SUMMARY
Positioning of centrosomes is vital for cell division and development. In metazoan cells, spindle positioning is controlled by a dynamic pool of subcortical actin that organizes in response to the position of retraction fibers. These actin “clouds” are proposed to generate pulling forces on centrosomes and mediate spindle orientation. However, the motors that pull astral microtubules toward these actin structures are not known. Here, we report that the unconventional myosin, Myo10, couples actin-dependent forces from retraction fibers and subcortical actin clouds to centrosomes. Myo10-mediated centrosome positioning requires its direct microtubule binding. Computational image analysis of large microtubule populations reveals a direct effect of Myo10 on microtubule dynamics and microtubule-cortex interactions. Myo10’s role in centrosome positioning is distinct from, but overlaps with, that of dynein. Thus, Myo10 plays a key role in integrating the actin and microtubule cytoskeletons to position centrosomes and mitotic spindles.
INTRODUCTION
The regulated positioning of centrosomes within cells has essential roles in tissue homeostasis, morphogenesis, and the specification of cell fate during development (McNally, 2013; Morin and Bellaïche, 2011). In symmetrically dividing cells, centrosome positioning centers the mitotic spindle, which is important for maintaining normal daughter cell size (Kiyomitsu and Cheeseman, 2013). Centrosome and spindle positioning is also important for asymmetric cell divisions, which control some cell fate decisions during development and are required for stem cell maintenance (McCaffrey and Macara, 2011; Morin and Bellaïche, 2011; Siller and Doe, 2009). Defects in spindle positioning are implicated in developmental defects and tumorigenesis (McCaffrey and Macara, 2011; Pease and Tirnauer, 2011).
Centrosome positioning is controlled by mechanisms that differ between cell types. In the simplest case, pushing forces from polymerizing microtubules can center asters in cell fragments (Rodionov and Borisy, 1998) and microfabricated chambers (Laan et al., 2008), or nuclei in the fission yeast, S. pombe (Chang and Martin, 2009).
Most commonly, pulling forces on astral microtubules that originate near the cell cortex or from within the adjacent cytoplasm are critical for centrosome positioning (Goshima and Scholey, 2010; McNally, 2013; Minc et al., 2011). Spindle-cortex interactions are best understood in budding yeast, where two mechanisms work in parallel to pull astral microtubules into the daughter cell (Pearson and Bloom, 2004; Siller and Doe, 2009). A first budding yeast mechanism involves a complex of proteins at the plus ends of astral microtubules that binds a type V myosin, which then transports the astral microtubule along polarized arrays of actin cables. A second mechanism is actin-independent and is mediated by the microtubule motor dynein.
In mammalian cells, dynein is also a major cortical force generator that mediates spindle orientation. The functional importance of dynein for spindle positioning is established in many studies (McNally, 2013; Morin and Bellaïche, 2011; Siller and Doe, 2009). During interphase, dynein can mediate end-on attachment of microtubules to the cell cortex, with force generation coupled to microtubule depolymerization (Laan et al., 2012; Yi et al., 2013). Dynein can also mediate lateral attachment of microtubules to the mitotic cell cortex that generates sliding of microtubule ends along the cortex (Adames and Cooper, 2000; Gusnowski and Srayko, 2011). The cortical distribution of dynein can be regulated by external cues (Morin and Bellaïche, 2011; Siller and Doe, 2009) or by signals from the spindle or the chromosomes (Kiyomitsu and Cheeseman, 2012).
Like in budding yeast, spindle positioning in mammalian cells requires the actin cytoskeleton (Kunda and Baum, 2009), but the underlying molecular mechanism, including possible roles for actin-based motors, is less well understood. An important effect of actin is indirect: F-actin is required to maintain cortical rigidity that prevents end-on microtubule attachments from pulling strands of plasma membrane into the cytoplasm (Kunda and Baum, 2009; Redemann et al., 2010). In some cell types, asymmetric contraction of the cortical actomyosin network may pull on attached astral microtubules, facilitating the positioning of mitotic centrosomes (Rosenblatt et al., 2004). Actin also indirectly affects dynein function by maintaining cortical localization of LGN, a cortical recruitment factor for dynein (Zheng et al., 2013). Finally, prior work has implicated the microtubule-binding myosin Myo10 in spindle positioning (Liu et al., 2012; Toyoshima and Nishida, 2007; Weber et al., 2004); however, whether Myo10 affects spindle orientation directly or indirectly has been unclear.
Although the molecular mechanisms by which the actin cytoskeleton controls spindle position are not well understood in mammalian cells, significant progress has been made in identifying the relevant actin structures. Mitotic actin-dependent pulling forces originate from retraction fibers, which are cytoplasmic extensions that link rounded mitotic cells to sites of strong cell matrix adhesion (Mitchison, 1992; Théry and Bornens, 2006). Because retraction fibers control spindle orientation, the position of retraction fibers can determine the plane of cell division. This has been demonstrated most elegantly by experiments in which fibronectin micropatterns are used to manipulate the position of retraction fibers (Fink et al., 2011; Théry et al., 2007). In cells with multiple centrosomes, retraction fibers either force cells into multipolar divisions or force the clustering of centrosomes to generate bipolar division, depending on the position of the retraction fibers (Kwon et al., 2008). Thus, mitotic cortical pulling forces concentrate near retraction fibers. Recent work suggests that the assembly of subcortical “actin clouds” is required for pulling forces on centrosomes toward retraction fibers. Actin clouds are migrating pools of subcortical actin that extend from the plasma membrane into the cytoplasm, concentrating near retraction fibers (Fink et al., 2011; Mitsushima et al., 2010). Together, these findings suggest a model where cortical regions with attached retraction fibers organize the adjacent cytoplasm by controlling a dynamic subcortical actin network. This network then concentrates force-generating molecules that pull on astral microtubules.
Here, we demonstrate that Myo10, an unconventional microtubule-binding myosin (Weber et al., 2004), is required to orient centrosomes and spindle poles toward actin clouds and retraction fibers. Our data indicate that direct binding to microtubules is critical for Myo10’s role in centrosome positioning. By computational image analysis, we demonstrate that Myo10 regulates astral microtubule dynamics and is required for end-on cortical microtubule interactions prior to anaphase. These Myo10 effects are distinct from those of dynein, which we find overlaps with Myo10 for spindle positioning in mammalian cells.
RESULTS
Myo10 Is Required to Orient Centrosomes toward Retraction Fibers
Force generators that orient centrosomes toward the mitotic retraction fiber regions should be present near retraction fibers and/or within the associated subcortical actin clouds; and they should be necessary for orienting centrosomes toward retraction fibers. Ideally, such proteins could also be motor proteins that directly or indirectly bind both actin and microtubules. Motivated by prior work and the results of our genome-wide RNAi screen (Kwon et al., 2008; Toyoshima and Nishida, 2007; Weber et al., 2004), we explore here whether the unconventional microtubule-binding myosin Myo10 is such a force generating protein.
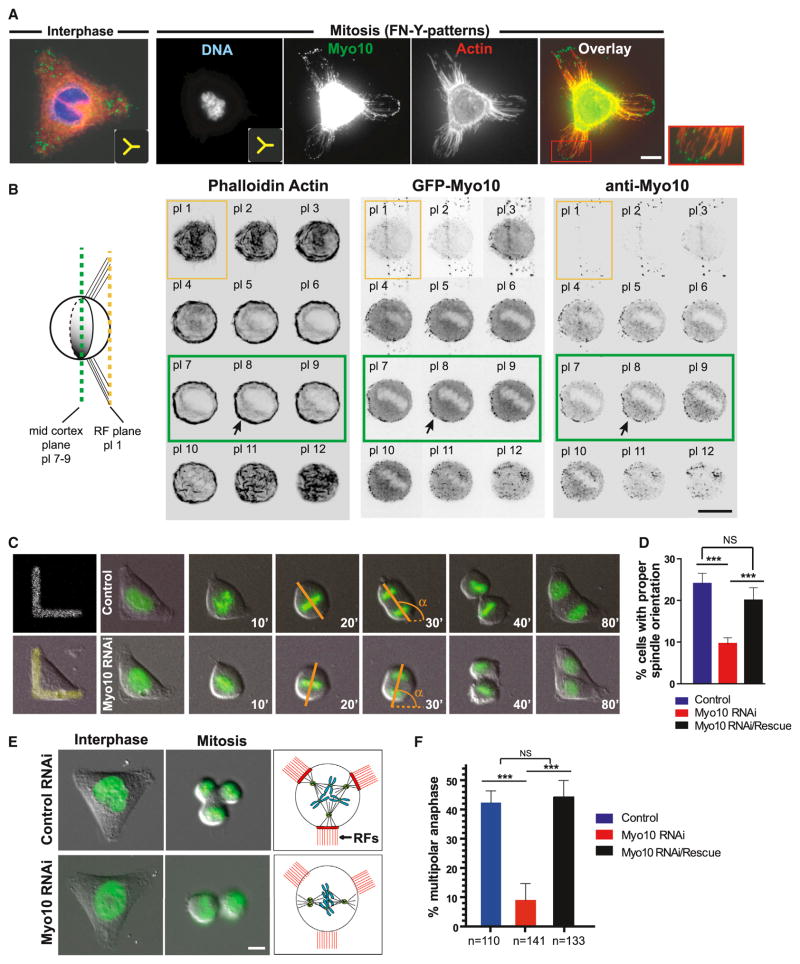
During interphase, Myo10 concentrated at sites of cell-matrix adhesion, often in filopodia (Figure 1A, left), as expected from previous studies (Kerber and Cheney, 2011). During mitosis, Myo10 concentrated in retraction fibers (Figure 1A, right), a finding that was independently confirmed by monitoring GFP-Myo10 localization in three different cell types (Figures 1B and S1A–S1C). In addition to its retraction fiber localization, Myo10 also accumulated asymmetrically at the mid-cortex where asymmetrically positioned subcortical actin clouds assembled (Figure 1B, see planes 7–9).
