence with high-order association areas in both chimpanzees and humans (25, 26). In humans, sulcal morphology shows a high degree of interindividual variability that is linked to differences in functional networks and long-range corticocortical connectivity (27), whereas lobe- or region-specific volumetric measures and cortical thickness have been shown to be less variable and highly heritable (28).
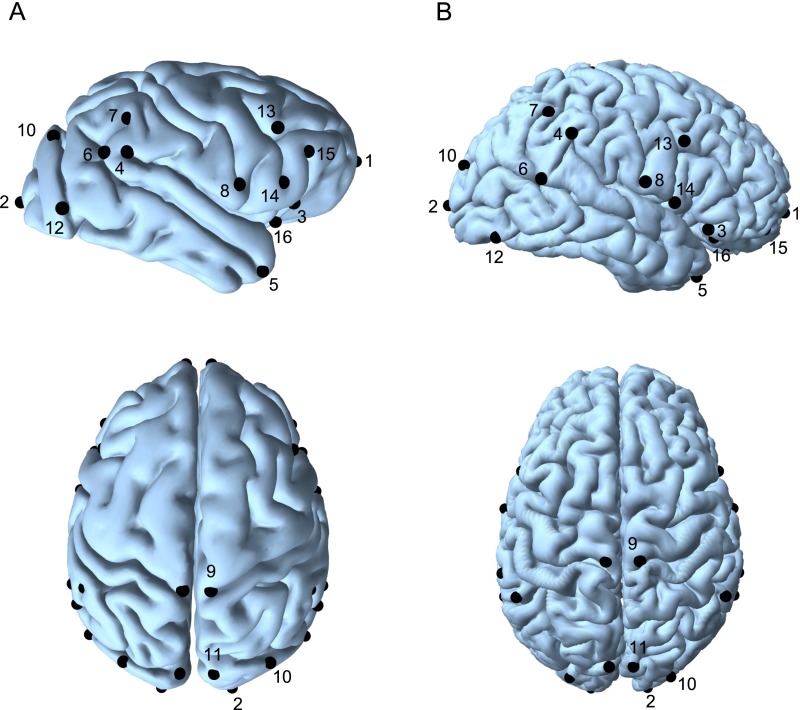
Fig. S1.
Anatomically homologous landmarks used in this study. (A) Anatomically homologous landmarks in chimpanzee brains in lateral view (Upper) and dorsal view (Lower). (B) Anatomically homologous landmarks in a representative human brain in lateral view (Upper) and dorsal view (Lower). Definitions are listed in Table S1.
Table S1.
Definition of anatomical landmarks
| Landmark | Definition |
| 1 | Frontal pole |
| 2 | Occipital pole |
| 3 | Anterior end of the Sylvian fissure (defined on the pars orbitalis in humans) |
| 4 | Posterior end of the Sylvian fissure (following the main course of the fissure when the terminal segment is divided) |
| 5 | Anterior end of the superior temporal sulcus (close to the temporal pole) |
| 6 | Inflection point between the horizontal segment and the ascending segment of the superior temporal sulcus |
| 7 | Most posterior and superior point of the superior temporal sulcus (located between the supramarginal gyrus and the angular gyrus) |
| 8 | Inferior termination of the central sulcus |
| 9 | Superior termination of the central sulcus (intersection between the central sulcus and the midline) |
| 10 | In chimpanzees: intersection between the intraparietal sulcus and the lunate sulcus |
| In humans: intersection between the intraparietal sulcus and the transverse occipital sulcus | |
| 11 | In chimpanzees: intersection of the lunate sulcus with the midline |
| In humans: intersection of the parieto-occipital sulcus with the midline | |
| 12 | In chimpanzees: most inferior and lateral point of the lunate sulcus |
| In humans: occipital notch | |
| 13 | Intersection of the inferior frontal sulcus with the precentral sulcus |
| 14 | Inferior end of the precentral sulcus |
| 15 | In chimpanzees: superior end of the fronto-orbital sulcus |
| In humans: anterior end of the latero-orbital sulcus | |
| 16 | In chimpanzees: inferior end of the fronto-orbital sulcus |
| In humans: posterior end of the latero-orbital sulcus |
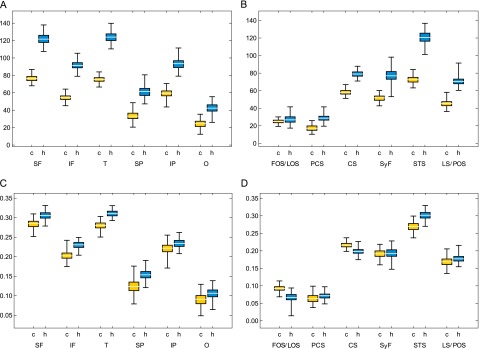
Fig. S2.
Boxplots showing variation in linear metrics. Data for chimpanzees (c) are shown in yellow, and data for humans (h) are shown in blue. (A) Variation in original lobe dimensions (in native space). (B) Variation in original sulcal dimensions. In A and B dimensions are provided in millimeters. (C) Variation in lobe dimensions measured in Procrustes-superimposed configurations of landmarks. (D) Variation in sulcal dimensions in Procrustes-superimposed configurations. In C and D linear dimensions were measured after scaling all individuals to a centroid size of 1. In A and C, IF, inferior frontal length; IP, inferior parietal length; O, occipital length; SF, superior frontal length; SP, superior parietal length; T, temporal length. In B and D, CS, central sulcus; FOS, fronto-orbital sulcus; LOS, latero-orbital sulcus; LS, lunate sulcus; PCS, precentral sulcus; POS, parieto-occipital sulcus; STS, superior temporal sulcus; SyF, Sylvian fissure.
Table S2.
Definitions of lobe and sulcal dimensions
| Variable | Defined between landmarks |
| Lobe dimensions | |
| Superior frontal length (SF) | 1 and 9 |
| Inferior frontal length (IF) | 1 and 8 |
| Temporal length (T) | 5 and 10 |
| Superior parietal length (SP) | 9 and 11 |
| Inferior parietal length (IP) | 8 and 10 |
| Occipital length (O) | 2 and 11 |
| Sulcal dimensions | |
| Fronto-orbital (FOS) or latero-orbital sulcus (LOS) | 15 and 16 |
| Precentral sulcus (PCS) | 13 and 14 |
| Central sulcus (CS) | 8 and 9 |
| Sylvian fissure (SyF) | 3 and 4 |
| Superior temporal sulcus (STS) | 5 and 6, plus 6 and 7 |
| Lunate (LS) or parieto-occipital sulcus (POS) | 10 and 11, plus 10 and 12 |
Results
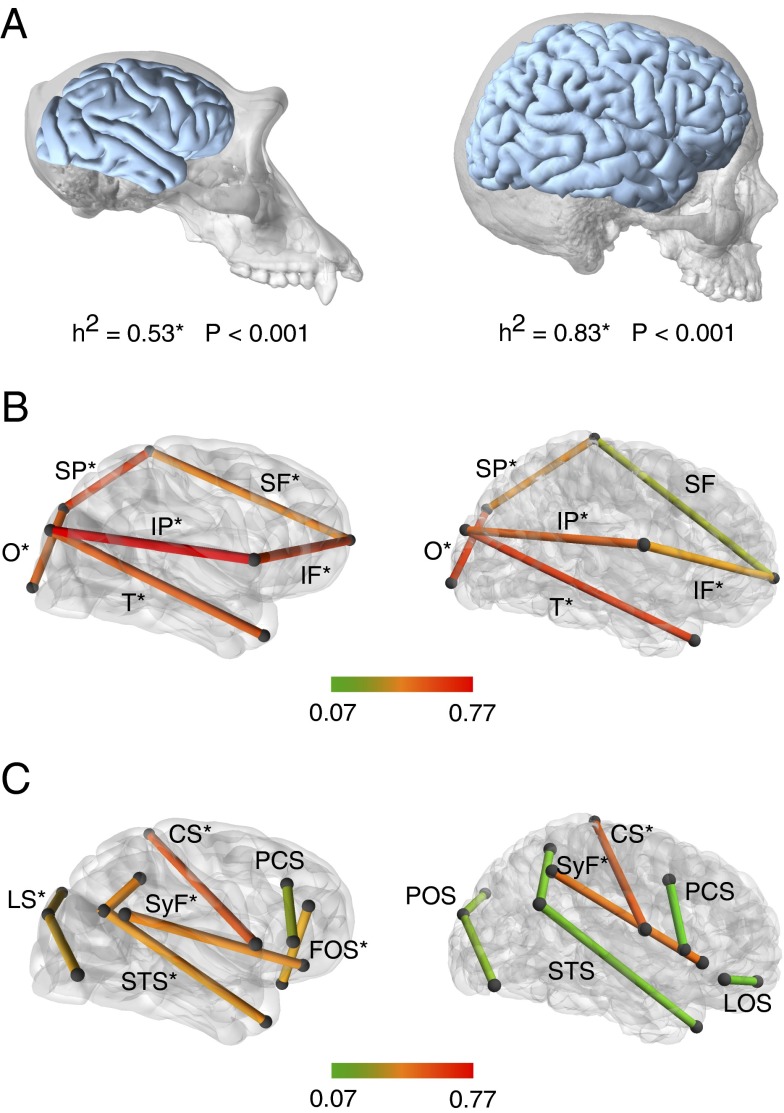
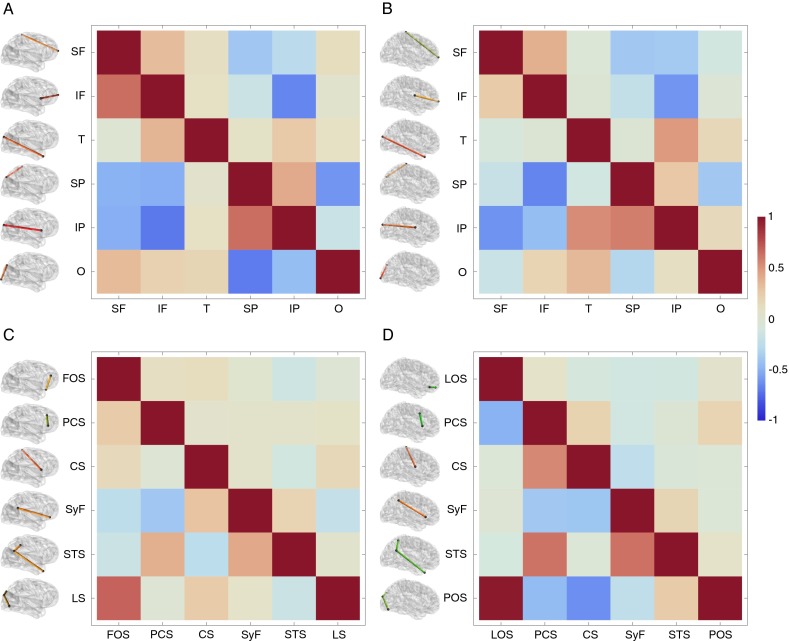
