Abstract
Global expression analyses demonstrate that alterations in microRNA (miRNA) levels correlate with a variety of metabolic diseases. miRNAs regulate central metabolic pathways and thus play vital roles in maintaining organismal energy balance and metabolic homeostasis. Here, we highlight novel sequencing technologies used to comprehensively define the target spectrum of miRNAs in metabolic disease that complement recent literature reporting physiologic roles for miRNAs in the regulation of glucose and lipid metabolism in peripheral tissues of animal models of metabolic dysfunction. These emerging technologies help decipher the complexity of the miRNA interactome and enrich our understanding of how miRNAs mediate physiologic effects by targeting a spectrum of gene transcripts simultaneously. miRNA-based therapeutics emerge as a viable strategy for treating metabolic diseases.
Keywords: insulin, microRNA, lipid metabolism, adipose tissue, sequencing
Introduction
Control of metabolic homeostasis in higher organisms is maintained by interwoven regulatory networks that sense and respond to environmental and physiologic stimuli. Multicellular organisms have evolved precise strategies to activate or repress metabolic pathways to rapidly satisfy the energetic needs of cells, tissues, and organs. A significant body of work suggests that precise and rapid modulation of target gene transcripts is a critical component in maintaining metabolic homeostasis.
miRNAs play a key role in metabolic homeostasis by governing the response to alterations in nutrients, or fluctuations in various metabolic substrates that include cholesterol, lipids, glucose, and insulin [1–5]. miR NAs are short (21–23 nucleotides) noncoding RNA molecules that primarily function to prevent mRNA translation or initiate mRNA degradation [6]. miRNA targeting is largely defined by the miRNA’s seed region consisting of nucleotides 2–8 of the miRNA [7–10], which undergoes complementary base-pairing with specific sequences in target mRNAs. This miRNA seed (see Glossary) is responsible for guiding the effector proteins that comprise the RNA-induced silencing complex (RISC) to the 3′ untranslated region (UTR) of target mRNAs (Box 1). Target prediction algorithms based on Watson-Crick base pairing and species conservation (e.g. TargetScan [11], PicTar [12], miRanda [13], miRWalk [14]) suggest that miRNAs have hundreds of mRNA targets, and miRNA target spectrums frequently overlap [10,15]. Conversely, mRNAs are predicted to be targets of many distinct miRNAs. In addition, single miRNAs can have multiple target sites in the 3′UTRs of a particular mRNA, likely increasing mRNA repression. Thus, miRNAs may rival transcriptional mechanisms in terms of regulatory output potential. Indeed, a few hundred conserved miRNAs are estimated to regulate ~30–80% of human genes [16]. However, miRNAs normally exert relatively modest effects on individual mRNA targets, acting as rheostats to finely regulate protein expression [6]. Given the large number of miRNAs, their broad range of targets, and the consequences of global miRNA deletion (Box 2), it is no surprise that miRNAs regulate energy metabolism, glucose homeostasis, and whole body insulin sensitivity.
Box 1. The AGO-RISC Complex.
Like other genes transcribed by RNA Polymerase II, miRNA expression is transcriptionally regulated by conventional transcription factors. A freshly transcribed miRNA is referred to as a pri-miRNA. Following transcription, the pri-miRNA is cleaved in the nucleus by the Drosha/DGCR8 microprocessor complex to form a pre-miRNA. The pre-miRNA is then exported to the cytoplasm by exportin-5 and is cleaved in the cytoplasm by DICER to form the mature miRNA duplex, which is subsequently loaded onto the Argonaute protein. The duplex strand with the less stable 5′end [73] is preferentially loaded, accounting for characteristic differences in total expression (non-dominant arms are frequently annotated as the star (*) strand). While one arm is typically dominant, recent efforts provide evidence that both strands are biologically active and arm dominance may be tissue specific. miRNA strands are more specifically termed 3p and 5p strands based on chromosomal location, and this nomenclature may be preferable due to its increased specificity and non-biased terminology. Potent biological activity of a * miRNA is demonstrated by miR-378-3p (miR-378*) in this review [64].
The AGO-miRNA complex forms the core of the miRNA induced silencing complex (RISC). If a miRNA maintains perfect complementarity with its target, RISC mediates catalytic cleavage of the target mRNA and subsequent transcript decay [74]. Perfect miRNA complementarity is uncommon in mammalian systems and, in mammals, only AGO2 contains an active nuclease [75].
Most mRNA targeting by RISC requires recruitment of the larger RISC complex. The other required component of this complex is the GW182 family of proteins. The GW182 protein family acts as a scaffold that bridges AGO binding with effector components of RISC [74]. These effector components include poly-A binding protein, which may stabilize miRNA-RISC binding [76], and the PAN2/3 and CCR4-NOT deadenylase complexes [74]. Canonically, these interactions can interfere with mRNA translation by preventing mRNA circularization, followed by deadenylation and shortening of polyA tails [77] which leads to decapping by DCP1/2 and subsequent mRNA decay.
Recent research has focused on functional regulation of RISC components as a means to globally regulate the miRNA pool. For example, oncogenic stress drives AGO phosphorylation to inhibit miRNA biogenesis by disrupting Dicer-AGO protein-protein interactions [78]. Other findings include Ser387 phosphorylation of AGO increasing its affinity for GW182 [30,79] and the discovery that GW182 protein degradation is modified through ubiquitination [31]. These potential forms of RISC complex regulation may be manifest in the ability of different tissues to form highly variable levels of the RISC complex [29], suggesting the quantity of miRNA activity in a cell may be just as important a contributor to miRNA function as the miRNA expression.
Box 2. Genetic disruption of miRNA processing in mice.
The miRNA processing factors Dicer, Dgcr8, Drosha, and Ago2 are essential for viability in mice [80–82]. KO mice individually lacking these central miRNA-processing genes die during early gestation with severe developmental defects. Nematodes have been useful in this regard as Dicer mutations confer reduced life span and stress tolerance [83]. To study the tissue-specific phenotypes of mature miRNA loss, Cre-inducible conditional KO mice lacking Dicer and Dgcr8 have been generated [66,82–87]. Studies in mice with conditional Dicer KO in adipose tissue [66,83,87] and pancreas [85,86] have shown that miRNAs are essential for maintaining normal glucose and lipid homeostasis.
The endoribonuclease Dicer is required to generate the majority of mature miRNAs [88]. The miRNA pool is largely inactive if Dicer is not present, and Dicer knockout (KO) mice die from severe developmental defects [80]. Deletion of Dicer in adipocytes, and perhaps other cells, using ap2-Cre transgenic mice display juvenile lethality and depletion of WAT [87]. Dicer KO in mature adipocytes using an Adipoq-Cre transgene shows profound lipodystrophy and severe insulin resistance [66]. Supporting a role for miRNAs in glucose homeostasis, the KO of miRNA processing in the adult pancreas established the requirement for miRNAs in the maintenance of glucose homeostasis and beta cell mass [86]. Although these KO studies clearly demonstrated the importance of miRNAs on specific developmental and metabolic processes, it is not clear which miRNAs are responsible for the observed phenotypes.
