Significance
Forgetting is an important aspect of memory and cognition, yet its underlying mechanisms remain poorly understood. In this work, we describe neural circuits involved in the forgetting of olfactory associative memory after odor–shock pairing in Drosophila. Our work establishes an anatomically well-defined example where forgetting is regulated by multiple neural pathways that impinge upon a memory center. We also show that forgetting over time and the acute forgetting of conflicting memory during reversal learning rely on separable neural circuits. Overall, these findings support the emerging view that forgetting is a biologically regulated process of the brain, and also open the door to studying the diverse circuit mechanisms of forgetting in different contexts.
Keywords: forgetting, Drosophila melanogaster, associative memory, mushroom body, reversal learning
Abstract
Recent studies have identified molecular pathways driving forgetting and supported the notion that forgetting is a biologically active process. The circuit mechanisms of forgetting, however, remain largely unknown. Here we report two sets of Drosophila neurons that account for the rapid forgetting of early olfactory aversive memory. We show that inactivating these neurons inhibits memory decay without altering learning, whereas activating them promotes forgetting. These neurons, including a cluster of dopaminergic neurons (PAM-β′1) and a pair of glutamatergic neurons (MBON-γ4>γ1γ2), terminate in distinct subdomains in the mushroom body and represent parallel neural pathways for regulating forgetting. Interestingly, although activity of these neurons is required for memory decay over time, they are not required for acute forgetting during reversal learning. Our results thus not only establish the presence of multiple neural pathways for forgetting in Drosophila but also suggest the existence of diverse circuit mechanisms of forgetting in different contexts.
Although forgetting commonly has a negative connotation, it is a functional process that shapes memory and cognition (1–4). Recent studies, including work in relatively simple invertebrate models, have started to reveal basic biological mechanisms underlying forgetting (5–15). In Drosophila, single-session Pavlovian conditioning by pairing an odor (conditioned stimulus, CS) with electric shock (unconditioned stimulus, US) induces aversive memories that are short-lasting (16). The memory performance of fruit flies is observed to drop to a negligible level within 24 h, decaying rapidly early after training and slowing down thereafter (17). Memory decay or forgetting requires the activation of the small G protein Rac, a signaling protein involved in actin remodeling, in the mushroom body (MB) intrinsic neurons (6). These so-called Kenyon cells (KCs) are the neurons that integrate CS–US information (18, 19) and support aversive memory formation and retrieval (20–22). In addition to Rac, forgetting also requires the DAMB dopamine receptor (7), which has highly enriched expression in the MB (23). Evidence suggests that the dopamine-mediated forgetting signal is conveyed to the MB by dopamine neurons (DANs) in the protocerebral posterior lateral 1 (PPL1) cluster (7, 24). Therefore, forgetting of olfactory aversive memory in Drosophila depends on a particular set of intracellular molecular pathways within KCs, involving Rac, DAMB, and possibly others (25), and also receives modulation from extrinsic neurons. Although important cellular evidence supporting the hypothesis that memory traces are erased under these circumstances is still lacking, these findings lend support to the notion that forgetting is an active, biologically regulated process (17, 26).
Although existing studies point to the MB circuit as essential for forgetting, several questions remain to be answered. First, whereas the molecular pathways for learning and forgetting of olfactory aversive memory are distinct and separable (6, 7), the neural circuits seem to overlap. Rac-mediated forgetting has been localized to a large population of KCs (6), including the γ-subset, which is also critical for initial memory formation (21, 27). The site of action of DAMB for forgetting has yet to be established; however, the subgroups of PPL1-DANs implicated in forgetting are the same as those that signal aversive reinforcement and are required for learning (28–30). It leaves open the question of whether the brain circuitry underlying forgetting and learning is dissociable, or whether forgetting and learning share the same circuit but are driven by distinct activity patterns and molecular machinery (26). Second, shock reinforcement elicits multiple memory traces through at least three dopamine pathways to different subdomains in the MB lobes (28, 29). Functional imaging studies have also revealed Ca2+-based memory traces in different KC populations (31). It is poorly understood how forgetting of these memory traces differs, and it remains unknown whether there are multiple regulatory neural pathways. Notably, when PPL1-DANs are inactivated, forgetting still occurs, albeit at a lower rate (7). This incomplete block suggests the existence of an additional pathway(s) that conveys forgetting signals to the MB. Third, other than memory decay over time, forgetting is also observed through interference (32, 33), when new learning or reversal learning is introduced after training (6, 34, 35). Time-based and interference-based forgetting shares a similar dependence on Rac and DAMB (6, 7). However, it is not known whether distinct circuits underlie forgetting in these different contexts.
In the current study, we focus on the diverse set of MB extrinsic neurons (MBENs) that interconnect the MB lobes with other brain regions, which include 34 MB output neurons (MBONs) of 21 types and ∼130 dopaminergic neurons of 20 types in the PPL1 and protocerebral anterior medial (PAM) clusters (36, 37). These neurons have been intensively studied in olfactory memory formation, consolidation, and retrieval in recent years (e.g., 24, 28–30, 38–48); however, their roles in forgetting have not been characterized except for the aforementioned PPL1-DANs. In a functional screen, we unexpectedly found that several Gal4 driver lines of MBENs showed significantly better 3-h memory retention when the Gal4-expressing cells were inactivated. The screen has thus led us to identify two types of MBENs that are not involved in initial learning but play important and additive roles in mediating memory decay. Furthermore, neither of these MBEN types is required for reversal learning, supporting the notion that there is a diversity of neural circuits that drive different forms of forgetting.
Results
Screen for MBENs Involved in Memory Decay.
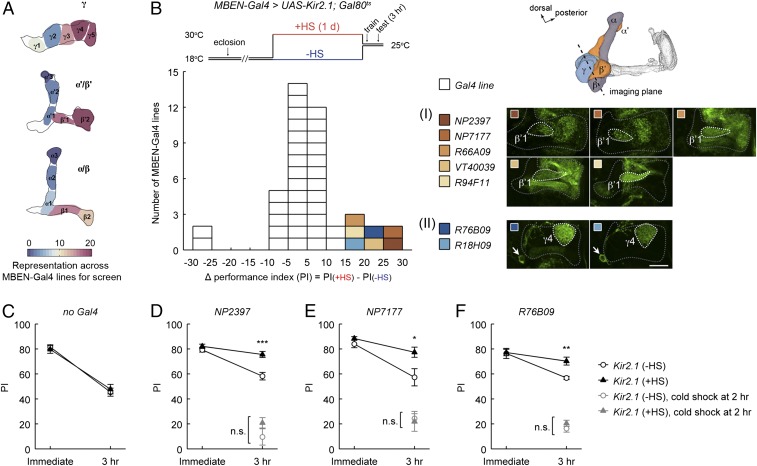
We collected 47 Gal4 driver lines (37, 49, 50) that had expression in putative MBENs (SI Appendix, Fig. S1 and Table S1). Different types of MBENs have stereotyped innervation in distinct subdomains in the MB lobes, which defines 15 compartments (36, 37) along the axons of the three KC subtypes, KCγ, KCα/β, and KCα′/β′. Based on lobe compartment coverage in the expression patterns of the collected Gal4 drivers (Fig. 1A), we estimated that our screen included most MBEN types connected to the MB horizontal (medial) lobes and also a few connected to the vertical lobes.
Fig. 1.
Neuronal inactivation screening has identified two types of MB extrinsic neurons as candidates required for early memory decay. (A) The screen used 47 Gal4 driver lines that covered a broad variety of MBEN types, which have stereotyped innervation of distinct compartments on the MB lobes. Pseudocolors in the schematic reflect the number of times of the appearance of each lobe compartment in the expression patterns of the screened Gal4 drivers. (B) The MBEN-Gal4 lines were combined with the Kir2.1 potassium channel and screened for 3-h olfactory aversive memory after single-session training. HS regimen for Kir2.1 induction is shown. The distribution plot (Bottom) summarizes the screen data (SI Appendix, Table S1), with each square representing one Gal4 line and the x axis indicating a 3-h memory difference between the +HS and −HS groups. Colored squares highlight the seven Gal4 lines showing better memory retention with Kir2.1 inactivation. These lines converged on two MBEN types: (I) PAM-DANs innervating the β′1-compartment (PAM-β′1); and (II) a γ4-output neuron (MBON-γ4>γ1γ2) (SI Appendix, Fig. S2). Gal4 expression patterns are single confocal sections of the brain region including the MB horizontal lobes (gray dashed lines). The β′1- and γ4-compartments are marked, and the MBON-γ4>γ1γ2 soma is indicated by arrows. (Scale bar, 20 μm.) (C–F) Immediate and 3-h memory for the “no Gal4” control (UAS-Kir2.1/+; Gal80ts/+) and for UAS-Kir2.1; Gal80ts flies combined with two PAM-β′1 lines (NP2397 and NP7177) and one MBON-γ4>γ1γ2 line (R76B09) from the screen. For all these three tested Gal4 lines, HS-induced expression of Kir2.1 slowed memory decay without affecting initial learning, and the slower decay was blocked by subjecting flies to a cold-shock treatment at 2 h after training to eliminate the early labile memory components. All statistically significant differences (t test) are marked. Data are means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001; n.s., not significant; n ≥ 6. Three-hour memory data in D–F are an independent replication of the screen results.
Our initial screen for candidate lines simply involved looking at 3-h memory performance, which should be enhanced when the forgetting process is disrupted. Further characterization of specific lines was subsequently used to establish that the decay of memory is specifically affected (see below). In the screen, we electrically silenced Gal4-expressing cells using the UAS-driven Kir2.1 inwardly rectifying potassium channel (51). To minimize undesirable effects from developmental perturbation and chronic neuronal inactivation, Kir2.1 expression throughout this study was temporally controlled by Gal80ts (Tubp-Gal80ts) (52). To induce Kir2.1 expression, 2- to 6-day-old adult flies were subjected to heat shock (HS) at 30 °C for 1 d, following which we tested memory formation and retention (Fig. 1B). The performance index (PI) of the “+HS” groups of flies was compared with that of control groups kept at 18 °C (“−HS”), which did not have Kir2.1 expression but were otherwise genetically identical.
We found seven MBEN-Gal4 lines that showed significantly higher 3-h memory when driving HS-induced expression of Kir2.1 (Fig. 1B and SI Appendix, Table S1). To rule out nonspecific effects due to the HS-induction procedure, we tested control flies lacking a Gal4 driver (UAS-Kir2.1/+; Gal80ts/+) and did not observe HS-dependent effects on memory performance (Fig. 1C). Moreover, and to further support screen validity, we found lower memory in two MBEN-Gal4 lines, one of which (NP2758; SI Appendix, Table S1) was expressed in the MP1 subgroup of PPL1-DANs, and the memory defect was expected from its impaired learning (28).