Consistent with previous studies (28), our findings demonstrate that humans show very high heritability for brain size (Fig. 1A and Table S3). Chimpanzees show significant heritability for brain size although substantially lower than in humans (Fig. 1A and Table S3). Several reviews and meta-analyses have demonstrated that twin-based studies inflate heritability estimates compared with family- or pedigree-based studies (28); such differences are likely related to the higher heritability for brain size observed in the human sample (SI Text). Cerebral lobe dimensions show significant and relatively high heritability in both chimpanzees and humans (Fig. 1B and Table S4). In both species the sulci that demarcate cerebral lobe subdivisions, such as the central sulcus and the Sylvian fissure, also have significant heritability (Fig. 1C and Table S4), pointing to strong genetic control of lobar organization. However, other sulci within cortical association regions show significant heritability only in chimpanzees, but not in humans (Fig. 1C and Table S4). Low heritabilities in human sulci within higher-order association regions suggest a greater degree of plasticity in brain architecture that is not observed in chimpanzees. Genetic correlations between variables tend to be low in both species, although there are exceptions that include some lobe dimensions (Fig. S3 and Tables S5 and S6). Those correlations reflect the inverse relationship between relative proportions of cerebral lobes.
Fig. 1.
Heritability for brain size and lobe and sulcal dimensions. (A) Heritability for brain size (brain volume including white and gray matter but not ventricular spaces) for chimpanzees (Left) and humans (Right). (B) Heritability for cerebral lobe dimensions in chimpanzees (Left) and humans (Right). IF, inferior frontal length; IP, inferior parietal length; O, occipital length; SF, superior frontal length; SP, superior parietal length; T, temporal length. (C) Heritability for sulcal lengths in chimpanzees (Left) and humans (Right). CS, central sulcus; FOS, fronto-orbital sulcus; LOS, latero-orbital sulcus; LS, lunate sulcus; PCS, precentral sulcus; POS, parieto-occipital sulcus; STS, superior temporal sulcus; SyF, Sylvian fissure. In B and C lobe dimensions and sulci are color-coded according to heritability values as indicated in the color scale bars. Lobe dimensions and sulci marked with an asterisk show significant heritability after a false-discovery rate approach was used to control for multiple comparisons. Detailed heritabilities, SEs, and P values are listed in Tables S3 and S4. In B and C chimpanzee and human brains are not to scale.
Table S3.
Heritability for brain size
| Variable | Heritability | SE | P value | Significant covariates | Var |
| Chimpanzee | |||||
| Volume | 0.53 | 0.16 | <0.001 | Sex, age | 0.12 |
| Centroid size | 0.52 | 0.17 | 0.002 | Sex, age | 0.14 |
| Human | |||||
| Volume | 0.83 | 0.05 | <0.001 | Sex | 0.44 |
| Centroid size | 0.86 | 0.05 | <0.001 | Sex | 0.43 |
Centroid size, landmark-based measure of size defined as the squared root of the sum of the squared distances between each landmark and the centroid of the configuration; Var, proportion of variance explained by significant covariates; Volume, brain volume including white and gray matter, but not ventricular spaces.
Table S4.
Heritability for lobe and sulcal dimensions (after Procrustes superimposition)
| Variable | Heritability | SE | P value | Significant covariates | Var |
| Chimpanzee | |||||
| Superior frontal length (SF) | 0.49 | 0.14 | <0.001 | Scan | 0.05 |
| Inferior frontal length (IF) | 0.64 | 0.17 | <0.001 | Age, sex*age, size | 0.05 |
| Temporal length (T) | 0.58 | 0.17 | <0.001 | Size | 0.02 |
| Superior parietal length (SP) | 0.63 | 0.13 | <0.001 | Sex | 0.04 |
| Inferior parietal length (IP) | 0.77 | 0.13 | <0.001 | ||
| Occipital length (O) | 0.52 | 0.16 | <0.001 | Sex, size | 0.13 |
| Fronto-orbital sulcus (FOS) | 0.35 | 0.15 | 0.004 | ||
| Precentral sulcus (PCS) | 0.24 | 0.19 | 0.089 | ||
| Central sulcus (CS) | 0.59 | 0.15 | <0.001 | Size, colony | 0.07 |
| Sylvian fissure (SyF) | 0.45 | 0.13 | <0.001 | Colony | 0.03 |
| Superior temporal sulcus (STS) | 0.42 | 0.18 | 0.007 | ||
| Lunate sulcus (LS) | 0.34 | 0.16 | 0.009 | Scan | 0.13 |
| Human | |||||
| Superior frontal length (SF) | 0.25 | 0.18 | 0.088 | ||
| Inferior frontal length (IF) | 0.38 | 0.15 | 0.009 | Sex | 0.03 |
| Temporal length (T) | 0.65 | 0.11 | <0.001 | Sex | 0.01 |
| Superior parietal length (SP) | 0.43 | 0.14 | 0.003 | Size | 0.01 |
| Inferior parietal length (IP) | 0.53 | 0.13 | <0.001 | Sex, age | 0.07 |
| Occipital length (O) | 0.65 | 0.14 | <0.001 | ||
| Latero-orbital sulcus (LOS) | 0.10 | 0.14 | 0.241 | Age | 0.02 |
| Precentral sulcus (PCS) | 0.07 | 0.16 | 0.328 | Sex, size | 0.06 |
| Central sulcus (CS) | 0.58 | 0.10 | <0.001 | Size | 0.07 |
| Sylvian fissure (SyF) | 0.51 | 0.12 | <0.001 | Sex*age, size | 0.04 |
| Superior temporal sulcus (STS) | 0.14 | 0.14 | 0.151 | ||
| Parieto-occipital sulcus (POS) | 0.19 | 0.18 | 0.132 | ||
Sex*age, interaction between age and sex; Var, proportion of variance explained by significant covariates.
