Significance
Plant cytoplasmic male sterility (CMS) and sterility restoration by nuclear restorer-of-fertility (Rf) genes provide a crucial breeding tool to harness hybrid vigor in many crops. Here, we identify RF6 as a novel pentatricopeptide repeat family protein that restores the fertility of Honglian CMS (HL-CMS), a major type of rice CMS used in breeding. We demonstrated that RF6 and hexokinase 6 function together in mitochondria to promote the processing of the aberrant CMS-associated transcript atp6-orfH79, thereby restoring the fertility of HL-CMS rice. This study links CMS restoration to hexokinase, providing novel insights into the mechanisms of CMS restoration, pollen development, mitochondria metabolism, and nuclear-cytoplasmic communication. In addition, the present study offers opportunities to further exploit rice heterosis in hybrid rice production.
Keywords: cytoplasmic male sterility, pentatricopeptide repeat, restorer-of-fertility, RF6, hexokinase 6
Abstract
Cytoplasmic male sterility (CMS) has been extensively used for hybrid seed production in many major crops. Honglian CMS (HL-CMS) is one of the three major types of CMS in rice and has contributed greatly to food security worldwide. The HL-CMS trait is associated with an aberrant chimeric mitochondrial transcript, atp6-orfH79, which causes pollen sterility and can be rescued by two nonallelic restorer-of-fertility (Rf) genes, Rf5 or Rf6. Here, we report the identification of Rf6, which encodes a novel pentatricopeptide repeat (PPR) family protein with a characteristic duplication of PPR motifs 3–5. RF6 is targeted to mitochondria, where it physically associates with hexokinase 6 (OsHXK6) and promotes the processing of the aberrant CMS-associated transcript atp6-orfH79 at nucleotide 1238, which ensures normal pollen development and restores fertility. The duplicated motif 3 of RF6 is essential for RF6-OsHXK6 interactions, processing of the aberrant transcript, and restoration of fertility. Furthermore, reductions in the level of OsHXK6 result in atp6-orfH79 transcript accumulation and male sterility. Together these results reveal a novel mechanism for CMS restoration by which RF6 functions with OsHXK6 to restore HL-CMS fertility. The present study also provides insight into the function of hexokinase 6 in regulating mitochondrial RNA metabolism and may facilitate further exploitation of heterosis in rice.
Plant cytoplasmic male sterility (CMS) is a trait characterized by a lack of functional pollen grains in plants, although female gametes are still viable (1). CMS has been widely used to produce hybrid seeds to improve plant yield, improve resistance to diseases or stresses, or enhance taste (2). Plant CMS results from genomic incompatibility between mitochondria and nuclei and is typically associated with an aberrant chimeric gene in the mitochondria (3–6). A specific set of nuclear genes called restorer-of-fertility (Rf) genes, which primarily belong to the pentatricopeptide repeat (PPR) family, repress CMS through the promotion of the cleavage, degradation, or editing of CMS-associated aberrant transcripts or the inhibition of the translation of these genes (7–9). PPR proteins regulate RNA metabolism at multiple levels, from RNA editing, stability, processing, and splicing to translation (10, 11). PPR proteins can be classified into PLS and P families based on the structure of PPR motifs (11). PLS family members generally exhibit direct RNA binding (12–14) or editing activity (15–17), whereas P-family proteins often lack sites for binding to RNA, particularly mitochondrial CMS-related RNA (18, 19). Most of the known PPR proteins implicated in restoration of fertility (RF) are P-family members. Thus, RF-related PPR proteins may require additional partners, such as RNA-binding or splicing factors to process CMS-related transcripts.
Rice CMS is categorized into three major types: Wild Abortive (WA), originating from a wild abortive rice line; Honglian (HL), originating from red-awned wild rice; and BT, originating from ChinsurahBoro II rice (20). These three types of CMS contribute to ∼60% of the global hybrid rice production (21, 22). HL-CMS/Rf rice has been planted on over 6 million hectares in China and Southeast Asia in the past 2 decades, significantly contributing to food security worldwide (23). At the molecular level, WA-CMS is associated with the mitochondrial gene WA352. Pollen abortion in these rice plants occurs at the uninucleate stage and is suppressed by the nuclear genes Rf3 and Rf4 (24). BT-CMS involves the mitochondrial chimeric gene atp6-orf79 (25, 26). The pollen of these plants aborts at the trinucleate stage and can be suppressed by Rf1A and Rf1B (26). For HL-CMS, pollen aborts at the dinucleate stage and exhibits a spherical shape and negative staining with iodine-potassium iodide (I2-KI) solution (20). The HL-CMS trait is associated with orfH79, a chimeric mitochondrial gene located downstream of atp6. The ORFH79 protein interacts with P61, a subunit of mitochondrial complex III, and impairs the activity of this complex. This inhibition leads to dysfunctional energy metabolism and elevated oxidative stresses in mitochondria, eventually causing pollen sterility (27–30). The HL-CMS phenotype can be rescued by two nonallelic Rf genes, Rf5 or Rf6 (31). F1 hybrids carrying a single restorer gene display ∼50% normal pollen grains, whereas those harboring both Rf genes generate 75% fertile pollen and a higher seed set rate under environmental stress conditions (31), thus indicating that both Rf genes are equally important for maintaining fertility and the seed set rate. Recently, we have reported that Rf5 encodes a protein with 16 putative PPR motifs and that RF5 functions with GRP162, an RNA-binding protein, to promote the cleavage of the CMS-associated transcript orfH79 (18, 32). In the present study, we identified Rf6 as a restorer gene for HL-CMS rice using a map-based cloning strategy. Mechanistically, we revealed that RF6 acts with hexokinase 6 (OsHXK6) in mitochondria to reduce the accumulation of the CMS-associated transcript atp6-orfH79 through a mechanism distinct from that of RF5.
