Significance
Many different types of plankton bloom in the coastal ocean. Some produce harmful toxins, whereas others are considered important contributors to primary production and ecosystem dynamics. Routine ocean color sensors can reveal a dense bloom or “red tide” from space but cannot differentiate types of plankton. The Hyperspectral Imager for the Coastal Ocean on the International Space Station allowed for new “hyperspectral” approaches to identify pigments and assess biophysical forcing associated with different blooms. Consistent with field and molecular observations, imagery revealed dense surface patches of Mesodinium rubrum, a ciliate that uses the chloroplasts from its cryptophyte algal prey to photosynthesize. For the first time, to our knowledge, the distribution and abundance were mapped based on the unique yellow fluorescence properties of its signature pigment phycoerythrin.
Keywords: remote sensing, hyperspectral, ocean color, red tide, Mesodinium ciliate
Abstract
Mesodinium rubrum is a globally distributed nontoxic ciliate that is known to produce intense red-colored blooms using enslaved chloroplasts from its algal prey. Although frequent enough to have been observed by Darwin, blooms of M. rubrum are notoriously difficult to quantify because M. rubrum can aggregate into massive clouds of rusty-red water in a very short time due to its high growth rates and rapid swimming behavior and can disaggregate just as quickly by vertical or horizontal dispersion. A September 2012 hyperspectral image from the Hyperspectral Imager for the Coastal Ocean sensor aboard the International Space Station captured a dense red tide of M. rubrum (106 cells per liter) in surface waters of western Long Island Sound. Genetic data confirmed the identity of the chloroplast as a cryptophyte that was actively photosynthesizing. Microscopy indicated extremely high abundance of its yellow fluorescing signature pigment phycoerythrin. Spectral absorption and fluorescence features were related to ancillary photosynthetic pigments unique to this organism that cannot be observed with traditional satellites. Cell abundance was estimated at a resolution of 100 m using an algorithm based on the distinctive yellow fluorescence of phycoerythrin. Future development of hyperspectral satellites will allow for better enumeration of bloom-forming coastal plankton, the associated physical mechanisms, and contributions to marine productivity.
Massive red-colored blooms of Mesodinium rubrum (syn. Myrionecta rubra) up to 106 cells per liter have been reported in coastal waters, sometimes in association with upwelling and eastern boundary currents (1) (Fig. 1 A and B). During the voyage of the HMS Beagle, Darwin (1839) observed enormous patches of red water off Chile and associated them with a small ciliate he observed in the microscope, writing: “Some of the water placed in a glass was of a pale reddish tint; and examined under a microscope, was seen to swarm with minute animalcula darting about, and often exploding. Their shape is oval and contracted in the middle by a ring of vibrating curved ciliae.” (2). His description of the size, shape, color, swimming motion, and fragility of the ciliate suggests strongly that it was M. rubrum. Even though considered a microzooplankton, its photosynthetic contribution can represent up to 70% of total primary productivity (1), making it an occasionally dominant member of the phytoplankton community. However, because it is a mixotroph and difficult to enumerate (3), M. rubrum is regularly excluded from reports on both phytoplankton and zooplankton.
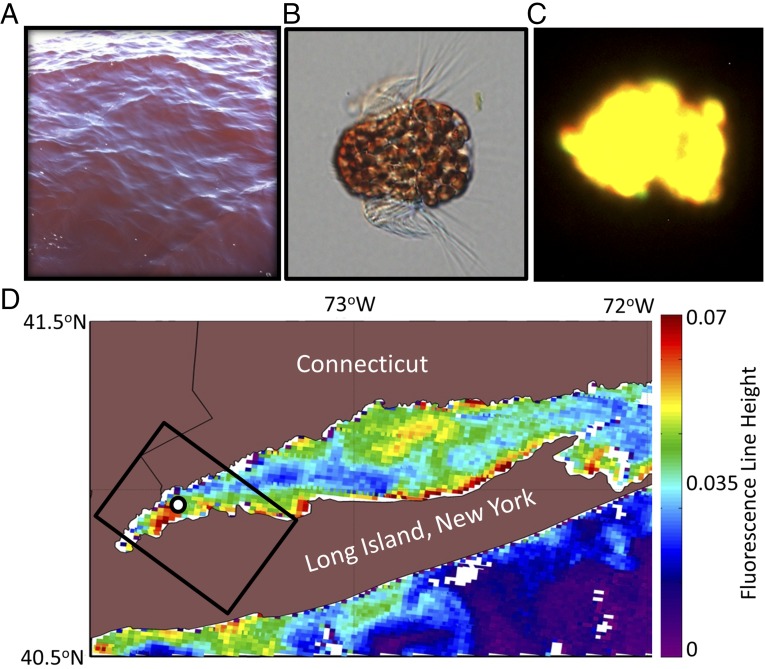
Fig. 1.