Fig. S3.
Phenotypic and genetic correlations between lobe and sulcal dimensions. (A) Lobe dimensions in chimpanzees. (B) Lobe dimensions in humans. In A and B, IF, inferior frontal length; IP, inferior parietal length; O, occipital length; SF, superior frontal length; SP, superior parietal length; T, temporal length. (C) Sulcal dimensions in chimpanzees. (D) Sulcal dimensions in humans. In C and D, CS, central sulcus; FOS, fronto-orbital sulcus; LOS, latero-orbital sulcus; LS, lunate sulcus; PCS, precentral sulcus; POS, parieto-occipital sulcus; STS, superior temporal sulcus; SyF, Sylvian fissure. Phenotypic correlations are represented above the diagonal, and genetic correlations are represented below the diagonal.
Table S5.
Genetic correlations (below diagonal) and phenotypic correlations (above diagonal) between lobe dimensions
| Variable | SF | IF | T | SP | IP | O |
| Chimpanzee | ||||||
| SF | 0.33 | 0.11 | −0.39 | −0.26 | 0.13 | |
| P < 0.001 | P = 0.165 | P < 0.001 | P < 0.001 | P = 0.088 | ||
| IF | 0.65 (0.20) | 0.10 | −0.16 | −0.65 | 0.04 | |
| P = 0.006 | P = 0.182 | P = 0.036 | P < 0.001 | P = 0.589 | ||
| T | 0.01 (0.25) | 0.37 (0.21) | 0.09 | 0.27 | 0.10 | |
| P = 0.949 | P = 0.095 | P = 0.221 | P < 0.001 | P = 0.173 | ||
| SP | −0.49 (0.17) | −0.50 (0.20) | 0.05 (0.21) | 0.42 | −0.60 | |
| P = 0.022 | P = 0.016 | P = 0.816 | P < 0.001 | P < 0.001 | ||
| IP | −0.50 (0.21) | −0.69 (0.11) | 0.10 (0.21) | 0.65 (0.15) | −0.17 | |
| P = 0.021 | P = 0.001 | P = 0.628 | P < 0.001 | P = 0.022 | ||
| O | 0.34 (0.24) | 0.22 (0.24) | 0.19 (0.25) | −0.68 (0.14) | −0.44 (0.24) | |
| P = 0.173 | P = 0.355 | P = 0.459 | P = 0.001 | P = 0.059 | ||
| Human | ||||||
| SF | 0.40 | −0.03 | −0.38 | −0.37 | −0.11 | |
| P < 0.001 | P = 0.650 | P < 0.001 | P < 0.001 | P = 0.126 | ||
| IF | 0.25 (0.36) | −0.01 | −0.21 | −0.60 | −0.02 | |
| P = 0.573 | P = 0.866 | P = 0.002 | P < 0.001 | P = 0.764 | ||
| T | −0.06 (0.30) | −0.01 (0.22) | −0.01 | 0.49 | 0.17 | |
| P = 0.847 | P = 0.958 | P = 0.874 | P < 0.001 | P = 0.018 | ||
| SP | −0.19 (0.34) | −0.66 (0.28) | −0.12 (0.21) | 0.28 | −0.38 | |
| P = 0.608 | P = 0.023 | P = 0.607 | P < 0.001 | P < 0.001 | ||
| IP | −0.61 (0.28) | −0.44 (0.19) | 0.54 (0.13) | 0.59 (0.22) | 0.15 | |
| P = 0.080 | P = 0.139 | P = 0.009 | P = 0.016 | P = 0.039 | ||
| O | −0.17 (0.31) | 0.20 (0.27) | 0 0.35 (0.17) | −0.29 (0.21) | 0.11 (0.19) | |
| P = 0.593 | P = 0.421 | P = 0.044 | P = 0.264 | P = 0.584 | ||
For genetic correlations, associated SEs are provided in parentheses. IF, inferior frontal length; IP, inferior parietal length; O, occipital length; SF, superior frontal length; SP, superior parietal length; T, temporal length.
Table S6.
Genetic correlations (below diagonal) and phenotypic correlations (above diagonal) between sulcal dimensions
| Variable | FOS/LOS | PCS | CS | SyF | STS | LS/POS |
| Chimpanzee | ||||||
| FOS | 0.11 | 0.13 | 0.03 | −0.14 | 0.00 | |
| P = 0.105 | P = 0.091 | P = 0.695 | P = 0.060 | P = 0.989 | ||
| PCS | 0.25 (0.42) | 0.05 | 0.06 | 0.06 | 0.09 | |
| P = 0.563 | P = 0.500 | P = 0.433 | P = 0.373 | P = 0.205 | ||
| CS | 0.15 (0.28) | 0.00 (0.37) | 0.06 | −0.11 | 0.17 | |
| P = 0.582 | P = 0.998 | P = 0.382 | P = 0.134 | P = 0.018 | ||
| SyF | −0.24 (0.29) | −0.39 (0.38) | 0.30 (0.23) | 0.20 | −0.20 | |
| P = 0.393 | P = 0.258 | P = 0.190 | P = 0.004 | P = 0.006 | ||
| STS | −0.16 (0.33) | 0.40 (0.73) | −0.25 (0.26) | 0.43 (0.25) | 0.04 | |
| P = 0.657 | P = 0.500 | P = 0.368 | P = 0.118 | P = 0.608 | ||
| LS | 0.69 (0.40) | Not computable | 0.25 (0.27) | 0.08 (0.29) | −0.16 (0.34) | |
| P = 0.064 | P = 0.391 | P = 0.789 | P = 0.649 | |||
| Human | ||||||
| LOS | 0.08 | −0.08 | −0.12 | −0.11 | 0.05 | |
| P = 0.210 | P = 0.218 | P = 0.080 | P = 0.098 | P = 0.477 | ||
| PCS | −0.49 (1.53) | 0.21 | −0.12 | −0.01 | 0.20 | |
| P = 0.702 | P = 0.001 | P = 0.074 | P = 0.910 | P = 0.004 | ||
| CS | −0.03 (0.44) | 0.55 (0.93) | −0.23 | −0.04 | −0.03 | |
| P = 0.949 | P = 0.354 | P = 0.001 | P = 0.572 | P = 0.624 | ||
| SyF | Not computable | −0.38 (0.58) | −0.40 (0.15) | 0.19 | −0.03 | |
| P = 0.489 | P = 0.027 | P = 0.006 | P = 0.695 | |||
| STS | −0.09 (0.88) | 0.63 (1.00) | −0.03 (0.35) | 0.64 (0.36) | 0.08 | |
| P = 0.919 | P = 0.492 | P = 0.941 | P = 0.082 | P = 0.219 | ||
| POS | 0.98 (1.34) | −0.45 (1.58) | −0.62 (0.48) | −0.22 (0.38) | 0.25 (0.66) | |
| P = 0.299 | P = 0.701 | P = 0.078 | P = 0.376 | P = 0.716 | ||
For genetic correlations, associated SEs are provided in parentheses. Some genetic correlations corresponding to sulci with very low genetic variance are not computable. CS, central sulcus; FOS, fronto-orbital sulcus; LOS, latero-orbital sulcus; LS, lunate sulcus; PCS, precentral sulcus; POS, parieto-occipital sulcus; STS, superior temporal sulcus; SyF, Sylvian fissure.