Results
Identification of Rf6 as a Restorer Gene for HL-CMS Rice.
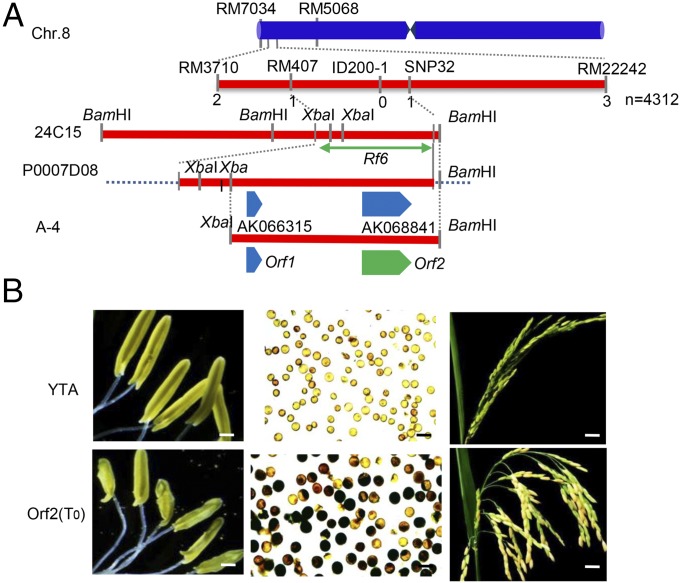
We aimed to identify the other HL-CMS restorer gene, Rf6, using a map-based cloning strategy. In a previous study, we showed that Rf6 cosegregates with the marker RM407 and associates with a region of ∼200 kb flanked by the markers RM3710 and RM22242 on chromosome 8 (31) (Fig. 1A). To narrow down the location of Rf6, we selected an individual plant, (R-2), from the F2 population obtained by crossing the restorer line (9311) with the HL-CMS abortive line (Yuetai A, or YTA) (31). R-2 contains YTA cytoplasm and is homozygous at the Rf6 locus (Rf6Rf6), and 100% of the pollen grains generated from this plant exhibited black staining with 1% I2-KI solution. R-2 was subsequently backcrossed to YTA, generating a BC1F2 population containing 4,312 plants. Genetic linkage analysis identified two recombinant plants with the RM3710 marker and three recombinant plants carrying the RM22242 marker from this population (Fig. 1A). To obtain a high-resolution map, we selected three polymorphic markers, RM407, ID200-1, and SNP32 (SI Appendix, Table S1), to screen for individual recombinants. The markers RM407 and SNP32 each identified a single recombinant plant, whereas the marker ID200-1 did not (Fig. 1A). Thus, the Rf6 locus was delimited to a 16.8-kb region between RM407 and SNP32 and cosegregated with the marker ID200-1. In this region, there are two putative genes, AK066315 and AK068841, in the P0007D08 Nipponbare PAC clone (Fig. 1A). To obtain the corresponding genomic sequence for the restorer line, we constructed a 9311 BAC library (SI Appendix, Fig. S1) and screened this library with the markers RM407 and SNP32. A positive clone, 24C15, containing two putative ORFs, was identified (Fig. 1A) and subcloned for sequencing. Both ORFs (Orf1 and Orf2) were individually cloned into a vector under the control of the ubiquitin promoter of Zea mays and transformed into the YTA sterile line for complementation analyses. All 17 of the Orf1 transformants showed the same sterile phenotype as YTA. Conversely, all 19 of the transgenic T0 plants containing Orf2 displayed a normal fertile phenotype (Fig. 1B). Moreover, we observed that ∼50% of the pollen grains were fertile, as shown by I2-KI staining, and the seed-setting rate of these plants was 80.5% on average (Fig. 1B and SI Appendix, Fig. S2). To confirm these results, three Orf2 T0 plants were self-crossed to produce T1-generation plants. We observed a 1:1 ratio for plants with 100% normal pollen compared with those with 50% abortive pollen in the T1 population (SI Appendix, Table S2), which indicates that Orf2 restores the fertility of HL-CMS in a gametophytic manner. Thus, we demonstrated that Orf2 is the Rf6 gene responsible for restoring HL-CMS fertility.
Fig. 1.
Rf6 restores the fertility of HL-CMS rice. (A) Schematic diagram showing the mapping and location of Rf6. Clone number and restriction sites are indicated. (B) Functional complementation analysis of Rf6 in rescuing HL-CMS sterility. Orf2 was transformed into the sterile line YTA (rf6rf6) by Agrobacterium tumefaciens-mediated methods. The anthers, 1% I2-KI–stained pollen grains, and seed set rate for YTA and the transgenic plants carrying Orf2 are shown. (Scale bars: 1 mm, 50 µm, and 500 mm in anthers, pollen grains, and seeds, respectively.)
Duplicated 3–5 PPR Motifs Are Essential for Restoring HL-CMS Fertility.