Figure 1. Myo10 Is a Retraction Fiber Protein Required for Centrosome Positioning.
(A) Immunolocalization of Myo10 in HeLa cells plated on FN patterns (insets show Y pattern geometry). The red box shows a higher magnification view of Myo10 puncta along retraction fibers. The cell body signal of Myo10 is over-exposed to enable visualization of Myo10 signal at retraction fibers.
(B) Co-enrichment of Myo10 and subcortical actin at the mid-cortex. The left image shows a schematic of a cell plated on a FN-bar pattern, side view. The right image shows a Z-focal plane series (0.5 μm steps) from HeLa cells labeled to detect actin (left), GFP-Myo10 (middle), and Myo10 (right). The yellow and green boxes indicate the planes of retraction fiber (RF) and the mid-cortex, respectively.
(C–F) Requirement of Myo10 for spindle orientation (C and D) and the positioning of multiple centrosomes (E and F).
(C) Time-lapse series of control and Myo10-depleted U2OS cells expressing GFP-H2B on FN-L patterns. Time is the minutes from nuclear envelope breakdown (NEBD, t = 0) and α is the angle of the anaphase cell division axis.
(D) Percentage of cells that divide within ±10 degrees of the median angle of 135 degrees, obtained from Figure S1D (α = 135 ± 10, ***p < 0.003, non-parametric Student’s t test, and three experiments).
(E) Images from a time-lapse series in U2OS cells expressing GFP-H2B that contain extra centrosomes after transient overexpression of Plk4 (Movies S1 and S2): interphase, mitosis, and cartoon of experimental findings (red: RFs).
(F) The percentage of multipolar anaphases in the indicated conditions (***p < 0.003, non-parametric Student’s t test, and three experiments). The scale bars represent 10 μm. See also Figure S1. All error bars represent mean ± SEM.
Experiments using fibronectin (FN) micropatterns demonstrated that Myo10 is required to position centrosomes toward retraction fibers. It is known that retraction fibers control mitotic centrosome positioning in cells with a normal number of centrosomes (Théry et al., 2007). On L-shaped FN patterns, as expected, control U2OS cells oriented their spindles such that the poles were typically positioned between the adhesive regions of the pattern, at an ~135° degree angle with respect to the horizontal portion of the “L” (α, Figure 1C). By contrast, depletion of Myo10 led to significant impairment of spindle orientation with respect to the adhesive pattern (Figures 1C, 1D, and S1D). Ectopic expression of small interfering (si)RNA-resistant GFP-Myo10 rescued this defect, demonstrating the specificity of the knockdown effect (Figures 1D and S1D).
In addition to their role in normal spindle positioning, retraction fibers also control the clustering of multiple centrosomes in cancer cells with centrosome amplification (Kwon et al., 2008). Some cancer cells have the capacity to cluster extra centrosomes (e.g., U2OS cells). In permissive cell types, the clustering of multiple centrosomes provides an independent assay for retraction fiber-mediated centrosome positioning. We therefore induced extra centrosomes in U2OS cells by transient overexpression of the kinase Plk4 (Kleylein-Sohn et al., 2007); these cells undergo bipolar or multipolar divisions, depending (in part) on the position of their retraction fibers (Kwon et al., 2008). When such cells are directly plated onto coverslips, without manipulating the position of retraction fibers, most cells with extra centrosomes cluster their centrosomes and assemble pseudo-bipolar spindles. We found that Myo10 is required for this centrosome clustering, consistent with the hypothesis that it could mediate forces from retraction fiber regions on centrosomes (Figures S1E–S1G).
Clustering of multiple centrosomes, and the consequent pseudo-bipolar division, can be overridden by directly manipulating the position of retraction fibers. On a Y-shaped FN pattern, cortical forces pulled centrosomes toward the retraction fibers at the tips of the “Y,” resulting in tripolar cell division ~45% of the time (Figure 1E, upper, and Movie S1). In this case, interfering with the actin-based pulling forces on centrosomes releases tension, allows pseudo-bipolar spindles to form, and increases the fraction of cells that undergo bipolar divisions. Indeed, after knockdown of Myo10, the cell division axis was decoupled from the position of retraction fibers (Figure 1E, bottom, and Movie S2), and most cells underwent bipolar division (Figure 1F). We note that the requirement of Myo10 for centrosome positioning in mammalian cells cannot be explained by the effect of Myo10 on centrosome number or spindle pole integrity (Figures S1H and S1I) (Kwon et al., 2008; Liu et al., 2012; Toyoshima and Nishida, 2007), unlike what is reported for amphibians (Weber et al., 2004; Woolner et al., 2008). Thus, multiple assays suggest that Myo10 is required for cortical pulling forces to position centrosomes at or near retraction fibers.
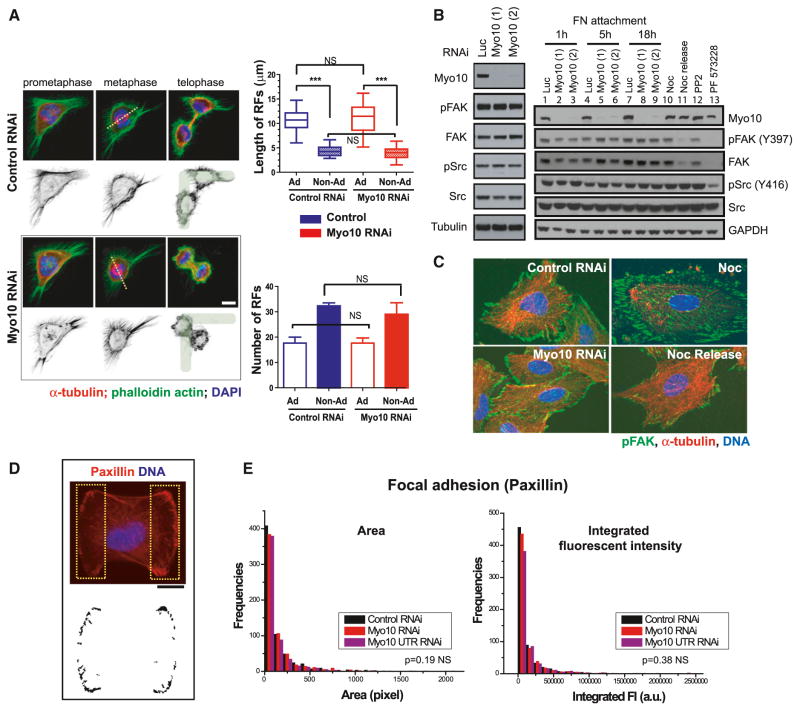
Myo10 Is Not Required for Normal Cell-Matrix Adhesion or for Retraction Fiber Assembly
The function of Myo10 in mitotic centrosome positioning is independent of the requirement for Myo10 in retraction fiber assembly, mitotic cell rounding (Figures 2A and S2A–S2D), and assembly of subcortical actin (Figures 4, S4, 5, and S5; see next section). Myo10 depletion led to no detectable defects in mitotic cell rounding (Figure S2D), in contrast with one previous report (Toyoshima and Nishida, 2007), and no detectable defects in the morphology or organization of subcortical actin.
Figure 2. Myo10 Inhibition Does Not Disrupt Retraction Fiber Formation or Cell Adhesion.
(A) The left image has immunofluorescence images showing normal retraction fiber (RF) morphology during prometaphase, metaphase, and telophase of control or Myo10-depleted HeLa cells plated on L-patterns. The cells were stained for microtubules, actin, and DNA. The dotted lines show the pole-to-pole spindle axis. The right image shows no difference in length (top) and number (bottom) of RFs in both adhesive (Ad) and non-adhesive (Non-Ad) regions upon control or Myo10 RNAi (n = 20 cells from three experiments, ***p < 0.0001, and non-parametric Student’s t test, not significant: NS) (mean ± SEM).
(B and C) Comparable level of activation and distribution of downstream effectors of integrin-mediated adhesion signaling after Myo10 depletion in RPE-1 cells. The two independent Myo10 siRNAs were used.
(B) Western blots showing steady-state Src and FAK activation (left, 24 hr post-attachment) and the kinetics of activation (right, 1, 5, and 18 hr post-attachment). The positive controls show decreased FAK and Src activation after treatment with Src (PP2) or FAK (PF573228) inhibitors or by microtubule-regrowth-mediated focal adhesion disassembly, Nocodazole (Noc) release (Ezratty et al., 2005).
(C) Immunofluorescence images of pFAK (Y397), α-tubulin, and DNA staining in control or Myo10-depleted cells. The Noc treatment (microtubule-depolymerization-induced focal adhesion assembly) and 1 hr post-Noc release (microtubule-regrowth-mediated focal adhesion disassembly) serve as positive and negative controls for FAK activation.
(D and E) No defects in focal adhesion protein distribution in control or Myo10-depleted RPE-1 cells plated on FN-H patterns.
(D) Immunofluorescence image of a cell stained for paxillin and DNA 5 hr post-attachment. The two yellow dotted boxes corresponding to the adhesive surfaces of cells on the H pattern were segmented (bottom image) for paxillin signal.
(E) Corresponding quantitation of the area and integrated fluorescence intensity of paxillin islands (D) in the indicated conditions (n = 15 cells per condition from two experiments). The scale bar represents 10 μm. See also Figure S2.