The seven MBEN-Gal4 lines showing higher memory fell into two categories based on their labeled MBEN types. Five lines shared the expression in a subset of dopaminergic neurons from the PAM-DAN cluster (PAM-β′1), which innervate the β′1-compartment. The remaining two lines had common expression in a pair of neurons from the MB output neuron population (MBON-γ4>γ1γ2), which are connected to the γ4-compartment. This categorization was supported by examining Gal4-driven reporter expression in the MB horizontal lobes (Fig. 1B), and was further validated by colocalizing Gal4-expressing cells with R24E12-LexA, a genetic landmark of both PAM-β′1 and MBON-γ4>γ1γ2 (SI Appendix, Fig. S2). Finding the convergence of hits into two categories, affecting either PAM-β′1 or MBON-γ4>γ1γ2, suggests that the increased 3-h memory retention is attributable to the inactivation of these two MBEN types. Indeed, we have validated this notion using refined genetic manipulation, which we present later. In the next section, we describe experiments that used the Gal4 lines showing the strongest screen phenotype to first confirm an effect on memory decay.
Bidirectional Regulation of Early Memory Decay.
We chose two Gal4 lines labeling PAM-β′1 (NP2397 and NP7177) and one Gal4 line labeling MBON-γ4>γ1γ2 (R76B09) to perform further behavioral tests. It is worth mentioning that NP2397 and NP7177 both carry P{Gal4} enhancer traps inserted in amnesiac (amn) on the X chromosome (SI Appendix, Fig. S3A). The expression patterns of these two drivers are highly similar (SI Appendix, Fig. S1); we therefore mainly used NP2397, whereas including NP7177 only in initial studies. amn encodes a putative neuropeptide precursor (53), which is expressed in dorsal paired medial (DPM) neurons to regulate olfactory memory (54). We did not detect expression in the DPM neurons for NP2397 and NP7177 (SI Appendix, Fig. S3A, legend) and, to limit confounding effects due to any possible perturbation of amn, we only used heterozygous females of NP2397 and NP7177 as Gal4 drivers (Materials and Methods). We observed increased 3-h memory retention in an HS-dependent manner in these females (Gal4/+; UAS-Kir2.1/+; Gal80ts/+) in comparison with simultaneously assayed male siblings that lacked the Gal4 driver (+/Y; UAS-Kir2.1/+; Gal80ts/+) (SI Appendix, Fig. S3 B and C). This result, together with the observation that the female heterozygotes of Gal4 drivers alone showed normal 3-h memory irrespective of HS (SI Appendix, Fig. S3 D and E), rules out nonspecific effects due to genetic background. Note that the Gal4 line used to manipulate MBON-γ4>γ1γ2, R76B09, has its insertion in a prechosen attP2 site on the third chromosome (55), which is the same as most other Gal4 lines used in the screen and is less likely to confound data interpretation.
For all three Gal4 lines, we found that the slope of memory decay was decreased with Kir2.1 inactivation (Fig. 1 D–F). These data directly show that the initial memory is not affected in these flies, and that the effect is in memory retention. To assess whether task-relevant behaviors other than learning are altered, we tested sensorimotor abilities of these flies and found normal olfactory acuity and shock reactivity (SI Appendix, Table S2). After a single training session, the kinetics of memory decay are primarily influenced by the rapid forgetting of early, anesthesia-sensitive memory components and the gradual consolidation of memory into a late, more stable form that is anesthesia-resistant (56). To test whether the decreased slope of memory decay arises because early memories are protected from forgetting or because memory consolidation is accelerated, we cold-anesthetized flies at 2 h after training, a treatment that eliminates early labile memory components but does not affect consolidated memories (57). We found that cold shock blocked the increase of 3-h memory retention for all three Gal4 lines (Fig. 1 D–F), suggesting that the extra memory is labile and sensitive to cold anesthesia. This is consistent with the idea that inactivating Gal4-expressing cells does not increase memory consolidation but instead protects early memories from decay by impairing forgetting processes.
Subtle effects on initial memory acquisition might not be observed in regular training, which induced near-saturated learning performance (SI Appendix, Fig. S5 B and C). To investigate whether our neuronal inactivation alters memory acquisition, we tested NP2397 and R76B09 flies using a weaker training regimen where we reduced the overall duration of CS–US pairing and decreased the number of shock pulses delivered (SI Appendix, Fig. S5A). The results of weaker training showed that the levels of memory acquisition with Kir2.1 inactivation were comparable to those of the controls (SI Appendix, Fig. S5 B and C). Note that, for NP2397, the experimental group showed higher learning performance than the −HS groups under the training condition with two shock pulses. However, the difference under this circumstance might be attributed to nonspecific effects of HS, because the performance of the experimental group was similar to that of the HS-treated male siblings under all training conditions [compare “NP2397/+; UAS-Kir2.1/+; Gal80ts/+ (+HS)” and “+/Y; UAS-Kir2.1/+; Gal80ts/+ (+HS)”; SI Appendix, Fig. S5B]. The fact that memory acquisition is not significantly altered by inactivating neurons in these drivers supports the notion that the increased memory retention is due to slower memory decay rather than an elevation of initial memory.
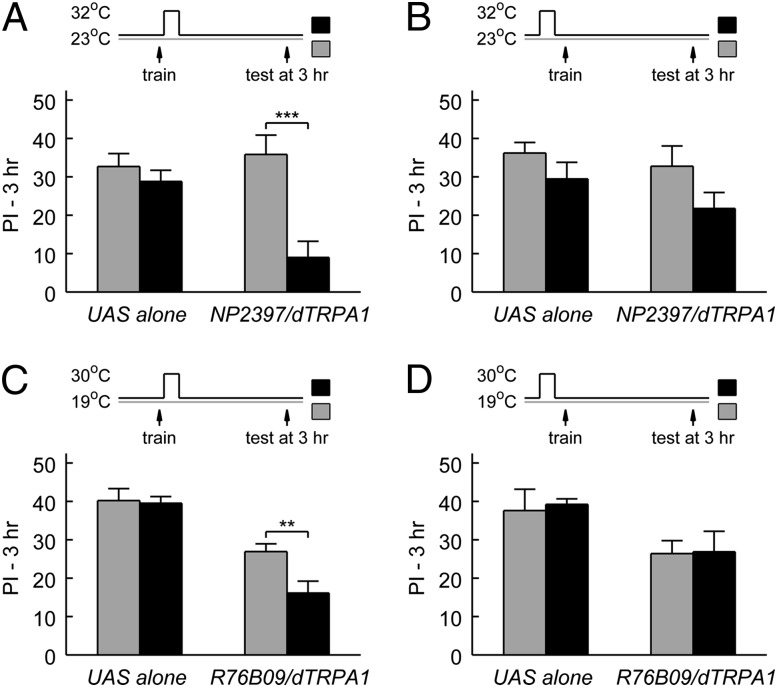
We reasoned that if Gal4-expressing cells have an active role in driving forgetting, then activating these cells would facilitate decay and decrease memory retention. We conducted neuronal activation experiments by using the two Gal4 drivers NP2397 and R76B09 to express UAS-dTRPA1, which encodes a heat-activated cation channel (58) that stimulates neurons at a moderately warm temperature (>25 °C). Indeed, we found decreased 3-h memory scores when we shifted flies to a higher temperature for 20 min immediately after training, in comparison with flies left at the lower temperature (Fig. 2 A and C). This decrease in 3-h memory was not caused by nonspecific consequences of temperature shift, because no significant effects were observed when the dTRPA1-expressing flies were shifted to the higher temperature before training (Fig. 2 B and D). Furthermore, the same temperature shift did not affect 3-h memory of the UAS-dTRPA1/+ control flies (“UAS alone”; Fig. 2). Together, these behavioral data using Kir2.1 and dTRPA1 to bidirectionally modulate neuronal activity confirm that the Gal4-expressing cells in the two categories of screened MBEN-Gal4 lines are involved in memory decay.
Fig. 2.
Neuronal activation after training decreases 3-h memory retention. One PAM-β′1 line (NP2397) and one MBON-γ4>γ1γ2 line (R76B09) from the screen were used to drive the expression of dTRPA1, a heat-activated cation channel. For acute neuronal activation, flies were moved to a higher temperature for 20 min starting either immediately after training or at 35 min before training (Materials and Methods). The groups experiencing a temperature shift (black) were compared (t test) with the control groups that stayed at the lower temperature (gray). (A and B) Activating NP2397 neurons after but not before training decreased 3-h memory retention. For NP2397/+; UAS-dTRPA1/+, “after training,” P < 0.001, n = 12; “before training,” P = 0.12, n = 8. For the male siblings that were identically treated but lacked the Gal4 driver (+/Y; UAS-dTRPA1/+), after training, P = 0.39, n = 12; before training, P = 0.21, n = 8. (C and D) Activating R76B09 neurons after but not before training decreased 3-h memory retention. For UAS-dTRPA1/+; R76B09/+, after training, P = 0.007, n = 14; before training, P = 0.94, n = 6. For the UAS-alone parental control (UAS-dTRPA1/+), after training, P = 0.85, n = 14; before training, P = 0.79, n = 6. Data are means ± SEM. **P < 0.01, ***P < 0.001.
A Subset of Dopaminergic Neurons Is Required for Forgetting.
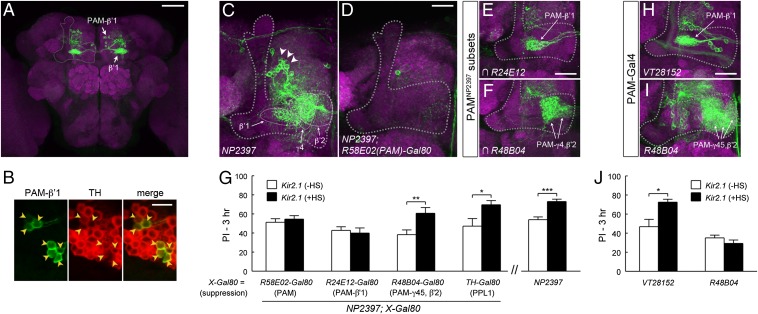
As mentioned above (Fig. 1B), NP2397 and four other Gal4 lines identified from the screen labeled PAM-β′1, a group of ∼13 dopaminergic neurons in the PAM cluster that specifically innervate the β′1-compartment (36). In the anterior inferior medial protocerebrum (aimpr), where PAM-β′1 somas are located, each of these Gal4 lines showed overlap in 11–12 cells with R24E12-LexA (SI Appendix, Fig. S2E), which in this brain area preferentially labeled PAM-β′1 (SI Appendix, Fig. S2 D and E). In addition, PAM-β′1 appeared to be the only MBEN type that was significantly shared by all five Gal4 lines we identified in this category, including the driver labeling the fewest cells in the aimpr (R94F11; SI Appendix, Fig. S2E). Consistent with the labeling of PAM-β′1, an intersectional Gal4 driver (59, 60) constructed from R94F11 and R24E12 (Materials and Methods) specifically visualized 13 ± 1 PAM-β′1 neurons in each hemisphere (mean ± SEM, n = 6 hemispheres) (Fig. 3A, see Fig. 5A and Movie S1 for more information). Tyrosine hydroxylase (TH) is the rate-limiting enzyme for dopamine biosynthesis and is used as a marker for dopaminergic neurons. We observed TH immunoreactivity in 112 cells in the PAM cluster, and all PAM-β′1 cells labeled by the intersectional Gal4 were immunopositive to TH (n = 2 hemispheres; Fig. 3B), suggesting that PAM-β′1 is dopaminergic.