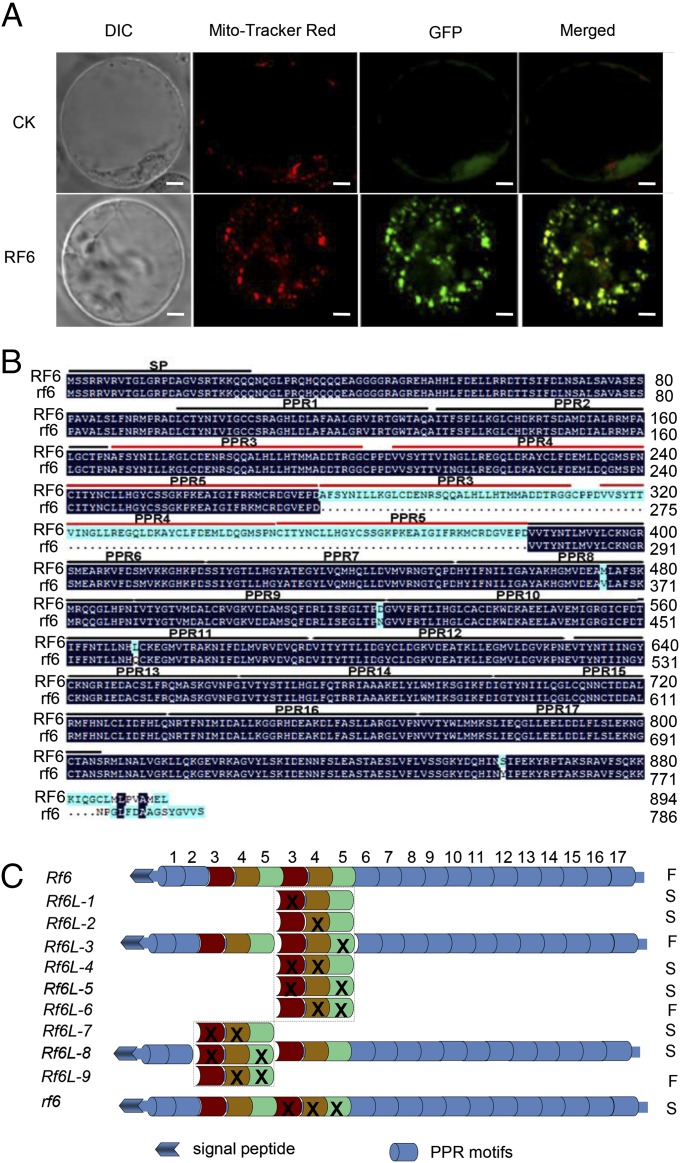
Next, we used RT-PCR to examine the expression patterns of the Rf6 gene in R-2 plants and the recessive allele, rf6, in the YTA abortive line. Both alleles exhibited similar expression profiles in a variety of organs (SI Appendix, Fig. S3), implying that the expression of these genes is not tissue-specific. Rf6 contains a mitochondrial transit signal at the N terminus (https://urgi.versailles.inra.fr/predotar/predotar.html). Indeed, we observed that a GFP-fused RF6 protein expressed from a plasmid driven by the CaMV 35S promoter in rice protoplasts was localized to mitochondria (Fig. 2A). Sequence analysis revealed that Rf6 encodes a novel PPR protein of 894 amino acids with 20 PRR motifs (Fig. 2B). Notably, the RF6 protein contained a duplication of motifs 3–5, which are absent in rf6 or other known RF proteins (18, 33–38) (Fig. 2B and SI Appendix, Fig. S4). Outside this region, RF6 and rf6 are nearly identical in amino acid sequences (Fig. 2B), suggesting that this region is associated with the restoration of fertility. To evaluate the importance of each of the three PPR motifs, we synthesized a series of truncated Rf6 genes lacking these PPR motifs (Fig. 2C, Rf6Ls and Rf6L-1 to Rf6L-9) and transformed these genes into the YTA sterile line to examine their effects on fertility restoration (SI Appendix, Fig. S5A). We found that the deletion of motif 3 alone or the deletion of motifs 3 and 4 or 3 and 5 resulted in sterility in Rf6L-1, Rf6L-4, Rf6L-5, Rf6L-7, or Rf6L-8 transgenic plants. However, the simultaneous disruption of motifs 4 and 5 and retention of motif 3 (Rf6L-6 or Rf6L-9) did not impair fertility (Fig. 2C and SI Appendix, Fig. S5B and Table S2). Thus, the duplication of motif 3, but not 4 or 5, is essential and sufficient for the function of Rf6 in fertility restoration. Furthermore, motif order plays an important role in this process because the direct fusion of motifs 5–3 (Rf6L-2, deletion of motif 4) resulted in sterility (Fig. 2C and SI Appendix, Fig. S5B and Table S2), as motif 4 alone is not required for fertility restoration. These three motifs do not share a high degree of homology and are clearly classified into distinct phylogenetic branches (SI Appendix, Fig. S6).
Fig. 2.
The duplicated PPR motifs 3–5 in RF6 are crucial for restoring fertility. (A) Localization of Rf6 in mitochondria. Protoplasts transformed with 35S:GFP:nos (CK) and 35S:Rf6-GFP: nos (Rf6) constructs were analyzed using fluorescence microscopy. The dye Mito-Tracker Red was used as a mitochondrial marker. (Scale bars: 10 µm.) (B) Comparison of the amino acid sequences for the RF6 and rf6 proteins. Sequence alignment was performed using DNAMAN6.0 software. The PPR motifs are indicated with solid lines. SP: signal peptide. (C) Schematic diagrams showing wild-type Rf6 and engineered Rf6L constructs (Rf6L-1–Rf6L-9). The crosses involved the deletion of the corresponding PPR motifs. F: fertility; S: sterility.
Furthermore, to address whether this duplication could influence the structure of RF6, we used the crystal structure of maize chloroplast PPR10 (39) as a template to perform homology-based modeling of RF6, rf6, and RF6L-6 proteins. PPR10 belongs to the P-class of PPR family members and shares homology with RF6 in PPR motifs (SI Appendix, Fig. S7A). The duplicated motifs form a loosely extended structure connecting both N- and C-terminal domains of the RF6 protein and become shorter in the RF6L-6 and, particularly, the rf6 protein (SI Appendix, Fig. S7B). We speculated that this region may aid in establishing an optimal RF6 conformation or serve as an interface for protein–protein interactions. To examine whether the duplicated region also appears in other rice species, we amplified and analyzed Rf6 homologs from a variety of cultivars and their ancestors, the wild rice species with AA genomes (SI Appendix, Table S3). We observed that the duplicated region also appeared in multiple indica cultivars and in several wild rice species (SI Appendix, Fig. S8). Thus, the duplicated region in RF6 likely originated from a duplication event that occurred in wild rice species.
