Abstract
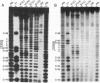
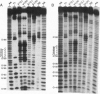
Signal recognition particle (SRP) RNA exhibits significant primary sequence conservation only in domain IV, a bulged hairpin capped by a GNRA (N, any nucleotide; R, purine) tetranucleotide loop except in plant homologs. Tetraloops conforming to this sequence or to the consensus UNCG enhance the stability of synthetic RNA hairpins and have strikingly similar three-dimensional structures. To determine the biological relevance of this similarity, as well as to assess the relative contributions of sequence and structure to the function of the domain IV tetraloop, we replaced the GAAA sequence in fission yeast SRP RNA with UUCG. Haploid strains harboring this substitution are viable, providing experimental evidence for the functional equivalence of the two tetraloops. We next tested the two sequences found in plant SRP RNAs at this location for function in the context of the Schizosaccharomyces pombe RNA. While substitution of CUUC does not allow growth, a viable strain results from replacing GAAA with UUUC. Although the viable tetraloop substitution mutants exhibit wild-type growth under normal conditions, all three express conditional defects. To determine whether this might be a consequence of structural perturbations, we performed enzymatic probing. The results indicate that RNAs containing tetraloop substitutions exhibit subtle differences from the wild type not only in the tetraloop itself, but also in the 3-base pair adjoining stem. To directly assess the importance of the latter structure, we disrupted it partially or completely and made the compensatory mutations to restore the helix. Surprisingly, mutant RNAs with as little as one Watson-Crick base pair can support growth.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andreazzoli M., Gerbi S. A. Changes in 7SL RNA conformation during the signal recognition particle cycle. EMBO J. 1991 Apr;10(4):767–777. doi: 10.1002/j.1460-2075.1991.tb08008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antao V. P., Lai S. Y., Tinoco I., Jr A thermodynamic study of unusually stable RNA and DNA hairpins. Nucleic Acids Res. 1991 Nov 11;19(21):5901–5905. doi: 10.1093/nar/19.21.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennwald P., Liao X., Holm K., Porter G., Wise J. A. Identification of an essential Schizosaccharomyces pombe RNA homologous to the 7SL component of signal recognition particle. Mol Cell Biol. 1988 Apr;8(4):1580–1590. doi: 10.1128/mcb.8.4.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong C., Varani G., Tinoco I., Jr Solution structure of an unusually stable RNA hairpin, 5'GGAC(UUCG)GUCC. Nature. 1990 Aug 16;346(6285):680–682. doi: 10.1038/346680a0. [DOI] [PubMed] [Google Scholar]
- Glück A., Endo Y., Wool I. G. Ribosomal RNA identity elements for ricin A-chain recognition and catalysis. Analysis with tetraloop mutants. J Mol Biol. 1992 Jul 20;226(2):411–424. doi: 10.1016/0022-2836(92)90956-k. [DOI] [PubMed] [Google Scholar]
- Heus H. A., Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991 Jul 12;253(5016):191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- Holbrook S. R., Cheong C., Tinoco I., Jr, Kim S. H. Crystal structure of an RNA double helix incorporating a track of non-Watson-Crick base pairs. Nature. 1991 Oct 10;353(6344):579–581. doi: 10.1038/353579a0. [DOI] [PubMed] [Google Scholar]
- Knapp G. Enzymatic approaches to probing of RNA secondary and tertiary structure. Methods Enzymol. 1989;180:192–212. doi: 10.1016/0076-6879(89)80102-8. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen N., Zwieb C. SRP-RNA sequence alignment and secondary structure. Nucleic Acids Res. 1991 Jan 25;19(2):209–215. doi: 10.1093/nar/19.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X. B., Brennwald P., Wise J. A. Genetic analysis of Schizosaccharomyces pombe 7SL RNA: a structural motif that includes a conserved tetranucleotide loop is important for function. Proc Natl Acad Sci U S A. 1989 Jun;86(11):4137–4141. doi: 10.1073/pnas.86.11.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X., Selinger D., Althoff S., Chiang A., Hamilton D., Ma M., Wise J. A. Random mutagenesis of Schizosaccharomyces pombe SRP RNA: lethal and conditional lesions cluster in presumptive protein binding sites. Nucleic Acids Res. 1992 Apr 11;20(7):1607–1615. doi: 10.1093/nar/20.7.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poritz M. A., Siegel V., Hansen W., Walter P. Small ribonucleoproteins in Schizosaccharomyces pombe and Yarrowia lipolytica homologous to signal recognition particle. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4315–4319. doi: 10.1073/pnas.85.12.4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SantaLucia J., Jr, Kierzek R., Turner D. H. Context dependence of hydrogen bond free energy revealed by substitutions in an RNA hairpin. Science. 1992 Apr 10;256(5054):217–219. doi: 10.1126/science.1373521. [DOI] [PubMed] [Google Scholar]
- Selinger D., Brennwald P., Liao X., Wise J. A. Identification of RNA sequences and structural elements required for assembly of fission yeast SRP54 protein with signal recognition particle RNA. Mol Cell Biol. 1993 Mar;13(3):1353–1362. doi: 10.1128/mcb.13.3.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel V., Walter P. Binding sites of the 19-kDa and 68/72-kDa signal recognition particle (SRP) proteins on SRP RNA as determined in protein-RNA "footprinting". Proc Natl Acad Sci U S A. 1988 Mar;85(6):1801–1805. doi: 10.1073/pnas.85.6.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. W., Ott J., Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C., Gauss P., Thermes C., Groebe D. R., Gayle M., Guild N., Stormo G., d'Aubenton-Carafa Y., Uhlenbeck O. C., Tinoco I., Jr CUUCGG hairpins: extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1364–1368. doi: 10.1073/pnas.85.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenbeck O. C. Tetraloops and RNA folding. Nature. 1990 Aug 16;346(6285):613–614. doi: 10.1038/346613a0. [DOI] [PubMed] [Google Scholar]
- Walter P., Blobel G. Purification of a membrane-associated protein complex required for protein translocation across the endoplasmic reticulum. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7112–7116. doi: 10.1073/pnas.77.12.7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., Lingappa V. R. Mechanism of protein translocation across the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1986;2:499–516. doi: 10.1146/annurev.cb.02.110186.002435. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Winker S., Gutell R. R. Architecture of ribosomal RNA: constraints on the sequence of "tetra-loops". Proc Natl Acad Sci U S A. 1990 Nov;87(21):8467–8471. doi: 10.1073/pnas.87.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood H., Luirink J., Tollervey D. Evolutionary conserved nucleotides within the E.coli 4.5S RNA are required for association with P48 in vitro and for optimal function in vivo. Nucleic Acids Res. 1992 Nov 25;20(22):5919–5925. doi: 10.1093/nar/20.22.5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C. Recognition of a tetranucleotide loop of signal recognition particle RNA by protein SRP19. J Biol Chem. 1992 Aug 5;267(22):15650–15656. [PubMed] [Google Scholar]