Fig. 3.
PAM-β′1 subset of dopaminergic neurons is involved in forgetting. (A) PAM-β′1 neurons in the R94F11∩R24E12 split-Gal4 driver (R94F11-Gal4DBD; R24E12-p65AD). Throughout the figures, gray dashed lines indicate MB lobes. (B) Colocalization of PAM-β′1 with tyrosine hydroxylase, a dopamine marker. Arrowheads indicate PAM-β′1 somas visualized by a membrane-tagged reporter (myr::GFP) driven by R94F11∩R24E12. Representative images are single confocal sections. (C and D) Projection of the brain region including the MB lobes. NP2397 was expressed in putative PAM-DANs (arrowheads) whose processes innervated β′1, β′2, and γ4 (white dashed lines). The expression was substantially removed by R58E02-Gal80, which suppressed Gal4 activity in PAM-DANs. (E and F) Magnified views of the MB horizontal lobes. Intersection of NP2397 with R24E12-LexA and R48B04-LexA revealed PAM-DANs that innervated distinct lobe compartments (Materials and Methods). (G) Suppressing the expression of NP2397 in PAM-β′1 restored memory decay at 3 h. UAS-Kir2.1; Gal80ts flies were crossed to flies carrying NP2397 in combination with the indicated Gal80 transgene. Rescue of the 3-h memory increment was observed with R58E02-Gal80 (P = 0.54, n = 10, t test) and R24E12-Gal80 (P = 0.68, n = 8, t test) but not with TH-Gal80 (P = 0.03, n = 6, t test) and R48B04-Gal80 (P = 0.009, n = 12, t test). (H and I) VT28152 was selective for PAM-β′1, whereas R48B04 was mainly expressed in PAM-DANs innervating γ4, γ5, and β′2. (J) Kir2.1 inactivation of VT28152 neurons (P = 0.01, n = 6, t test) but not R48B04 neurons (P = 0.22, n = 9, t test) led to increased 3-h memory retention. Data in G and J are means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. [Scale bars, 50 μm (A), 10 μm (B), and 20 μm (C–F, H, and I).]
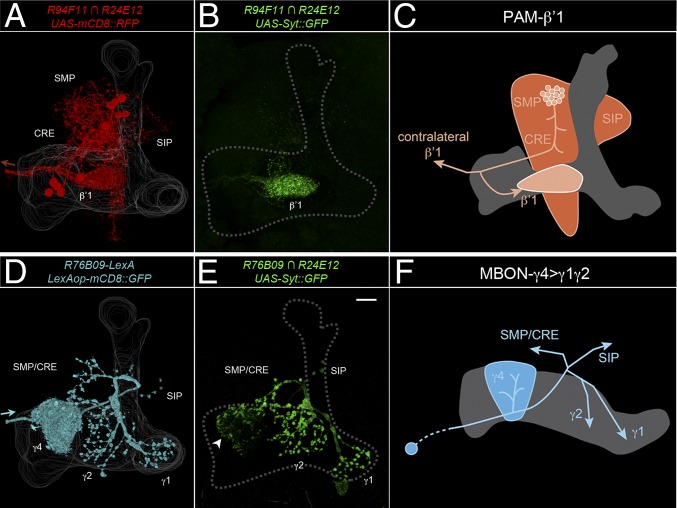
Fig. 5.
PAM-β′1 and MBON-γ4>γ1γ2 neurons have presynaptic terminals in the projections to the MB lobes. (A) Three-dimensional reconstruction of PAM-β′1 on one hemisphere visualized by a membrane-tagged reporter (mcd8::RFP). PAM-β′1 neurons ramified in the superior medial protocerebrum (SMP), superior intermediate protocerebrum (SIP), and crepine (CRE), an area encircling the horizontal lobes, and innervated ipsilateral and contralateral β′1 on the MB lobes. Arrow indicates neurites extending to the contralateral side. The MB landmark was based on nc82 counterstaining. (B) The presynaptic marker Syt::GFP in PAM-β′1 was enriched in β′1. PAM-β′1 somas obscured the Syt::GFP distribution pattern and were artificially removed. (C) Schematic of PAM-β′1. The gray area is the α′/β′-lobe. Arrows indicate axonal projections. (D) Three-dimensional reconstruction of MBON-γ4>γ1γ2 visualized by a membrane-tagged reporter (mCD8::GFP). MBON-γ4>γ1γ2 neurons sent neurites only to the contralateral MB, densely innervating γ4 en route and then projecting to more proximal γ1 and γ2 with minor branches sent outside the MB to SMP/CRE and SIP. Arrow indicates that the origination of the cell body fiber from contralateral MBON-γ4>γ1γ2. For a clear presentation, the ipsilateral soma and cell body fiber, as well as reporter signals from other R76B09-expressing neurons, were artificially removed. (E) Syt::GFP signal in MBON-γ4>γ1γ2 was distributed in multiple areas, which on the MB lobes included γ1 and γ2 and the periphery of γ4 (arrowhead). (Scale bar, 20 μm.) (F) Schematic of MBON-γ4>γ1γ2. The gray area is the γ-lobe. Arrows indicate axonal projections.
Given the overlap in PAM-β′1, we sought to determine whether PAM-β′1 is specifically responsible for the memory decay phenotype. We focused on the above-characterized NP2397 driver and used a panel of tissue-specific Gal80 lines to refine the Gal4 expression patterns. As described in an earlier study (37), NP2397 labeled ∼50 neurons in the aimpr, whose processes innervated multiple horizontal lobe compartments, primarily β′1, β′2, and γ4 (Fig. 3C). The majority of these neurons were removed by R58E02-Gal80 (Fig. 3D), which suppresses Gal4 expression in PAM-DANs (41). This suggests that NP2397 labeled multiple subsets of PAM-DANs, which included but were not limited to PAM-β′1. We examined whether removing PAM-DANs in NP2397 restored the behavioral effects. Indeed, we found that combining R58E02-Gal80 with NP2397 rescued the 3-h memory increment observed with Kir2.1 inactivation (Fig. 3G). In contrast, suppressing Gal4 expression in PPL1-DANs by TH-Gal80 (61) or in glutamatergic neurons by VGLUT-Gal80 (62) did not significantly alter the expression of NP2397 in PAM-DANs, and also failed to rescue the memory phenotype (see Fig. 3G and SI Appendix, Fig. S6 for data of TH-Gal80). These results indicate that the expression of NP2397 in PAM-DANs underlies the forgetting phenotype. To further distinguish different subsets of PAM-DANs, we took advantage of Gal80 lines derived from R24E12 and R48B04, two drivers that overlapped with NP2397 in PAM-β′1 and in other PAM-DANs, respectively (Fig. 3 E and F). Note that the R24E12 driver had expression in both PAM-β′1 and MBON-γ4>γ1γ2 (SI Appendix, Fig. S2A); we therefore used the R24E12-Gal80 transgene to suppress PAM-β′1 in this experiment and also to suppress MBON-γ4>γ1γ2 in an experiment described later (Fig. 4D). Removing PAM-β′1 in NP2397 using R24E12-Gal80 (SI Appendix, Fig. S6B) rescued the 3-h memory increment (Fig. 3G), whereas in a reciprocal experiment, removing PAM subsets other than PAM-β′1 using R48B04-Gal80 (SI Appendix, Fig. S6B) left the memory phenotype unaffected (Fig. 3G). For all of the Gal80 lines tested, the expression of NP2397 in areas outside the MB was not apparently affected (SI Appendix, Fig. S6A). These results suggest that PAM-β′1 neurons are a strong candidate for the behavioral phenotype.
Fig. 4.
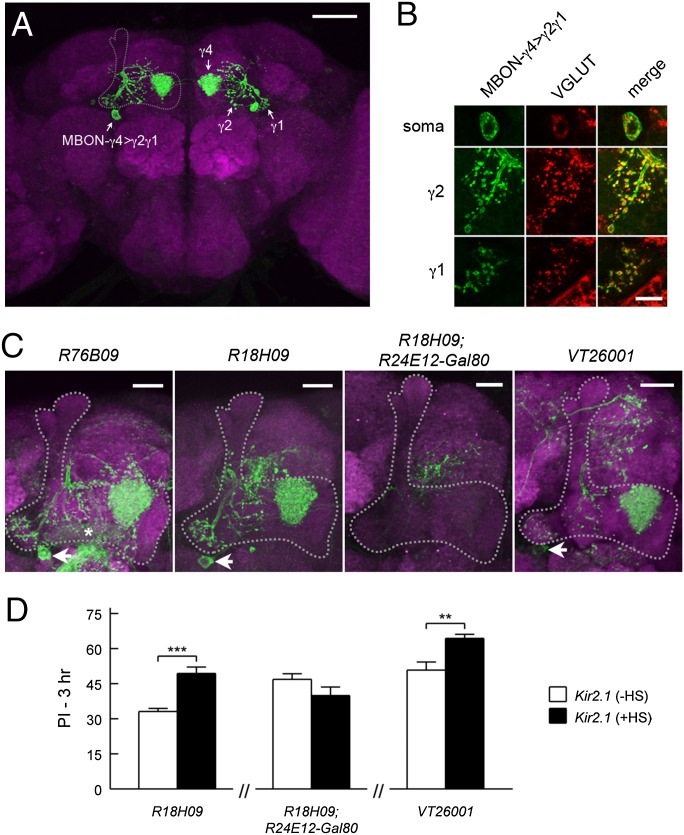
Pair of glutamatergic MBON-γ4>γ1γ2 neurons are involved in forgetting. (A) Visualization of MBON-γ4>γ1γ2 using the R76B09∩R24E12 split-Gal4 (R76B09-Gal4DBD; R24E12-p65AD). The soma on one hemisphere and the projection to the contralateral MB lobes are indicated. (B) MBON-γ4>γ1γ2 soma and the terminals at γ1 and γ2 were immunopositive for Drosophila vesicular glutamate transporter, a glutamate marker. MBON-γ4>γ1γ2 were visualized by myr::GFP driven by R76B09. (C) Projection of the brain region including the MB lobes. Arrows indicate MBON-γ4>γ1γ2 in the two lines identified from the screen (R76B09 and R18H09) and in a Gal4 line for validation (VT26001). R76B09 has weak expression in MBON-β1>α that innervates β1 (asterisk). The combination of R18H09 with R24E12-Gal80 removed the expression in MBON-γ4>γ1γ2. (D) Inactivating MBON-γ4>γ1γ2 neurons led to increased 3-h memory retention. UAS-Kir2.1; Gal80ts were combined with the indicated Gal4 drivers. The +HS group had better memory retention than the −HS group for R18H09 (P < 0.001, n = 6, t test) and VT26001 (P = 0.006, n = 6, t test) but not for R18H09, R24E12-Gal80 (P = 0.13, n = 8, t test). Data are means ± SEM. **P < 0.01, ***P < 0.001. [Scale bars, 50 μm (A), 5 μm (B), and 20 μm (C).]