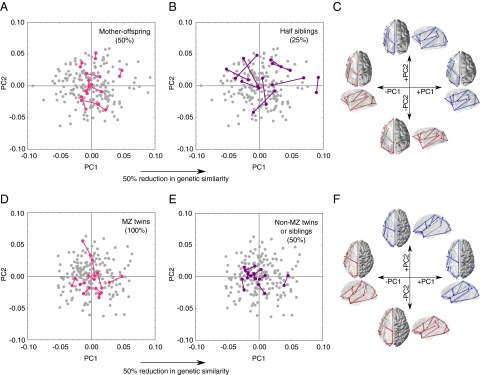
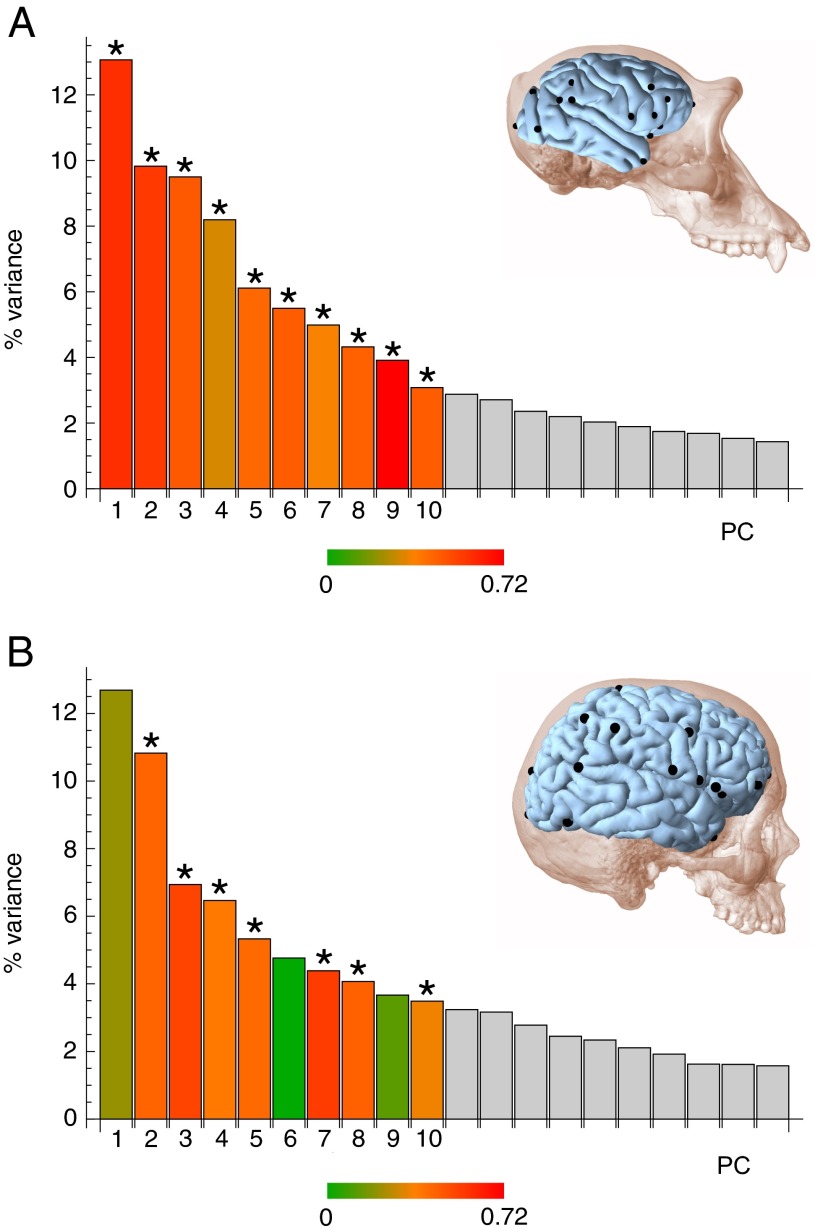
Principal components analyses (PCAs) of shape variation within each species show different patterns of divergence with respect to genetic similarity in chimpanzees and humans. In chimpanzees, mother–offspring pairs, which share 50% genetic similarity, show less shape divergence than half-sibling pairs, which share 25% genetic similarity on average (Fig. 2 A and B). In humans, however, a 50% decrease in genetic similarity is not associated with an increase in shape differences: monozygotic twins, who share 100% genetic similarity, show the same degree of shape variation as nonmonozygotic twins and nontwin siblings, who share on average 50% genetic similarity (Fig. 2 D and E). These differences are reflected further in the substantially higher heritabilities observed in chimpanzees for principal components (PCs) of shape variation than in humans. Chimpanzees show significant heritability in the first 10 PCs, which correspond to the main patterns of shape variation (Fig. 3A and Table S7). The main pattern of variation in chimpanzee brains, summarized by PC1, corresponds to differences in the general proportions of the brain, which vary from long and narrow to short and broad (Fig. 2C). This component of anatomical variation shows a highly significant heritability of 0.59 (P < 0.001) (Fig. 3A and Table S7). Subsequent PCs also show significant and relatively high heritabilities, with a weighted mean (weighted by the proportion of variance explained by each PC) of 0.48 (Fig. 3A and Table S7). Notably, the heritabilities for shape variation approach the same degree of heritability for overall brain size in chimpanzees. Humans, however, show nonsignificant heritabilities in several of these components, including PC1 (Fig. 3B and Table S7). In the human sample, the main pattern of shape variation corresponds to differences within perisylvian areas that involve reorientation of the Sylvian fissure and reorganization of the superior temporal sulcus (Fig. 2F). This pattern of variation has a nonsignificant heritability of 0.21 (P = 0.142) (Fig. 3B and Table S7), indicating that this aspect of interindividual variability in sulcal morphology of humans is under relaxed genetic control. The weighted mean heritability for the first 10 PCs of cortical shape variation in humans is 0.35 (Fig. 3B and Table S7), less than half the heritability for brain size. The temporal and inferior parietal regions, the variation of which is associated with the lowest heritability values, are involved in cognitive functions in humans that include language, attention, and memory (29). Our findings highlight the importance of cortical plasticity as a foundation for the emergence of high-order cognitive functions (29) because environmental influence on areas dedicated to these functions is substantially greater in humans than in chimpanzees.
Fig. 2.
PCAs of shape variation in chimpanzee and human brains. (A and B) PCA of shape variation in chimpanzee brains showing the 10 closest mother–offspring pairs (A, pink links), which share 50% genetic similarity, and the 10 closest pairs of half siblings (B, purple links), which share 25% genetic similarity on average. (C) Brain models showing shape variation corresponding to positive and negative extremes of PC1 and PC2 in chimpanzees. (D and E) PCA of shape variation in human brains showing the 10 closest pairs of monozygotic twins (D, pink links), who share 100% genetic similarity, and the 10 closest pairs of nonmonozygotic twins or nontwin siblings (E, purple links), who share 50% genetic similarity on average. (F) Brain models showing shape variation corresponding to positive and negative extremes of PC1 and PC2 in humans. C and F include dorsal and lateral views, with right hemispheres represented as opaque models with landmarks and left hemispheres represented as transparent models with overlaid schematic representations of landmark variation. Red is used in brain models to show variation corresponding to negative extremes on PC1 and PC2, and blue is used to show variation corresponding to positive extremes in those PCs.
Fig. 3.
Distribution of variance and heritability of phenotypic shape variation. (A) Scree plot showing the distribution of shape variance in chimpanzee brains. (B) Scree plot corresponding to shape variation in human brains. Heritabilities for the first 10 PCs are represented using a color code. PCs marked with an asterisk show significant heritability after a false-discovery rate was applied to control for multiple comparisons. Only the first 20 PCs are represented; heritabilities of PC11–PC20 have not been estimated because they account for very minor proportions of variance. Detailed heritabilities, SEs, and P values are listed in Table S7.
Table S7.