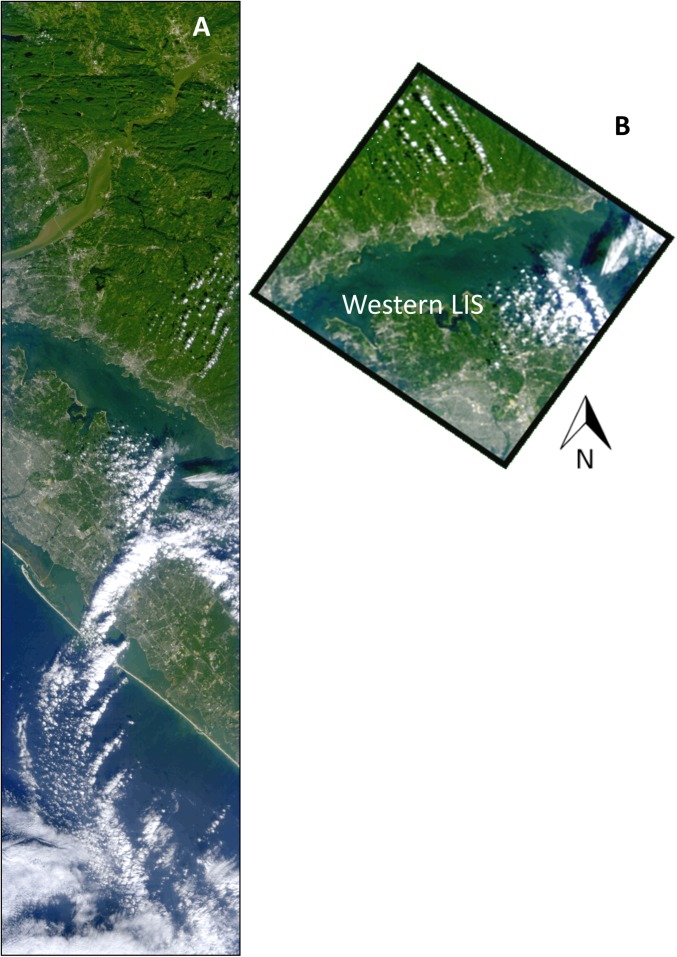
(A) Photograph of red water in WLIS during the observed M. rubrum bloom. Photo courtesy of Kay Howard-Strobel (University of Connecticut, Groton, CT). (B) Micrograph of the ciliate M. rubrum with enslaved cryptophyte chloroplasts. Micrograph courtesy of National Oceanic and Atmospheric Administration Phytoplankton Monitoring Network. (C) Fluorescent microscopy under green light excitation showing characteristic yellow fluorescence (565–570 nm) (26) of M. rubrum due to high amounts of phycoerythrin pigment. (D) Satellite image of Chl a fluorescence line height from the MODIS Terra sensor on September 23, 2012 shows a dense bloom in WLIS, where M. rubrum was sampled at 1 × 106 cells per liter (black circle). The black rectangle outlines the swath of the coincident HICO imagery obtained from the International Space Station.
The nature of the relationship between the litostome ciliate and its cryptophyte organelles has been the subject of considerable study. Although it has been described as a symbiosis, it seems clear that the cryptophyte never survives ingestion and the arrangement is similar to the practice of chloroplast enslavement known from other ciliates and some sacoglossan molluscs (4–6). Unlike the latter cases, however, cryptophyte nuclei remain transcriptionally active within M. rubrum (7). Retained chloroplasts can also reproduce, allowing the ciliate to survive without food for prolonged periods in the light (8). Nevertheless, the arrangement is obligate for M. rubrum, which cannot grow in the dark, even with abundant food.
Generally, M. rubrum is more abundant in lower salinity estuarine water (9, 10), but the causes of bloom initiation and demise are not well known (11). In western Long Island Sound (WLIS), approximately monthly monitoring in all seasons over a 12-y span found M. rubrum to be present in 80% of 572 samples at a mean concentration of 354 cells per liter and a maximum of 2.6 × 104 cells per liter (Table S1). Coarse monthly sampling, however, is not sufficient to ensure quantification of episodic blooms (Fig. S1), in part, due to the extreme motility and phototactic behavior of the organism (12). The ciliate can migrate vertically tens of meters per day, aggregate in thin subsurface layers at specific periods of the day, and move up and downstream with flood and ebb tides (10, 13). Hence, abundances can rapidly increase to bloom conditions due to near-surface aggregation and disappear quickly with migration vertically and dispersion horizontally. Better methods are needed to capture these blooms and differentiate them from other types of red tides.
Table S1.
Summary of M. rubrum abundance from microscopic counts of Long Island Sound monitoring samples from 2002 to 2013
| Metric | Abundance (cells L−1) | Biomass (μg L−1) |
| Maximum | 25,671 | 39 |
| Minimum | 0 | 0 |
| Median | 110 | 0.12 |
| Mean | 354 | 0.59 |
| N | 572 | 572 |
| SD | 1,363 | 2.18 |
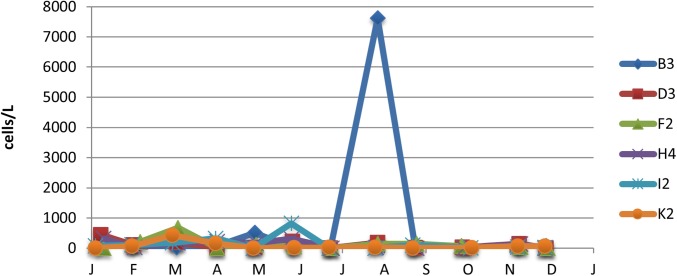
Fig. S1.
Abundance of Mesodinium spp. at six stations in Long Island Sound during monthly sampling in 2012. Stations B3 to K2 are aligned along the main axis of the sound, alphabetically, from west to east. The monthly sampling on July 31 at the westernmost station B3 revealed a high amount of Mesodinium (>7,000 cells per liter). Routine monthly sampling on October 2, 2012, however, missed the intense bloom (>1 × 106 cells per liter) observed on September 24, 2012.