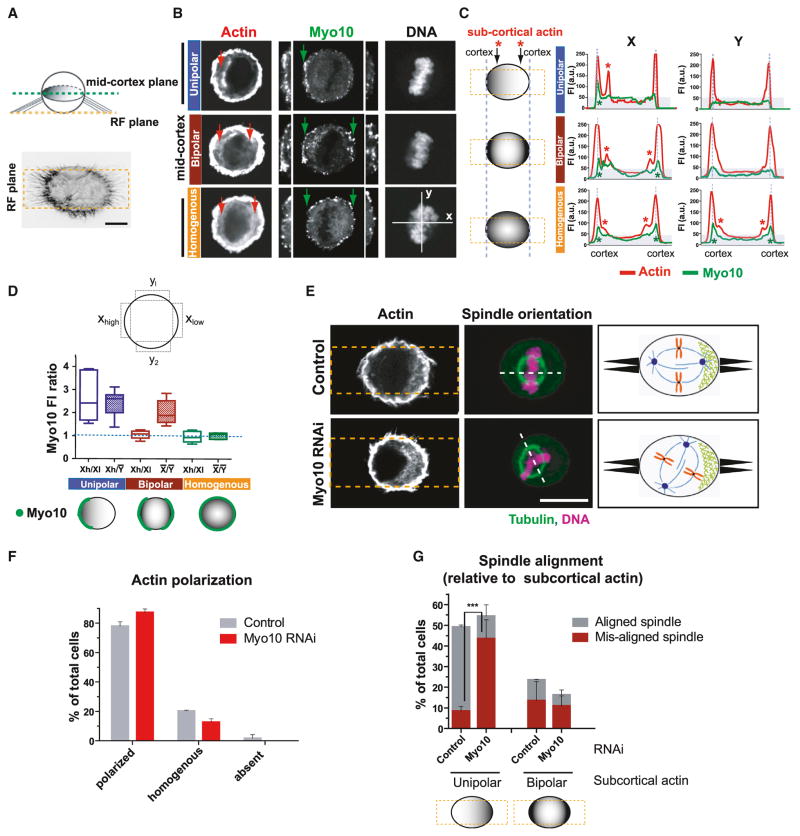
Figure 4. Myo10 Is Required for Spindle Orientation toward Polarized Subcortical Actin.
(A–D) Asymmetric accumulation of Myo10 at the mid-cortical plane of the mitotic cortex is correlated with the asymmetric distribution of subcortical actin.
(A) The top image shows a schematic of a cell plated on FN-bar pattern from a side view. The bottom image shows the phalloidin actin staining of a HeLa cell from the retraction fiber (RF) focal plane.
(B) Representative images of cells where actin and Myo10 are concentrated on one side of the cell (unipolar), both sides (bipolar), or homogeneously distributed. Note that Myo10 accumulates near the cell boundary, at the base of the retraction fibers (see green arrows and insets with enlarged view of both sides of the cell).
(C) Fluorescence line scans of actin (red) and Myo10 (green) along x axis (left) or y axis (right) of the bar pattern from (B). The red and green asterisks show sites of high accumulation of subcortical actin and Myo10, respectively (see also corresponding arrows in B).
(D) Schematic for the Myo10 fluorescent intensity (FI) quantitation shown below. The plot shows FI ratios of Myo10 in the indicated regions from cells classified based on the distribution of subcortical actin in (B) and (C). The open boxes measure asymmetry along the x axis of the pattern; hatched boxes measure the asymmetry of x over y axis of the pattern (n = ~15 line scans per category from two experiments). The bottom image shows a cartoon summarizing observations.
(E–G) Myo10 does not affect the assembly or distribution of subcortical actin clouds, but is required for spindle alignment relative to subcortical actin.
(E) Immunofluorescence images of control or Myo10-depleted cells plated on FN-bar patterns (yellow dashed boxes) and stained for actin, tubulin, and DNA. The schematic summary is shown in the right box: retraction fibers (black protrusions) and subcortical actin (green mesh). The scale bar represents 10 μm.
(F) Percentage of cells with the indicated distribution of subcortical actin with control or Myo10 RNAi treatment (mean ± SEM).
(G) Percentage of cells exhibiting aligned or misaligned spindle with unipolar or bipolar actin clouds (n = ~100 cells from three experiments, ***p < 0.001, and non-parametric Student’s t test. See Figure S4C for data including detailed category of actin cloud organization (mean ± SEM). See also Figure S4.
Because Myo10 transports α5β1 integrin along filopodia via its FERM domain (Zhang et al., 2004), we examined the effect of Myo10 depletion on integrin signaling (Figures 2B and 2C). Both the Src and FAK kinases were fully activated in multiple cell lines after Myo10 depletion (Figures 2B, 2C, and S2E), although a small, but reproducible, delay in FAK activation was evident at the 1 hr time point (Zhang et al., 2004). Likewise, in cells plated on FN micropatterns, recruitment of paxillin to focal adhesions was also unaffected by Myo10 depletion (Figures 2D and 2E). Thus, Myo10 depletion does not detectably affect retraction fiber assembly, mitotic rounding, or integrin signaling.
Microtubule Binding by Myo10 Is Required for Centrosome Positioning
Myo10 is a processive actin motor that binds microtubules through the MyTH4 domain located in its tail (Kerber and Cheney, 2011; Weber et al., 2004) (Figure S1B). Although the motor domain of Myo10 (Myo10-HMM) was sufficient for its localization to retraction fibers (Figures S1B and S1C), expression of the isolated motor domain did not rescue the centrosome-clustering defect in Myo10 knockdown cells (Figure S1E). Likewise, the Myo10 tail domain (PH-MyTH4-FERM) on its own was not able to complement the depletion of Myo10 (Figure S1E). Centrosome positioning by Myo10, therefore, requires both its actin-binding motor and microtubule-binding tail domains.
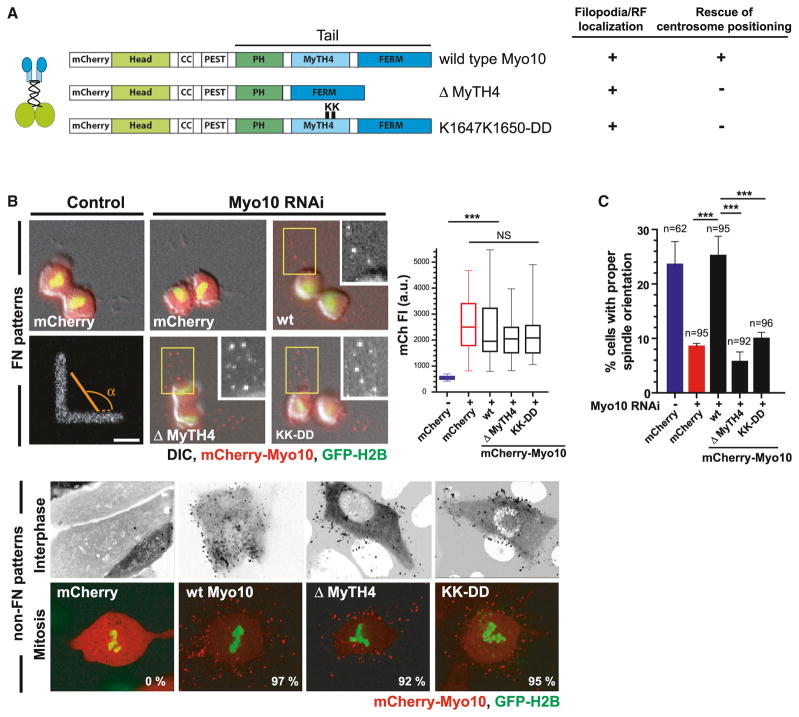
Other interaction partners of the Myo10 tail could also be important for centrosome positioning (Kerber and Cheney, 2011; Liu et al., 2008; Woolner and Bement, 2009). This not only includes FERM domain interactions with integrins, but also microtubule regulators (Katanin and TPX2), actin regulators (Ena and VASP), and regulators of cell polarity (aPKC). Previous studies were not able to discriminate the effects of microtubule-binding from the effects of these other interactions (Woolner and Bement, 2009) (Figure S1E). However, a recent X-ray structure identified specific residues (K1647 and K1650) within the MyTH4 domain that are essential for direct binding to the acidic carboxy-terminal tubulin tail (Hirano et al., 2011; Wei et al., 2011; Wu et al., 2011) (Figure S3A, asterisks in the red box). This enabled us to generate the Myo10-KK-DD point mutation construct (Figures 3A and S3A) that specifically disrupts microtubule lattice-binding without altering the interaction of Myo10 with FERM domain-binding cargo (Hirano et al., 2011).
Figure 3. Direct Binding of Myo10 to Microtubules Is Required for Centrosome Positioning.
(A) Schematic of mCherry-tagged Myo10 constructs used in siRNA-rescue experiments.
(B) Expression and localization of the indicated mCherry-tagged Myo10 constructs in cells expressing GFP-H2B. The top panels are from time-lapse images of control and Myo10-depleted U2OS cells on FN-L patterns. The insets have enlarged views showing retraction fiber localization of Myo10 constructs in yellow boxed regions. The right graph shows the corresponding mCherry fluorescence quantitation. The bottom image shows the normal localization of the indicated Myo10 constructs to filopodia (top: inverted images) and retraction fibers (bottom) in the cells with extra centrosomes. The percentage of cells with proper Myo10 localization is indicated.
(C) Requirement of Myo10 microtubule-binding, but not its cargo-binding, for spindle orientation on FN-L patterns. The spindle orientation analysis was done as in Figures 1C and 1D (***p < 0.001, non-parametric Student’s t test, and three experiments). The scale bar represents 10 μm (mean ± SEM). See also Figure S3.