RF6 Promotes Processing of the Chimeric Transcript atp6-orfH79 at Nucleotide 1238.
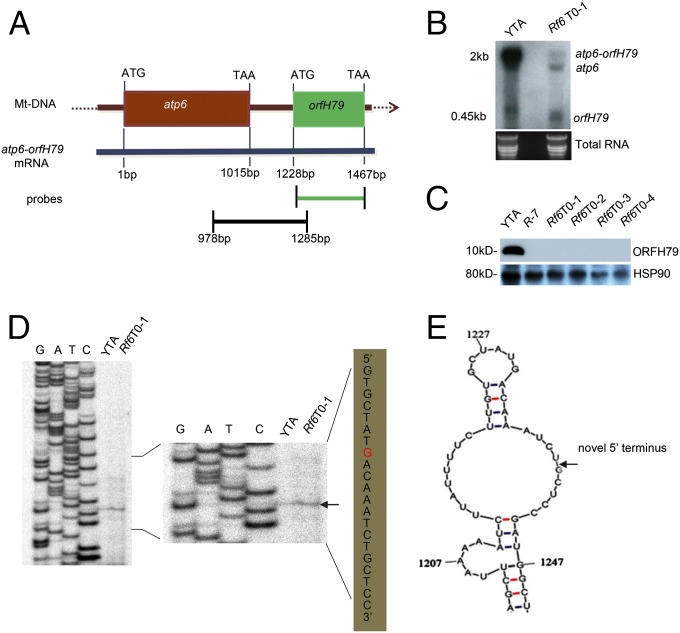
Based on previous studies, the sterility of HL-CMS rice is caused by the presence of the ORFH79 protein product from the chimeric transcript atp6-orfH79 in mitochondria. ORFH79 impairs the function of the mitochondrial electron transport chain complex III through an association with P61 (27–30). The restorer protein RF5 and its cofactor, GRP162, promote the cleavage of this aberrant transcript into two fragments, atp6 and orfH79, leading to the disruption of the ORFH79 protein and fertility restoration (18). Thus, we examined whether Rf6 restores the fertility of abortive lines through the same mechanism. The atp6-orfH79 transcript was largely replaced by two separate fragments, atp6 and orfH79, in transgenic plants carrying Rf6, whereas this transcript remained intact in YTA, which carries rf6 (Fig. 3 A and B and SI Appendix, Fig. S9A). Consequently, the ORFH79 protein was not detected in the presence of Rf6 (Fig. 3C). Furthermore, we observed the accumulation of the atp6-orfH79 transcript and OFRH79 protein in transgenic plants without motif 3 duplications (SI Appendix, Table S2 and Fig. S9B). These observations demonstrated that Rf6 and motif 3 are crucial for preventing the accumulation of the abnormal transcript. Next, we further defined the transcript changes associated with fertility restoration. We performed a primer extension assay using Rf6 T0 transgenic plants and observed that a specific band appeared 12 nucleotides (at the G residue) downstream from the start codon of orfH79 but was not detected in YTA (Fig. 3D). This observation likely reflects the cleavage of the chimeric transcript by RNA-processing factors. However, we cannot fully exclude the possibility that the restorer complex may protect the transcript against exonuclease processing or block the translation of the transcript. This processing site is located at nucleotide 1238 on a single-stranded loop in the atp6-orfH79 transcript (Fig. 3E) and obviously differs from the one generated by RF5, which lies in the junction region between atp6 and orfH79 at nucleotide 1169 of the same transcript (18). Thus, RF6 likely differs from RF5 mechanistically in processing the atp6-orfH79 transcript.
Fig. 3.
RF6 promotes the processing of the atp6-orfH79 transcript at nucleotide 1238. (A) The schematic diagram of the atp6-orfH79 chimeric gene and transcript. (B) Northern blot analysis showing the level of the atp6-orfH79 transcript in YTA and the transgenic plant with Rf6. The orfH79-coding sequence was used as a probe. (Lower) A gel stained with ethidium bromide to confirm that equal amounts of RNA were loaded in each lane. (C) A Western blot showing the protein level of ORF79 in transgenic plants with Rf6. YTA and R-7 were used as negative and positive controls, respectively. HSP90 protein was used as a loading control. (D) A primer extension assay mapping the processing site in the atp6-orfH79 transcript in T0 transgenic plants. The processing site G (red) is indicated with an arrow. (E) The secondary structure of the atp6-orfH79 transcript was predicted by the Mfold Web Server (unafold.rna.albany.edu). The processing site at nucleotide 1238 of the atp6-orfH79 transcript is indicated with an arrow. The RNA and protein for these experiments were prepared from plant leaves.
RF6 Does Not Bind the atp6-orfH79 Transcript or GRP162 Protein.
To understand how Rf6 may process the aberrant atp6-orfH79 transcript, we examined whether Rf6 can directly bind to the transcript. We purified the RF6 protein using an maltose-binding protein tag (SI Appendix, Fig. S10A) and incubated this fusion protein with a biotin-labeled RNA probe containing the linker region between the atp6- and orfH79- coding sequences (Fig. 3A). We did not observe any shift in electrophoretic mobility shift assays (EMSAs), even when RF6 was added at a concentration of 10 nM (SI Appendix, Fig. S10B), which suggests that RF6 does not directly bind to the atp6-orfH79 transcript. RF5 functions through an association with GRP162, a glycine-rich protein that binds to atp6-orfH79 mRNA, to promote the processing of the aberrant transcript (18). Therefore, we examined whether RF6 interacts with GRP162 by a yeast two-hybrid assay. However, we did not observe any signs of interaction between these proteins (SI Appendix, Fig. S11), suggesting that RF6 does not bind to this RNA-processing factor. Thus, RF6 likely requires other types of cofactors for binding and processing the atp6-orfH79 transcript.