Results and Discussion
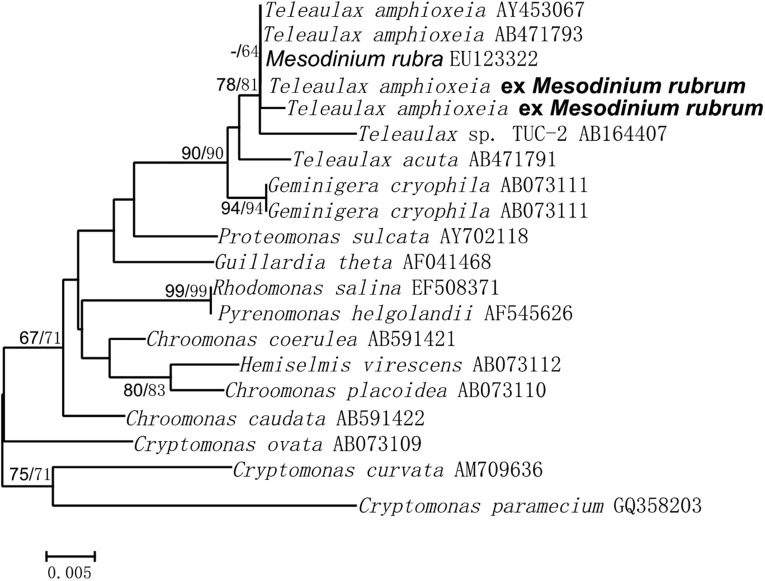
On September 24, 2012, a dense aggregation of M. rubrum was observed in WLIS manifest as intensely red-colored water (40.9°N, 73.6°W) (Fig. 1B). Conditions were calm with wind speeds averaging 3 m⋅s−1. A water sample revealed cell counts as high as 1.03 × 106 cells per liter of M. rubrum, higher than ever measured during 12 y of routine sampling (Table S1). Plastid 16S rDNA analysis of the bloom showed that M. rubrum harbored plastids of the cryptophyte Teleaulax amphioxeia (Fig. S2). Transcriptome data obtained from the bloom show active expression of the plastid protein synthesis apparatus (16S rRNA) and functional proteins (e.g., light harvesting protein, carbon fixing enzyme Rubisco) (Table S2), suggesting that the chloroplasts were actively photosynthesizing. The high expression level of the phycoerythrin gene is consistent with the extremely abundant green-light harvesting pigment phycoerythrin (14) observed microscopically as strong orange-yellow fluorescence (Fig. 1C). The intense red water color was caused by high absorption of blue and green photons, and a consequent shift of reflected light to wavelengths >550 nm (15). The extent and duration of the bloom, however, could not be deduced from ship sampling.
Fig. S2.
Plastid 16S rDNA phylogenetic tree indicating that M. rubrum in the bloom harbored a plastid of T. amphioxeia (bold type indicates sequences obtained in this study). Values at nodes are bootstrap values from maximum likelihood/neighbor-joining analysis. The scale bar shows the substitution rate per nucleotide site.
Table S2.
Expression of chloroplast genes detected from the bloom
| Gene | Transcript abundance, FPKM |
| 16S rRNA | Detected but not quantified* |
| Rubisco | 11.7 |
| Phycoerythrin synthesis gene (pe) | 39,153 |
Only genes relevant to this paper are shown; more complete transcriptome data will be published elsewhere. FPKM, fragments per kilobases per million mapped reads.
Not quantified because it is not a protein-coding gene, and hence not represented in the transcriptomic data; it was detected by PCR with random primer-based cDNA as a template.
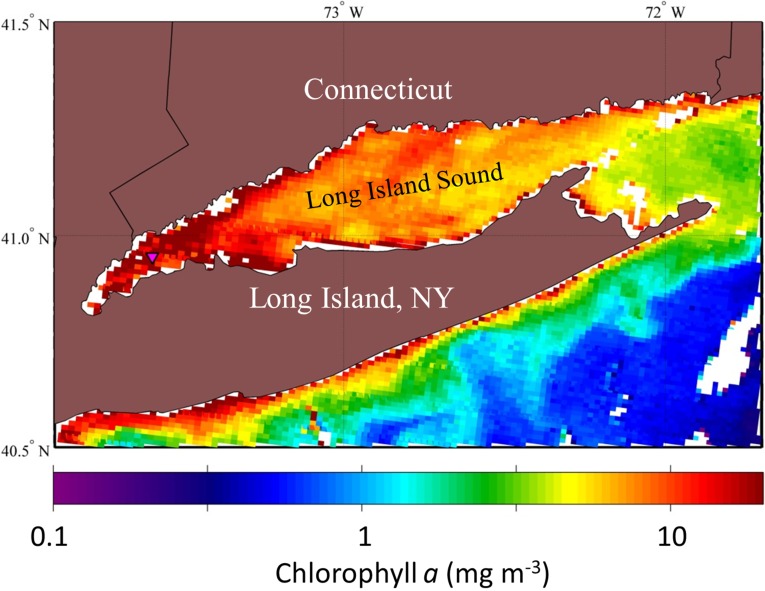
Remote sensing affords the opportunity to examine the spatial and temporal changes in water color. The standard chlorophyll a (Chl a) algorithm, however, fails in the optically complex coastal waters of WLIS due to rich concentrations of suspended minerals and colored dissolved organic matter (CDOM) (16–18). These blue-absorbing and scattering constituents can mimic the water color produced by phytoplankton and result in overestimates of their abundance, as shown by the high Chl a estimates throughout the sound (Fig. S3). However, Chl a, which is found in all phytoplankton, fluoresces in far red wavelengths of ∼678 nm. Although not contributing significantly to the color observed by the human eye (15), the amount of natural fluorescence and backscattering at far red wavelengths can be used to detect surface blooms of phytoplankton without contamination by CDOM and minerals (19, 20). Measurements at a resolution of 1 km from the Moderate Resolution Imaging Spectrometer (MODIS) Terra sensor on September 23, 2012 revealed the heterogeneous distribution of Chl a in the sound and, in particular, a patchy ∼20-km2 bloom covering our field station in WLIS (Fig. 1D).
Fig. S3.
MODIS Terra image of Chl a retrieved at 11:25 AM Eastern Standard Time on September 23, 2012. The Chl a concentration using the standard algorithm is overestimated in the complex waters of Long Island Sound, with high amounts of colored dissolved and suspended matter.