Dysregulation of specific miRNAs in response to genetic or environmental factors can contribute to aberrant gene expression patterns underlying metabolic dysfunction and cancer [15]. As discussed below, miRNAs often affect multiple targets in linear signaling pathways or interconnected nodes in regulatory networks, exerting a significant cumulative effect. In this review, we discuss the use of high throughput sequencing technologies, specifically Argonaute crosslinking immunoprecipitation coupled to high throughput sequencing (AGO-CLIP-seq) [17,18], to elucidate miRNA targeting and the role of miRNAs in controlling metabolic gene networks. We also discuss recent studies demonstrating the capacity of miRNAs to alter measures of whole body metabolism, including effects on glucose and lipid metabolism. Collectively, the existing data suggest that miRNA-based therapeutics are a novel strategy for treating metabolic diseases.
High throughput approaches delineate the spectrum of miRNA targets
Comprehensive understanding of miRNA function requires identification of the entire spectrum of a given miRNA’s targets. Traditional prediction methods utilize recognition of a specific miRNA seed complement on the 3′UTR of an mRNA target, evolutionary conservation of these complements, and the potential free energy of interaction between the miRNA seed-complement pair [8,10,11,19].
Methods that combine UV crosslinking and immunoprecipitation (CLIP) of RNA binding proteins (RBPs) with high throughput sequencing (Seq) allow broad mapping of interactions between RBPs and their RNA target sites [20]. In contrast to RNA immunoprecipitation coupled to high throughput sequencing (RIP-Seq), CLIP-seq uses ultraviolet light to reversibly crosslink RBPs to nearby RNAs. Recently, CLIP methods have been adapted to identify miRNA-mRNA interactions in a genome-wide manner through purification of Argonaute (AGO)-associated RNAs, which includes miRNAs bound to their respective gene targets [17,18]. In particular, immunoprecipitation of the Argonaute protein coupled to high throughput sequencing (AGO-CLIP-seq) defines points of AGO-mRNA interactions, which allows definition of underlying miRNA seed complements. AGO Photoactivatable-Ribonucleoside-Enhanced CLIP (AGO-PAR-CLIP) [18] includes an added step where nucleotide analogues such as 4-thiouridine are introduced before crosslinking. These nucleotide analogues, when crosslinked, undergo T→C transitions during the reverse-transcription step of the AGO-CLIP experiment [18], allowing precise identification of the point of RNA–protein interaction. After sequencing, AGO-CLIP sequence reads are mapped onto the genome and segregated into read “clusters”. Read clusters are then statistically analyzed using a number of available methods to define their statistical likelihood of existing above background. Clusters above threshold values are designated as true bound targets of AGO. In AGO-PAR-CLIP the presence of T→C transitions in a read cluster additionally increases the statistical probability of true positives by defining the point of physical linkage between the AGO protein and bound transcript.
Global analyses of miRNA-mRNA interactions by AGO-CLIP-seq have revealed frequent miRNA targeting of genomic regions other than the 3′UTR, which may represent >30% of all miRNA binding sites. Because miRNA target prediction algorithms rely on binding site conservation across species, miRNA interactions within coding regions or 5′-UTRs are rarely predicted because the higher conservation of these regions obscures the conservation signature of miRNA target sites. Therefore, AGO-CLIP-seq methods are the only method currently available to perform comprehensive, unbiased, and tissue-specific mapping of miRNA target spectra.
Intronic and Polycistronic miRNAs
miRNA transcriptional regulation is frequently physically coupled to other genes or miRNAs [21]. While single, intergenic miRNAs under control of their own promoter are common, many miRNAs are linked to other transcripts. One-third to one-half of mammalian miRNAs may be located within the introns of coding or noncoding host genes [22]. Most of these intronic miRNAs are transcriptionally linked to their host gene, though up to one third of intronic miRNAs may have their own promoter [22]. Canonical intronic miRNAs are located in large introns and are cleaved by Drosha/DGCR8 (Box 1) during transcription and before splicing. In contrast, non-canonical ‘miRtrons’, derive from small, hairpin, intronic loci, and do not require Drosha-DGCR8 cleavage [21]. Intronic miRNA function is often coupled with the host gene and may promote host gene action in a synergistic manner, or provide negative feedback by antagonizing host gene action [22]. Intronic miRNAs exemplified in this review include miR-33 and miR-378, which act in concert with their host genes to modulate hepatic lipid and cholesterol metabolism.
Polycistronic miRNA transcripts are evolutionarily conserved and can derive from tandem duplications of the same miRNA leading to common regulation of highly similar miRNAs that often have the same seed sequence [23]. However, polycistronic transcripts consisting of many different miRNAs also exist [21]. Similar to intronic miRNAs and their host genes, polycistronic miRNAs have tightly coupled expression because they typically derive from the same promoter as a coupled pri-miRNA polycistronic transcriptional unit [24]. Coupled transcription and seed sequence similarity allows polycistronic transcripts to target overlapping gene programs. Polycistronic and clustered miRNA loci exemplified in this review include the hepatic miR-96/182/183 locus [25] and the pancreatic DLK1-MEG3 miRNA cluster [26].
Stress and the miRNA interactome
Cellular stress is a key mechanism leading to metabolic dysregulation. Recent studies by Karginov et. al. sought to consider how induced cell stress affects the miRNA targetome [27]. Using AGO-CLIP analysis, the authors noted global increases in AGO2 occupancy on the 3′UTR upon stress induction. These changes were next corroborated with polyribosomal sequencing, demonstrating that cellular stress may not only alter mRNA and miRNA expression changes, but may globally enhance miRNA binding to the mRNA. Such differential activation could provide flexible intensity of miRNA activity in order to re-calibrate intensity of miRNA mediated mRNA decay.
In muscle, recent work demonstrated AGO2 protein and miR-1 are present in mitochondria [28]. CLIP-based target profiling of C2C12 myoblasts defined mitochondrial transcripts NADH dehydrogenase 1 (ND1), cytochrome c oxidase subunit 1 (COX1), ATP synthase 8 (ATP8), and cytochrome c oxidase subunit 3 (COX3) as targets of miR-1. Following target identification, miR-1 activity on some mitochondrial mRNAs such as ND1 and COX1 stimulated, rather than suppressed, translational capacity of the genes and supported increased mitochondrial protein synthesis during muscle cell differentiation. Together, this work demonstrated the capacity of the miRNA targetome to undergo state-dependent flux, and unique, non-canonical enhancement of mRNA translation via miRNA binding.