Heritability for PCs of shape variation
| Variable | Heritability | SE | P value | Significant covariates | Var |
| Chimpanzee | |||||
| PC1 | 0.59 | 0.14 | <0.001 | Sex*age | 0.01 |
| PC2 | 0.56 | 0.17 | <0.001 | Sex, size, scan | 0.19 |
| PC3 | 0.47 | 0.14 | <0.001 | Age, sex*age | 0.04 |
| PC4 | 0.30 | 0.17 | 0.020 | Sex | 0.01 |
| PC5 | 0.43 | 0.16 | 0.002 | ||
| PC6 | 0.46 | 0.16 | 0.001 | Age, sex*age | 0.01 |
| PC7 | 0.35 | 0.17 | 0.009 | Size, scan | 0.11 |
| PC8 | 0.45 | 0.14 | <0.001 | Sex | 0.05 |
| PC9 | 0.72 | 0.17 | <0.001 | Sex, sex*age | 0.01 |
| PC10 | 0.46 | 0.15 | <0.001 | Sex, size | 0.05 |
| Human | |||||
| PC1 | 0.21 | 0.20 | 0.142 | Sex*age, size | 0.03 |
| PC2 | 0.44 | 0.14 | 0.003 | Size | 0.07 |
| PC3 | 0.53 | 0.11 | <0.001 | Sex | 0.05 |
| PC4 | 0.38 | 0.14 | 0.004 | ||
| PC5 | 0.42 | 0.14 | 0.004 | ||
| PC6 | 0.00 | 0.500 | Size | 0.02 | |
| PC7 | 0.55 | 0.10 | <0.001 | Sex | 0.03 |
| PC8 | 0.45 | 0.16 | 0.004 | ||
| PC9 | 0.13 | 0.16 | 0.217 | Size | 0.08 |
| PC10 | 0.34 | 0.18 | 0.032 | Age | 0.04 |
Sex*age, interaction between age and sex; Var, proportion of variance explained by significant covariates.
Discussion
Differences in the population structure of the chimpanzee and human samples make it necessary to restrict comparisons of heritability values within each species separately. Furthermore, heritability values are characteristic of given populations and particular environmental conditions, requiring caution in making cross-species and cross-study comparisons. Nonetheless, within-species differences are marked in our analyses. Specifically, chimpanzees are characterized by similar heritability levels for brain size and cortical morphology, whereas humans show a much higher heritability for brain size than for cortical organization, indicating elevated plasticity in our species for the latter. This interpretation is further supported by previous findings demonstrating that, compared with chimpanzees, human brains exhibit a higher level of fluctuating asymmetry in cortical association areas (30). The lack of clear homology between humans and chimpanzees in some sulci of the inferior frontal and occipital lobes (SI Text) is notable and reflects the higher variability of the human brain. Because humans do not have clear fronto-orbital and lunate sulci, our analyses in the human sample focused on alternate sulci and the landmarks that can be identified most reliably in the inferior frontal region and in the parieto-occipital boundary. Those sulcal dimensions still show substantially lower heritability in humans than do the developmentally and evolutionarily primary sulci such as the central sulcus and the Sylvian fissure.
Studies of cortical development in humans have shown differential regional enlargement, which has been suggested to reflect extended maturation and complexity of dendritic and synaptic architecture in association areas (24). Lateral temporal, lateral parietal, and dorsal and medial prefrontal regions show the greatest degree of expansion from birth to adulthood, and it has been suggested that cortical circuits in these regions may be more sensitive to postnatal experience (24). Heritability patterns observed in chimpanzees and humans in the present study are consistent with the proposition that humans have evolved relaxed genetic control on cortical organization, especially in areas related to higher-order cognitive functions. Although particular plastic changes are not themselves heritable, the level of developmental plasticity in different traits can have a genetic basis and therefore can be evolvable, and it has been demonstrated that plasticity levels may respond to both artificial and natural selection (31–33). A high level of cortical plasticity means that neural circuits that are responsible for behavior are formed under a complex array of environmental influences that directly shape those networks, thus providing a neurobiological basis for socially and culturally mediated behavioral evolution.
A causal factor driving the highly plastic nature of the human brain is likely the underdeveloped or altricial condition of humans at birth (34), which requires that a relatively larger fraction of brain maturation occurs postnatally. Humans have evolved a secondary altricial pattern of development compared to the more precocial pattern that characterizes other living primates (34); this developmental pattern might be related to obstetrical (35) or metabolic constraints (36). Regardless of the initial causal factor, an altricial pattern of development, once established, may have provided fundamental selective advantages through the opportunity for postnatal maturation and associated increased learning abilities to allow human offspring to incorporate cultural information through social-transmission mechanisms.
The increase in brain size that is observed during hominin evolution may have created the opportunity for a more extended postnatal period of brain maturation, thus promoting a synergistic interaction between an increased computational capacity [larger brains (3) with expanded neocortices (1) and more neurons (37)] and the ability to form connections in a plastic, environment-dependent manner. This model therefore predicts that hominin species with a large brain size and modern body proportions also likely had an altricial pattern of development, a prolonged postnatal period of brain maturation, and an increased level of cerebral cortical plasticity. Although several studies of brain growth in Homo erectus, the first hominin species characterized by these anatomical traits, indicate that this species likely had a pattern of brain development intermediate between those of chimpanzees and modern humans (11, 38), other analyses have suggested that H. erectus and Homo sapiens shared similar developmental patterns (39). In either case, secondary altriciality seems to have been characteristic of different species of the genus Homo and to have evolved at least in the last common ancestor of Neanderthals and modern humans (11). If so, despite the differences in the evolution and development of endocranial shape between Neanderthals and modern humans (6, 12), they may have shared the anatomical bases for social learning and cultural accumulation that are related to human cognitive evolution. Our results showing relaxed genetic control of cortical anatomy in human brains compared with chimpanzees point to the fundamental role of developmental plasticity in increasing learning abilities and allowing behavioral flexibility in later hominins, thus providing a link between biological evolution and cultural evolution.
Materials and Methods
Samples and MRI Scans.
A sample of 206 chimpanzee (79 males, 127 females, age range 8–53 y) and 218 human (87 males, 131 females, age range 22–30 y) MRI scans was used. The number of human individuals was chosen to match approximately the number of available chimpanzee scans. Chimpanzees used in this study were housed at the Yerkes National Primate Research Center (YNPRC) in Atlanta, and at the University of Texas MD Anderson Cancer Center (UTMDACC) in Bastrop, TX. They were scanned using a 3-T scanner (Siemens Trio; Siemens Medical Solutions) or a 1.5-T scanner (Phillips; Model 51, Philips Medical Systems). Technical details regarding scanning procedures and processing can be found in ref. 40. Scanning procedures in chimpanzees were approved by the Institutional Animal Care and Use Committees at YNPRC and UTMDACC and also followed the guidelines of the Institute of Medicine on the use of chimpanzees in research. No paternity tests were conducted for the purposes of this study, but well-documented pedigrees, including information on mother, father, and offspring identity for many individuals, are available for these chimpanzees. This chimpanzee population has been used previously in quantitative genetic studies of behavioral phenotypes (41, 42). Human MRI scans were obtained from the Human Connectome Project (HCP) database (43). Individuals were scanned with a Siemens Skyra 3-T scanner. Technical details of scanning procedures and processing in human subjects can be found in refs. 43 and 44. Consent from human participants was obtained in the context of the HCP, and terms for the use of open and restricted data were accepted and observed as per the HCP requirement (45). The secondary use of these scans is considered exempt from approval by the Institutional Review Board of The George Washington University. The HCP database includes monozygotic twins, nonmonozygotic twins, and nontwin siblings. To minimize interpopulation variability resulting from genetic ancestry, which recently has been proposed to correlate with general brain anatomy (46), all human subjects included in this study have the same ancestry (as self-reported).
3D Reconstructions and Landmarks.
3D models of the cortical surface were reconstructed from MRI scans using BrainVisa software (47) for chimpanzees and FreeSurfer software (48) for humans (for the human sample, 3D models were obtained directly from the HCP database). Thirty-two anatomically homologous landmarks (16 bilateral landmarks) were placed on the intersections and extreme points of the most constant sulci in the chimpanzee cortical surface (Fig. S1 and Table S1) (30, 49). The same sulci were used to identify equivalent anatomically homologous landmarks in human brains (but see SI Text). Because the anatomical complexity of the human cortical surface makes it difficult to identify some sulci, landmark placement was aided by a comparison with automatically parcellated models. These parcellated models, obtained with FreeSurfer software version 5.3.0 according to the Desikan surface atlas (50), are provided in the HCP database. As compared with other studies of heritability in brain structure, our study can be considered a minimal-processing approach. The use of anatomically homologous landmarks makes our study reliant on anatomical criteria rather than on processing steps that have been demonstrated to have a significant effect on the evaluated phenotypes (23).