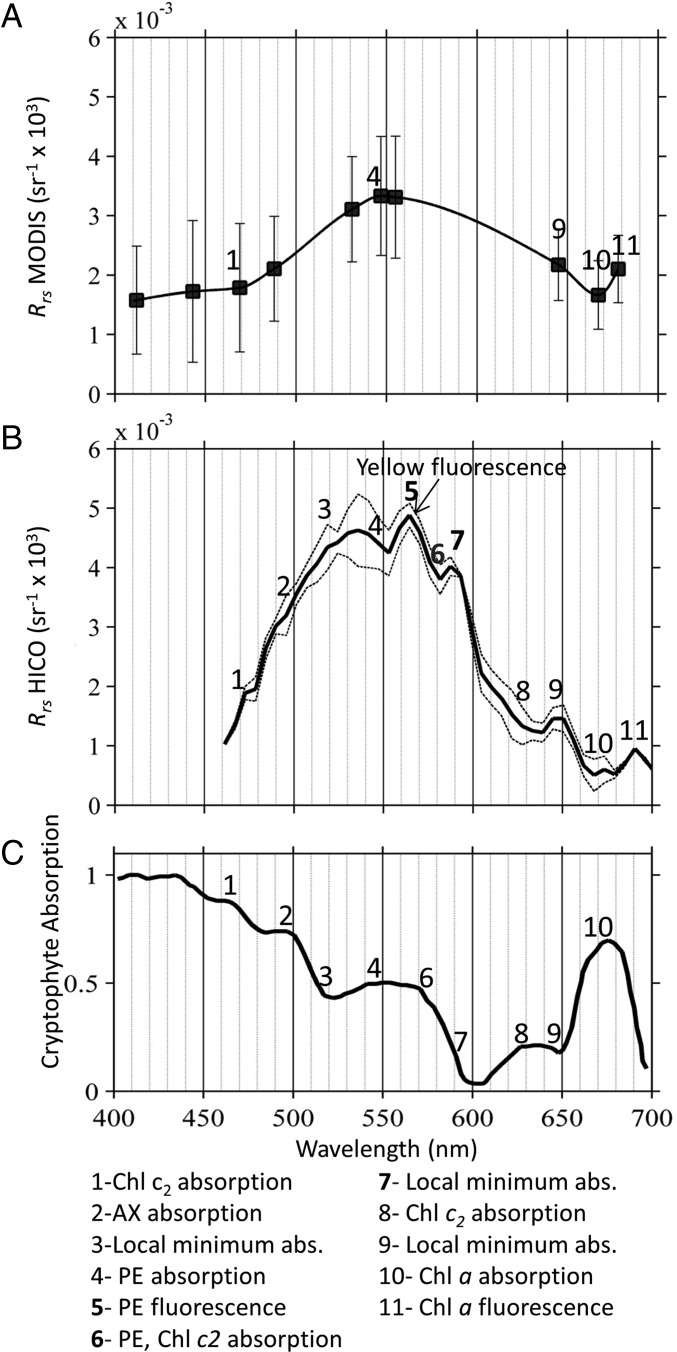
Progress has been made in identifying types of phytoplankton from space; however, the multichannel sensors currently in orbit do not have the spectral resolution necessary to differentiate complex coastal assemblages without prior knowledge and regional tuning. The MODIS Terra reflectance spectrum shows differences primarily in magnitude between bloom and nonbloom pixels in the sound due to enhanced green absorbing pigments, but there are large spectral gaps where no information is known (e.g., 555–665 nm; Fig. 2A). Tuned to open ocean conditions (16), the limited spectral information prevents differentiation of an M. rubrum bloom from other types of phytoplankton known to occur in this region (e.g., diatoms, dinoflagellates) (17).
Fig. 2.
Compared with the limited remote sensing reflectance (Rrs) data available on the ocean color sensor MODIS (A), the spectral information available from the hyperspectral HICO sensor shows many peaks and dips (B) related to the pigment absorption (abs.) features from phycoerythrin (PE), alloxanthin (AX), Chl a, and Chl c2 contained within the enslaved cryptophytes in the ciliate M. rubrum (C). Fluorescence features are also visible, and the yellow PE fluorescence peak at 565 nm compared with absorption dip at 575 nm was used to quantify the concentration of M. rubrum cells in the HICO imagery.
Such discrimination is possible with the Hyperspectral Imager for the Coastal Ocean (HICO) proof-of-concept imaging spectrometer located aboard the International Space Station (21, 22). A spectacular HICO image of WLIS on September 23, 2012 captured the water color during the M. rubrum bloom at a resolution of 110 m (Fig. S4). With 100 channels in the visible region, the “hyperspectral” capabilities of the sensor reveal fine dips and peaks in the reflectance spectrum that are not evident from the multichannel satellite imagery (Fig. 2B). M. rubrum contains a suite of photosynthetic pigments from the enslaved cryptophytes, including phycoerythrin and Chl c2 (23). A cryptophyte absorption spectrum shows the spectral regions where the ancillary pigments are known to absorb in vivo (1, 24) (Fig. 2C). Enhanced absorption produces dips in the HICO reflectance corresponding to pigment-specific enhancements in absorption (Fig. 2 and Fig. S5).
Fig. S4.
(A) HICO image from the International Space Station collected on September 23, 2012 shown in pseudo-true color. White features represent cloud cover. (B) Highlight of the portion of the HICO swath covering WLIS shows the color of the water containing M. rubrum.
Fig. S5.