We further confirmed the involvement of PAM-β′1 with an independent Gal4 line, VT28152 (SI Appendix, Fig. S7A). Note that the intersectional driver R94F11∩R24E12 (Fig. 3A) was constructed from the Gal80-incompatible p65AD (60), which prohibited use with UAS-Kir2.1/Gal80ts for testing the specificity to PAM-β′1. We therefore used the VT28152 driver. PAM-DANs labeled in this Gal4 belonged exclusively to the PAM-β′1 subset (Fig. 3H and SI Appendix, Fig. S7 B and C). Again, we observed increased 3-h memory retention in VT28152 with Kir2.1 inactivation (Fig. 3J). In comparison, and consistent with the functional heterogeneity of MB-innervating dopaminergic neurons, we did not detect a memory phenotype when either Kir2.1 (Fig. 3J) or tetanus toxin light chain (SI Appendix, Fig. S2B) was expressed with R48B04, which labeled ∼60 PAM-DANs (38, 43) that were nonoverlapping with PAM-β′1 (Fig. 3I and SI Appendix, Fig. S2E). In summary, multiple lines of evidence, including screen convergence, Gal80 refinement of the NP2397 driver, and post hoc validation using an independent Gal4 line, support a distinctive role of PAM-β′1 neurons in regulating memory decay.
A Pair of Glutamatergic Output Neurons Involved in Forgetting.
As mentioned above (Fig. 1B and SI Appendix, Fig. S2F), R76B09 and another Gal4 line identified in the screen, R18H09, labeled a pair of γ-lobe output neurons termed MBON-γ4>γ1γ2 (36). The somas of MBON-γ4>γ1γ2 were located underneath the junction of vertical and horizontal lobes; the processes densely innervated the γ4-compartment and projected further to γ1 and γ2, as well as small areas outside the MB (Fig. 4A, see Fig. 5D and Movie S2 for more information). Immunostaining of vesicular glutamate transporter (VGLUT) suggests that MBON-γ4>γ1γ2 is glutamatergic (Fig. 4B).
We found that the Gal4 expression of R76B09 and R18H09 was relatively specific to MBON-γ4>γ1γ2 in the adult brain (Fig. 4C and SI Appendix, Fig. S8). Although R76B09 also had weak expression in another MBEN type, MB-MV2 (MBON-β1>α), inactivating MB-MV2 with stronger Gal4 lines did not show a phenotype in 3-h memory (e.g., NP0242; SI Appendix, Table S1). These observations suggest that MBON-γ4>γ1γ2 is a strong candidate for the memory decay phenotype. For further confirmation, we used R24E12-Gal80 to remove the expression of R18H09 in MBON-γ4>γ1γ2. Accompanying the specific suppression of MBON-γ4>γ1γ2 expression (Fig. 4C and SI Appendix, Figs. S2F and S8), R24E12-Gal80 also rescued the 3-h memory increment with Kir2.1 inactivation (Fig. 4D) (note that we were unable to perform a similar rescue of R76B09, because neither R24E12-Gal80 nor VGLUT-Gal80 was able to fully suppress the expression of R76B09 in MBON-γ4>γ1γ2). As further evidence of the involvement of MBON-γ4>γ1γ2, we identified an independent Gal4 line (VT26001) that was specific to MBON-γ4>γ1γ2 (Fig. 4C and SI Appendix, Figs. S2F and S8). Inactivating MBON-γ4>γ1γ2 neurons with VT26001 showed increased 3-h memory retention (Fig. 4D), consistent with those observed in R76B09 and R18H09. Taken together, MBON-γ4>γ1γ2 is a second MBEN type identified from the screen that regulates memory decay.
Identified Neurons Relay Information to the MB.
We determined the direction of information flow in PAM-β′1 and MBON-γ4>γ1γ2 using genetically encoded neuronal polarity markers. For both groups of neurons, the presynaptic marker Syt::GFP (63) was found in the projections to the MB lobes. PAM-β′1 showed enriched Syt::GFP signal in the β′1-compartment (Fig. 5B), consistent with a role as modulatory afferent neurons to the β′-lobe. MBON-γ4>γ1γ2 is one of the few types of MBONs that interconnect multiple lobe compartments (36). The dendritic marker DenMark (64) was confined to the γ4-compartment (SI Appendix, Fig. S9B), whereas Syt::GFP was distributed in multiple projection areas, including the proximal compartments of the γ-lobe, γ1 and γ2 (Fig. 5E). Thus, both PAM-β′1 and MBON-γ4>γ1γ2 send axons to the MB lobes (Fig. 5 C and F). The β′- and γ-lobes bearing the projections of these neurons have important functions in early memory processing (65–67), which is in line with the idea that these neurons supply signals to modulate forgetting in memory-relevant sites.
Parallel Forgetting Pathways in Early Memory Decay.
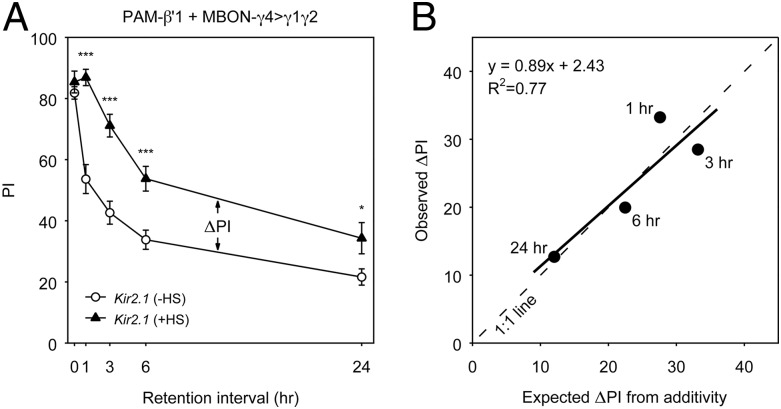
Given the observation that PAM-β′1 and MBON-γ4>γ1γ2 projected axons to distinct subdomains in the MB lobes, we next sought to determine whether they regulate memory decay in an additive manner. We noticed that when PAM-β′1 (NP2397) and MBON-γ4>γ1γ2 (R76B09) were individually inactivated, memory decay was slowed down but significant effects were observed primarily at early time points after training: up to 6 h but not at 24 h for PAM-β′1, and up to 3 h but marginal at later time points for MBON-γ4>γ1γ2 (SI Appendix, Fig. S10 B and C). We then examined the effects of inactivating both MBEN types simultaneously using a double Gal4 driver, NP2397; R76B09. Inactivating PAM-β′1 and MBON-γ4>γ1γ2 in combination produced a stronger effect, which largely prevented memory decline within 3 h and led to notably slower decay, lasting out to 24 h (Fig. 6A). To facilitate the comparison of combinatorial and individual inactivation, we calculated the differences in memory performance (ΔPI) between the +HS and −HS groups. This showed a greater effect on memory retention when both neuron types were inactivated simultaneously rather than individually (SI Appendix, Fig. S10E). When the effect of inactivating both PAM-β′1 and MBON-γ4>γ1γ2 was compared with what was predicted from a summation of the effects of inactivating PAM-β′1 and MBON-γ4>γ1γ2 individually, we observed a relationship close to a 1:1 match (Fig. 6B). Taken together, these results confirm important roles of PAM-β′1 and MBON-γ4>γ1γ2 in early memory decay and suggest an additive or near-additive effect from these two neural pathways.
Fig. 6.
Combinatorial inactivation of PAM-β′1 and MBON-γ4>γ1γ2 has an additive effect on memory decay. (A) Memory decay curves of flies bearing UAS-Kir2.1; Gal80ts combined with the double Gal4 driver NP2397; R76B09 labeling both PAM-β′1 and MBON-γ4>γ1γ2. The memory retention of the +HS and −HS groups was compared at different time points after training. All statistically significant differences (t test) are marked. Data are means ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001; n ≥ 6. (B) Relationship between the observed effect and the predicted effect under the assumption of simple additivity. The performance index difference between the +HS and −HS groups (ΔPI) observed for combinatorial inactivation in A was plotted against what is expected from the summation of individual inactivation of PAM-β′1 (NP2397; SI Appendix, Fig. S10B) and MBON-γ4>γ1γ2 (R76B09; SI Appendix, Fig. S10C). The dashed line is a 1:1 relationship, and the solid line is a regression fit.
Distinct Circuit Requirement for Reversal Learning.