RF6 Physically Interacts with OsHXK6 Both in Vitro and in Vivo.
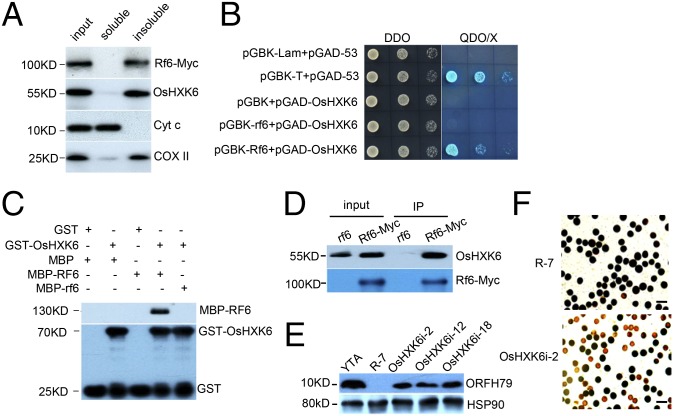
To identify potential targets that interact with RF6, we generated a yeast two-hybrid library for F1 hybrids and screened this library using RF6 protein as bait. Among the proteins identified (SI Appendix, Table S4), OsHXK6, a putative rice hexokinase localized to the mitochondria and nucleus (40), was detected. We observed that OsHXK6 displayed an expression pattern similar to that of RF6 and was primarily distributed within the insoluble fraction of mitochondria (Fig. 4A and SI Appendix, Fig. S13A). Importantly, OsHXK6 exhibited a strong specific interaction with RF6 but not rf6 in the yeast two-hybrid assay (Fig. 4B). This result was confirmed by a pull-down assay with purified GST-OsHXK6 and MBP-RF6 or MBP-rf6 proteins (Fig. 4C and SI Appendix, Fig. S10A). Furthermore, we performed a coimmunoprecipitation assay using purified mitochondria from plants with Myc-tagged RF6 and observed a strong interaction between OsHXK6 and RF6 in mitochondria (Fig. 4D). Thus, we demonstrated that RF6 directly interacts with OsHXK6 both in vitro and in vivo and that the duplicated motifs are essential for this interaction.
Fig. 4.
OsHXK6 functions with RF6 to promote the restoration of HL-CMS fertility. (A) A Western blot showing the coexistence of the RF6 and OsHXK6 proteins in insoluble mitochondria fractions. (B) Yeast two-hybrid analysis results showing the interaction between OsHXK6 and RF6 proteins. The 53-Lam and 53-T interactions were used as negative and positive controls, respectively. DDO: SD-Leu/-Trp medium; QDO: SD/-Leu/-Trp/-His/-Ade medium; X: X-αIL. (C) A pull-down assay revealing the direct physical interaction between OsHXK6 and RF6 but not rf6 in vitro. A “+” and a “−” indicate the presence and absence of corresponding components in the reactions, respectively. (D) Coimmunoprecipitation of the RF6 and OsHXK6 proteins from mitochondria. (E) Western blot analysis of the ORFH79 protein level in indicated plants. YTA and R-7 were used as negative and positive controls, respectively. HSP90 protein was used as a loading control. (F) Microscopic analysis of pollen grains stained with 1% I2-KI solution in the indicated plants. (Scale bars: 50 µm.)
C terminus of RF6 Mediates the RF6–OsHXK6 Interaction.
Next, we sought to identify the RF6 domains that directly mediate the association with OsHXK6. We examined the association of OsHXK6 with truncated RF6 proteins by a yeast two-hybrid assay. Surprisingly, the region that directly interacted with OsHXK6 was located at the C terminus of RF6, from residue 386 to 894, whereas the duplicated motifs at the N terminus did not interact with OsHXK6 (SI Appendix, Fig. S12A). Thus, these results suggest that duplicated motifs are needed to establish an optimal conformation appropriate for OsHXK6 association rather than serving as a direct docking site.
Association with RF6 Does Not Depend on the Catalytic Motif of OsHXK6.
Two mutations in the Gly (G112) residue to Asp (D) and in a Ser (S185) residue to Ala (A) at the N terminus of OsHXK6 result in the loss of sugar-kinase catalytic activity in rice (40). Thus, we examined whether the catalytic domain of OsHXK6 is required for association with RF6. To this end, we performed a yeast two-hybrid assay using full-length RF6 as bait and truncated OsHXK6 as prey. We observed strong interactions between RF6 and truncated OsHXK6, encompassing residues 386–506. However, we did not observe a direct association between RF6 and the OsHXK6 N-terminal domain, which included the sugar-kinase catalytic motif (SI Appendix, Fig. S12B). This result suggests that the association with RF6 does not involve the catalytic domain of OsHXK6.
OsHXK6 Promotes the Processing of the atp6-orfH79 Transcript and Restoration of Fertility.
The above results suggest that RF6 and OsHXK6 may function together to process the atp6-orfH79 transcript and restore fertility. To examine the role of OsHXK6 in this process, we used RNA interference to down-regulate the expression of the OsHXK6 gene in R-7 F3 plants, which are homozygous at the Rf6 locus and exhibit 100% normal pollen grains. We obtained three transgenic lines with reduced levels of OsHXK6 (SI Appendix, Fig. S13B). As expected, the atp6-orfH79 transcript accumulated in OsHXK6i plants (SI Appendix, Table S2), and ORFH79 protein also accumulated (Fig. 4E). As a result, we observed that ∼50% of pollen grains were abortive in these plants (Fig. 4F and SI Appendix, Fig. S13 C and D), a proportion equivalent to that in the F1 hybrids. Therefore, we concluded that RF6 functions with OsHXK6 in mitochondria to reduce the accumulation of the atp6-orfH79 transcript, thus leading to the restoration of HL-CMS fertility. Furthermore, we observed that the expression of the Rf6 gene in OsHXK6i transgenic lines remains unaffected (SI Appendix, Fig. S14), suggesting that the roles of OsHXK6 in restoring fertility are not through impacting the expression of Rf6.