Imaging experiments demonstrated that the Myo10 microtubule-binding site mutations, including deletions that completely lack the MyTH4 domain, had no effect on Myo10’s localization to filopodia or retraction fibers (Figures 3A and 3B). However, complementation with siRNA-resistant constructs demonstrated that Myo10 microtubule-binding is critical for centrosome positioning (Figures 3C and S3B–S3D). In cells plated on FN-L patterns, neither MyTH4 deletion nor the Myo10-KK-DD point mutant rescued the spindle orientation defect resulting from Myo10 knockdown (Figures 3C and S3C). The failure of the mutants to complement cannot be explained by defects in expression or localization (Figures 3B, upper, and S3B). Similar results were obtained for the microtubule-binding deficient mutants in the multiple centrosome-clustering assay (Figures 3B, bottom, and S3D). Collectively, these data suggest that direct microtubule lattice-binding by Myo10 is critical for its ability to position centrosomes in response to cortical forces (Figure S3E).
Myo10 Is Required for Spindle Orientation Relative to Subcortical Actin Clouds
Although localization of subcortical actin clouds is strongly correlated with centrosome positioning (Fink et al., 2011), the molecular connection between the actin clouds and the astral microtubules and/or centrosomes is not known. To test the idea that Myo10 is this link, we first quantified its localization relative to the position of the actin clouds. The density of Myo10 at the cell cortex corresponded to the distribution of the underlying subcortical actin clouds (Figures 4A–4D). This was clearly apparent in HeLa cells plated on bar-shaped FN patterns that accentuate the asymmetrical accumulation of the actin clouds at the mid-cortex (Figures 4A–4D).
Next, we investigated whether Myo10 is required to form the subcortical actin clouds. F-actin was visualized by phalloidin staining in cells plated on FN-bar patterns (Figures 4E–4G). As expected (Fink et al., 2011), subcortical actin concentrated on one (unipolar distribution) or both (bipolar) sides of the FN bar in the majority (~80%) of control cells, but was homogenously distributed in the remaining 10%–20% of the cells (Figures 4E, 4F, S4A, and S4B). Importantly, depletion of Myo10 had no effect on either the formation or polarization of subcortical actin (Figures 4F and S4D), but led to a ~4-fold decrease in the ability of cells to align their spindles with unipolar subcortical actin (Figures 4G and S4C). Thus, Myo10 is essential to orient centrosomes with subcortical actin, but is not required to assemble these actin structures (Figure 4E).
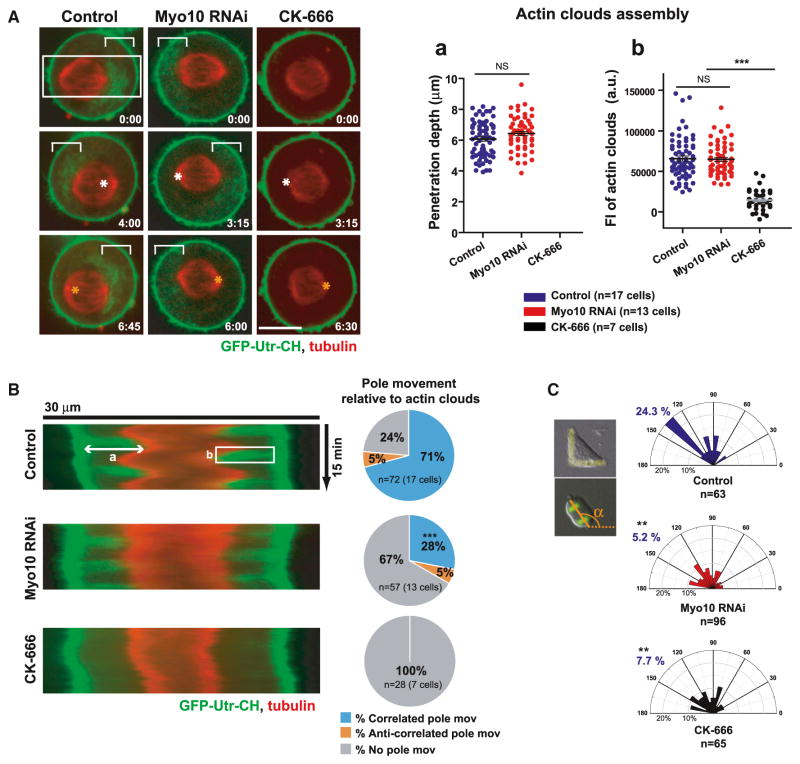
Live-cell imaging confirmed a key role for Myo10 in coordinating centrosome movement with the local positioning of actin clouds (Figures 5 and S5). Subcortical actin dynamics were visualized by GFP-tagged calponin homology (CH) domain of utrophin (GFP-Utr-CH), a fluorescent F-actin binding protein (Mitsushima et al., 2010; Woolner et al., 2008). Imaging of cells expressing GFP-Utr-CH and mCherry-tubulin showed that, during oscillatory spindle movements, spindle poles move toward subcortical actin clouds after a time lag (Figure 5A, left, and Movie S3), as reported (Fink et al., 2011). The correlated pole movement toward actin clouds was confirmed in kymographs (Figures 5B and S5). Strikingly, spindle pole movement toward actin clouds was largely dependent upon Myo10 (Figures 5A, 5B, and S5), consistent with the above analysis of fixed cells (Figures 4E–4G and S4). Although Myo10 depleted cells had no detectable defects in the polarization and movement of subcortical actin clouds (Figure 5A), they did show a significant decrease in the frequency and velocity of pole movement toward actin clouds (Figures 5B, middle, S5B, and S5C and Movie S3). This effect of Myo10 depletion was mirrored by that of Arp2/3 inhibition, which completely disrupts actin clouds (Figures 5A, 5B, S5B, and S5C and Movie S3) without affecting retraction fibers (Mitsushima et al., 2010). Actin clouds and Myo10 are also required for spindle orientation in cells plated on FN-L patterns (Figure 5C). Thus, Myo10 is not essential for actin cloud assembly, but is vital for forces that move centrosomes toward actin clouds during spindle oscillations and spindle orientation.
Figure 5. Myo10 Is Not Required for the Assembly or Dynamics of Actin Clouds but Is Required to Pull Centrosomes toward Subcortical Actin Clouds.
(A) Time-lapse images of control, Myo10-depleted, or CK-666 (100 μM, Arp2/3 inhibitor)-treated HeLa cells expressing GFP-Utr-CH and Cherry-tubulin (see Movie S3). Equatorial planes were imaged at 15 s intervals during metaphase spindle oscillations (time, min: sec). The white and orange asterisks (middle and bottom) show the positions of spindle pole at the previous time point (top and middle). The white brackets show actin clouds. The right graphs show the penetration depth (a) and integrated fluorescent intensity (b) of actin structures quantified from kymographs, as shown in (B) (***p < 0.0001, non-parametric Student’s t test, and three experiments). The scale bar represents 10 μm.
(B) A decrease in the coordinated spindle pole movements toward actin clouds in Myo10-depleted cells. The left image shows kymographs of actin clouds (green) and spindle poles (red) generated from the region indicated by the white boxes (70 pixel in width shown in A) along the spindle axis in the indicated conditions. Pie charts at right show the spindle pole movements relative to actin cloud accumulation that were classified as correlated (blue, pole moves toward actin clouds), anti-correlated (orange, pole moves against actin clouds), or no (gray) movement, as described in Figures S5A and S5B. The percentage of pole movements in each category is shown in the indicated conditions (***p < 0.003, non-parametric Student’s t test, and three experiments).
(C) Requirement of Myo10 and actin clouds for spindle orientation in HeLa cells plated on FN-L patterns. The imaging and analysis were done as in Figures 1C and 1D (**p < 0.01, non-parametric Student’s t test, and two experiments ±SEM). See also Figure S5.
The Effect of Actin and Myo10 on Astral Microtubule Dynamics
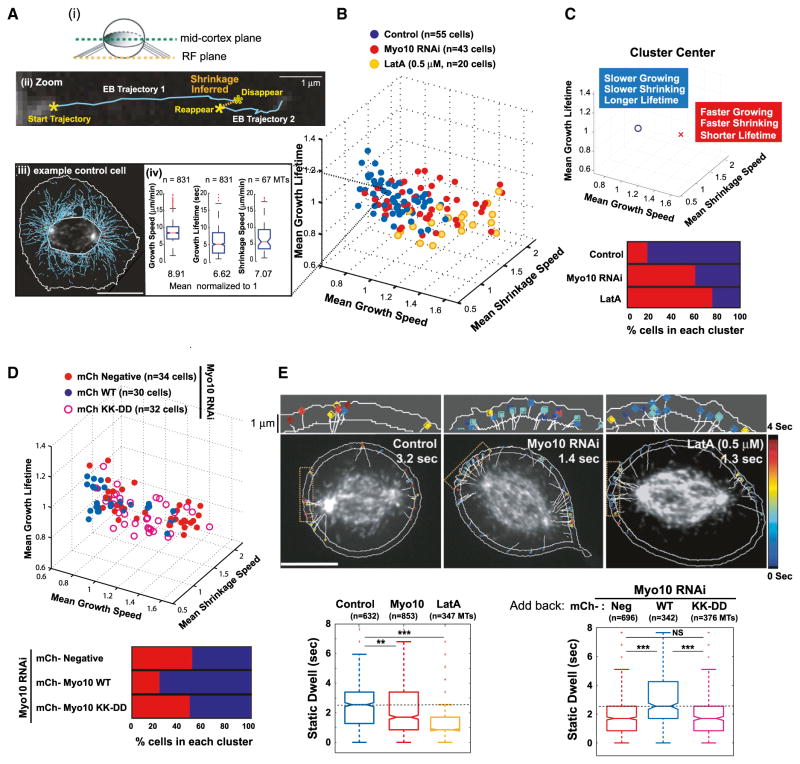
Despite numerous studies on the role of the actin cytoskeleton in spindle orientation (Kunda and Baum, 2009; Sandquist et al., 2011), how the actin cytoskeleton affects astral microtubule dynamics during mitosis has not been systematically examined. To determine the effect of actin disassembly or Myo10 knockdown on microtubule dynamics, we used spinning disc confocal microscopy of cells expressing GFP-EB3, a microtubule plus end binding protein that forms comets on growing microtubules (Figures 6 and S6). Comet trajectories were recorded using the plusTipTracker software (Applegate et al., 2011; Matov et al., 2010), enabling unbiased analyses of tens of thousands of microtubules for a given condition. The method yields direct measurements of microtubule growth speeds and growth lifetimes and includes an algorithm to infer shrinkage events from spatially collinear growth events (Figure 6A and Movie S4). To achieve high spatio-temporal resolution, we employed single focal plane imaging at the mid-cortex in Z near the spindle poles, where actin clouds accumulate. Because Myo10 is essential for mitotic centrosome positioning in multiple cell lines (Figures 1C–1E, S2A–S2C, 3, 4E–4G, and 5), we selected RPE-1 cells, whose flat morphology enabled robust automated analysis of microtubule dynamics.