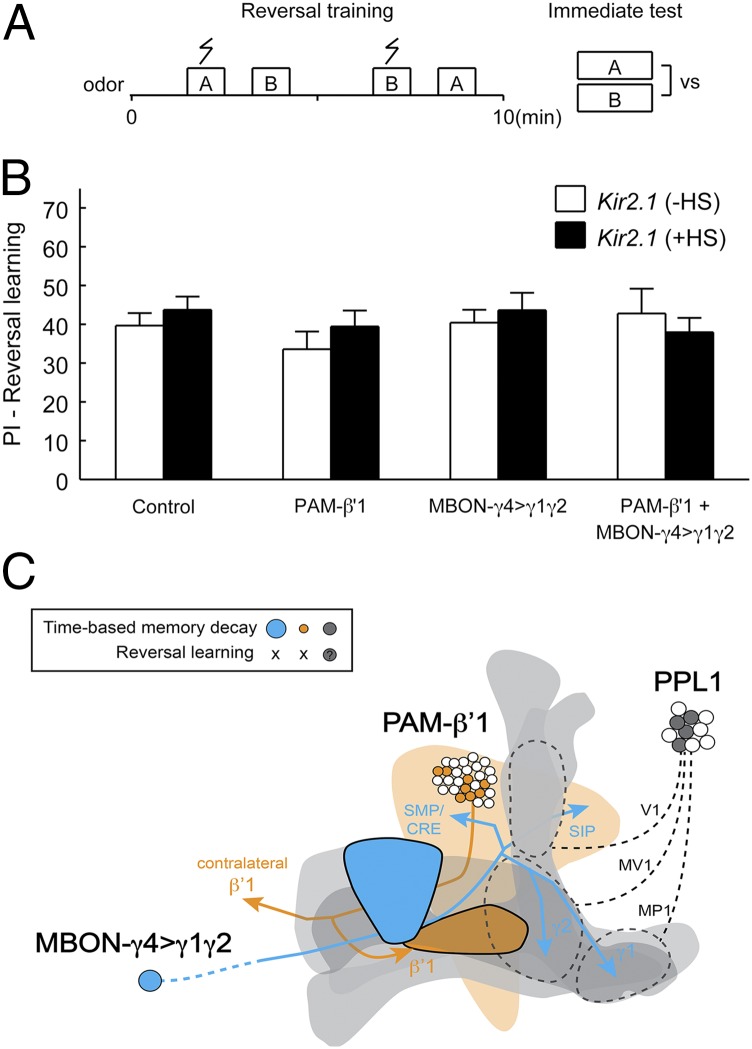
The ability to forget confers adaptive advantages to animals living in a changing environment (68, 69); for example, memory inconsistent with the current circumstance needs to be quickly removed or suppressed, which otherwise may endanger one’s survival. We here used a simple reversal learning paradigm (Fig. 7A), where a previously “safe” odor becomes “punishing,” whereas the punishing odor now becomes safe. Flies are readily able to learn such a reversal; when given a choice between the two trained odors, they selectively avoid the odor last paired with punishment (70). This flexible behavior is made possible by either acute forgetting or suppression of the memory acquired in the initial training session in a Rac-dependent manner (6). We tested whether PAM-β′1 and MBON-γ4>γ1γ2 also play a role in forgetting in this context. Interestingly, flies in which we inactivated PAM-β′1, MBON-γ4>γ1γ2, or even both in combination showed normal reversal learning performance (Fig. 7B), implying that their ability to unlearn or suppress the previously acquired conflicting information is intact. Therefore, despite their significant roles in time-based memory decay, PAM-β′1 and MBON-γ4>γ1γ2 are not required for the acute forgetting during reversal learning.
Fig. 7.
Kir2.1 inactivation of PAM-β′1 and MBON-γ4>γ1γ2 does not affect reversal learning. (A) Schematic of reversal learning. Immediately after a regular training session, flies were retrained but with the contingency between shock and the two trained odors reversed. Positive reversal learning PI indicates selective avoidance of the odor last paired with shock (Materials and Methods). (B) For all tested genotypes, no differences were found between the +HS and −HS groups in reversal learning. For “Control” (no Gal4), P = 0.39, n = 12; for “PAM-β′1” (NP2397), P = 0.35, n = 12; for “MBON-γ4>γ1γ2” (R76B09), P = 0.66, n = 6; for “PAM-β′1 + MBON-γ4>γ1γ2” (NP2397; R76B09), P = 0.60, n = 6; t test. Data are means ± SEM. (C) Three groups of MBENs are involved in forgetting of olfactory aversive memory. The schematic shows the PAM-β′1 and MBON-γ4>γ1γ2 neurons identified in this study. Their axonal projections are indicated by arrows, and the MB lobes are shown in gray. The schematic also illustrates the PPL1-DANs previously reported to be involved in forgetting, which may include three subgroups that project to the heel/peduncle, junction/lower-stalk, and upper-stalk regions of the MB neuropil (7). Memory decay over time requires these three groups of extrinsic neurons: PAM-β′1, MBON-γ4>γ1γ2, or PPL1-DANs. Blocking each of these neuronal groups inhibits, but does not fully stop, memory decay. Forgetting in reversal learning may recruit distinct circuit mechanisms, which are at least independent of PAM-β′1 and MBON-γ4>γ1γ2.
Discussion
From a neuronal inactivation screen, we identified two sets of MBENs that regulate forgetting of early olfactory aversive memory in Drosophila. The cellular organization of these neurons and the additive effect on memory decay by inactivating both sets of neurons simultaneously support the notion that these neurons represent parallel neural pathways that supply forgetting signals to the MB. Our results thus establish an anatomically well-defined example where forgetting is regulated by multiple neural pathways that impinge upon a memory center.
The identified MBENs include a subgroup of dopaminergic neurons in the PAM cluster whose axons innervate the β′1-compartment of the horizontal lobes (PAM-β′1) and a pair of glutamatergic output neurons with dendrites in the γ4- and synaptic terminals in the γ1- and γ2-compartments (MBON-γ4>γ1γ2). For both PAM-β′1 and MBON-γ4>γ1γ2, blocking their functions slows memory decay over a 3-h time period without affecting memory acquisition, memory consolidation, or task-relevant sensorimotor abilities. These findings, together with the fact that activating these neurons after but not before training decreases 3-h memory retention, support the idea that PAM-β′1 and MBON-γ4>γ1γ2 are neural components that contribute to an active process of forgetting.
Forgetting of olfactory aversive memory, therefore, involves at least three groups of MBENs: PAM-β′1, MBON-γ4>γ1γ2, and the previously reported PPL1-DANs (7) that innervate the heel/peduncle (MP1), lower stalk/junction (MV1), and upper stalk (V1) of the MB neuropil (Fig. 7C). These MBENs project axons to a broad area of the MB lobes and have access to all three KC subtypes. Conceivably, such circuit organization renders these neural pathways for forgetting well-positioned to counter memory distributed in the MB neuronal network (65–67). There exist notable differences among these neural pathways. In addition to their role in forgetting, PPL1-DANs are also required for learning and serve as the neural pathways that convey aversive reinforcement to the MB. In contrast, the inactivation of PAM-β′1 and MBON-γ4>γ1γ2 has a trivial effect (if any at all) on aversive learning (SI Appendix, Fig. S5). The unimportance of PAM-β′1 and MBON-γ4>γ1γ2 in learning demonstrates that at least part of the brain circuitry underlying learning and forgetting is dissociable, and their difference from PPL1-DANs also indicates that the multiple forgetting neural pathways may receive divergent inputs and undergo differential regulation.
The mechanisms by which the identified neurons cause forgetting await further investigation. PAM-β′1 is putatively dopaminergic, as suggested by TH immunoreactivity (Fig. 3B). Dopamine is a major neuromodulator in the MB circuit (36, 71), and plays a dual role in learning and forgetting by signaling through two different dopamine receptors, dDA1 and DAMB, respectively (7). The DAMB receptor mediating forgetting is expressed broadly within the MB but at higher levels in the α′/β′-lobe (72). It would be of considerable interest to test whether PAM-β′1, in addition to PPL1-DANs (7), functions through DAMB signaling to affect forgetting.
In our study, two PAM-β′1 Gal4 drivers, NP2397 and NP7177, are enhancer traps inserted in amn (SI Appendix, Fig. S3A). This potentially suggests that the putative AMN neuropeptide (53) is expressed in PAM-β′1 and is perhaps released in parallel with dopamine. We observed an increment of 3-h memory retention when the synaptic output of NP2397 neurons was blocked by expressing tetanus toxin light chain but not by expressing Shibirets1 (SI Appendix, Fig. S4). The replication of the Kir2.1 phenotype with tetanus toxin but not Shibirets1 is slightly consistent with a role of the AMN neuropeptide in PAM-β′1, because Shibirets1, a temperature-sensitive mutant of dynamin, blocks fast synaptic transmission but is presumably ineffective in blocking neuropeptide release (73). However, alternative explanations, such as insufficient expression level of UAS-Shibirets1 transgene, are equally valid to account for the absence of phenotype with Shibirets1. It is as yet unclear whether the AMN neuropeptide plays a role in the forgetting function mediated by PAM-β′1.
The other identified neuron group (MBON-γ4>γ1γ2) is glutamatergic, supporting the notion that forgetting regulation involves multiple neurotransmitter systems. In Drosophila, glutamatergic synapses are distributed only sparsely in the MB, albeit widely across other adult brain neuropils (74). MBON-γ4>γ1γ2 is among the few neurons (36) that supply glutamatergic inputs to the MB lobes. At present, we cannot distinguish whether MBON-γ4>γ1γ2 provides presynaptic inputs onto KCs, the MB intrinsic neurons. A recent study (36) found that the axonal terminals of MBON-γ4>γ1γ2 overlapped with the dendrites of a type of dopaminergic extrinsic neuron (PAM-γ4<γ1γ2), raising the possibility that MBON-γ4>γ1γ2 may also target other MBENs innervating the γ1- and γ2-lobe compartments. In rodents, it has been shown that NMDA receptor antagonists sustain long-term potentiation (LTP) and spatial memory when administered after LTP induction or following training (14). Here, the receptor signaling (75, 76) activated by MBON-γ4>γ1γ2 remains to be defined. Furthermore, it might be noteworthy that MBON-γ4>γ1γ2 and the MP1/MV1 subsets of PPL1-DANs both project axons to the γ1- and γ2-compartments. It is possible that the glutamate- and dopamine-mediated forgetting signals cross-talk in these compartments.
Apart from forgetting described here, several behavioral functions also depend in part on MBON-γ4>γ1γ2 in a recent study that screened through a set of behavioral tasks (39). For example, the activation of MBON-γ4>γ1γ2 elicits aversion, and a more chronic activation promotes wakefulness. The involvement of MBON-γ4>γ1γ2 in pleiotropic behavioral functions might be attributed to the influence of MBON-γ4>γ1γ2 on these behaviors at different time courses, or might be accounted for by its interaction with different sets of neurons, given the projection of MBON-γ4>γ1γ2 to multiple lobe compartments. Alternatively, it has been proposed that the ensemble of MBONs conveys an abstract representation of valence and internal state (39). In this regard, it would be an interesting possibility that the activity of MBON-γ4>γ1γ2 is linked with certain internal states, for example alertness or stress levels, which coordinate diverse behaviors, including how fast an aversive memory is to be forgotten.