Both of the above studies were able to track wide scale changes in miRNA binding patterns that extended to the global changes on miRNA occupancy of the 3′UTR. The intersection of variable miRNA activity within metabolic stress pathways and the overall capacity of miRNAs to modulate the metabolic gene program is an intriguing area for future study, especially in stress-dependent disease states such as type 2 diabetes. These findings are even more relevant in light of recent evidence showing that the miRNA pool may require activation in many tissues [29], and that GW182 and Argonaute may be heavily regulated by post-translational modifications [30,31]. These observations also inform other studies and provide clues to explain why certain miR-dependent phenotypes only emerge in specific contexts or under significant cellular stress [32].
Pancreatic miRNA circuitry regulates insulin secretion
miRNAs have been implicated in controlling pancreatic islet development, beta cell differentiation, and glucose stimulated insulin secretion (GSIS) [33–35]. As aberrant beta cell function and insulin secretion leads to insulin resistance and diabetes, a better understanding of how miRNAs integrate signaling pathways in the pancreas is critically important.
Recent pioneering studies combined high throughput sequencing of small RNAs with AGO-CLIP-seq to define the miRNA interactome in human diabetic and non-diabetic islets [26]. In one study, Kameswaran and colleagues demonstrated the DLK1-MEG3 cluster of miRNAs on chromosome 14q32 was specifically expressed in pancreatic beta cells [26]. Using high throughput sequencing of small RNAs, the authors identified seven miRNAs located within the DLK1-MEG3 locus that were significantly downregulated in pancreatic islets isolated from donors with type 2 diabetes. The maternally imprinted DLK1-MEG3 cluster exhibited MEG3 promoter hypermethylation in diabetic islets, which corresponded to the reduced expression of miRNAs within the cluster. By performing AGO-CLIP on islets isolated from human cadaveric pancreas, 996 mRNA targets of 38 miRNAs encoded within the DLK-MEG3 cluster were identified, including several known to induce beta cell apoptosis such as islet amyloid polypeptide (IAPP) and p53-induced nuclear protein 1 (TP53INP1). Although the DLK1-MEG3 cluster is susceptible to epigenetic events that suppress anti-apoptotic miRNAs in pancreatic islets, it is interesting to note that the full extent of this regulation remains incomplete, owing to the large number of miRNAs and targets discovered in the analysis.
Additional studies have identified a number of miRNAs required for proper pancreatic function. Amongst the earliest studies of miRNA function in mice, miR-375 was cloned from pancreatic islets and shown to negatively regulate glucose stimulated insulin secretion [36]. Further studies established miR-375 stimulates beta cell proliferation by modulating genes governing cell growth and proliferation [35]. Deletion of miR-375 in mice resulted in reduced beta cell mass, pancreatic failure, hyperglycemia, increased hepatic gluconeogenesis and type 2 diabetes. Microarray profiling of pancreatic islets isolated from miR-375 KO mice identified 55 transcripts with significantly increased expression, that contained a conserved miR-375 seed motif, demonstrating the broad effects miRNAs can have on the genome [35]. Similar to miR-375, miR-7 is a negative regulator of GSIS in beta cells in mice. miR-7 interacts with and directly regulates genes that control the insulin exocytotic machinery, which are necessary for exocytosis of insulin granules from beta cells [34].
Initial studies showed expression of miR-200 is increased in islets isolated from diabetic mice and, specifically, miR-200b promotes pancreatic cell apoptosis by targeting Zeb1 [37]. More recent work has shown that genetic ablation of the polycistronic miR-141/200c cluster in mice prevents p53-dependent beta cell apoptosis in response to metabolic stress [33]. The ability of miR-141/200c to maintain physiologic insulin sensitivity only occurs under conditions of cellular stress, and no apparent phenotype is observed in unstressed mice. The finding that a specific miRNA family or cluster [26,33,37] only mediates a phenotypic response in pathological conditions appears to be an emerging theme in miRNA research.
Collectively, these studies highlight the critical roles of miRNAs in pancreatic beta cell proliferation, insulin granule exocytosis, and beta cell survival and demonstrate how, by targeting multiple transcripts involved in a variety of cellular processes, individual miRNA and miRNA families modulate key metabolic outputs.
Hepatic miRNAs regulate lipid and cholesterol metabolism
Hepatic insulin resistance and type 2 diabetes are primarily caused by ectopic lipid accumulation in the liver, which inhibits insulin signaling and promotes hepatic glucose production [38]. Transcriptional regulation of de novo lipid synthesis is partially mediated by insulin activation and feedback inhibition of sterol regulatory binding proteins (SREBPs) [39]. The human SREBF1 (SREBP-1) regulates core lipogenic genes including fatty acid synthase (FASN) and stearoyl-CoA desaturase (SCD1). The SREBPF2 (SREBP-2) gene preferentially controls expression of cholesterol synthesis genes, including HMGCR and LDLR. Both genes are host genes for a highly conserved intronic miRNA. SREBF1 on chromosome 17 harbors miR-33b in intron 17, and SREBF2 on chromosome 22 contains miR-33a in intron 16. Mature miR-33a and miR-33b are highly homologous with a largely overlapping predicted target spectrum [3,4,40]. The integration of miR-33 expression concurrently with SREBP transcripts allows co-regulation of these miRNAs and the SREBP proteins during insulin and lipid signaling. In agreement with this concept, miR-33a and miR-33b expression is especially abundant in the liver [3,4].
Antisense inhibition in mouse and human cell lines showed that miR-33a and miR-33b sustain lipid and cholesterol synthesis programs by repressing genes important for fatty acid beta oxidation and cholesterol efflux, particularly the cholesterol efflux pump ABCA1. miR-33a and miR-33b mediate an elegant negative regulatory loop with SREBP wherein depleted cholesterol levels increase SREBP and miR-33ab levels to restore cholesterol biosynthesis and reduce cholesterol efflux through miR-33-dependent silencing of ABCA1 [3,4,40].
Another miRNA involved in lipid and cholesterol metabolism is miR-122, which accounts for ~5% of all miRNA expression in the adult liver [41]. miR-122 inhibition in vivo improves lipid and cholesterol profiles in wild type and mice with diet-induced obesity (DIO) [42]. Although direct target genes of miR-122 have not been comprehensively identified until recently [41], antisense inhibition of miR-122 blocks the expression of lipogenic and cholesterol synthesis genes in the liver including SREBF1 (SREBP-1), FASN, SCD1, ACACA (ACC1), ACACB (ACC2), CD36, HMGCR, and LDLR [42]. Other miRNAs that increase lipid and cholesterol synthesis include the polycistronic miR-96/182/183 locus, which is directly regulated by dietary cholesterol and functions to amplify the lipogenic activity of SREBP-2 [25]. More recently, Huang and colleagues established that miR-26, which exhibited reduced expression in the liver of overweight individuals, protects against hepatic steatosis and insulin resistance by co-targeting critical genes for gluconeogenesis and lipid biosynthesis [43]. Together, these studies reveal an extensive network of miRNA signaling in the liver that functions to modulate lipid and cholesterol metabolism. miRNAs discussed in this review with significant physiological effects in mice are summarized in Table 1.