Figure 6. Actin or Myo10 Suppresses Microtubule Dynamicity near the Cell Cortex prior to Anaphase.
(A–C) Myo10 depletion increases microtubule growth rates, shrinkage rates, and numbers of short-lived microtubules (RPE-1 cells expressing GFP-EB3).
(A) (i) In the schematic, the images were taken from the mid-cortical plane of a rounded mitotic cell. (ii) The zoomed image of a microtubule shrinkage event (orange dotted line) inferred from the linkage of two collinear EB3 comet trajectories (Movie S4). (iii) Representative image of a control cell with EB3 comet trajectories (solid cyan lines). (iv) Distributions of the indicated microtubule parameters extracted from the representative control cell shown in (iii). Data were obtained from cells with a metaphase chromosome configuration as judged by DIC.
(B) 3D plot of microtubule growth speed, shrinkage speed, and growth lifetime obtained from control cells (blue), cells depleted of Myo10 (red), or cells treated with LatA (yellow). Each data point represents the average microtubule parameters quantified from the entire astral microtubule population per cell from five experiments. The per cell values for each parameter were normalized by the mean value of the per cell control distribution for a given experimental day. The mean control values were normalized to 1 from measured values of 9.6 ± 1.0 μm/min (growth speed), 6.8 ± 1.0 μm/min (shrinkage speed), and 6.1 ± 0.5 s (growth lifetime).
(C) Both actin disruption and Myo10 depletion result in increased microtubule dynamicity. The top graph shows that K-means clustering of data shown in (B) distinguishes two separate populations of cells that possess different characteristic microtubule dynamics (see Supplemental Information for details). Each cell belongs to a respective cluster, “O” (less dynamic) or “X” (highly dynamic). The bottom graph shows the percentage of cells possessing highly dynamic (red, cluster center X) or less dynamic (blue, cluster center O) microtubules after the indicated treatment from (B).
(D) Requirement of Myo10 microtubule-binding to regulate astral microtubule dynamics. The top graph shows a 3D plot of indicated microtubule parameters obtained from Myo10-depleted cells expressing mCherry (mCh)-tagged wild-type Myo10 or the KK-DD mutant. The mCh-negative Myo10-depleted cells (closed red circles) serve as the negative control. The mean wild-type Myo10 rescue values were normalized to 1. The bottom graph shows the data presented as in Figure 6C (two experiments).
(E) Myo10-dependent cortical interactions of astral microtubules. The top image shows the automated measurements of static dwell time within the 1 μm cortical band. The microtubule tracks are shown as white lines and the dwell time is presented as colors from a heatmap. The bottom box plots show the static dwell time (sec) of astral microtubules (MTs) in the indicated conditions from five (left) and two (right) experiments (***p < 0.00001, **p < 0.006, and permutation test of the medians). The scale bars represent 10 μm. See also Figure S6.
After actin depolymerization with Latrunculin A (LatA), astral microtubules grew and shrank significantly faster, and had shorter growth lifetimes, than in control cells (Figures 6B and S6A; permutation t test, ***p < 0.0001, and **p < 0.02). This effect was also evident when pooling the data for control and experimental conditions, followed by unbiased segmentation using K-means clustering of populations of cells with highly dynamic (fast-growing/shrinking and short lifetime-high frequency switching), and less dynamic (slow-growing/shrinking with longer lifetime-low frequency switching) microtubule behavior (Figure 6C). The increase in microtubule dynamics was dependent on the dose of LatA (Figure S6B). These findings thus reveal that interactions with the actin cytoskeleton suppress the dynamicity of astral microtubules during mitosis.
Next, we determined the effect of Myo10 depletion on astral microtubule dynamics (Figures 6B, 6C, and S6A). Overall, Myo10 knockdown (red) increased astral microtubule dynamics in a manner that was similar to that of LatA treatment (yellow), although to a slightly lesser extent. By contrast, treatment of cells with blebbistatin (green), a specific myosin II ATPase inhibitor, had little effect on microtubule growth and shrinkage speeds, despite the decrease in microtubule growth lifetime (Figures S6A and S6B). Thus, the distinct effect of Myo10 on microtubule dynamics cannot be simply explained by a general defect in actomyosin contractility.
The effect of Myo10 on astral microtubule dynamics requires microtubule-binding. After Myo10 knockdown, add-back of the Myo10 point mutant (KK-DD, open red circles), which is deficient in microtubule-binding, had an effect on astral microtubule dynamics that was similar to that of the Myo10 knockdown (Figures 6D and S6C). Moreover, Myo10 knockdown predominantly affected astral microtubule dynamics (Figure S6D): Kymograph analysis of spindle microtubules, where single particle tracking of comets is inaccurate, showed no detectable effect of Myo10 knockdown on microtubule growth. These results suggest that Myo10 directly mediates interactions of astral microtubules with the actin network in vivo and modulates their dynamics.
The Effect of Actin and Myo10 on Astral Microtubule Cortical Interactions
Next, we determined whether Myo10 is required for astral microtubule ends to interact with the cell cortex. We developed software to make quantitative measures of cortical-microtubule interactions during mitosis. We analyzed the lifetime of EB comets in a 1 μm wide band from the cell boundary (Figure 6E; see Supplemental Information for details). The microtubule cortical dwell time was measured as the time that the EB3 comet persisted within a 200 nm radius around each microtubule trajectory end point. Actin disruption or Myo10 depletion diminished the cortical dwell time during metaphase (Figure 6E, bottom left). This effect was not due to faster dissociation of EB3 from microtubule plus ends (Figure S6E; see Supplemental Information for details), validating the use of EB3 to visualize microtubule ends and measure cortical dwell time. LatA-treated or Myo10-depleted cells also exhibited significantly higher frequency of contact between astral microtubules and the cell cortex relative to control cells (Figures 6E, upper, and S6F), consistent with the increased microtubule dynamicity in these cells (Figure 6C). Importantly, cells that express the microtubule-binding deficient point mutants (Myo10-KK-DD) displayed similar microtubule-cortical interactions as LatA-treated or Myo10 knockdown cells (Figure 6E, bottom right). Together, these results show that the microtubule-binding activity of Myo10 facilitates interactions between astral microtubules and the cell cortex.
Overlapping Roles of Myo10 and Dynein
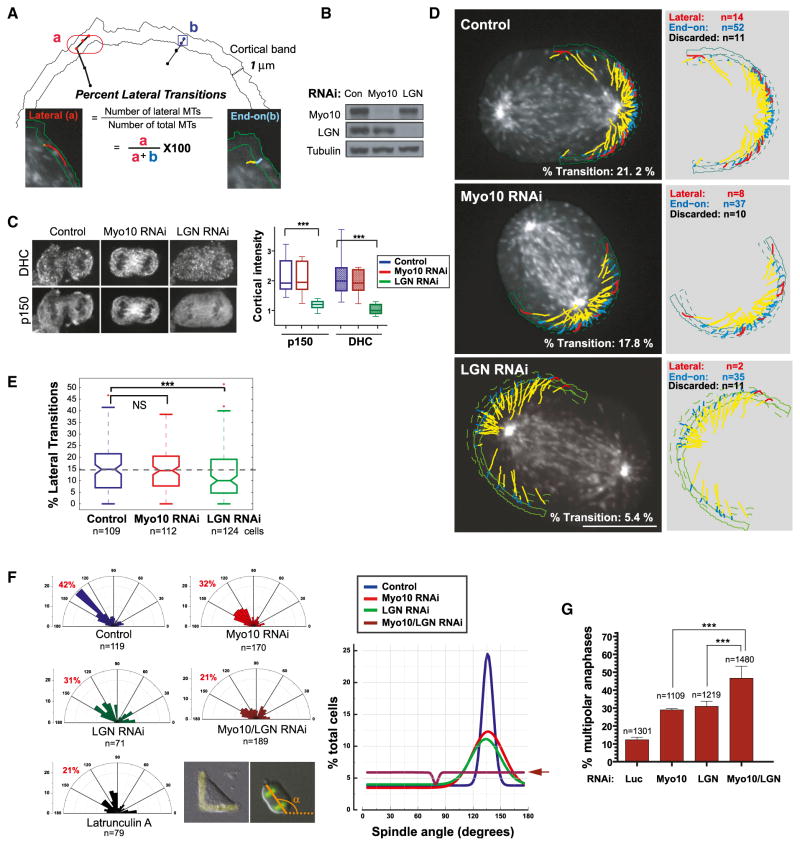
We observed two modes of astral microtubule interactions with the cell cortex that are broadly similar to findings previously reported in budding yeast (Adames and Cooper, 2000; Pearson and Bloom, 2004). Some astral microtubules approached the cell edge directly, before stalling or undergoing catastrophe (hereafter referred to as “end-on microtubules,” Figure 7A, blue), whereas others appeared to slide along the cell edge (hereafter referred to as “lateral sliding microtubules,” red). We developed software that distinguishes between these two types of microtubules (Figures 7A and S7A and Movie S5). A cortical microtubule was defined as undergoing lateral sliding when it grew for >0.7 μm within the 1 μm wide cortical band and maintained a terminal direction of motion roughly parallel to the cell edge (Figure S7A; see Supplemental Information for details). All other cortical microtubules that traveled a significant distance within the cortical band (>0.3 μm) were defined as end-on interacting. Consistent with observations in C. elegans (Gusnowski and Srayko, 2011), we observed few lateral sliding events in pre-anaphase cells, when the spindle pole is relatively far from the cell cortex; however, lateral sliding became relatively frequent post-anaphase, when the poles are in close proximity to the cortex. To directly compare the impact of Myo10 and dynein inhibition on these different modes of cortical-microtubule contact, we examined pre- and post-anaphase cells, in the latter case, our analysis was restricted to cells that had a comparable pole-to-cortex distance (3–4 μm).