Among the open questions arising from the current study are the different circuit mechanisms underlying forgetting in time-based memory decay and reversal learning. Rooted respectively in the concepts of decay and interference (32, 77), these two forms of forgetting have notable and interesting distinctions. Forgetting in memory decay proceeds at a timescale of hours and occurs without the overt presence of a triggering event, whereas forgetting in reversal learning takes place within minutes (or maybe instantly) and is evoked by reversing the contingency of the sensory cues that drive learning. Both rely on Rac activity in KCs, but Rac activation was found at distinct time domains, namely 1 h after training in memory decay but immediately in reversal learning (6). In this study, PAM-β′1 and MBON-γ4>γ1γ2 play important roles in memory decay but are dispensable in reversal learning. This raises the likelihood that the complex Rac activation kinetics might be coupled to the involvement of different forgetting neural pathways. It remains to be tested whether PAM-β′1 and MBON-γ4>γ1γ2 support a slow Rac activation and signal forgetting in time-based memory decay and, of equal importance, whether in reversal learning PPL1-DANs or other neurons (78) support a rapid Rac activation and guide acute forgetting. Deepening this line of research, we may approach an understanding of how the brain taps into mechanisms at multiple levels to achieve forgetting in different contexts.
Materials and Methods
Fly Strains and Reagents.
Drosophila melanogaster stocks were raised on a standard cornmeal medium and maintained at 25 °C at 60% relative humidity under a 16/8-h light/dark cycle with lights on at 9:00 AM. w1118 (isoCJ1) was wild type. Tubp-Gal80ts, UAS-dTRPA1, and UAS-Shibirets1 were extant stock in the laboratory. Gal4 lines for the screen (SI Appendix, Table S1) were mostly obtained from Bloomington Drosophila Stock Center (BDSC), Vienna Drosophila Resource Center (VDRC), and Japan Drosophila Genetic Resource Center in Kyoto (DGRC-Kyoto). VT28152 and VT26001 for validation were obtained from the VDRC. Additional flies from the BDSC included 10xUAS-IVS-mCD8::RFP [attP18], 13xLexAop2-mCD8::GFP [su(Hw)attP8] (32229), LexAop-2xmRFP.nls (29956), 10xUAS-IVS-myr::GFP [su(Hw)attP5] (32199), UAS-TNT (28838) (79), UAS-IMPTNT-VA2 (28840), and UAS-DenMark, UAS-Syt::GFP (33065). UAS-Kir2.1 was a gift from Gero Miesenboeck, University of Oxford, Oxford; 10xUAS-IVS-myr::GFP [attP40] and 13xLexAop2-IVS-GFP-p10 [Su(Hw)attP5] were from Gerald Rubin, Janelia Research Campus, Ashburn, VA; VT999036 was from Yoshinori Aso and Barry Dickson, Janelia Research Campus, Ashburn, VA; 8xLexAop2-Flpl [attP40]; UAS>stop>myr::GFP [attP1] (80) was from Yufeng Pan, Janelia Research Campus, Ashburn, VA; and R58E02, R58E02-Gal80, and TH-Gal80 were from Hiromu Tanimoto, Tohoku University, Sendai, Japan. VGLUT-Gal80 was shared generously by Leslie Vosshall, Rockefeller University, New York before publication. VGLUT antibody was from Aaron DiAntonio, Washington University in St. Louis, St. Louis.
Transgenes.
Enhancer fragments were amplified from genomic DNA of wild-type flies with Platinum Pfx high-fidelity DNA polymerase (Invitrogen). Sequences of PCR primers for R24E12, R94F11, R76B09, and R48B04 (55) were obtained from the Janelia FlyLight website. PCR products were cloned into pENTR/D-TOPO (Invitrogen) and sequenced for verification. The entry clone was then recombined in Gateway LR reactions (Invitrogen) with appropriate vectors (60) obtained from Addgene, including pBPnlsLexA::p65Uw, pBPZpGAL4DBDUw, pBPp65ADZpUw, and pBPG80Uw-6. The resulting clones were confirmed by PCR and sent for injection to make transgenic flies by BestGene. The attP sites and inserted chromosomes were as follows: R24E12-LexA (VK00027, III), R48B04-LexA (attP2, III), R76B09-LexA (VK00027, III), R24E12-Gal80 (VK00027, III), R48B04-Gal80 (attP40, II), R76B09-Gal80 (VK00027, III), R24E12-p65AD (VK00027, III), R94F11-Gal4DBD (attP40, II), and R76B09-Gal4DBD (attP40, II).
Fly crosses.
The combination of transgenes was performed using standard genetic crosses. In all crosses for behavioral experiments, male flies bearing Gal4 drivers were crossed to female flies carrying various UAS-driven effectors, including Kir2.1 (UAS-Kir2.1; Gal80ts), TNT (UAS-TNT, Gal80ts), IMPTNT (UAS-IMPTNT; Gal80ts), dTRPA1 (UAS-dTRPA1), and Shibirets1 (UAS-Shibirets1;;UAS-Shibirets1). For experiments involving the X chromosome-located NP2397 and NP7177, the male progeny did not carry the Gal4 driver and were used as controls bearing UAS-driven effectors alone (Figs. 2 A and B and 7B and SI Appendix, Figs. S3 B and C, S4 A–C, S5B, and S10A). They were trained and tested together with their female counterparts but were segregated after the memory test and scored separately. A similar strategy was applied to the combined driver NP2397; R76B09, except that the male progeny carried R76B09. These males generated behavioral results that were consistent with the fly progeny (males and females not segregated) from the crosses of UAS-driven effectors to R76B09. The data were thus combined and presented as a single group (MBON-γ4>γ1γ2; Fig. 7B and SI Appendix, Fig. S10C).
Behavioral assays.
Behavioral experiments were performed between 9:00 AM and 11:00 PM in an environmental room with 70% relative humidity. The temperature was set at 25 °C except for dTRPA1 and Shibirets1 experiments. The relevant fly groups were assayed side-by-side in a balanced manner and the assignment of flies to experimental groups was randomized. The conditions of the experiments were not blind to experimenters. Pavlovian olfactory aversive conditioning was performed as described previously (16). Briefly, around 100 flies were exposed sequentially to two aversive odors [3-octanol (OCT; Sigma-Aldrich) and 4-methylcyclohexanol (MCH; Sigma-Aldrich), 1.5 × 10−3 and 1 × 10−3 dilutions in heavy mineral oil (Fluka), respectively] for 60 s with a 45-s flush of fresh air after each odor. Flies received a 60-V foot shock (US) during the presence of the first odor (CS+) but not the second (CS−). To test memory performance, flies were given a choice between the two trained odors in a T maze for 120 s. PI was calculated as the fraction of flies avoiding CS+ minus the fraction avoiding CS− and finally multiplied by 100. A PI of 0 indicates an equal distribution; a PI of 100 indicates that all flies avoid the negatively reinforced CS+. To eliminate odor bias, each PI (n = 1) was averaged over two reciprocally trained groups, one associating shock with OCT, the other with MCH. Learning performance was tested immediately (within 3 min) after training. For memory retention, flies were placed in food vials for a given retention interval (at 25 °C for retention up to 3 h and at 18 °C for that longer than 3 h) before they were transferred back to a T maze for testing at 25 °C. Flies were tested once and killed. Different time points on memory decay curves were from different groups of flies. Reversal learning was performed as in our previous study (6). The odor (either OCT or MCH) paired with shock in the first session was not paired with shock in the second session and vice versa. Flies were immediately tested for a choice between the two reversely trained odors. PI was calculated as in normal learning, except that the odor last paired with shock was taken as “CS+.” Task-relevant sensorimotor responses were assayed as described previously (70). Olfactory acuity was quantified by exposing groups of 100 untrained flies to an odor (either OCT or MCH, 1.5 × 10−3 and 1 × 10−3 dilutions, respectively) versus fresh air in a T maze. Shock reactivity was tested by exposing groups of 100 untrained flies to two T-maze arms with a 60-V foot shock delivered to one arm but not the other. In both tests, flies were allowed to make a choice between the two T-maze arms for 120 s, after which they were trapped, anesthetized, and counted. PI was then calculated from the distribution of flies as in memory test. Each experiment also consisted of two reciprocal groups to control for a potential side bias of the T maze.
Gal4/Gal80ts induction.
Crosses were raised at 18 °C. Two- to 6-d-old adult progeny were collected and divided into two groups. The +HS group was transferred to a 30-°C incubator for 1 d, whereas the −HS group was kept continuously at 18 °C for the same period. Occasionally in the screen, flies receiving HS for up to 3 d were still used. Both the +HS and −HS groups were acclimated to the environmental room (25 °C) for about 1 h and then subjected to behavioral experiments under the same condition.
Cold-shock treatment.
At 2 h after training, flies were transferred to precooled empty vials and immersed in ice water. The treatment immobilized and anesthetized flies within a few seconds. After 2 min, flies were transferred back to fresh food vials and allowed to recover for 1 h before 3-h memory was tested.
dTRPA1 activation.
Crosses were raised at 18 °C to avoid unintended dTRPA1 activation. Two- to 6-d-old adult progeny were collected and moved to an environmental room at around 1 h before behavioral assays. The environmental room was set at a cool temperature below the activation threshold (∼25 °C) of dTRPA1, 23 °C for NP2397 and 19 °C for R76B09. The latter was set lower because we initially observed a below-average performance of R76B09/UAS-dTRPA1 flies at 23 °C, and were concerned that it might be caused by leaky activation of dTRPA1 at this temperature. Control groups were kept in the cool environmental room throughout the behavioral experiment. Activation groups were subjected to a brief temperature shift at the indicated time windows starting either immediately after training or at 35 min before the beginning of training. These flies were transferred to preheated food vials and kept for 20 min in a second environmental room set at a warm temperature sufficient to activate dTRPA1, 32 °C for NP2397 and 30 °C for R76B09. After that, they were returned to the cool environmental room to test 3-h memory.
Gal4–LexA intersection.
The Gal4–LexA intersectional strategy took advantage of a LexAop-driven flippase to remove the FRT-flanked stop codon in a UAS-fluorescence reporter (80). In the experiments, NP2397; LexAop2-Flpl; UAS>stop>myr::GFP flies were crossed to R24E12-LexA or R48B04-LexA flies and the adult progeny were examined for GFP reporter expression in the brain. Expected intersectional patterns were observed, but in a stochastic manner. A portion of flies showed the specific patterns as in the representative images in Fig. 3 E and F (4 in 31 for R24E12-LexA and 4 in 15 for R48B04-LexA). For the rest, a few showed no or very sparse labeling of neurons, likely attributable to the stochastic nature of flipout events, but the majority were reminiscent of the original NP2397 pattern, which might be caused by the less specific LexA driver expression earlier in development.
Immunostaining.