Table 1.
miRNAs involved in metabolic homeostasis
| miRNA | Tissue | Mechanism of Action | Target(s) | Ref |
|---|---|---|---|---|
| miR-33ab | liver | Tail vein injection of miR-33a/b is sufficient to increase whole body HDL. miR-33 antagonists lower VLDL triglycerides | ABCA1 | [3,4,40] |
| miR-103/107 | liver, White adipose | Promotes insulin resistance and increased serum glucose levels | CAV1 | [29] |
| miR-143 | liver | Transgenic overexpression in the liver impairs glucose homeostasis. | ORP8 | [30] |
| miR-802 | liver | Inhibits hepatic insulin sensitivity and impairs glucose tolerance | HNF1b/TCF2 | [31] |
| miR-378 | brown adipose, skeletal muscle, white adipose, liver | Adipose-specific overexpression has an anti-obesity effect. Deletion confers resistance to diet-induced obesity and exhibit increased energy expenditure | CRAT, MED13, Pde1b | [41, 54] |
| miR-133 | brown adipose | Inhibits brown adipocyte differentiation and thermogenic capacity. | PRDM16 | [47, 49, 51] |
| Let-7 | skeletal muscle, liver, pancreas | Inhibits insulin sensitivity, insulin secretion | IGF1R, INSR, IRS2, HMGA2 | [27, 28] |
| miR-375 | pancreas | Increases beta cell mass | Multiple genes | [26] |
| miR-7 | pancreas | Reduces insulin secretion | SCNA | [25] |
| miR-200 | pancreas | Induces beta-cell apoptosis | DNAJC3, JAZF1, RPS6KB1, XIAP, ZEB1 | [33,37] |
| DLK1-MEG3 microRNA cluster | pancreas | Epigenetically silenced in diabetes, suppresses mediators of beta cell apoptosis. | >900 targets including IAPP and TP53INP1 | [57] |
| miR-155 | brown adipose | reduces brown adipose tissue mass and thermogenic capacity | CEBPB | [53] |
| miR-30b/c | brown adipose | agomirs promote thermogenesis in brown adipose tissue | RIP140 | [42] |
| mR-196a | brown adipose, white adipose | aP2-driven transgenic expression improves insulin sensitivity and energy expenditure | HOXC8 | [50] |
| miR-96/182 cluster | liver | Activated by SREBP2 to increase cholesterol and lipid synthesis | Fbxw7, INSIG2 | [23] |
miRNAs coordinate essential steps in glucose metabolism and insulin action
In addition to modulating insulin secretion by impacting beta cell function, miRNAs coordinate downstream insulin signaling pathways. While some miRNAs exhibit tissue-restricted expression (e.g. miR-375), many are broadly expressed and likely serve a more general role in maintaining metabolic homeostasis.
The let-7 miRNA family regulates multiple aspects of glucose metabolism in the pancreas, liver, and muscle [44,45]. Let-7 antagonism improves glucose tolerance in DIO mice, an effect partially mediated by improving liver and muscle insulin sensitivity. Conversely, let-7 overexpression in the pancreas or skeletal muscle promotes insulin resistance and impairs glucose tolerance in mice [44]. Let-7 inhibits the canonical Akt-mediated insulin signaling cascade by suppressing expression of the insulin receptor, insulin growth factor-1 receptor (IGF1R), and insulin receptor substrate-2 (IRS2) [45]. By simultaneously targeting multiple substrates along the insulin-signaling cascade, let-7 coordinately regulates glucose metabolism and insulin action in a nuanced manner.
Several miRNAs play roles in metabolic disorders associated with an aberrant response to insulin signaling. For example, miR-103/miR-107 are upregulated in the liver of mice with genetic or diet-induced insulin resistance. Antisense inhibition of these miRNAs leads to improved insulin action by stimulating caveolin-1 expression to increase insulin receptor availability [5]. Forced miR-103/107 expression is sufficient to induce impaired glucose homeostasis by decreasing insulin receptor availability due to reduced caveolin-mediated endocytosis of the receptor.
Similar to miR-103/107, miR-143 [1] and miR-802 [2] (table 1) are overexpressed in the liver of leptin-deficient (ob/ob) and DIO mice. Transgenic overexpression of miR-143 reduces insulin sensitivity, presumably by targeting an obligate target of Akt [1], oxysterol-binding protein-related protein 8 (ORP8). miR-802 is also capable of causing impaired glucose tolerance and attenuated insulin sensitivity in mice [2]. HNF1b is a target of miR-802, and the loss of HNF1b results in mature onset diabetes of the young (MODY), providing a possible mechanism for miR-802 pro-diabetic activity. The common thread shared between miR-103/107, miR-143, and miR-802 is metabolic stress that alters the expression of these miRNAs in the liver, resulting in insulin resistance and demonstrating the diverse effects exerted by these miRNAs on organismal glucose metabolism (Figure 1).
Figure 1. miRNAs target the insulin pathway at multiple levels.
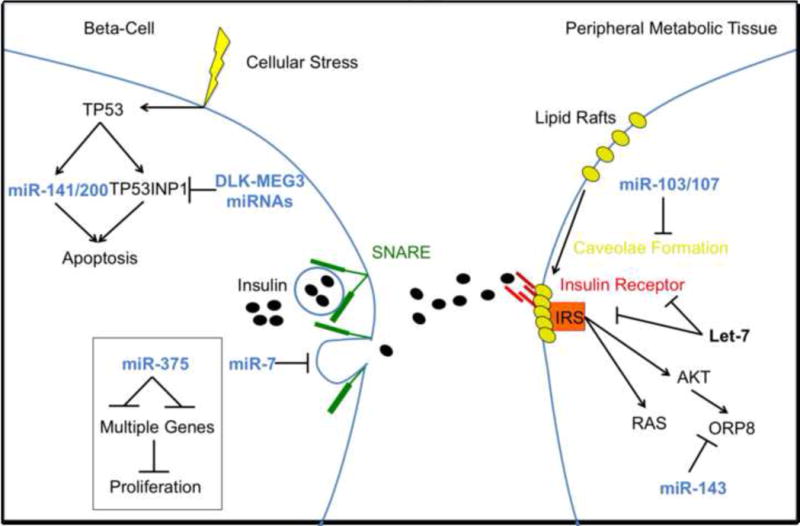
miRNA targeting of insulin signaling occurs centrally at the beta cell and peripherally in target tissues. Several miRNAs, such as the miR-141/200c family and the DLK1-MEG3 cluster, directly regulate cellular response to stress and are active in facilitating (miR-141/200c) or disrupting (DLK1-MEG3 cluster) beta cell apoptosis in response to metabolic stress. miR-375 is important for beta cell proliferation and maintenance of beta cell mass while miR-7 regulates innsulin granule release by inhibiting the SNARE complex. Peripherally, miRNAs are capable of disrupting insulin signaling at almost every point along its axis. miR-103/107 disrupt caveolae formation, impacting insulin receptor and insulin-like growth factor receptor expression at the membrane. The let-7 miRNA family directly targets high-level insulin signaling by reducing insulin receptor (IR) and insulin receptor substrate (IRS) expression. Hnf1b, a transcription factor important for maintaining insulin signaling, is targeted by miR-802. Finally, miR-143 inhibits insulin activity by targeting effector genes downstream of AKT such as ORP8.