Measurement of Brain Volume and Linear Distances.
Brain volumes were obtained from the HCP database for humans, which in turn were obtained from the FreeSurfer structural pipeline (48). In chimpanzees, brain volumes were obtained from BrainVisa (47) masks. Potential differences in the values obtained from the two approaches do not impact our results because the two species were not compared with each other in the same analyses.
Linear distances between landmarks were calculated in Mathematica (Wolfram Research). Euclidean distances between landmarks were measured after Procrustes superimposition [which entails a translation, scaling, and rotation of configurations until distances between homologous landmarks are minimized following a least squares criterion (51)] to remove differences in general size. Individuals were scaled so that centroid size, defined as the squared root of the sum of the squared distances between each landmark and the centroid of the configuration, was 1 in all individuals. Variations in original dimensions (as measured in individuals’ native space before Procrustes superimposition) and in dimensions obtained after Procrustes superimposition are shown in Fig. S2. Asymmetric variation was removed by averaging left and right values because our aim was to assess general patterns of heritability in cortical anatomy without regard to side-specific differences. Interlandmark linear distances included two types of variables. The first corresponded to the dimensions of the major cerebral lobes, which were defined as superior and inferior frontal lengths, superior and inferior parietal lengths, and temporal and occipital lengths (Table S2). The second group of variables corresponded to linear approximations of the lengths of major cortical sulci, including the central sulcus, Sylvian fissure, fronto-orbital sulcus (latero-orbital sulcus in humans), precentral sulcus, superior temporal sulcus, and lunate sulcus (parieto-occipital sulcus in humans). Potential concerns regarding the homology of some of these sulci between chimpanzees and humans are discussed in SI Text. The first group of linear distances (lobe dimensions) describes the general proportions of cerebral lobes. The second group of linear distances (sulcal dimensions) describes more detailed aspects of cortical organization.
Geometric Morphometrics.
Asymmetric variation was removed by mirror-imaging and averaging the original and mirrored configurations of landmarks for each specimen (52). As indicated above, variation corresponding to position, orientation, and size of individuals in the digitized space was removed through Procrustes superimposition (51). No further affine or nonaffine registration was performed to maintain and analyze all shape variation in analyses. Procrustes-superimposed landmark coordinates were subjected to separate PCs analyses for each species because our major interest was to understand the genetic bases of shape variation within each species. Each PC corresponds to a set of phenotypically correlated changes in the position of certain landmarks across the whole population; the genetic bases of these shape variations were estimated later. Patterns of shape differences between kin-related individuals were visualized by highlighting pairs of individuals with different degrees of genetic similarity in the morphospace formed by PC1 and PC2. In chimpanzees, the 10 closest mother–offspring pairs (which share 50% genetic similarity) were represented and compared with the 10 closest pairs of half siblings (who share 25% genetic similarity on average). In humans, the 10 closest pairs of monozygotic twins (who share 100% genetic similarity) were represented and compared with the 10 closest pairs of nonmonozygotic twins or nontwin siblings (who share 50% genetic similarity on average). Patterns of shape variation corresponding to extreme values on PC1 and PC2 for each species are represented as well.
Quantitative Genetics.
A maximum likelihood approach was used to estimate the components of variance of the different evaluated variables as implemented in SOLAR software (53). Narrow-sense heritabilities were estimated, and their significance was tested using likelihood ratio tests. Following other quantitative genetic studies of brain, endocranial, and cranial anatomy in humans and nonhuman primates, age, sex, and the interaction between age and sex were used as covariates (54–56). Overall brain size also was tested as a covariate in analyses of linear distances and PCs of shape. Because linear metrics were obtained and geometric morphometric analyses were performed after all individuals were scaled to a common size through Procrustes superimposition, overall brain size was not expected to have a consistent significant effect. However, it still was tested as a covariate to explore potential allometric trends. Chimpanzees in the sample belong to different colonies, and they were scanned with two different types of scanner, but there is not a complete correspondence between these two variables. For these reasons, these variables (colony and scanner type) were used as covariates in analyses of chimpanzees. When covariates were significant at the P < 0.10 level, they were retained in final models to calculate heritabilities; they were excluded when not significant. Variables were inverse-normalized before analyses to force normality and to avoid high residual kurtosis (56).
Heritability for the first 10 PCs of shape variation, which correspond to the 10 major patterns of phenotypic variation within each sample, was estimated using the same methodological approach and the same covariates. A clear drop in eigenvalues is observed in PC5 for chimpanzees and in PC3 for humans, but the distribution of variance in general is very homogeneous in both species (Fig. 3). Because these first PCs explain a rather minor proportion of variance in both species (40.6% in chimpanzees for PC1–PC4 and 23.5% in humans for PC1 and PC2), analyses of heritability were extended to the first 10 PCs, which explain 68.5% of shape variance in chimpanzees and 62.6% in humans. Subsequent PCs were not included because they explain very minor proportions of shape variation. Univariate estimates of heritability in these PCs were preferred over a fully multivariate approach as described in refs. 57 and 58 because of the limited size of our samples. Although these sample sizes are large enough to estimate heritabilities, they do not allow a reliable calculation of genetic correlations and covariances.
Genetic Correlations Between Lobe and Sulcal Dimensions.
Genetic correlations between linear measures were estimated using bivariate models in which significant covariates for each variable were retained. The genetic correlation between two traits is defined as the association between those traits resulting from the correlation between the loci controlling both traits. These correlations can arise through linkage disequilibrium or pleiotropy (59), and they usually are considered to constrain evolution and reduce evolvability. As stated above, the reliable estimation of genetic correlations requires very large sample sizes that exceed the number of individuals available to our study. For this reason, the genetic correlations between lobe dimensions and between sulcal dimensions provided in Fig. S3 and Tables S5 and S6 should be taken with caution.
Representation of Heritabilities.
For heritability in linear measures, linear distances were represented and overlaid on 3D models of a representative chimpanzee and human brain. Linear dimensions were color-coded according to their heritability values. Differences between chimpanzees and humans were maximized by rescaling the color gradient to the minimum and maximum heritabilities observed in our study (0.07 and 0.77, respectively), instead of using the whole range of possible heritabilities (0–1). Heritabilities in patterns of shape variation (PCs) also were color-coded and overlaid on scree plots representing the distribution of variance within each species (the heritability range for the color code is 0–0.72). Heritabilities that remained significant after correcting for multiple comparisons using a false-discovery rate procedure (60, 61) are marked in Figs. 1 and 3. Original P values obtained for all analyses are listed in Tables S3, S4, and S7.
SI Text
Landmarks and Sulci.
The location and definition of landmarks used in this study are provided in Fig. S1 and Table S1. This configuration includes some of the landmarks defined in refs. 30 and 49. All except two of the landmark-defining sulci we used have a clear homology between chimpanzees and humans. The fronto-orbital sulcus used to demarcate the anterior aspect of Broca’s area homolog in chimpanzees (26, 62–64) does not have a clear correspondence in human brains. According to Connolly (65), the opercular expansion of the frontal lobe during hominin evolution made this sulcus shift caudally, where it became part of the anterior limiting sulcus of the insula (66). We used the latero-orbital sulcus in humans, despite its lack of homology with the chimpanzee fronto-orbital sulcus, to characterize shape variation in the orbito-frontal region. The second area of questionable homology between chimpanzees and humans is the lunate sulcus region (4, 5, 67, 68). Although clearly identifiable in chimpanzees, the posterior displacement and disappearance of the lunate sulcus is assumed to be a hominin character that is maximally expressed in H. sapiens (4, 5). For this reason, we selected three landmarks in the human sample that demarcate the occipital lobe: the intersection of the parieto-occipital sulcus with the midline, the intersection of the intraparietal sulcus with the transverse occipital sulcus, and the occipital notch (69). The use of these landmarks despite their questionable anatomical homology in chimpanzees and humans is an attempt to circumvent the lack of clearly homologous structures in the two species by using a more functional criterion (70).