Figure 7. Cooperation between Myo10 and Dynein-Mediated Mechanisms.
(A–E) Cortical microtubule (MT) behaviors requiring Myo10 or dynein in RPE-1 cells expressing GFP-EB3.
(A) Schematic classification of lateral (a) and end-on (b) MTs used to determine the percentage of MTs undergoing lateral sliding (% Transition). All of the cortical astral MTs reaching a 1 μm cortical band are classified into lateral (red) or end-on (blue) based on the their displacement and orientation relative to the cell edge (see Figure S7A; Supplemental Information for details).
(B) Western blotting showing depletion of Myo10 and LGN protein by siRNA.
(C) Immunofluorescence images showing decreased cortical dynein and p150 localization after LGN RNAi (left), with corresponding fluorescence quantitation (right, n = 20 line scans from ten cells per condition, ***p < 0.0001, and non-parametric Student’s t test). Note that in RPE-1 cells, LGN knockdown leads to significantly reduced cortical dynein or p150Glued during anaphase, unlike HeLa cells, where it has a more modest effect, due to parallel 4.1G/R-dependent cortical dynein recruitment (Kiyomitsu and Cheeseman, 2013).
(D) Overlay images of lateral (red) and end-on (blue) MTs from GFP-EB3 movies in indicated conditions (Movie S5) (right, without the overlay). The yellow indicates the MT trajectory prior to entry into the cortical band, whereas the blue/red correspond to trajectories within the cortical region classified as in (A). The outlines of the cell edge in the first and last frame (solid and dashed green lines) of the movie are shown. The scale bar represents 10 μm.
(E) Quantitation showing decreased lateral transitions after LGN knockdown (n = >100 spindles poles, each of which had ~2,500–4,000 MTs, per condition from four experiments, permutation t test, and ***p < 0.0003).
(F and G) Additive defects in spindle orientation (F) and centrosome clustering (G) after combinatorial inhibition of Myo10 and cortical dynein.
(F) Imaging of U2OS cells on FN-L patterns and analysis as in Figure 1C. The right graph shows a Gaussian curve that was fitted to the data for each treatment shown in the left panel.
(G) The percentage of multipolar divisions in U2OS cells with extra centrosomes that express GFP-H2B for the indicated RNAi treatments. Approximately 70% of the cells have extra centrosomes after the transient overexpression of Plk4, as confirmed by centrin staining or spindle multipolarity that was induced by HSET RNAi (Kwon et al., 2008) (mean ± SEM). See also Figure S7.
Unlike Myo10 (Figure 6E), the cortical pool of dynein did not affect the cortical dwell time of astral microtubules during metaphase (not significant [NS], permutation test of the medians from four experiments; control cells: n = 34 cells and 354 microtubules; and LGN RNAi: 23 cells and 240 microtubules), consistent with a previous report (Samora et al., 2011). In this experiment, the cortical pool of dynein was inhibited by knockdown of LGN, a cortical recruitment factor for dynein (Figure 7B) (Kiyomitsu and Cheeseman, 2012; Morin and Bellaïche, 2011). In this condition, we also did not detect any decrease in cortical dwell times during anaphase (Figure S7D). However, we did find that cortical dynein was required for anaphase lateral sliding events (Figures 7C–7E and Movie S5), as documented in other systems (Adames and Cooper, 2000; Pearson and Bloom, 2004). The conclusion that dynein is specifically required for promoting lateral sliding is robust to small-scale alterations of the lateral sliding classification parameters in our automated analyses (Figures S7B and S7C). Importantly, this effect cannot be explained by a decrease in the density of cortical EB3 comets (Figure S7E). Unlike dynein, Myo10-depletion did not impair end-on or lateral sliding microtubule attachments during anaphase (Figures 7D, 7E, and S7D and Movie S5). Thus, Myo10 and dynein have distinct effects on microtubule-cortical interactions: Myo10 is required for end-on microtubule attachment prior to anaphase (Figure 6), and dynein is required for lateral sliding of microtubules during anaphase.
To determine if the Myo10 and dynein mechanisms act in parallel, we examined the individual and combined effects of Myo10 and LGN knockdown on spindle orientation (Figure 7F) and on the clustering of centrosomes in cells with centrosome amplification (Figure 7G). Consistent with parallel action, double knockdown of Myo10 and LGN had an additive effect, randomizing spindle orientation, as compared to the individual knockdowns (Figure 7F, right, compare brown line with arrow to red or green lines). An additive effect of Myo10 and LGN knockdown was also observed using the centrosome-clustering assay (Figures 7G and S7F). Finally, Myo10 knockdown did not impair dynein cortical localization, and LGN knockdown similarly did not alter the cortical distribution of Myo10 (Figures 7C, S7G, and S7H). These results define two overlapping pathways for positioning centrosomes during mitosis in human cells: a Myo10-dependent pathway and an LGN/dynein/dynactin-dependent pathway.
DISCUSSION
Requirement of Myosin-10 for Spindle Pole Movement toward Retraction Fibers and Subcortical Actin Clouds
Here, we document that the microtubule-binding myosin Myo10 is required for actin-dependent forces on the centrosome in human cells during mitosis. Our data suggest that Myo10 is the major actin-based force generator that pulls astral microtubules toward retraction fibers and subcortical actin clouds. First, Myo10 concentrates in the cortical region around these actin structures. Second, Myo10 is functionally important to position spindle poles relative to these structures. The fact that Myo10 is not required for the assembly or dynamics of retraction fibers and actin clouds suggests that Myo10 is specifically required for the generation of pulling forces toward these structures. Because retraction fibers control the polarization of actin clouds, we propose that these structures form a continuous actin network, through which Myo10 motors orient astral microtubules. Indeed, Myo10 is a processive motor capable of walking on individual actin filaments and on actin bundles, that also has the capacity to switch between filament tracks (Kerber et al., 2009; Nagy et al., 2008; Nagy and Rock, 2010; Sun et al., 2010). Thus, Myo10 is well suited for transporting microtubules through variously shaped actin structures.
We can envision several ways in which the Myo10 biochemical activities may be harnessed to position centrosomes near retraction fibers. The simplest model would be direct transport of astral microtubules through the subcortical actin clouds toward the base of the retraction fibers, driven by the interaction of Myo10 with the microtubule lattice. This model requires that the subcortical actin clouds have a net polarity, which has not been tested. This is, however, plausible, as the actin clouds are assembled in a manner that requires the Arp2/3 complex (Mitsushima et al., 2010), in common with polarized actin in lamellipodia (Pollard and Borisy, 2003). In sea urchin eggs, cytoplasmic pulling forces on astral microtubules increase with increasing microtubule length (Minc et al., 2011). By binding to the microtubule lattice, in principal, the numbers of bound Myo10 molecules could be proportional to microtubule length, thus generating length-dependent pulling forces on centrosomes. Finally, Myo10 may facilitate the action of dynein or other, unknown, force generators, by anchoring microtubules to the actin structures associated with retraction fibers.
The Requirement for Microtubule Binding by Myo10
Our data demonstrate that the mitotic function of Myo10 in centrosome positioning depends, almost completely, on its ability to bind the microtubule lattice. Several studies suggest that Myo10 links actin and microtubules, based on deletions that encompass the MyTH4-FERM domain (Weber et al., 2004; Woolner et al., 2008). However, this prior work could not distinguish compromised microtubule-binding from the loss of other cargo that binds the FERM domain. Here, we make this distinction by utilizing the Myo10 point mutations that specifically disrupt microtubule-binding without altering other known cargo binding (Figure 3) (Hirano et al., 2011; Wei et al., 2011).
We also considered the possibility that Myo10 interacts indirectly with microtubules through EB-family microtubule plus end-binding proteins. Drosophila myosin XV, a related MyTH4-FERM domain myosin, binds to EB1 through its MyTH4-FERM domain and other microtubule regulators through its FERM domain (Liu et al., 2008). We also noticed that many Myo10 homologs contain several potential EB1 binding, SxIP-like motifs (Honnappa et al., 2009) within the MyTH4 domain (Figure S3A). However, a construct containing point mutations in all of the Myo10 SxIP motifs nonetheless rescued the centrosome-positioning defect in Myo10 knockdown cells to near completion (~18% as compared with 15% multipolar spindles for the controls, as in Figure S3D). Although we do not exclude a contribution from the EB protein binding to Myo10, our analysis indicates that microtubule lattice-binding is the primary mechanism underlying Myo10-dependent centrosome positioning.