Adipose tissue miRNAs remodel energy expenditure
Adipose tissue is an endocrine organ that stores energy in the form of lipids and regulates whole body energy homeostasis via secretion of metabolites and adipokines [46]. It is well established that chronic low-grade inflammation in white adipose tissue (WAT) is a hallmark of obesity and type 2 diabetes [47]. Several miRNAs identified in obese human WAT were found to control inflammation by directly or indirectly modulating pro-inflammatory cytokine release from adipocytes and macrophages [48,49].
Brown adipose tissue (BAT) functions to catabolize free fatty acids and dissipate energy in the form of heat [50]. A newly described pool of brown-like adipocytes, referred to as beige adipocytes, are located within and derived from subcutaneous WAT depots [51]. Similar to brown adipocytes, beige adipocytes undergo inducible thermogenesis. Activation of both depots is thought to protect against insulin resistance and co-morbidities associated with type 2 diabetes [52].
Adipose tissue miRNAs influence metabolism by regulating adipocyte differentiation states or directly modulating specific metabolic and endocrine functions. Over 40 miRNAs correlate with human obesity and type 2 diabetes [53] and numerous miRNAs affect adipocyte differentiation, including miR-103/107 [5], miR-193b-365 [54], miR-378 [55], and miR-30b/c [56]. Studies in mice have also shown that miRNAs alter brown adipogenesis [54,55,57] or stimulate a white-to-brown fat transition [56,58–60]. The miR-193b-365 [54] cluster of miRNAs and miR-133 [57,61] regulate brown adipogenesis by acting on PRDM16, a factor required for the development of brown adipocytes [62]. Likewise, miR-143 [1] and miR-103/107 [5] suppress white adipocyte differentiation, thereby promoting insulin resistance. In contrast, a screen for inhibitors of brown adipocyte differentiation identified miR-155, which impairs brown and beige adipocyte development by inhibiting the expression of C/EBPβ in a bidirectional transcriptional feedback loop [63]. Genetic or pharmacologic methods to increase the expression of miRNAs specific to BAT (miR-196a) or those induced by cold exposure (miR-30b/c) enhance browning of WAT by disrupting the expression of conventional transcriptional activators of white fat genes [56,60].
A recent study showed that the first intron of the PPARGC1B gene encodes miR-378, which counterbalance the metabolic actions of PGC1β [55,64]. Genetic deletion of miR-378 confers resistance to DIO and insulin resistance, while enhancing energy expenditure and mitochondrial fatty acid metabolism [64]. Additional effects of miR-378 on adipocyte metabolism were shown in mice overexpressing miR-378 under the aP2 promoter. In these mice, miR-378 drives BAT expansion, resulting in protection from genetic and diet induced insulin resistance [55]. Thus, it appears that the targets of miR-378 are contextually dependent and specific to certain tissues or cell types. In support of this notion, none of the known miR-378 target genes identified in other tissues, including IGFR (cardiomyocytes), CRAT and MED13 (liver), and ERRG (cancer cells), were significantly suppressed following miR-378 overexpression in BAT [55]. This finding is consistent with the idea that a single miRNA may target different mRNAs in different contexts, with distinct and tissue-specific functional outcomes.
Collectively, these findings highlight the roles miRNAs play in adipocyte gene expression. In addition, miRNAs might have pathological actions that are associated with impaired brown and white adipocyte differentiation, WAT inflammation, energy balance, and insulin resistance [65–67]. No studies have been published to date that directly examine the role of miRNAs in human BAT. Because miRNAs act as key regulators of adipocyte and BAT gene expression, they may be therapeutically leveraged to ‘brown’ white fat, thus improving insulin sensitivity and systemic metabolism.
miRNAs and metabolism: a bidirectional link
A hallmark of the models discussed above is that dysregulation of a single miRNA in one or more core metabolic tissues is sufficient to perturb global metabolic homeostasis, indicating that some miRNAs affect major metabolic pathways. Using Circos [68] to illustrate genomic interactions between miRNAs and their targets, let-7a, miR-143, miR-33a, and miR-33b [1,44,45,69,70] affect both common and discrete components of insulin action (Key figure, figure 2), including genes such as ONECUT2 that are critical for pancreatic beta cell function [71]. Other metabolic pathways are clearly co-targeted by one or more microRNAs, including central glucose metabolism and energy sensing nodes like hexokinase 2 (HK2), lactate dehydrogenase A (LDHA), and the alpha subunit of AMP kinase (PRKAA2). Such co-targeting between miRNAs and targets is a common mechanism for establishing redundant miRNA activity [15]. These overlapping but independent target spectrums are likely to act in concert to decrease insulin action at multiple nodes in the insulin signaling cascade [45] as well as the metabolism of glucose and lipids in the liver [43]. Therefore, the relationship between miRNAs and metabolism is a bidirectional link. miRNAs impinge on metabolism by interacting with the production and signaling of key players involved in metabolic regulation. Similarly, miRNA expression is modulated by major metabolic stimuli, including dietary stress.
KEY FIGURE, Figure 2. miRNAs exert physiologic function by targeting multiple genes and gene networks.