Age Distribution of Chimpanzee and Human Samples.
Different studies of heritability in brain size and structure have used individuals of different ages. A review of several of these studies demonstrated that heritability for brain variables decreases from early to late adulthood in humans (71). Therefore, the difference in age range between the chimpanzee sample (8–53 y old) and the human sample (22–30 y old) may be involved in the higher heritability observed in humans for brain size. However, this observation is likely to be true for many different brain phenotypes, which are expected to show lower heritability as individuals age and environmental influences increase. Therefore, the low heritabilities for cortical organization observed in humans can be considered an overestimate in comparison with the values that potentially could be obtained in a population with a larger range of variation in age.
Other Supporting Information Files
Supplementary Material
Acknowledgments
Three-dimensional models of chimpanzee and human skulls were provided by José Manuel de la Cuétara. This work was supported by NIH Grants NS42867, NS73134, and NS092988 and by Grant 220020293 from the James S. McDonnell Foundation. Chimpanzees at UTMDACC were supported by NIH Cooperative Agreement U42 OD-011197. Human data were provided by the Human Connectome Project, Washington University–University of Minnesota Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research and by the McDonnell Center for Systems Neuroscience at Washington University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. L.A.K. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1512646112/-/DCSupplemental.
References
- 1.Rilling JK, Insel TR. The primate neocortex in comparative perspective using magnetic resonance imaging. J Hum Evol. 1999;37(2):191–223. doi: 10.1006/jhev.1999.0313. [DOI] [PubMed] [Google Scholar]
- 2.Smaers JB, Soligo C. 2013 Brain reorganization, not relative brain size, primarily characterizes anthropoid brain evolution. Proc R Soc B Biol Sci 280(1759):20130269. [Google Scholar]
- 3.Striedter GF. Principles of brain evolution. Sinauer Associates; Sunderland: 2005. [Google Scholar]
- 4.Holloway RL, Clarke RJ, Tobias PV. Posterior lunate sulcus in Australopithecus africanus: Was Dart right? C R Palevol. 2004;3(4):287–293. [Google Scholar]
- 5.Falk D. Hominin paleoneurology: Where are we now? Prog Brain Res. 2012;195:255–272. doi: 10.1016/B978-0-444-53860-4.00012-X. [DOI] [PubMed] [Google Scholar]
- 6.Bruner E, Manzi G, Arsuaga JL. Encephalization and allometric trajectories in the genus Homo: Evidence from the Neandertal and modern lineages. Proc Natl Acad Sci USA. 2003;100(26):15335–15340. doi: 10.1073/pnas.2536671100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balzeau A, Holloway RL, Grimaud-Hervé D. Variations and asymmetries in regional brain surface in the genus Homo. J Hum Evol. 2012;62(6):696–706. doi: 10.1016/j.jhevol.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 8.Carlson KJ, et al. The endocast of MH1, Australopithecus sediba. Science. 2011;333(6048):1402–1407. doi: 10.1126/science.1203922. [DOI] [PubMed] [Google Scholar]
- 9.Spoor F, et al. Reconstructed Homo habilis type OH 7 suggests deep-rooted species diversity in early Homo. Nature. 2015;519(7541):83–86. doi: 10.1038/nature14224. [DOI] [PubMed] [Google Scholar]
- 10.Leigh SR. Brain growth, life history, and cognition in primate and human evolution. Am J Primatol. 2004;62(3):139–164. doi: 10.1002/ajp.20012. [DOI] [PubMed] [Google Scholar]
- 11.Coqueugniot H, Hublin J-J, Veillon F, Houët F, Jacob T. Early brain growth in Homo erectus and implications for cognitive ability. Nature. 2004;431(7006):299–302. doi: 10.1038/nature02852. [DOI] [PubMed] [Google Scholar]
- 12.Gunz P, Neubauer S, Maureille B, Hublin J-J. Brain development after birth differs between Neanderthals and modern humans. Curr Biol. 2010;20(21):R921–R922. doi: 10.1016/j.cub.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 13.Hublin J-J, Neubauer S, Gunz P. Brain ontogeny and life history in Pleistocene hominins. Philos Trans R Soc Lond B Biol Sci. 2015;370(1663):20140062. doi: 10.1098/rstb.2014.0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Florio M, et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science. 2015;347(6229):1465–1470. doi: 10.1126/science.aaa1975. [DOI] [PubMed] [Google Scholar]
- 15.Prüfer K, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505(7481):43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomasello M. The Cultural Origins of Human Cognition. Harvard Univ Press; Cambridge, MA: 1999. [Google Scholar]
- 17.Herrmann E, Call J, Hernàndez-Lloreda MV, Hare B, Tomasello M. Humans have evolved specialized skills of social cognition: The cultural intelligence hypothesis. Science. 2007;317(5843):1360–1366. doi: 10.1126/science.1146282. [DOI] [PubMed] [Google Scholar]
- 18.Boyd R, Richerson PJ, Henrich J. The cultural niche: Why social learning is essential for human adaptation. Proc Natl Acad Sci USA. 2011;108(Suppl 2):10918–10925. doi: 10.1073/pnas.1100290108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Russell JL, Lyn H, Schaeffer JA, Hopkins WD. The role of socio-communicative rearing environments in the development of social and physical cognition in apes. Dev Sci. 2011;14(6):1459–1470. doi: 10.1111/j.1467-7687.2011.01090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lendvai B, Stern EA, Chen B, Svoboda K. Experience-dependent plasticity of dendritic spines in the developing rat barrel cortex in vivo. Nature. 2000;404(6780):876–881. doi: 10.1038/35009107. [DOI] [PubMed] [Google Scholar]
- 21.Cáceres M, Suwyn C, Maddox M, Thomas JW, Preuss TM. Increased cortical expression of two synaptogenic thrombospondins in human brain evolution. Cereb Cortex. 2007;17(10):2312–2321. doi: 10.1093/cercor/bhl140. [DOI] [PubMed] [Google Scholar]
- 22.Enard W, et al. A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell. 2009;137(5):961–971. doi: 10.1016/j.cell.2009.03.041. [DOI] [PubMed] [Google Scholar]
- 23.Gronenschild EHBM, et al. The effects of FreeSurfer version, workstation type, and Macintosh operating system version on anatomical volume and cortical thickness measurements. PLoS One. 2012;7(6):e38234. doi: 10.1371/journal.pone.0038234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hill J, et al. Similar patterns of cortical expansion during human development and evolution. Proc Natl Acad Sci USA. 2010;107(29):13135–13140. doi: 10.1073/pnas.1001229107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischl B, et al. Cortical folding patterns and predicting cytoarchitecture. Cereb Cortex. 2008;18(8):1973–1980. doi: 10.1093/cercor/bhm225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherwood CC, Broadfield DC, Holloway RL, Gannon PJ, Hof PR. Variability of Broca’s area homologue in African great apes: Implications for language evolution. Anat Rec A Discov Mol Cell Evol Biol. 2003;271(2):276–285. doi: 10.1002/ar.a.10046. [DOI] [PubMed] [Google Scholar]
- 27.Mueller S, et al. Individual variability in functional connectivity architecture of the human brain. Neuron. 2013;77(3):586–595. doi: 10.1016/j.neuron.2012.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peper JS, Brouwer RM, Boomsma DI, Kahn RS, Hulshoff Pol HE. Genetic influences on human brain structure: A review of brain imaging studies in twins. Hum Brain Mapp. 2007;28(6):464–473. doi: 10.1002/hbm.20398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherwood CC, Subiaul F, Zawidzki TW. A natural history of the human mind: Tracing evolutionary changes in brain and cognition. J Anat. 2008;212(4):426–454. doi: 10.1111/j.1469-7580.2008.00868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gómez-Robles A, Hopkins WD, Sherwood CC. Increased morphological asymmetry, evolvability and plasticity in human brain evolution. Proc R Soc B Biol Sci. 2013;280(1761):20130575. doi: 10.1098/rspb.2013.0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pigliucci M. Evolution of phenotypic plasticity: where are we going now? Trends Ecol Evol. 2005;20(9):481–486. doi: 10.1016/j.tree.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 32.Scheiner SM. Genetics and evolution of phenotypic plasticity. Annu Rev Ecol Syst. 1993;24:35–68. [Google Scholar]
- 33.West-Eberhard MJ. Developmental plasticity and the origin of species differences. Proc Natl Acad Sci USA. 2005;102(Suppl 1):6543–6549. doi: 10.1073/pnas.0501844102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Portmann A. 1969. Biologische Fragmente zu einer Lehre vom Menschen (Benno Schwabe, Basel); trans Schaefer J (1990) (Columbia Univ Press, New York). German.