Fourth derivative is often used to discriminate spectral features due to absorption (14, 35, 36) and reflectance spectra (37). The dips in the reflectance spectrum can be observed as negative peaks in the fourth derivative and relate to absorption by alloxanthin (479 nm), phycoerythrin (552–580 nm), Chl c2 (581 and 639 nm), and Chl a (667 nm). The peaks in the reflectance spectrum appear as positive peaks in the fourth derivative and correspond to fluorescence of phycoerythrin (565 nm) and Chl a (673 nm), as well as local minima in absorption. Max, maximum; Min, minimum.
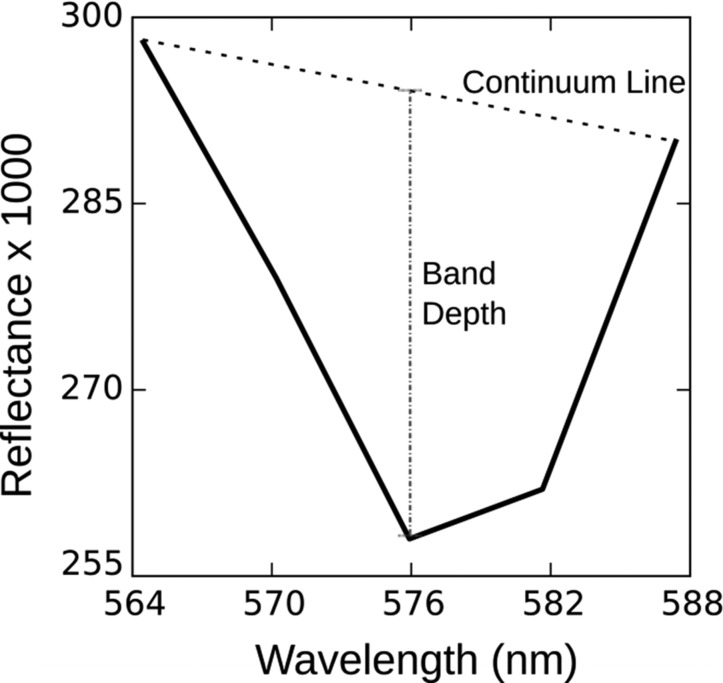
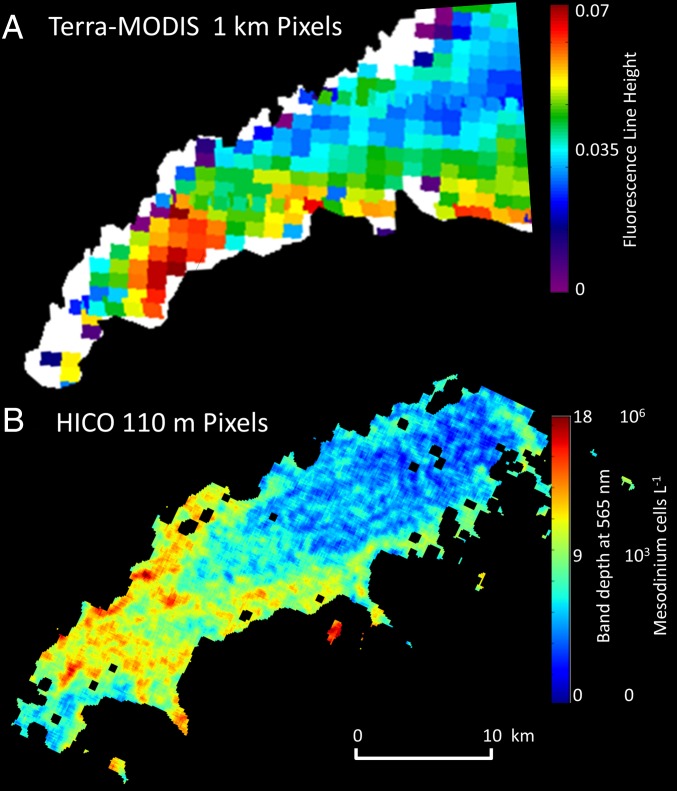
In contrast, peaks in the reflectance spectrum are due to local minima in absorption or production of light through fluorescence. The HICO spectrum in WLIS contained a unique “yellow” peak at 565 nm associated with phycoerythrin fluorescence (25–27). A band depth algorithm approach was used to quantify pixels in the HICO with enhanced yellow fluorescence based on the reflectance signal at three wavelengths: 564.5 nm, 575 nm, and 587 nm (Fig. S6). Compared with the 1-km MODIS image, the higher spatial resolution (110 m) of the HICO image revealed intense small patches of yellow fluorescing Mesodinium in WLIS (Fig. 3). If we conservatively assume the highest values correspond to our measured 1 × 106 cells per liter, then the M. rubrum concentration can be roughly estimated across the image. The extremely patchy nature of the bloom revealed here is consistent with the irregular patches or clouds of red water reported at the sea surface during calm conditions (8).
Fig. S6.
Band depth algorithm used to identify the presence of phycoerythrin from the Lambertian equivalent water-leaving reflectance peak at 565 nm due to yellow fluorescence and the dip at 575 nm due to absorption. The band depth was roughly related to the concentration of M. rubrum cells such that 0.0018 was comparable to 1 × 106 cells per liter.
Fig. 3.
(A) Image of WLIS at a resolution of 1 km from the MODIS Terra sensor shows an elevated Chl a fluorescence patch on September 23, 2012, but the type of bloom cannot be distinguished from the limited spectral bands. (B) In contrast, hyperspectral HICO imagery from the International Space Station reveals characteristic yellow fluorescence due to phycoerythrin pigment within the enslaved chloroplasts of the ciliate M. rubrum. Dense and patchy near-surface blooms of this motile and actively photosynthesizing mixotrophic marine protist (>1 × 106 cells per liter) periodically dominate primary productivity in the region.