Discussion
In the present study, we report the identification and characterization of a novel PPR family restorer gene, Rf6, and demonstrate that RF6 protein works with OsHXK6 in mitochondria to stimulate the processing of the HL-CMS–associated transcript, atp6-orfH79, leading to normal pollen development and fertility restoration. The results suggest that RF6 and RF5 function through distinct mechanisms to rescue the sterility of HL-CMS. Unlike RF5, RF6 does not interact with the RNA-processing factor, GRP162 (SI Appendix, Fig. S11). Additionally, RF6 likely promotes the cleavage of the atp6-orfH79 transcript at a novel position, as shown in the model (Fig. 5). To our knowledge, this study is the first to link restoration of CMS fertility to the hexokinase OsHXK6, providing novel insights into the mechanisms of CMS restoration, pollen development, and hexokinase 6 function in regulating mitochondrial RNA metabolism.
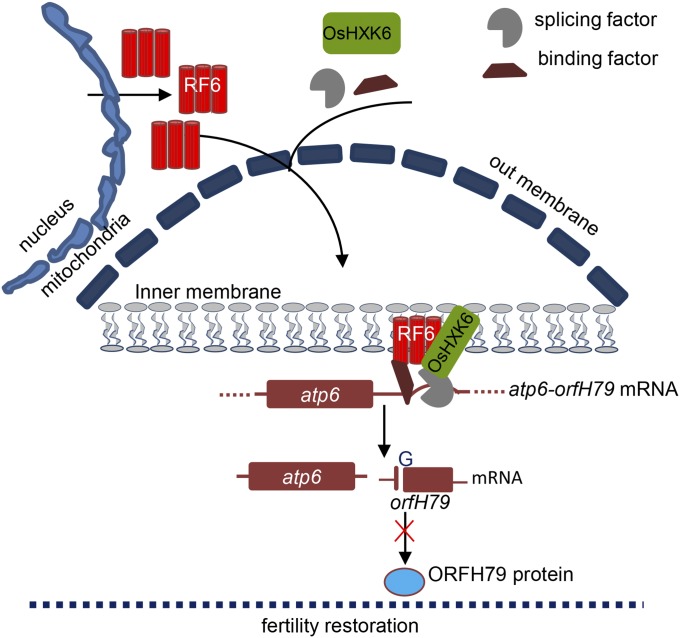
Fig. 5.
A working model showing the mechanism by which RF6 restores the fertility of HL-CMS. RF6 and OsHXK6 proteins are targeted to mitochondria, forming a complex with other proteins, including RNA-binding and processing factors, likely to cleave the aberrant atp6-orfH79 transcript, thus disrupting orfH79 mRNA and preventing its translation. Consequently, this processing ensures normal pollen development and leads to the restoration of CMS fertility.
The PPR protein family is one of the most perplexing protein families in plants such as maize, rice, sorghum, rapeseed, petunia, radish, and sunflower (9). PPR proteins are tandem arrays of 2–27 repeats of PPR motifs, and each motif contains 35 amino acids (7). Structurally, PPR motifs consist of two distinct antiparallel alpha helices, helices A and B, closely resembling the tetratricopeptide repeat motif, which is involved in mediating protein–protein interactions (7, 41). We observed that RF6 contains a unique duplication of PPR motifs, which are not present in rf6 or other known RF proteins. This duplication does not interact with OsHXK6 but is essential for enabling RF6 to interact with OsHXK6 to process the atp6-orfH79 transcript. Thus, this duplication likely establishes an optimal conformation appropriate for OsHXK6 association or is associated with an unknown mediator protein in the restoration complex.
The HL-CMS restorer gene, Rf5, which encodes a P-family member of PPR family proteins, is identical to the BT-CMS restorer gene, Rf1A (18). However, Rf1A directly binds to and processes the CMS-associated transcript atp6-orf79 in the BT-CMS line (42), whereas Rf5 apparently requires additional factors, including GRP162, to process the aberrant transcript atp6-orfH79 in HL-CMS lines (18), suggesting that RF5 protein binds both protein and RNA molecules, depending on the specific situation in a given CMS system. Importantly, we observed that Rf6 also restored the fertility of BT-CMS rice. Therefore, it will be necessary to test whether Rf6 functions through the same mechanism in the two CMS systems and whether OsHXK6 is needed to rescue BT-CMS sterility. Regardless, a broad role for Rf6 in rescuing both HL- and BT-CMS sterility should greatly facilitate the further exploitation of heterosis in rice breeding.
In addition to functioning as glucose sensors, hexokinase proteins phosphorylate hexose to form hexose-6-P, thereby regulating many physiological processes, such as germination, flowering, senescence, and pathogen defense responses to biotic and abiotic stresses in plants (43–45). OsHXK6 belongs to a hexokinase family comprising 10 members in rice (46). Among these members, OsHXK6 was closely related to AtHXK1, which functions in a complex involved in regulating gene expression or RNA metabolism in the nucleus (47). OsHXK6 can be exported into the nucleus, and the nuclear localization of this protein may be critical for its sugar-mediated signaling functions in rice (40). In contrast, little is known regarding the function of OsHXK6 in mitochondria. Here, we identified OsHXK6 as a RF6 partner that participates in the processing of a mitochondrial RNA transcript. The sugar-kinase catalytic domain is not involved in interactions with RF6, suggesting that the function of OsHXK6 in fertility restoration is likely independent of its role in sugar sensing. However, the precise role of OsHXK6 in this process is unclear. Although both RF6 and OsHXK6 are essential for processing the chimeric transcript, neither protein can directly bind to RNA (SI Appendix, Figs. S10B and S15), implying that additional proteins capable of binding or splicing RNA should be involved (Fig. 5). Identifying and characterizing the functions of these factors will be an important task necessary for fully elucidating the mechanism of CMS and fertility restoration. In addition, this study provides information for further studies on the function of hexokinases in mitochondria metabolism and nuclear-cytoplasm communication.