Two- to 6-d-old female adult brains were dissected in PBS and then fixed in 4% (wt/vol) paraformaldehyde (Electron Microscopy Sciences) in PBS on ice for 1 h. After three washes of 15 min each in 2% (vol/vol) Triton X-100 (Sigma-Aldrich) in PBS, the samples were blocked in 10% (vol/vol) normal goat serum (NGS; Jackson ImmunoResearch) in 2% (vol/vol) Triton X-100 (Sigma-Aldrich) in PBS for 1 h. They were then incubated with primary antibody diluted in 1% NGS in 0.2% Triton X-100 in PBS at 4 °C for 2 d. After three washes of 15 min each in 2% (vol/vol) Triton X-100 in PBS, the samples were incubated with secondary antibody diluted in 1% NGS in 0.2% Triton X-100 at 4 °C for 2 d. After two washes of 15 min each in 2% (vol/vol) Triton X-100 in PBS and one wash of 15 min in PBS, they were mounted in VECTASHIELD Mounting Medium (Vector Laboratories). For VGLUT staining, samples were fixed in Bouin’s solution (Sigma-Aldrich) on ice for 1 h. Primary antibodies used were mouse monoclonal nc82 [1:25; Developmental Studies Hybridoma Bank (DSHB)], rabbit polyclonal to dsRed (1:500; Clontech), chicken polyclonal IgY fraction to GFP (1:1,000; Aves Labs), mouse monoclonal to TH (1:500; ImmunoStar), rabbit polyclonal to VGLUT (1:500; Aaron DiAntonio, Washington University in St. Louis, St. Louis, MO), and rabbit polyclonal to GFP (1:1,000; Invitrogen). Secondary antibodies were Alexa Fluor-conjugated (1:250; Invitrogen).
Imaging.
Data were acquired on a Zeiss LSM 510 confocal microscope using a 20× dry objective (N.A. 0.75) or a 40× water-immersion objective (N.A. 1.2). The volume resolution (xyz) was 0.62 × 0.62 × 2 μm for whole-brain expression, 0.22 × 0.22 × 1 μm for MB lobe expression and soma colocalization, and 0.12 × 0.12 × 0.44 μm for 3D reconstruction. For each genotype, at least two brains were examined. Maximal intensity projections of Z stacks were processed in Fiji/ImageJ (NIH). Images were cropped and adjusted for brightness and contrast in Photoshop CS5 (Adobe). Three-dimensional reconstruction was realized in Vaa3D (81). The MB landmark in the 3D reconstruction was based on nc82 counterstaining. It was synthesized from a mask generated using the “Segmentation Editor” plugin in Fiji/ImageJ (fiji.sc/Fiji).
Statistics.
Statistical analyses were performed in MATLAB (MathWorks). All data satisfied the assumption of normal distribution (one-sample Kolmogorov–Smirnov test). Data involving comparison between two groups were analyzed by two-tailed unpaired t test. Those violating the homogeneity of variance assumption (F test) were subjected to two-tailed unpaired t test without assuming equal variances. Data involving comparison among multiple groups were analyzed by one-way ANOVA followed by multiple-comparisons t tests with Bonferroni’s correction, except that the data in SI Appendix, Fig. S10E were analyzed by two-way ANOVA followed by multiple comparisons of population marginal means. Significance level was set at P < 0.05. The sample sizes were similar to those in previous publications (6, 16).
Supplementary Material
Acknowledgments
We thank G. Miesenboeck (University of Oxford), G. Rubin, B. Dickson, and Y. Pan (Janelia Research Campus), H. Tanimoto (Tohoku University), L. Vosshall, N. Yapici, and J. Bussell (Rockefeller University), A. DiAntonio (Washington University in St. Louis), VDRC, BDSC, DGRC-Kyoto, and DSHB for flies and reagents. We are grateful to Y. Aso (Janelia Research Campus) for generous information sharing and helpful communication and G. Turner, T. Hige, J. Beshel, M. M. Shih (Cold Spring Harbor Laboratory), and Y. Hu (Tsinghua University) for critical reading of the manuscript. The research was supported by National Institutes of Health Grant R01DC013071 (to Y.Z.) and a William and Joyce O’Neil Fellowship (to Y.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1512792112/-/DCSupplemental.
References
- 1.Hardt O, Nader K, Nadel L. Decay happens: The role of active forgetting in memory. Trends Cogn Sci. 2013;17(3):111–120. doi: 10.1016/j.tics.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Schacter DL. The Seven Sins of Memory: How the Mind Forgets and Remembers. Houghton Mifflin; Boston: 2001. [Google Scholar]
- 3.Anderson MC, Hanslmayr S. Neural mechanisms of motivated forgetting. Trends Cogn Sci. 2014;18(6):279–292. doi: 10.1016/j.tics.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson JR, Schooler LJ. Reflections of the environment in memory. Psychol Sci. 1991;2(6):396–408. [Google Scholar]
- 5.Shuai Y, Hu Y, Qin H, Campbell RA, Zhong Y. Distinct molecular underpinnings of Drosophila olfactory trace conditioning. Proc Natl Acad Sci USA. 2011;108(50):20201–20206. doi: 10.1073/pnas.1107489109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shuai Y, et al. Forgetting is regulated through Rac activity in Drosophila. Cell. 2010;140(4):579–589. doi: 10.1016/j.cell.2009.12.044. [DOI] [PubMed] [Google Scholar]
- 7.Berry JA, Cervantes-Sandoval I, Nicholas EP, Davis RL. Dopamine is required for learning and forgetting in Drosophila. Neuron. 2012;74(3):530–542. doi: 10.1016/j.neuron.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hadziselimovic N, et al. Forgetting is regulated via Musashi-mediated translational control of the Arp2/3 complex. Cell. 2014;156(6):1153–1166. doi: 10.1016/j.cell.2014.01.054. [DOI] [PubMed] [Google Scholar]
- 9.Inoue A, et al. Forgetting in C. elegans is accelerated by neuronal communication via the TIR-1/JNK-1 pathway. Cell Reports. 2013;3(3):808–819. doi: 10.1016/j.celrep.2013.02.019. [DOI] [PubMed] [Google Scholar]
- 10.Jiang L, et al. Inhibition of Rac1 activity in the hippocampus impairs the forgetting of contextual fear memory. Mol Neurobiol. 2015 doi: 10.1007/s12035-015-9093-6. [DOI] [PubMed] [Google Scholar]
- 11.Frankland PW, Köhler S, Josselyn SA. Hippocampal neurogenesis and forgetting. Trends Neurosci. 2013;36(9):497–503. doi: 10.1016/j.tins.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Genoux D, et al. Protein phosphatase 1 is a molecular constraint on learning and memory. Nature. 2002;418(6901):970–975. doi: 10.1038/nature00928. [DOI] [PubMed] [Google Scholar]
- 13.Knezevic B, Dalesman S, Karnik V, Byzitter J, Lukowiak K. Low external environmental calcium levels prevent forgetting in Lymnaea. J Exp Biol. 2011;214(Pt 12):2118–2124. doi: 10.1242/jeb.054635. [DOI] [PubMed] [Google Scholar]
- 14.Villarreal DM, Do V, Haddad E, Derrick BE. NMDA receptor antagonists sustain LTP and spatial memory: Active processes mediate LTP decay. Nat Neurosci. 2002;5(1):48–52. doi: 10.1038/nn776. [DOI] [PubMed] [Google Scholar]
- 15.Berry JA, Cervantes-Sandoval I, Chakraborty M, Davis RL. Sleep facilitates memory by blocking dopamine neuron-mediated forgetting. Cell. 2015;161(7):1656–1667. doi: 10.1016/j.cell.2015.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tully T, Preat T, Boynton SC, Del Vecchio M. Genetic dissection of consolidated memory in Drosophila. Cell. 1994;79(1):35–47. doi: 10.1016/0092-8674(94)90398-0. [DOI] [PubMed] [Google Scholar]
- 17.Shuai Y, Zhong Y. Forgetting and small G protein Rac. Protein Cell. 2010;1(6):503–506. doi: 10.1007/s13238-010-0077-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heisenberg M. Mushroom body memoir: From maps to models. Nat Rev Neurosci. 2003;4(4):266–275. doi: 10.1038/nrn1074. [DOI] [PubMed] [Google Scholar]
- 19.Campbell RA, Turner GC. The mushroom body. Curr Biol. 2010;20(1):R11–R12. doi: 10.1016/j.cub.2009.10.031. [DOI] [PubMed] [Google Scholar]
- 20.Dubnau J, Grady L, Kitamoto T, Tully T. Disruption of neurotransmission in Drosophila mushroom body blocks retrieval but not acquisition of memory. Nature. 2001;411(6836):476–480. doi: 10.1038/35078077. [DOI] [PubMed] [Google Scholar]
- 21.Zars T, Fischer M, Schulz R, Heisenberg M. Localization of a short-term memory in Drosophila. Science. 2000;288(5466):672–675. doi: 10.1126/science.288.5466.672. [DOI] [PubMed] [Google Scholar]
- 22.McGuire SE, Le PT, Davis RL. The role of Drosophila mushroom body signaling in olfactory memory. Science. 2001;293(5533):1330–1333. doi: 10.1126/science.1062622. [DOI] [PubMed] [Google Scholar]
- 23.Han KA, Millar NS, Grotewiel MS, Davis RL. DAMB, a novel dopamine receptor expressed specifically in Drosophila mushroom bodies. Neuron. 1996;16(6):1127–1135. doi: 10.1016/s0896-6273(00)80139-7. [DOI] [PubMed] [Google Scholar]
- 24.Plaçais PY, et al. Slow oscillations in two pairs of dopaminergic neurons gate long-term memory formation in Drosophila. Nat Neurosci. 2012;15(4):592–599. doi: 10.1038/nn.3055. [DOI] [PubMed] [Google Scholar]
- 25.Walkinshaw E, et al. Identification of genes that promote or inhibit olfactory memory formation in Drosophila. Genetics. 2015;199(4):1173–1182. doi: 10.1534/genetics.114.173575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berry JA, Davis RL. Active forgetting of olfactory memories in Drosophila. Prog Brain Res. 2014;208:39–62. doi: 10.1016/B978-0-444-63350-7.00002-4. [DOI] [PubMed] [Google Scholar]
- 27.Qin H, et al. Gamma neurons mediate dopaminergic input during aversive olfactory memory formation in Drosophila. Curr Biol. 2012;22(7):608–614. doi: 10.1016/j.cub.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aso Y, et al. Three dopamine pathways induce aversive odor memories with different stability. PLoS Genet. 2012;8(7):e1002768. doi: 10.1371/journal.pgen.1002768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aso Y, et al. Specific dopaminergic neurons for the formation of labile aversive memory. Curr Biol. 2010;20(16):1445–1451. doi: 10.1016/j.cub.2010.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Claridge-Chang A, et al. Writing memories with light-addressable reinforcement circuitry. Cell. 2009;139(2):405–415. doi: 10.1016/j.cell.2009.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis RL. Traces of Drosophila memory. Neuron. 2011;70(1):8–19. doi: 10.1016/j.neuron.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wixted JT. The psychology and neuroscience of forgetting. Annu Rev Psychol. 2004;55:235–269. doi: 10.1146/annurev.psych.55.090902.141555. [DOI] [PubMed] [Google Scholar]