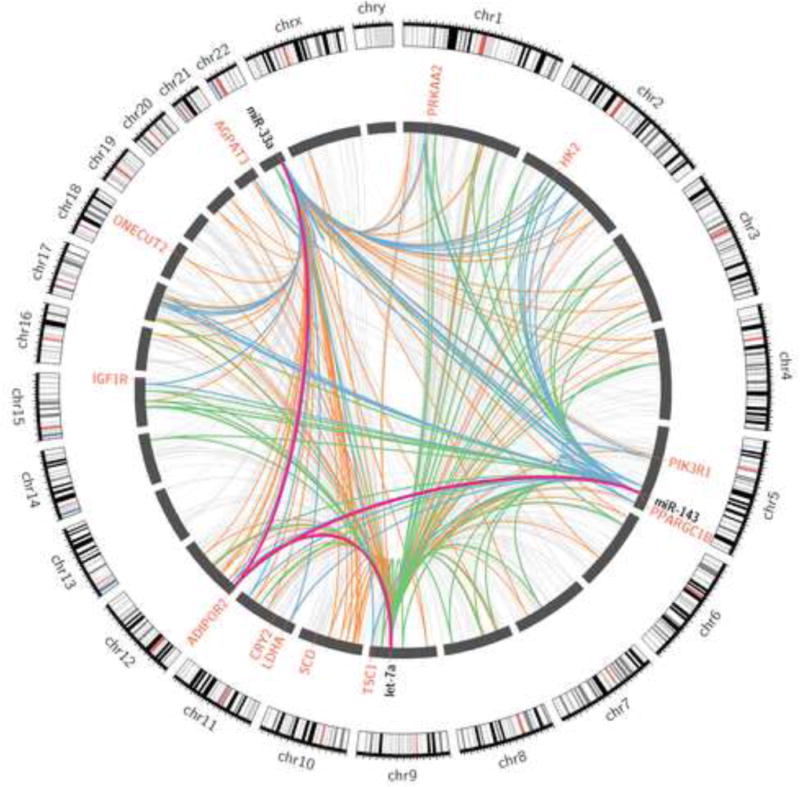
Though miRNA research often focuses on defining a few important targets of specific miRNAs that are relevant to the system or pathology in question, it is understood that the majority of miRNA targeting remains undescribed. In order to illustrate the number of potential targeting and co-targeting interactions that may occur in any given system, we defined targets of metabolically relevant miRNAs miR-33a, let-7 and miR-143 from all available human AGO-CLIP datasets. Individual miRNA-target interactions are illustrated as individual links. Targets of a single miRNA are illustrated as light grey links, co-targeted genes of miR-143 and let-7 are illustrated as green links, co-targets of miR-33 and miR-143 are illustrated as blue links, co-targets of miR-33 and let-7 are illustrated as orange links, and co-targets of all three genes are illustrated as purple links. Critical metabolic target genes are labeled in red. This figure demonstrates the possible breadth and complexity of miRNA targeting, as well as the ability of miRNAs to form complex, redundant, co-targeting networks.
Concluding remarks and future perspectives
Metabolic miRNA research has progressed rapidly in recent years. However, despite these advances, we currently have identified only a handful of miRNAs capable of modulating whole body insulin sensitivity in animal models. Additionally, for these miRNAs, only a few important targets are clearly defined. Although changes in miRNA expression in human insulin resistance and type 2 diabetes exist, the limited available data do not draw a clear picture. Important pitfalls must be considered, including uniformity in miRNA measurement techniques, and confounding factors related to age, body mass index, and sex differences. However, recent studies have found that miRNAs can be readily detected in human plasma, suggesting possibilities for novel disease biomarker discovery. The detection of circulating miRNAs represents an important advance for biomarker discovery, and altered circulating miRNA profiles have been linked to metabolic diseases, including type 2 diabetes Whether or not miRNAs act as signaling molecules that affect gene expression in distant target organs and cells is still an unanswered question.
Future research will require additional effort to define novel pathways altered by currently undescribed miRNAs or those processed through non-canonical biogenesis mechanisms, in addition to more comprehensively defining the multiple pathways and targets through which known miRNAs operate. Finally, the arising evidence in mammals for the existence of intracellular miRNA sponges such as competing endogenous RNAs will affect the efficiency of miRNA-mRNA targeting [72] by physically sequestering miRNAs from their target genes. New approaches such as AGO-CLIP-seq, mass spectrometry proteomics, and polysome profiling techniques will help decipher the composition and regulation of miRNA networks to elucidate how these miRNA networks are coordinated with mRNA and protein turnover. Limitations of high throughput CLIP-based profiling techniques include limited specificity of cross-linking, dependency on presence or absence of specific miRNAs or regulatory proteins, and the detection limit of individual sequence reads. Although technological limitations exist, AGO-CLIP-seq is currently the only method that allows comprehensive, unbiased, and tissue-specific mapping of miRNA interactions with target mRNAs. Understanding how these miRNAs function globally, how they mediate their metabolic effects, and what, if any, “off-target” effects they may have in a therapeutic context, will be necessary prior to effective clinical translation.
In sum, while the incredible regulatory potential of miRNAs in human metabolic disease is currently apparent, additional efforts to define the scope and mechanisms of this control are clearly necessary.
Box 3. Non-canonical miRNA biogenesis.
Various alternative mechanisms can generate miRNAs or small miRNA-like RNAs [89,90]. Deep sequencing of small RNAs from cells deficient in miRNA processing machinery uncovered unconventional microRNAs produced through mRNA splicing [91]. Conventional miRNA processing is also bypassed by miRNAs produced from endogenous short-hairpin RNAs, which are transcribed by RNA Polymerase II and III [92]. Although most alternative miRNA pathways require Dicer, miRNA-451 biogenesis does not require Dicer and instead involves AGO2 catalytic activity [90,93,94]. In this case, the pri-miRNA is too short to be processed by Dicer and directly loads onto AGO2. The loading onto AGO2 leads to slicing of miRNA-451 in the middle of its 3′ strand, which yields a 30-nucleotide intermediate species that is subsequently trimmed by a ribonuclease to a 22 nucleotide. Interestingly, the 30 nucleotide and 22 nucleotide intermediates for miRNA-451 show identical repressive activity. Physiologic roles for miRNAs produced by non-canonical biogenesis have not been established, though miRNA-451 expression is regulated by extracellular glucose levels, leading to direct modulation of AMPK activation [95]. Although only 1% of conserved miRNAs are produced independently of the microprocessing machinery in vertebrates, the existence of alternative pathways reflects the evolutionary flexibility of miRNA biogenesis.
Outstanding questions.
How does the external environment regulate miRNA expression? What diurnal or circadian factors stimulate or repress miRNA production and/or function?
Given the likelihood that many miRNAs may regulate several metabolic signaling nodes simultaneously, how is the target selectively controlled by natural variations in gene and protein expression?
As miRNAs seem to support stress resistance and maintain metabolic homeostasis, what are the determining molecular factors that regulate AGO loading and miRNA targeting?
What is the spectrum of miRNA activity on non 3-UTR regions in response to various environmental of metabolic cues?
Is there a physiological role for miRNAs produced through non-canonical biogenesis pathways in metabolic disease?
Trends Box.
miRNAs regulate target mRNA expression by inducing mRNA degradation or translational inhibition.
miRNAs mediate adipocyte differentiation, control beta cell mass and insulin secretion, and co-target multiple nodes of the insulin signaling pathway to globally promote or suppress diabetic phenotypes in the adipose tissue, pancreas, and liver.