Materials and Methods
Information on the plant materials used in this study is included in SI Appendix, Materials and Methods. Total RNA was extracted using TRIzol reagent (Invitrogen). The yeast two-hybrid (Clontech) and EMSA (Thermo Scientific) assays were performed according to the manufacturer’s instructions. Detailed experimental procedures and reagents used in this study are described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Jianming Wan, Dr. Haiyang Wang, Dr. William Terzaghi, Dr. Marenda Wilson, and Dr. Mengxiang Sun for critical comments on the manuscript and Dr. Meizhong Luo for providing BAC vectors. This work was supported by grants from the National Natural Science Foundation of China (Grant 31371236) and the National High Technology Research and Development Program of China (Grant 2011AA10A101).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.R.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1511748112/-/DCSupplemental.
References
- 1.Chen L, Liu YG. Male sterility and fertility restoration in crops. Annu Rev Plant Biol. 2014;65:579–606. doi: 10.1146/annurev-arplant-050213-040119. [DOI] [PubMed] [Google Scholar]
- 2.Havey M. The use of cytoplasmic male sterility for hybrid seed production. In: Daniell H, Chase CD, editors. Molecular Biology and Biotechnology of Plant Organelles. Springer; Berlin: 2004. pp. 623–634. [Google Scholar]
- 3.Levings CS., III Thoughts on cytoplasmic male sterility in cms-T maize. Plant Cell. 1993;5(10):1285–1290. doi: 10.1105/tpc.5.10.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grelon M, Budar F, Bonhomme S, Pelletier G. Ogura cytoplasmic male-sterility (CMS)-associated orf138 is translated into a mitochondrial membrane polypeptide in male-sterile Brassica cybrids. Mol Gen Genet. 1994;243(5):540–547. doi: 10.1007/BF00284202. [DOI] [PubMed] [Google Scholar]
- 5.Schnable PS, Wise RP. The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 1998;3:175–180. [Google Scholar]
- 6.Balk J, Leaver CJ. The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell. 2001;13(8):1803–1818. doi: 10.1105/TPC.010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Small ID, Peeters N. The PPR motif: A TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci. 2000;25(2):46–47. doi: 10.1016/s0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- 8.Dahan J, Mireau H. The Rf and Rf-like PPR in higher plants, a fast-evolving subclass of PPR genes. RNA Biol. 2013;10(9):1469–1476. doi: 10.4161/rna.25568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barkan A, Small I. Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol. 2014;65:415–442. doi: 10.1146/annurev-arplant-050213-040159. [DOI] [PubMed] [Google Scholar]
- 10.Lurin C, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16(8):2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: A socket set for organelle gene expression. Trends Plant Sci. 2008;13(12):663–670. doi: 10.1016/j.tplants.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 12.Schmitz-Linneweber C, Williams-Carrier R, Barkan A. RNA immunoprecipitation and microarray analysis show a chloroplast Pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell. 2005;17(10):2791–2804. doi: 10.1105/tpc.105.034454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beick S, Schmitz-Linneweber C, Williams-Carrier R, Jensen B, Barkan A. The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol Cell Biol. 2008;28(17):5337–5347. doi: 10.1128/MCB.00563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfalz J, Bayraktar OA, Prikryl J, Barkan A. Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 2009;28(14):2042–2052. doi: 10.1038/emboj.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kotera E, Tasaka M, Shikanai T. A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature. 2005;433(7023):326–330. doi: 10.1038/nature03229. [DOI] [PubMed] [Google Scholar]
- 16.Okuda K, et al. The pentatricopeptide repeat protein OTP82 is required for RNA editing of plastid ndhB and ndhG transcripts. Plant J. 2010;61(2):339–349. doi: 10.1111/j.1365-313X.2009.04059.x. [DOI] [PubMed] [Google Scholar]
- 17.Robbins JC, Heller WP, Hanson MR. A comparative genomics approach identifies a PPR-DYW protein that is essential for C-to-U editing of the Arabidopsis chloroplast accD transcript. RNA. 2009;15(6):1142–1153. doi: 10.1261/rna.1533909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu J, et al. The rice pentatricopeptide repeat protein RF5 restores fertility in Hong-Lian cytoplasmic male-sterile lines via a complex with the glycine-rich protein GRP162. Plant Cell. 2012;24(1):109–122. doi: 10.1105/tpc.111.093211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillman JD, Bentolila S, Hanson MR. The petunia restorer of fertility protein is part of a large mitochondrial complex that interacts with transcripts of the CMS-associated locus. Plant J. 2007;49(2):217–227. doi: 10.1111/j.1365-313X.2006.02953.x. [DOI] [PubMed] [Google Scholar]
- 20.Li SQ, Yang DC, Zhu YG. Characterization and use of male sterility in hybrid rice breeding. J Integr Plant Biol. 2007;49:791–804. [Google Scholar]
- 21.Virmani SS. Advances in hybrid rice research and development in the tropics. In Proceedings of the 4th International Symposium on Hybrid Rice. In: Virmani SS, Mao CX, Hardy B, editors. International Rice Research Institute; Manila, Philippines: 2003. pp. 7–20. [Google Scholar]