Actin and Myo10 Regulate Dynamics and Cortical Attachment of Microtubules during Mitosis
Using computational image analysis, we show that actin and Myo10 decrease microtubule dynamicity and promote end-on cortical-microtubule interactions prior to anaphase (Figures 6 and S6). The similarity in the effect of LatA treatment and Myo10 knockdown, together with the fact that Myo10 does not detectably alter subcortical actin structures, indicates that Myo10 is an important mediator of actin’s effect on microtubule dynamics. The abrogation of this effect by point mutations in the Myo10 microtubule-binding domain (Figures 6D and S6C) strongly suggests a direct mechanism.
The analytical tools that we developed have enabled us to acquire the first in depth, large-scale analysis of cortical microtubule interactions in human cells (Figures 6, 7A–7E, and S7) and to uncover distinct, but overlapping, roles for Myo10 and dynein. Prior to anaphase, Myo10 plays an important role in mediating end-on microtubule cortical interactions (Figure 6E). Once cells enter anaphase, Myo10 becomes less important, with the microtubule motor dynein playing the dominant role in promoting lateral sliding of microtubules along the cortex (Figures 7D, 7E, and S7D). This is consistent with dynein’s role in spindle positioning in other systems (Adames and Cooper, 2000; Pearson and Bloom, 2004). The absence of a dynein effect on lateral sliding during metaphase may be explained by the known antagonism of dynein by MAP4 during this stage of mitosis (Samora et al., 2011).
Together, our data demonstrate that Myo10 and dynein are the major motors that position centrosomes and spindles in somatic mammalian cells and that they largely act in parallel. This conclusion is supported by the following evidence. First, Myo10’s role in spindle positioning requires its direct microtubule-binding capacity (Figures 3, S3, 6D, 6E, and S6C). Second, the localization of LGN or dynein is independent of Myo10 and vice versa (Figures 7C, S7G, and S7H). Third, Myo10 and dynein inhibition have distinct effects on microtubule dynamics and microtubule-cortical interactions (Figures 6, S6, 7D, 7E, and S7D). Finally, combined inhibition of Myo10 and dynein results in an additive defect in centrosome positioning (Figures 7F and 7G).
The mechanisms that underlie spindle orientation in human cells, as defined in the present study, have interesting parallels to spindle positioning mechanisms previously described in budding yeast (Adames and Cooper, 2000; Pearson and Bloom, 2004). The Bim1-Kar9-myosin V mechanism in yeast promotes end-on cortical-microtubule attachment, whereas the dynein mechanism promotes lateral sliding. Similar to what we now demonstrate for Myo10 and dynein in mammalian cells, the actin-dependent mechanism in yeast is dominant prior to anaphase, whereas dynein dominates during anaphase. Myo10, therefore, appears to have amalgamated functions for centrosome positioning accomplished by Bim1, Kar9, and myosin V in budding yeast. Thus, evolutionarily divergent cell types have reached convergent solutions to integrate the actin and microtubule cytoskeletons for centrosome and spindle positioning.
Unlike yeast, spindle positioning in animal cells is heavily influenced by the tissue microenvironment through cell matrix attachments and the position of retraction fibers (McCaffrey and Macara, 2011; Morin and Bellaïche, 2011; Théry and Bornens, 2006). Abnormalities in cell-matrix adhesion and tissue rigidity are commonly observed in disease states such as cancer (McCaffrey and Macara, 2011; Pease and Tirnauer, 2011). The current work defines the major motors required for centrosome and spindle positioning in human cells and provides a foundation for future work on this process in the context of development or disease.
EXPERIMENTAL PROCEDURES
Detailed descriptions of cell lines, culture conditions, plasmid construction, immunofluorescence microscopy, live-cell imaging, quantitation, automated image analysis, statistical analysis of the data, and antibodies used in this study can be found in the Supplemental Information.
Quantitation of Paxillin
Cells plated on FN-H patterns for 6 hr were fixed and stained for paxillin, phalloidin actin, and DNA. For measurements of focal adhesion area and total fluorescence intensity, paxillin images were subjected to background subtraction by the same image after Gaussian blur and discrete paxillin-positive structures were obtained across all conditions. There were two rectangular regions (75 × 255 pixels) along the adhesive surface of a FN-H pattern that were used for paxillin quantitation: images were applied for threshold and the subsequent objects selected were quantified using the “analyze particles” function in Image J software. Data presentation for histograms was done using Origin (Origin Lab).
Cell Division on Patterned Substrates
Cells were trypsinized, seeded on CYTOO chips, and assembled to the CYTOO chamber according to the manufacturer’s instructions (CYTOO) 6–8 hr prior to fixation or live imaging. In some experiments, 20 μM of MG132 (Calbiochem) was used to enrich mitotic cells before fixation. For live imaging, the cell division plane was determined by the long axis of elongating anaphase cells at the time frame with the first appearance of a cleavage furrow and was measured using NIS elements software (Nikon Instruments). The angular distribution of spindle orientation was displayed using Origin (OriginLab).
Quantitation of Mitotic Cell Rounding
RPE-1 cells expressing RFP-H2B were plated on FN-L micropatterns and imaged at 10 min intervals by differential interference contrast (DIC) and fluorescence imaging. For the kinetics of mitotic cell rounding and post-mitotic cell spreading (re-adhesion), cell surface areas were measured using NIS elements software (Nikon Instruments) at the indicated time points.
Quantitation of Subcortical Actin Clouds and Spindle Pole Movements
HeLa cells expressing GFP-Utr-CH and mCherry-tubulin were imaged at a single confocal plane where both poles are visible (mid-cortical plane) at 15 s intervals for 15 min using a 60× objective (see Supplemental Information for details). To quantify subcortical actin clouds and spindle pole movements, kymographs were generated from a 70 pixel-width line along the major axis of the spindle. Cells in metaphase, as judged by chromosome configuration from DIC images, were analyzed. The penetration depth of the actin clouds into cytoplasm and their integrated fluorescence intensity were measured, as described (Fink et al., 2011). To quantify spindle pole movements relative to the actin cloud position (Figure S5A), the time intervals corresponding to each peak of GFP-Utr-CH were determined by line scans along the y axis of the kymographs. Pole velocity was obtained by quantifying the average slope of spindle pole position (both poles, determined by mCherry-tubulin signal) at each time interval (relative to the peak GFP-Utr-CH signal) using MetaMorph software.
Statistical Methods
To determine the significance of differences between conditions, a Student’s t test (GraphPad Software) was used, unless otherwise specified. For Figure 2E, a one-way ANOVA and Tukey’s multiple comparison tests were used to compare mean differences for all pairs. For all microtubule dynamics measurements, a permutation test was used to calculate statistical significance of changes in either the mean (when calculated per cell) or median (when pooled for multiple cells) of the distribution between conditions (see Supplemental Information for details).
Automated EB3 Comet Detection, Tracking, and Analysis of Astral EB3 Trajectories
The EB3 fluorescent comets were detected and tracked using a modified version of the plusTipTracker package (Applegate et al., 2011; Matov et al., 2010). Modifications were made to the watershed detection scheme in order to accommodate the large variance in fluorescence intensities observed in EB3-GFP mitotic cells and new methods for post-processing of the resulting EB3 trajectories were developed. Previously unpublished analytic schemes include the K-means clustering analysis, the cortical dwell time analysis, and the automated end-on versus lateral classifications. A discussion of the detection modifications, a listing of all detection/tracking parameters employed, and details regarding all newly developed EB3 trajectory analyses are provided in the Supplemental Information. All EB3 comet analyses were implemented in MATLAB (MathWorks).
Supplementary Material
Highlights.
Myo10 orients centrosomes toward retraction fibers and subcortical actin clouds
Spindle positioning by Myo10 requires its direct microtubule binding
Myo10 and cortical dynein have distinct effects on cortical-microtubule interactions
Myo10 and cortical dynein overlap to promote spindle positioning in mammalian cells
Acknowledgments
We thank M. Thery, B. Akins, H. Arellano-Santoyo, S. Jhaveri-Schneider, and N. Umbreit for comments on the manuscript; L. Ding for sharing computer code; the Nikon Imaging Center at Harvard Medical School and H. Elliott from the Image and Data Analysis Core at Harvard Medical School for their help with kymograph analysis of microtubule dynamics; and R. Cheney, W. Krek, E. Nigg, I. Cheeseman, T. Wittmann, and W. Bement for reagents. D.P. was supported by the Howard Hughes Medical Institute and the NIH (GM061345); M.K. was supported by a special fellow award from the Leukemia and Lymphoma Society and a Susan Komen grant; and M.B. and G.D. were funded by the NIH (GM067230).
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and five movies and can be found with this article online at http://dx.doi.org/10.1016/j.devcel.2015.06.013.