- 33.Dudai Y. The neurobiology of consolidations, or, how stable is the engram? Annu Rev Psychol. 2004;55:51–86. doi: 10.1146/annurev.psych.55.090902.142050. [DOI] [PubMed] [Google Scholar]
- 34.Reaume CJ, Sokolowski MB, Mery F. A natural genetic polymorphism affects retroactive interference in Drosophila melanogaster. Proc Biol Sci. 2011;278(1702):91–98. doi: 10.1098/rspb.2010.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tully T, et al. Genetic dissection of memory formation in Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1990;55:203–211. doi: 10.1101/sqb.1990.055.01.022. [DOI] [PubMed] [Google Scholar]
- 36.Aso Y, et al. The neuronal architecture of the mushroom body provides a logic for associative learning. eLife. 2014;3:e04577. doi: 10.7554/eLife.04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka NK, Tanimoto H, Ito K. Neuronal assemblies of the Drosophila mushroom body. J Comp Neurol. 2008;508(5):711–755. doi: 10.1002/cne.21692. [DOI] [PubMed] [Google Scholar]
- 38.Yamagata N, et al. Distinct dopamine neurons mediate reward signals for short- and long-term memories. Proc Natl Acad Sci USA. 2015;112(2):578–583. doi: 10.1073/pnas.1421930112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aso Y, et al. Mushroom body output neurons encode valence and guide memory-based action selection in Drosophila. eLife. 2014;3:e04580. doi: 10.7554/eLife.04580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plaçais PY, Trannoy S, Friedrich AB, Tanimoto H, Preat T. Two pairs of mushroom body efferent neurons are required for appetitive long-term memory retrieval in Drosophila. Cell Reports. 2013;5(3):769–780. doi: 10.1016/j.celrep.2013.09.032. [DOI] [PubMed] [Google Scholar]
- 41.Liu C, et al. A subset of dopamine neurons signals reward for odour memory in Drosophila. Nature. 2012;488(7412):512–516. doi: 10.1038/nature11304. [DOI] [PubMed] [Google Scholar]
- 42.Owald D, et al. Activity of defined mushroom body output neurons underlies learned olfactory behavior in Drosophila. Neuron. 2015;86(2):417–427. doi: 10.1016/j.neuron.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin S, et al. Neural correlates of water reward in thirsty Drosophila. Nat Neurosci. 2014;17(11):1536–1542. doi: 10.1038/nn.3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perisse E, et al. Different Kenyon cell populations drive learned approach and avoidance in Drosophila. Neuron. 2013;79(5):945–956. doi: 10.1016/j.neuron.2013.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Burke CJ, et al. Layered reward signalling through octopamine and dopamine in Drosophila. Nature. 2012;492(7429):433–437. doi: 10.1038/nature11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krashes MJ, et al. A neural circuit mechanism integrating motivational state with memory expression in Drosophila. Cell. 2009;139(2):416–427. doi: 10.1016/j.cell.2009.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pai TP, et al. Drosophila ORB protein in two mushroom body output neurons is necessary for long-term memory formation. Proc Natl Acad Sci USA. 2013;110(19):7898–7903. doi: 10.1073/pnas.1216336110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bouzaiane E, Trannoy S, Scheunemann L, Plaçais PY, Preat T. Two independent mushroom body output circuits retrieve the six discrete components of Drosophila aversive memory. Cell Reports. 2015;11(8):1280–1292. doi: 10.1016/j.celrep.2015.04.044. [DOI] [PubMed] [Google Scholar]
- 49.Jenett A, et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Reports. 2012;2(4):991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kvon EZ, et al. Genome-scale functional characterization of Drosophila developmental enhancers in vivo. Nature. 2014;512(7512):91–95. doi: 10.1038/nature13395. [DOI] [PubMed] [Google Scholar]
- 51.Baines RA, Uhler JP, Thompson A, Sweeney ST, Bate M. Altered electrical properties in Drosophila neurons developing without synaptic transmission. J Neurosci. 2001;21(5):1523–1531. doi: 10.1523/JNEUROSCI.21-05-01523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302(5651):1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- 53.Feany MB, Quinn WG. A neuropeptide gene defined by the Drosophila memory mutant amnesiac. Science. 1995;268(5212):869–873. doi: 10.1126/science.7754370. [DOI] [PubMed] [Google Scholar]
- 54.Waddell S, Armstrong JD, Kitamoto T, Kaiser K, Quinn WG. The amnesiac gene product is expressed in two neurons in the Drosophila brain that are critical for memory. Cell. 2000;103(5):805–813. doi: 10.1016/s0092-8674(00)00183-5. [DOI] [PubMed] [Google Scholar]
- 55.Pfeiffer BD, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci USA. 2008;105(28):9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Margulies C, Tully T, Dubnau J. Deconstructing memory in Drosophila. Curr Biol. 2005;15(17):R700–R713. doi: 10.1016/j.cub.2005.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quinn WG, Dudai Y. Memory phases in Drosophila. Nature. 1976;262(5569):576–577. doi: 10.1038/262576a0. [DOI] [PubMed] [Google Scholar]
- 58.Hamada FN, et al. An internal thermal sensor controlling temperature preference in Drosophila. Nature. 2008;454(7201):217–220. doi: 10.1038/nature07001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luan H, Peabody NC, Vinson CR, White BH. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron. 2006;52(3):425–436. doi: 10.1016/j.neuron.2006.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pfeiffer BD, et al. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186(2):735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sitaraman D, et al. Serotonin is necessary for place memory in Drosophila. Proc Natl Acad Sci USA. 2008;105(14):5579–5584. doi: 10.1073/pnas.0710168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bussell JJ, Yapici N, Zhang SX, Dickson BJ, Vosshall LB. Abdominal-B neurons control Drosophila virgin female receptivity. Curr Biol. 2014;24(14):1584–1595. doi: 10.1016/j.cub.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang YQ, Rodesch CK, Broadie K. Living synaptic vesicle marker: Synaptotagmin-GFP. Genesis. 2002;34(1-2):142–145. doi: 10.1002/gene.10144. [DOI] [PubMed] [Google Scholar]
- 64.Nicolaï LJ, et al. Genetically encoded dendritic marker sheds light on neuronal connectivity in Drosophila. Proc Natl Acad Sci USA. 2010;107(47):20553–20558. doi: 10.1073/pnas.1010198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guven-Ozkan T, Davis RL. Functional neuroanatomy of Drosophila olfactory memory formation. Learn Mem. 2014;21(10):519–526. doi: 10.1101/lm.034363.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perisse E, Burke C, Huetteroth W, Waddell S. Shocking revelations and saccharin sweetness in the study of Drosophila olfactory memory. Curr Biol. 2013;23(17):R752–R763. doi: 10.1016/j.cub.2013.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dubnau J, Chiang AS. Systems memory consolidation in Drosophila. Curr Opin Neurobiol. 2013;23(1):84–91. doi: 10.1016/j.conb.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 68.Kraemer PJ, Golding JM. Adaptive forgetting in animals. Psychon Bull Rev. 1997;4(4):480–491. [Google Scholar]
- 69.Brea J, Urbanczik R, Senn W. A normative theory of forgetting: Lessons from the fruit fly. PLOS Comput Biol. 2014;10(6):e1003640. doi: 10.1371/journal.pcbi.1003640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tully T, Quinn WG. Classical conditioning and retention in normal and mutant Drosophila melanogaster. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 1985;157(2):263–277. doi: 10.1007/BF01350033. [DOI] [PubMed] [Google Scholar]
- 71.Waddell S. Reinforcement signalling in Drosophila; dopamine does it all after all. Curr Opin Neurobiol. 2013;23(3):324–329. doi: 10.1016/j.conb.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Crittenden JR, Skoulakis EM, Han KA, Kalderon D, Davis RL. Tripartite mushroom body architecture revealed by antigenic markers. Learn Mem. 1998;5(1-2):38–51. [PMC free article] [PubMed] [Google Scholar]
- 73.Kitamoto T. Targeted expression of temperature-sensitive dynamin to study neural mechanisms of complex behavior in Drosophila. J Neurogenet. 2002;16(4):205–228. doi: 10.1080/01677060216295. [DOI] [PubMed] [Google Scholar]
- 74.Daniels RW, Gelfand MV, Collins CA, DiAntonio A. Visualizing glutamatergic cell bodies and synapses in Drosophila larval and adult CNS. J Comp Neurol. 2008;508(1):131–152. doi: 10.1002/cne.21670. [DOI] [PubMed] [Google Scholar]
- 75.Xia S, et al. NMDA receptors mediate olfactory learning and memory in Drosophila. Curr Biol. 2005;15(7):603–615. doi: 10.1016/j.cub.2005.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu WW, Wilson RI. Glutamate is an inhibitory neurotransmitter in the Drosophila olfactory system. Proc Natl Acad Sci USA. 2013;110(25):10294–10299. doi: 10.1073/pnas.1220560110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jonides J, et al. The mind and brain of short-term memory. Annu Rev Psychol. 2008;59:193–224. doi: 10.1146/annurev.psych.59.103006.093615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu Y, Ren Q, Li H, Guo A. The GABAergic anterior paired lateral neurons facilitate olfactory reversal learning in Drosophila. Learn Mem. 2012;19(10):478–486. doi: 10.1101/lm.025726.112. [DOI] [PubMed] [Google Scholar]
- 79.Sweeney ST, Broadie K, Keane J, Niemann H, O’Kane CJ. Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron. 1995;14(2):341–351. doi: 10.1016/0896-6273(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 80.Pan Y, Baker BS. Genetic identification and separation of innate and experience-dependent courtship behaviors in Drosophila. Cell. 2014;156(1-2):236–248. doi: 10.1016/j.cell.2013.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Peng H, Ruan Z, Long F, Simpson JH, Myers EW. V3D enables real-time 3D visualization and quantitative analysis of large-scale biological image data sets. Nat Biotechnol. 2010;28(4):348–353. doi: 10.1038/nbt.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.