Due to limitations in spectral resolution, prior remote sensing studies of M. rubrum have been based on either nonspecific markers of darkened water or on general absorption of green light (520–600 nm) (28). However, such markers are not specific to the cryptophytes, and enhanced green absorption in coastal waters may be due to other types of algal blooms. A 1984 study reported an M. rubrum bloom with a reflectance peak near 610 nm and a shift in fluorescence from 690 to 710 nm (29), but such results were not observed here. Use of yellow phycoerythrin fluorescence has also been proposed for quantifying blooms of the cyanobacteria Trichodesmium (25), although routine ocean color sensors have not had the required spectral resolution for implementation. Although our proposed approach would not necessarily differentiate M. rubrum from other phycoerythrin-containing blooms, other additional spectral features could be used. For example, the virtual absence of solar-induced fluorescence of Chl a and the optical brightness resulting from Trichodesmium gas vacuoles would also be visible in hyperspectral imagery (25). Hyperspectral inverse ocean color modeling has been applied to ship-based radiometry to differentiate M. rubrum from several other phytoplankton types based on its unique phycoerythrin absorption properties (30). Incorporation of fluorescence into such reflectance inversion models would provide even more power to assess complex assemblages of pigments from hyperspectral measurements.
Proposed future missions aim to be hyperspectral and provide a potential means to differentiate bloom-forming phototrophic and mixotrophic plankton in surface waters autonomously and routinely, as shown here. Such methods could prevent unnecessary shellfish and beach closures due to misidentification of coastal red tides. Additionally, application of new algorithms in coastal waters will allow for adaptive sampling of the vertical structure of the bloom that cannot be assessed remotely and better documentation of bloom initiation and termination, which may, in turn, lead to better understanding of how this particular plankter can escape normal population controls to produce such spectacular blooms.
Materials and Methods
Red-colored water was encountered in WLIS on September 24, 2012, as revealed in a photograph taken from the R/V Connecticut at 40.9502°N, 73.5955°W (Fig. S1). A water sample was collected for cell enumeration and genetic analysis.
Mesodinium sp. Enumeration.
M. rubrum and recently proposed cogeners (31) were quantified in routine monthly monitoring samples taken from six stations in the sound by the Connecticut Department of Energy and Environmental Protection. Whole-water samples from several depths at each station were pooled and preserved in Lugol’s iodine solution, settled in 100-mL aliquots, and counted on an inverted microscope. Biomass was estimated based on linear dimensions converted to cell volumes. Statistics from 572 counts from 2002 to 2012 are shown in Table S1. For 2012, monthly sampling revealed generally low concentrations of M. rubrum, with the exception of July 31 at the westernmost station B3, where abundance exceeded 7,000 cells per liter. The next subsequent monthly sampling at station B3 was done on October 2, when the abundance was <100 cells per liter (Fig. S1). When the bloom was fortuitously observed on September 24, 2012, a surface bucket sample was collected. M. rubrum was quantified microscopically using a Sedgwick–Rafter counting chamber. The cell concentration in that sample was 1.03 × 106 cells per liter.
Genetic Analysis.
M. rubrum cells were picked from the bloom water sample on an inverted microscope. The isolated cells were rinsed carefully with autoclaved filtered seawater. Twenty of these M. rubrum cells were resuspended in 0.5 mL of DNA lysis buffer [0.1 M EDTA (pH 8.0), 1% SDS, 200 mg⋅mL−1 proteinase K] and incubated for 48 h at 55 °C. DNA extraction, PCR amplification, cloning, and gene sequencing for the plastid 16S rRNA gene were carried out as described previously (32). The plastid 16S rDNA sequence was analyzed using a BLAST search against databases in GenBank. Sequences showing significant similarity in the BLAST search to the sequences obtained in this study were retrieved from the databases. These sequences were aligned using ClustalX, in which a neighbor-joining tree was inferred with 1,000 resamplings for bootstrap analysis. ModelTest was run to identify the most appropriate evolutionary model, and the general time-reversible model with a gamma distribution was selected for maximum likelihood analysis using PhyML3.0 aLRT (33). Categories of substitution rates were set at four, and other parameters were estimated based on the dataset.
We extracted RNA from the bloom sample and conducted high-throughput transcriptome sequencing using an Illumina HiSeq 2000 system. The resulting cDNA sequences were filtered to remove low-quality reads, assembled, and clustered (to remove redundancy) to yield unigenes using Trinity software (34). After functional annotation, we looked for photosynthesis-related genes and estimated their expression levels in terms of fragments per kilobase per million mapped reads (35).
Satellite Imagery.
MODIS Terra Level 2 Chl a and natural fluorescence line height imagery from September 23, 2015 at 11:25 AM Eastern Daylight Time was downloaded from the NASA Ocean Color Web (oceancolor.gsfc.nasa.gov/cms/). A HICO image incorporating WLIS at a 110-m pixel resolution from September 23, 2012 was downloaded from the ocean color website (H2012267133200.L1B_ISS). A pseudo-true color image shows the entire HICO swath covering a subset of WLIS (Fig. S4). Dark patches in the sound can be related to high light absorption, as well as some cloud shadows. Imagery from the HICO is not atmospherically corrected and our processing methods are presented below.
Atmospheric Correction.
The majority of the signal reaching a space-borne sensor is actually from sunlight scattered within the atmosphere and reflection off the sea surface that must be removed through the process of “atmospheric correction.” Uncertainties in HICO radiometric calibrations prevent the retrieval of aerosol models and optical depths from HICO data on a pixel-by-pixel basis using the ocean version of the atmospheric correction algorithm (36). Instead, a land version is used (37) in the present processing of the HICO imagery. A clear model atmosphere with a small optical depth of ∼0.08 at 550 nm is assumed for the removal of Rayleigh and aerosol scattering effects from HICO data. The atmospheric water vapor absorption effects in the complete HICO-covered wavelength range are removed on a pixel-by-pixel basis using information contained in the 725-nm and 825-nm water vapor bands in the HICO data. Because aerosol modeling was highly uncertain, the retrieved HICO spectra were higher than the MODIS imagery, and an adjustment of 0.0015 (steradian−1) was subtracted from each wavelength for visualization in Fig. 3B. This adjustment has no impact on the band depth algorithm developed below.