- 22.Cheng SH, Zhuang JY, Fan YY, Du JH, Cao LY. Progress in research and development on hybrid rice: A super-domesticate in China. Ann Bot (Lond) 2007;100(5):959–966. doi: 10.1093/aob/mcm121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang W, et al. Research and development of HL type hybrid rice. Sci China Life Sci. 2012;42(9):689–698. [Google Scholar]
- 24.Luo D, et al. A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nat Genet. 2013;45(5):573–577. doi: 10.1038/ng.2570. [DOI] [PubMed] [Google Scholar]
- 25.Akagi H, Sakamoto M, Shinjyo C, Shimada H, Fujimura T. A unique sequence located downstream from the rice mitochondrial atp6 may cause male sterility. Curr Genet. 1994;25(1):52–58. doi: 10.1007/BF00712968. [DOI] [PubMed] [Google Scholar]
- 26.Wang Z, et al. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell. 2006;18(3):676–687. doi: 10.1105/tpc.105.038240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yi P, Wang L, Sun Q, Zhu YG. Discovery of mitochondrial chimeric gene associated with male sterility of HL-rice. Chin Sci Bull. 2002;47(9):744–747. [Google Scholar]
- 28.Peng X, et al. The mitochondrial gene orfH79 plays a critical role in impairing both male gametophyte development and root growth in CMS-Honglian rice. BMC Plant Biol. 2010;10:125. doi: 10.1186/1471-2229-10-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu G, et al. Alterations of mitochondrial protein assembly and jasmonic acid biosynthesis pathway in Honglian (HL)-type cytoplasmic male sterility rice. J Biol Chem. 2012;287(47):40051–40060. doi: 10.1074/jbc.M112.382549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K, et al. ORFH79 impairs mitochondrial function via interaction with a subunit of electron transport chain complex III in Honglian cytoplasmic male sterile rice. New Phytol. 2013;198(2):408–418. doi: 10.1111/nph.12180. [DOI] [PubMed] [Google Scholar]
- 31.Huang W, et al. Two non-allelic nuclear genes restore fertility in a gametophytic pattern and enhance abiotic stress tolerance in the hybrid rice plant. Theor Appl Genet. 2012;124(5):799–807. doi: 10.1007/s00122-011-1755-9. [DOI] [PubMed] [Google Scholar]
- 32.Hu J, et al. The mechanism of ORFH79 suppression with the artificial restorer fertility gene Mt-GRP162. New Phytol. 2013;199(1):52–58. doi: 10.1111/nph.12310. [DOI] [PubMed] [Google Scholar]
- 33.Bentolila S, Alfonso AA, Hanson MR. A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc Natl Acad Sci USA. 2002;99(16):10887–10892. doi: 10.1073/pnas.102301599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown GG, et al. The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats. Plant J. 2003;35(2):262–272. doi: 10.1046/j.1365-313x.2003.01799.x. [DOI] [PubMed] [Google Scholar]
- 35.Akagi H, et al. Positional cloning of the rice Rf-1 gene, a restorer of BT-type cytoplasmic male sterility that encodes a mitochondria-targeting PPR protein. Theor Appl Genet. 2004;108(8):1449–1457. doi: 10.1007/s00122-004-1591-2. [DOI] [PubMed] [Google Scholar]
- 36.Komori T, et al. Map-based cloning of a fertility restorer gene, Rf-1, in rice (Oryza sativa L.) Plant J. 2004;37(3):315–325. doi: 10.1046/j.1365-313x.2003.01961.x. [DOI] [PubMed] [Google Scholar]
- 37.Klein RR, et al. Fertility restorer locus Rf1 [corrected] of sorghum (Sorghum bicolor L.) encodes a pentatricopeptide repeat protein not present in the colinear region of rice chromosome 12. Theor Appl Genet. 2005;111(6):994–1012. doi: 10.1007/s00122-005-2011-y. [DOI] [PubMed] [Google Scholar]
- 38.Tang H, et al. The rice restorer Rf4 for wild-abortive cytoplasmic male sterility encodes a mitochondrial-localized PPR protein that functions in reduction of WA352 transcripts. Mol Plant. 2014;7(9):1497–1500. doi: 10.1093/mp/ssu047. [DOI] [PubMed] [Google Scholar]
- 39.Yin P, et al. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature. 2013;504(7478):168–171. doi: 10.1038/nature12651. [DOI] [PubMed] [Google Scholar]
- 40.Cho J-I, et al. Role of the rice hexokinases OsHXK5 and OsHXK6 as glucose sensors. Plant Physiol. 2009;149(2):745–759. doi: 10.1104/pp.108.131227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Das AK, Cohen PW, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: Implications for TPR-mediated protein-protein interactions. EMBO J. 1998;17(5):1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kazama T, Nakamura T, Watanabe M, Sugita M, Toriyama K. Suppression mechanism of mitochondrial ORF79 accumulation by Rf1 protein in BT-type cytoplasmic male sterile rice. Plant J. 2008;55(4):619–628. doi: 10.1111/j.1365-313X.2008.03529.x. [DOI] [PubMed] [Google Scholar]
- 43.Jang JC, León P, Zhou L, Sheen J. Hexokinase as a sugar sensor in higher plants. Plant Cell. 1997;9(1):5–19. doi: 10.1105/tpc.9.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho J-I, Ryoo N, Hahn T-R, Jeon J-S. Evidence for a role of hexokinases as conserved glucose sensors in both monocot and dicot plant species. Plant Signal Behav. 2009;4(9):908–910. doi: 10.4161/psb.4.9.9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bolouri-Moghaddam MR, Le Roy K, Xiang L, Rolland F, Van den Ende W. Sugar signalling and antioxidant network connections in plant cells. FEBS J. 2010;277(9):2022–2037. doi: 10.1111/j.1742-4658.2010.07633.x. [DOI] [PubMed] [Google Scholar]
- 46.Cho JI, et al. Structure, expression, and functional analysis of the hexokinase gene family in rice (Oryza sativa L.) Planta. 2006a;224(3):598–611. doi: 10.1007/s00425-006-0251-y. [DOI] [PubMed] [Google Scholar]
- 47.Cho Y-H, Yoo S-D, Sheen J. Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell. 2006;127(3):579–589. doi: 10.1016/j.cell.2006.09.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.