Emerging genomic and sequencing technologies allow the definition of global miRNA-mRNA interactions that underpin metabolic regulation.
Currently most miRNAs have only few well-characterized targets, but are likely to work in complex, overlapping, signaling networks that target many genes in the same pathway.
Manipulation of anti-diabetic miRNAs in-vivo improves insulin sensitivity, suggesting miRNA-based therapeutics may represent a viable strategy for treating metabolic diseases.
Acknowledgments
This work was funded by NIH K01DK096093, American Heart Association Beginning Grant-in-Aid (15BGIA25850025), the Cancer Prevention Research Institute of Texas (CPRIT RP150232), the Alkek Center for Molecular Discovery (SM Hartig), the Baylor Research Advocates for Student Scientists (MP Hamilton), NIH 1F30CA196108-01(DA Bader), and the Caroline Weiss Law Foundation (SE McGuire). SE McGuire is a Dan L. Duncan Scholar and member of the Dan L. Duncan Cancer Center supported by National Cancer Institute Cancer Center Support Grant NIH P30CA125123. We acknowledge the joint participation by the Diana Helis Henry Medical Research Foundation through its direct engagement in the continuous active conduct of medical research.
Glossary
- Argonaute crosslinking-immunoprecipitation with high throughput sequencing (AGO-CLIP-seq)
An experimental method in which biological specimens undergo UV-crosslinking followed by immunoprecipitation of Argonaute, a central component of the RISC complex that is bound to both miRNAs and their mRNA targets. After processing and library preparation, the sample undergoes high throughput sequencing. Statistical methods are then applied to match each miRNA with its target mRNA(s)
- Crosslinking and immunoprecipitation (CLIP)
A method that combines UV crosslinking with immunoprecipitation in order to analyze protein interactions with RNA. CLIP-based techniques can be used to map RNA binding sites for a protein of interest on a genome-wide scale, thereby increasing the understanding of post-transcriptional regulatory networks. AGO-CLIP-seq is a variation of CLIP used to map AGO-RNA interactions
- Dicer
An endoribonuclease that cleaves immature pre-miRNA into mature 20–25 bp miRNA and assists in loading the mature miRNA onto Argonaute. Dicer is necessary for proper RISC function
- Host gene
A gene harboring a miRNA, often within an intron
- MicroRNA cluster
A genomic region containing two or more miRNAs in close proximity to one another
- miRtrons
miRNAs located in the introns of mRNA encoding host genes that arise from the spliced out introns
- miRNA seed region
A 6–8bp sequence on the 5′ end of a mature miRNA critical for defining the target spectrum of the miRNA
- Non-canonical miRNAs
Refers to non-canonical miRNA biogenesis through microprocessor or Dicer-independent processing of pri-miRNA
- Non-canonical miRNA targeting
Refers to microRNA targeting outside of the mRNA 3′UTR, such as the coding sequence, promoter, 5′UTR, intron, or on long non-coding RNAs
- Polycistronic
A single RNA transcript containing multiple discrete genetic elements (genes or miRNAs). In contrast, monocistronic transcripts contain one gene
- RNA-induced silencing complex (RISC)
A multiprotein, miRNA-activated complex that binds and cleaves (or translationally represses) mRNA transcripts complementary to the miRNA that activated RISC
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jordan SD, et al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat Cell Biol. 2011;13:434–446. doi: 10.1038/ncb2211. [DOI] [PubMed] [Google Scholar]
- 2.Kornfeld JW, et al. Obesity-induced overexpression of miR-802 impairs glucose metabolism through silencing of Hnf1b. Nature. 2013;494:111–115. doi: 10.1038/nature11793. [DOI] [PubMed] [Google Scholar]
- 3.Najafi-Shoushtari SH, et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rayner KJ, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trajkovski M, et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature. 2011;474:649–653. doi: 10.1038/nature10112. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai EC. MicroRNAs are complementary to 3′UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 8.Lewis BP, et al. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 9.Lewis BP, et al. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 10.Friedman RC, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grimson A, et al. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krek A, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 13.Betel D, et al. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dweep H, et al. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12:697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 15.Hamilton MP, et al. Identification of a pan-cancer oncogenic microRNA superfamily anchored by a central core seed motif. Nat Commun. 2013;4:13. doi: 10.1038/ncomms3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu J, et al. Impact of microRNA regulation on variation in human gene expression. Genome Res. 2012;22:1243–1254. doi: 10.1101/gr.132514.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chi SW, et al. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hafner M, et al. Transcriptome-wide Identification of RNA-Binding Protein and MicroRNA Target Sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia DM, et al. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ascano M, et al. Identification of RNA-protein interaction networks using PAR-CLIP. Wiley Interdiscip Rev RNA. 2012;3:159–177. doi: 10.1002/wrna.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marco A, et al. Multiple products from microRNA transcripts. Biochem Soc Trans. 2013;41:850–854. doi: 10.1042/BST20130035. [DOI] [PubMed] [Google Scholar]
- 22.Gao X, et al. Enemy or partner: relationship between intronic micrornas and their host genes. IUBMB life. 2012;64:835–840. doi: 10.1002/iub.1079. [DOI] [PubMed] [Google Scholar]
- 23.Tanzer A, et al. Molecular evolution of a microRNA cluster. J Mol Biol. 2004;339:327–335. doi: 10.1016/j.jmb.2004.03.065. [DOI] [PubMed] [Google Scholar]
- 24.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 25.Jeon TI, et al. An SREBP-responsive microRNA operon contributes to a regulatory loop for intracellular lipid homeostasis. Cell Metab. 2013;18:51–61. doi: 10.1016/j.cmet.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kameswaran V, et al. Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets. Cell Metab. 2014;19:135–145. doi: 10.1016/j.cmet.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karginov FV, et al. Remodeling of Ago2-mRNA interactions upon cellular stress reflects miRNA complementarity and correlates with altered translation rates. Genes Dev. 2013;27:1624–1632. doi: 10.1101/gad.215939.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, et al. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158:607–619. doi: 10.1016/j.cell.2014.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.La Rocca G, et al. In vivo, Argonaute-bound microRNAs exist predominantly in a reservoir of low molecular weight complexes not associated with mRNA. Proc Natl Acad Sci U S A. 2015;112:767–772. doi: 10.1073/pnas.1424217112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horman SR, et al. Akt-mediated phosphorylation of argonaute 2 downregulates cleavage and upregulates translational repression of MicroRNA targets. Mol Cell. 2013;50:356–367. doi: 10.1016/j.molcel.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li S, et al. TRIM65 regulates microRNA activity by ubiquitination of TNRC6. Proc Natl Acad Sci U S A. 2014;111:6970–6975. doi: 10.1073/pnas.1322545111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendell JT, et al. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belgardt BF, et al. The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nat Med. 2015;21:619–627. doi: 10.1038/nm.3862. [DOI] [PubMed] [Google Scholar]