Spectral Methods.
HICO imagery was georeferenced using a second-order polynomial transformation. Control points were derived from a Landsat 8 terrain-corrected image collected in September 2014. The spatial resolution of the georeferenced image was 110 m. A land and cloud mask was generated using an average visible-near infrared threshold such that any pixels with an average reflectance greater than 4% were masked. Mean remote sensing reflectance from the region of the water sample in WLIS revealed many spectral features related to absorption and fluorescence properties of cryptophytes (Fig. 3B). A fourth derivative was applied to the reflectance spectrum to highlight further the unique spectral features of the data (38, 39) (Fig. S5). To identify and enumerate the M. rubrum, a band depth algorithm was developed using reflectance features between 564.4 nm and 587.3 nm due to phycoerythrin fluorescence, phycoerythrin and Chl c absorption, and a local minimum in absorption at 587.3 nm (Fig. S6). The Chl c2 absorption at 570 nm may obscure phycoerythrin fluorescence at the same waveband and induce a spectral shift in peak fluorescence from 570 to 565 nm. The resulting band depth image (Fig. 3B) was smoothed using a 5 × 5 moving average window to remove noise.
Acknowledgments
We thank Kay Howard-Strobel (University of Connecticut) for collecting photographs and samples of the bloom, and the Hyperspectral Imager for the Coastal Ocean (HICO) Science Team and NASA Ocean Biology Distributed Active Archive Center for providing satellite imagery. H.D. and A.C. conducted the image analysis, B.-C.G. provided atmospheric correction of the HICO image, G.B.M. provided monitoring information on M. rubrum in Long Island Sound, and D.Q. and S.L. conducted cell count and genetic analyses for the bloom sample. This work was supported by NASA Ocean Biology and Biogeochemistry Award NNX15AC32G (to H.D.), the Connecticut Department of Energy & Environmental Protection (G.B.M. and S.L.), the Natural Science Foundation of China Grant 41129001 (to S.L.), and the University of Connecticut.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1512538112/-/DCSupplemental.
References
- 1.Crawford DW. Mesodinium rubrum: The phytoplankter that wasn’t. Mar Ecol Prog Ser. 1989;58(1):161–174. [Google Scholar]
- 2.Darwin C. A Naturalist’s Voyage Round the World: The Voyage of the Beagle. Skyhorse Publishing, Inc.; New York: 2014. [Google Scholar]
- 3.Flynn KJ, et al. Misuse of the phytoplankton–zooplankton dichotomy: The need to assign organisms as mixotrophs within plankton functional types. J Plankton Res. 2012;35(1):3–11. [Google Scholar]
- 4.McManus GB, Fuhrman JA. Bacterivory in seawater studied with the use of inert fluorescent particles1. Limnol Oceanogr. 1986;31(2):420–426. [Google Scholar]
- 5.McManus GB, Schoener DM, Haberlandt K. Chloroplast symbiosis in a marine ciliate: Ecophysiology and the risks and rewards of hosting foreign organelles. Front Microbiol. 2012;3:321. doi: 10.3389/fmicb.2012.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stoecker DK, Michaels AE, Davis LH. Large proportion of marine planktonic ciliates found to contain functional chloroplasts. Nature. 1987;326(6115):790–792. [Google Scholar]
- 7.Johnson MD, Oldach D, Delwiche CF, Stoecker DK. Retention of transcriptionally active cryptophyte nuclei by the ciliate Myrionecta rubra. Nature. 2007;445(7126):426–428. doi: 10.1038/nature05496. [DOI] [PubMed] [Google Scholar]
- 8.Johnson MD, Stoecker DK. Role of feeding in growth and photophysiology of Myrionecta rubra. Aquat Microb Ecol. 2005;39(3):303–312. [Google Scholar]
- 9.Johnson MD, Stoecker DK, Marshall HG. Seasonal dynamics of Mesodinium rubrum in Chesapeake Bay. J Plankton Res. 2013;39:303–312. [Google Scholar]
- 10.Yih W, et al. The red-tide ciliate Mesodinium rubrum in Korean coastal waters. Harmful Algae. 2013;30(Suppl 1):S53–S61. [Google Scholar]
- 11.Herfort L, Peterson TD, Campbell V, Futrell S, Zuber P. Myrionecta rubra (Mesodinium rubrum) bloom initiation in the Columbia River estuary. Estuar Coast Shelf Sci. 2011;95(4):440–446. [Google Scholar]
- 12.Fenchel T, Juel Hansen P. Motile behaviour of the bloom-forming ciliate Mesodinium rubrum. Mar Biol Res. 2006;2(1):33–40. [Google Scholar]
- 13.Sjöqvist CO, Lindholm TJ. Natural co-occurrence of Dinophysis acuminata (Dinoflagellata) and Mesodinium rubrum (Ciliophora) in thin layers in a coastal inlet. J Eukaryot Microbiol. 2011;58(4):365–372. doi: 10.1111/j.1550-7408.2011.00559.x. [DOI] [PubMed] [Google Scholar]
- 14.Kyewalyanga M, Sathyendranath S, Platt T. Effect of Mesodinium rubrum (= Myrionecta rubra) on the action and absorption spectra of phytoplankton in a coastal marine inlet. J Plankton Res. 2002;24(7):687–702. [Google Scholar]