- 35.Rosenberg KR. The evolution of modern human childbirth. Am J Phys Anthropol. 1992;35(S15):89–124. [Google Scholar]
- 36.Dunsworth HM, Warrener AG, Deacon T, Ellison PT, Pontzer H. Metabolic hypothesis for human altriciality. Proc Natl Acad Sci USA. 2012;109(38):15212–15216. doi: 10.1073/pnas.1205282109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herculano-Houzel S. The human brain in numbers: a linearly scaled-up primate brain. Front Hum Neurosci. 2009;3:31. doi: 10.3389/neuro.09.031.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Connell CA, DeSilva JM. Mojokerto revisited: evidence for an intermediate pattern of brain growth in Homo erectus. J Hum Evol. 2013;65(2):156–161. doi: 10.1016/j.jhevol.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 39.Leigh SR. Brain ontogeny and life history in Homo erectus. J Hum Evol. 2006;50(1):104–108, author reply 109–113. doi: 10.1016/j.jhevol.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 40.Bogart SL, et al. Cortical sulci asymmetries in chimpanzees and macaques: a new look at an old idea. Neuroimage. 2012;61(3):533–541. doi: 10.1016/j.neuroimage.2012.03.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hopkins WD, Russell JL, Schaeffer J. Chimpanzee intelligence is heritable. Curr Biol. 2014;24(14):1649–1652. doi: 10.1016/j.cub.2014.05.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hopkins WD, Reamer L, Mareno MC, Schapiro SJ. Genetic basis in motor skill and hand preference for tool use in chimpanzees (Pan troglodytes) Proc R Soc B Biol Sci. 2015;282(1800):20141223. doi: 10.1098/rspb.2014.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Essen DC, et al. WU-Minn HCP Consortium The Human Connectome Project: A data acquisition perspective. Neuroimage. 2012;62(4):2222–2231. doi: 10.1016/j.neuroimage.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glasser MF, et al. WU-Minn HCP Consortium The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage. 2013;80:105–124. doi: 10.1016/j.neuroimage.2013.04.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Essen DC, et al. WU-Minn HCP Consortium The WU-Minn Human Connectome Project: An overview. Neuroimage. 2013;80:62–79. doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fan CC, et al. Pediatric Imaging, Neurocognition, and Genetics Study Modeling the 3D geometry of the cortical surface with genetic ancestry. Curr Biol. 2015;25(15):1988–1992. doi: 10.1016/j.cub.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cointepas Y, Mangin J-F, Garnero L, Poline J-B, Benali H. BrainVISA: Software platform for visualization and analysis of multi-modality brain data. Neuroimage. 2001;13(6):S98. [Google Scholar]
- 48.Fischl B. FreeSurfer. Neuroimage. 2012;62(2):774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gómez-Robles A, Hopkins WD, Sherwood CC. Modular structure facilitates mosaic evolution of the brain in chimpanzees and humans. Nat Commun. 2014;5:4469. doi: 10.1038/ncomms5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Desikan RS, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31(3):968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 51.Rohlf FJ, Slice D. Extensions of the Procrustes method for the optimal superimposition of landmarks. Syst Zool. 1990;39(1):40–59. [Google Scholar]
- 52.Klingenberg CP, Barluenga M, Meyer A. Shape analysis of symmetric structures: Quantifying variation among individuals and asymmetry. Evolution. 2002;56(10):1909–1920. doi: 10.1111/j.0014-3820.2002.tb00117.x. [DOI] [PubMed] [Google Scholar]
- 53.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62(5):1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rogers J, et al. On the genetic architecture of cortical folding and brain volume in primates. Neuroimage. 2010;53(3):1103–1108. doi: 10.1016/j.neuroimage.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Atkinson EG, Rogers J, Mahaney MC, Cox LA, Cheverud JM. Cortical folding of the primate brain: An interdisciplinary examination of the genetic architecture, modularity, and evolvability of a significant neurological trait in pedigreed baboons (genus Papio) Genetics. 2015;200(2):651–665. doi: 10.1534/genetics.114.173443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martínez-Abadías N, et al. Heritability of human cranial dimensions: Comparing the evolvability of different cranial regions. J Anat. 2009;214(1):19–35. doi: 10.1111/j.1469-7580.2008.01015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martínez-Abadías N, et al. Pervasive genetic integration directs the evolution of human skull shape. Evolution. 2012;66(4):1010–1023. doi: 10.1111/j.1558-5646.2011.01496.x. [DOI] [PubMed] [Google Scholar]
- 58.Klingenberg CP, Debat V, Roff DA. Quantitative genetics of shape in cricket wings: Developmental integration in a functional structure. Evolution. 2010;64(10):2935–2951. doi: 10.1111/j.1558-5646.2010.01030.x. [DOI] [PubMed] [Google Scholar]
- 59.Roff DA. Evolutionary Quantitative Genetics. Chapman & Hall; New York: 1997. [Google Scholar]
- 60.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc, B. 1995;57(1):289–300. [Google Scholar]
- 61.Verhoeven KJF, Simonsen KL, McIntyre LM. Implementing false discovery rate control: Increasing your power. Oikos. 2005;108(3):643–647. [Google Scholar]
- 62.Keller SS, Roberts N, Hopkins W. A comparative magnetic resonance imaging study of the anatomy, variability, and asymmetry of Broca’s area in the human and chimpanzee brain. J Neurosci. 2009;29(46):14607–14616. doi: 10.1523/JNEUROSCI.2892-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Keller SS, Deppe M, Herbin M, Gilissen E. Variability and asymmetry of the sulcal contours defining Broca’s area homologue in the chimpanzee brain. J Comp Neurol. 2012;520(6):1165–1180. doi: 10.1002/cne.22747. [DOI] [PubMed] [Google Scholar]
- 64.Schenker NM, et al. Broca’s area homologue in chimpanzees (Pan troglodytes): Probabilistic mapping, asymmetry, and comparison to humans. Cereb Cortex. 2010;20(3):730–742. doi: 10.1093/cercor/bhp138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Connolly CJ. External Morphology of the Primate Brain. C. C. Thomas; Springfield: 1950. [Google Scholar]
- 66.de Sousa A, Cunha E. Hominins and the emergence of the modern human brain. Prog Brain Res. 2012;195:293–322. doi: 10.1016/B978-0-444-53860-4.00014-3. [DOI] [PubMed] [Google Scholar]
- 67.de Sousa AA, et al. Hominoid visual brain structure volumes and the position of the lunate sulcus. J Hum Evol. 2010;58(4):281–292. doi: 10.1016/j.jhevol.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 68.Allen JS, Bruss J, Damasio H. Looking for the lunate sulcus: A magnetic resonance imaging study in modern humans. Anat Rec A Discov Mol Cell Evol Biol. 2006;288(8):867–876. doi: 10.1002/ar.a.20362. [DOI] [PubMed] [Google Scholar]
- 69.Petrides M. The Human Cerebral Cortex: An MRI Atlas of the Sulci and Gyri in MNI Stereotaxic Space. Elsevier Science & Technology; New York: 2011. [Google Scholar]
- 70.Klingenberg CP. Novelty and “homology-free” morphometrics: What’s in a name? Evol Biol. 2008;35(3):186–190. [Google Scholar]
- 71.Batouli SAH, Trollor JN, Wen W, Sachdev PS. The heritability of volumes of brain structures and its relationship to age: A review of twin and family studies. Ageing Res Rev. 2014;13:1–9. doi: 10.1016/j.arr.2013.10.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.