References
- Adames NR, Cooper JA. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J Cell Biol. 2000;149:863–874. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applegate KT, Besson S, Matov A, Bagonis MH, Jaqaman K, Danuser G. plusTipTracker: Quantitative image analysis software for the measurement of microtubule dynamics. J Struct Biol. 2011;176:168–184. doi: 10.1016/j.jsb.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Martin SG. Shaping fission yeast with microtubules. Cold Spring Harb Perspect Biol. 2009;1:a001347. doi: 10.1101/cshperspect.a001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezratty EJ, Partridge MA, Gundersen GG. Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat Cell Biol. 2005;7:581–590. doi: 10.1038/ncb1262. [DOI] [PubMed] [Google Scholar]
- Fink J, Carpi N, Betz T, Bétard A, Chebah M, Azioune A, Bornens M, Sykes C, Fetler L, Cuvelier D, Piel M. External forces control mitotic spindle positioning. Nat Cell Biol. 2011;13:771–778. doi: 10.1038/ncb2269. [DOI] [PubMed] [Google Scholar]
- Goshima G, Scholey JM. Control of mitotic spindle length. Annu Rev Cell Dev Biol. 2010;26:21–57. doi: 10.1146/annurev-cellbio-100109-104006. [DOI] [PubMed] [Google Scholar]
- Gusnowski EM, Srayko M. Visualization of dynein-dependent microtubule gliding at the cell cortex: implications for spindle positioning. J Cell Biol. 2011;194:377–386. doi: 10.1083/jcb.201103128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano Y, Hatano T, Takahashi A, Toriyama M, Inagaki N, Hakoshima T. Structural basis of cargo recognition by the myosin-X MyTH4-FERM domain. EMBO J. 2011;30:2734–2747. doi: 10.1038/emboj.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa S, Gouveia SM, Weisbrich A, Damberger FF, Bhavesh NS, Jawhari H, Grigoriev I, van Rijssel FJ, Buey RM, Lawera A, et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- Kerber ML, Cheney RE. Myosin-X: a MyTH-FERM myosin at the tips of filopodia. J Cell Sci. 2011;124:3733–3741. doi: 10.1242/jcs.023549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerber ML, Jacobs DT, Campagnola L, Dunn BD, Yin T, Sousa AD, Quintero OA, Cheney RE. A novel form of motility in filopodia revealed by imaging myosin-X at the single-molecule level. Curr Biol. 2009;19:967–973. doi: 10.1016/j.cub.2009.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T, Cheeseman IM. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat Cell Biol. 2012;14:311–317. doi: 10.1038/ncb2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T, Cheeseman IM. Cortical dynein and asymmetric membrane elongation coordinately position the spindle in anaphase. Cell. 2013;154:391–402. doi: 10.1016/j.cell.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Kunda P, Baum B. The actin cytoskeleton in spindle assembly and positioning. Trends Cell Biol. 2009;19:174–179. doi: 10.1016/j.tcb.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, Thery M, Pellman D. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–2203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan L, Husson J, Munteanu EL, Kerssemakers JW, Dogterom M. Force-generation and dynamic instability of microtubule bundles. Proc Natl Acad Sci USA. 2008;105:8920–8925. doi: 10.1073/pnas.0710311105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laan L, Pavin N, Husson J, Romet-Lemonne G, van Duijn M, López MP, Vale RD, Jülicher F, Reck-Peterson SL, Dogterom M. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell. 2012;148:502–514. doi: 10.1016/j.cell.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Woolner S, Johndrow JE, Metzger D, Flores A, Parkhurst SM. Sisyphus, the Drosophila myosin XV homolog, traffics within filopodia transporting key sensory and adhesion cargos. Development. 2008;135:53–63. doi: 10.1242/dev.011437. [DOI] [PubMed] [Google Scholar]
- Liu KC, Jacobs DT, Dunn BD, Fanning AS, Cheney RE. Myosin-X functions in polarized epithelial cells. Mol Biol Cell. 2012;23:1675–1687. doi: 10.1091/mbc.E11-04-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matov A, Applegate K, Kumar P, Thoma C, Krek W, Danuser G, Wittmann T. Analysis of microtubule dynamic instability using a plus-end growth marker. Nat Methods. 2010;7:761–768. doi: 10.1038/nmeth.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey LM, Macara IG. Epithelial organization, cell polarity and tumorigenesis. Trends Cell Biol. 2011;21:727–735. doi: 10.1016/j.tcb.2011.06.005. [DOI] [PubMed] [Google Scholar]
- McNally FJ. Mechanisms of spindle positioning. J Cell Biol. 2013;200:131–140. doi: 10.1083/jcb.201210007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minc N, Burgess D, Chang F. Influence of cell geometry on division-plane positioning. Cell. 2011;144:414–426. doi: 10.1016/j.cell.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ. Actin based motility on retraction fibers in mitotic PtK2 cells. Cell Motil Cytoskeleton. 1992;22:135–151. doi: 10.1002/cm.970220207. [DOI] [PubMed] [Google Scholar]
- Mitsushima M, Aoki K, Ebisuya M, Matsumura S, Yamamoto T, Matsuda M, Toyoshima F, Nishida E. Revolving movement of a dynamic cluster of actin filaments during mitosis. J Cell Biol. 2010;191:453–462. doi: 10.1083/jcb.201007136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin X, Bellaïche Y. Mitotic spindle orientation in asymmetric and symmetric cell divisions during animal development. Dev Cell. 2011;21:102–119. doi: 10.1016/j.devcel.2011.06.012. [DOI] [PubMed] [Google Scholar]
- Nagy S, Rock RS. Structured post-IQ domain governs selectivity of myosin X for fascin-actin bundles. J Biol Chem. 2010;285:26608–26617. doi: 10.1074/jbc.M110.104661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy S, Ricca BL, Norstrom MF, Courson DS, Brawley CM, Smithback PA, Rock RS. A myosin motor that selects bundled actin for motility. Proc Natl Acad Sci USA. 2008;105:9616–9620. doi: 10.1073/pnas.0802592105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Bloom K. Dynamic microtubules lead the way for spindle positioning. Nat Rev Mol Cell Biol. 2004;5:481–492. doi: 10.1038/nrm1402. [DOI] [PubMed] [Google Scholar]
- Pease JC, Tirnauer JS. Mitotic spindle misorientation in cancer—out of alignment and into the fire. J Cell Sci. 2011;124:1007–1016. doi: 10.1242/jcs.081406. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Redemann S, Pecreaux J, Goehring NW, Khairy K, Stelzer EH, Hyman AA, Howard J. Membrane invaginations reveal cortical sites that pull on mitotic spindles in one-cell C. elegans embryos. PLoS ONE. 2010;5:e12301. doi: 10.1371/journal.pone.0012301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov VI, Borisy GG. Self-centering in cytoplasmic fragments of melanophores. Mol Biol Cell. 1998;9:1613–1615. doi: 10.1091/mbc.9.7.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt J, Cramer LP, Baum B, McGee KM. Myosin II-dependent cortical movement is required for centrosome separation and positioning during mitotic spindle assembly. Cell. 2004;117:361–372. doi: 10.1016/s0092-8674(04)00341-1. [DOI] [PubMed] [Google Scholar]
- Samora CP, Mogessie B, Conway L, Ross JL, Straube A, McAinsh AD. MAP4 and CLASP1 operate as a safety mechanism to maintain a stable spindle position in mitosis. Nat Cell Biol. 2011;13:1040–1050. doi: 10.1038/ncb2297. [DOI] [PubMed] [Google Scholar]
- Sandquist JC, Kita AM, Bement WM. And the dead shall rise: actin and myosin return to the spindle. Dev Cell. 2011;21:410–419. doi: 10.1016/j.devcel.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat Cell Biol. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- Sun Y, Sato O, Ruhnow F, Arsenault ME, Ikebe M, Goldman YE. Single-molecule stepping and structural dynamics of myosin X. Nat Struct Mol Biol. 2010;17:485–491. doi: 10.1038/nsmb.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry M, Bornens M. Cell shape and cell division. Curr Opin Cell Biol. 2006;18:648–657. doi: 10.1016/j.ceb.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Théry M, Jiménez-Dalmaroni A, Racine V, Bornens M, Jülicher F. Experimental and theoretical study of mitotic spindle orientation. Nature. 2007;447:493–496. doi: 10.1038/nature05786. [DOI] [PubMed] [Google Scholar]
- Toyoshima F, Nishida E. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 2007;26:1487–1498. doi: 10.1038/sj.emboj.7601599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber KL, Sokac AM, Berg JS, Cheney RE, Bement WM. A microtubule-binding myosin required for nuclear anchoring and spindle assembly. Nature. 2004;431:325–329. doi: 10.1038/nature02834. [DOI] [PubMed] [Google Scholar]
- Wei Z, Yan J, Lu Q, Pan L, Zhang M. Cargo recognition mechanism of myosin X revealed by the structure of its tail MyTH4-FERM tandem in complex with the DCC P3 domain. Proc Natl Acad Sci USA. 2011;108:3572–3577. doi: 10.1073/pnas.1016567108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolner S, Bement WM. Unconventional myosins acting unconventionally. Trends Cell Biol. 2009;19:245–252. doi: 10.1016/j.tcb.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolner S, O’Brien LL, Wiese C, Bement WM. Myosin-10 and actin filaments are essential for mitotic spindle function. J Cell Biol. 2008;182:77–88. doi: 10.1083/jcb.200804062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Pan L, Wei Z, Zhang M. Structure of MyTH4-FERM domains in myosin VIIa tail bound to cargo. Science. 2011;331:757–760. doi: 10.1126/science.1198848. [DOI] [PubMed] [Google Scholar]
- Yi J, Wu X, Chung AH, Chen JK, Kapoor TM, Hammer JA. Centrosome repositioning in T cells is biphasic and driven by microtubule end-on capture-shrinkage. J Cell Biol. 2013;202:779–792. doi: 10.1083/jcb.201301004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Berg JS, Li Z, Wang Y, Lång P, Sousa AD, Bhaskar A, Cheney RE, Strömblad S. Myosin-X provides a motor-based link between integrins and the cytoskeleton. Nat Cell Biol. 2004;6:523–531. doi: 10.1038/ncb1136. [DOI] [PubMed] [Google Scholar]
- Zheng Z, Wan Q, Liu J, Zhu H, Chu X, Du Q. Evidence for dynein and astral microtubule-mediated cortical release and transport of Gαi/LGN/NuMA complex in mitotic cells. Mol Biol Cell. 2013;24:901–913. doi: 10.1091/mbc.E12-06-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.