- 15.Dierssen HM, Kudela RM, Ryan JP, Zimmerman RC. Red and black tides: Quantitative analysis of water-leaving radiance and perceived color for phytoplankton, colored dissolved organic matter, and suspended sediments. Limnol Oceanogr. 2006;51(6):2646–2659. [Google Scholar]
- 16.Dierssen HM. Perspectives on empirical approaches for ocean color remote sensing of chlorophyll in a changing climate. Proc Natl Acad Sci USA. 2010;107(40):17073–17078. doi: 10.1073/pnas.0913800107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aurin DA, Dierssen HM. Advantages and limitations of ocean color remote sensing in CDOM-dominated, mineral-rich coastal and estuarine waters. Remote Sens Environ. 2012;125:181–197. [Google Scholar]
- 18.Aurin DA, Dierssen HM, Twardowski MS, Roesler CS. Optical complexity in Long Island Sound and implications for coastal ocean color remote sensing. J Geophys Res. 2010;115(C07011):1–11. [Google Scholar]
- 19.Smith RC, Baker KS. Optical classification of natural waters. Limnol Oceanogr. 1978;23:260–267. [Google Scholar]
- 20.Gower J, King S, Goncalves P. Global monitoring of plankton blooms using MERIS MCI. Int J Remote Sens. 2008;29(21):6209–6216. [Google Scholar]
- 21.Lucke RL, et al. Hyperspectral Imager for the Coastal Ocean: instrument description and first images. Appl Opt. 2011;50(11):1501–1516. doi: 10.1364/AO.50.001501. [DOI] [PubMed] [Google Scholar]
- 22.Corson MR, Davis CO. A new view of coastal oceans from the space station. Eos Transactions American Geophysical Union. 2011;92(19):161–162. [Google Scholar]
- 23.Rial P, Garrido JL, Jaén D, Rodríguez F. Pigment composition in three Dinophysis species (Dinophyceae) and the associated cultures of Mesodinium rubrum and Teleaulax amphioxeia. J Plankton Res. 2012;35(2):433–437. [Google Scholar]
- 24.Johnsen G, Samset O, Granskog L, Sakshaug E. In-vivo absorption characteristics in 10 classes of bloom-forming phytoplankton-taxonomic characteristics and responses to photoadaptation by means of discriminant and HPLC analysis. Mar Ecol Prog Ser. 1994;105(1-2):149–157. [Google Scholar]
- 25.Subramaniam A, Carpenter EJ, Karentz D, Falkowski PG. Bio-optical properties of the marine diazotrophic cyanobacteria Trichodesmium spp. I. Absorption and photosynthetic action spectra. Limnol Oceanogr. 1999;44(3):608–617. [Google Scholar]
- 26.McManus GB, Fuhrman JA. Photosynthetic pigments in the ciliate Laboea strobila from Long Island Sound, USA. J Plankton Res. 1986;8(2):317–327. [Google Scholar]
- 27.Seppälä J, Ylöstalo P, Kuosa H. Spectral absorption and fluorescence characteristics of phytoplankton in different size fractions across a salinity gradient in the Baltic Sea. Int J Remote Sens. 2005;26(2):387–414. [Google Scholar]
- 28.Garcia CA, Purdie DA, Robinson IS. Mapping a bloom of the photosynthetic ciliate Mesodinium rubrum in an estuary from airborne thematic mapper data. Estuar Coast Shelf Sci. 1993;37(3):287–298. [Google Scholar]
- 29.Gower JFR, Lin S, Borstad GA. The information content of different optical spectral ranges for remote chlorophyll estimation in coastal waters. Int J Remote Sens. 1984;5(2):349–364. [Google Scholar]
- 30.Roesler CS, Etheridge SM, Pitcher GC. Application of an ocean color algal taxa detection model to red tides in the Southern Benguela. In: Steidinger KA, Landsberg JH, Tomas CR, Vargo GA, editors. Harmful Algae 2002. Florida Fish and Wildlife Conservation Commission, Florida Institute of Oceanography, and Intergovernmental Oceanographic Commission of UNESCO; St. Petersburg, FL: 2004. pp. 303–305. [Google Scholar]
- 31.Garcia-Cuetos L, Moestrup Ø, Hansen PJ. Studies on the genus Mesodinium II. Ultrastructural and molecular investigations of five marine species help clarifying the taxonomy. J Eukaryot Microbiol. 2012;59(4):374–400. doi: 10.1111/j.1550-7408.2012.00630.x. [DOI] [PubMed] [Google Scholar]
- 32.Qiu D, Huang L, Liu S, Lin S. Nuclear, mitochondrial and plastid gene phylogenies of Dinophysis miles (Dinophyceae): Evidence of variable types of chloroplasts. PLoS One. 2011;6(12):e29398. doi: 10.1371/journal.pone.0029398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dereeper A, et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36(Web Server issue) Suppl 2:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 36.Gao BC, Montes MJ, Ahmad Z, Davis CO. Atmospheric correction algorithm for hyperspectral remote sensing of ocean color from space. Appl Opt. 2000;39(6):887–896. doi: 10.1364/ao.39.000887. [DOI] [PubMed] [Google Scholar]
- 37.Gao B-C, Heidebrecht KB, Goetz AF. Derivation of scaled surface reflectances from AVIRIS data. Remote Sens Environ. 1993;44(2):165–178. [Google Scholar]
- 38.Millie D, Schofield O, Kirkpatrick G, Johnsen G, Evens T. Using absorbance and fluorescence spectra to discriminate microalgae. Eur J Phycol. 2002;37(3):313–322. [Google Scholar]
- 39.Bidigare RR, Morrow JH, Kiefer DA. Derivative analysis of spectral absorption by photosynthetic pigments in the western Sargasso Sea. J Mar Res. 1989;47(2):323–341. [Google Scholar]