- 34.Latreille M, et al. MicroRNA-7a regulates pancreatic beta cell function. J Clin Invest. 2014;124:2722–2735. doi: 10.1172/JCI73066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poy MN, et al. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci U S A. 2009;106:5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poy MN, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 37.Filios SR, et al. MicroRNA-200 is induced by thioredoxin-interacting protein and regulates Zeb1 protein signaling and beta cell apoptosis. J Biol Chem. 2014;289:36275–36283. doi: 10.1074/jbc.M114.592360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shulman GI. Ectopic fat in insulin resistance, dyslipidemia, and cardiometabolic disease. N Engl J Med. 2014;371:2237–2238. doi: 10.1056/NEJMc1412427. [DOI] [PubMed] [Google Scholar]
- 39.Horton JD, et al. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rayner KJ, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478:404–407. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luna JM, et al. Hepatitis C virus RNA functionally sequesters miR-122. Cell. 2015;160:1099–1110. doi: 10.1016/j.cell.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Esau C, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 43.Fu X, et al. MicroRNA-26a regulates insulin sensitivity and metabolism of glucose and lipids. J Clin Invest. 2015;125:2947–2509. doi: 10.1172/JCI75438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frost RJ, et al. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci U S A. 2011;108:21075–21080. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu H, et al. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rutkowski JM, et al. The cell biology of fat expansion. J Cell Biol. 2015;208:501–512. doi: 10.1083/jcb.201409063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Glass CK, et al. Inflammation and lipid signaling in the etiology of insulin resistance. Cell Metab. 2012;15:635–645. doi: 10.1016/j.cmet.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arner E, et al. Adipose tissue microRNAs as regulators of CCL2 production in human obesity. Diabetes. 2012;61:1986–1993. doi: 10.2337/db11-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lorente-Cebrian S, et al. MicroRNAs regulate human adipocyte lipolysis: effects of miR-145 are linked to TNF-alpha. PLoS One. 2014;9:e86800. doi: 10.1371/journal.pone.0086800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cinti S. Between brown and white: novel aspects of adipocyte differentiation. Ann Med. 2011;43:104–115. doi: 10.3109/07853890.2010.535557. [DOI] [PubMed] [Google Scholar]
- 51.Wu J, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cohen P, et al. Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell. 2014;156:304–316. doi: 10.1016/j.cell.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arner P, et al. MicroRNA regulatory networks in human adipose tissue and obesity. Nat Rev Endocrinol. 2015;11:276–288. doi: 10.1038/nrendo.2015.25. [DOI] [PubMed] [Google Scholar]
- 54.Sun L, et al. Mir-193b-365 is essential for brown fat differentiation. Nat Cell Biol. 2011;13:958–964. doi: 10.1038/ncb2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pan D, et al. MicroRNA-378 controls classical brown fat expansion to counteract obesity. Nat Commun. 2014;5:4725. doi: 10.1038/ncomms5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu F, et al. miR-30 promotes thermogenesis and the development of beige fat by targeting RIP140. Diabetes. 2015;64:2056–2068. doi: 10.2337/db14-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trajkovski M, et al. MyomiR-133 regulates brown fat differentiation through Prdm16. Nat Cell Biol. 2012;14:1330–1335. doi: 10.1038/ncb2612. [DOI] [PubMed] [Google Scholar]
- 58.Kong X, et al. Glucocorticoids transcriptionally regulate miR-27b expression promoting body fat accumulation via suppressing the browning of white adipose tissue. Diabetes. 2015;64:393–404. doi: 10.2337/db14-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu W, et al. miR-133a regulates adipocyte browning in vivo. PLoS Genet. 2013;9:e1003626. doi: 10.1371/journal.pgen.1003626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mori M, et al. Essential role for miR-196a in brown adipogenesis of white fat progenitor cells. PLoS Biol. 2012;10:e1001314. doi: 10.1371/journal.pbio.1001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yin H, et al. MicroRNA-133 controls brown adipose determination in skeletal muscle satellite cells by targeting Prdm16. Cell Metab. 2013;17:210–224. doi: 10.1016/j.cmet.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seale P, et al. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest. 2011;121:96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen Y, et al. miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit. Nat Commun. 2013;4:1769. doi: 10.1038/ncomms2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carrer M, et al. Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378(star) Proc Natl Acad Sci U S A. 2012;109:15330–15335. doi: 10.1073/pnas.1207605109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim HJ, et al. MicroRNAs are required for the feature maintenance and differentiation of brown adipocytes. Diabetes. 2014;63:4045–4056. doi: 10.2337/db14-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mori MA, et al. Altered miRNA processing disrupts brown/white adipocyte determination and associates with lipodystrophy. J Clin Invest. 2014;124:3339–3351. doi: 10.1172/JCI73468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xie H, et al. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes. 2009;58:1050–1057. doi: 10.2337/db08-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davalos A, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A. 2011;108:9232–9237. doi: 10.1073/pnas.1102281108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ryu HS, et al. The induction of microRNA targeting IRS-1 is involved in the development of insulin resistance under conditions of mitochondrial dysfunction in hepatocytes. PLoS One. 2011;6:e17343. doi: 10.1371/journal.pone.0017343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roggli E, et al. Changes in microRNA expression contribute to pancreatic b-cell dysfunction in prediabetic NOD mice. Diabetes. 2012;61:1742–1751. doi: 10.2337/db11-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hansen TB, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 73.Okamura K, et al. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol. 2008;15:354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jonas S, et al. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet. 2015;16:421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 75.Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet. 2013;14:447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 76.Moretti F, et al. PABP and the poly(A) tail augment microRNA repression by facilitated miRISC binding. Nat Struct Mol Biol. 2012;19:603–608. doi: 10.1038/nsmb.2309. [DOI] [PubMed] [Google Scholar]
- 77.Subtelny AO, et al. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508:66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shen J, et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature. 2013;497:383–387. doi: 10.1038/nature12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rudel S, et al. Phosphorylation of human Argonaute proteins affects small RNA binding. Nucleic Acids Res. 2011;39:2330–2343. doi: 10.1093/nar/gkq1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bernstein E, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 81.Morita S, et al. One Argonaute family member, Eif2c2 (Ago2), is essential for development and appears not to be involved in DNA methylation. Genomics. 2007;89:687–696. doi: 10.1016/j.ygeno.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 82.Wang Y, et al. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mori MA, et al. Role of microRNA processing in adipose tissue in stress defense and longevity. Cell Metab. 2012;16:336–347. doi: 10.1016/j.cmet.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Harfe BD, et al. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lynn FC, et al. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes. 2007;56:2938–2945. doi: 10.2337/db07-0175. [DOI] [PubMed] [Google Scholar]
- 86.Martinez-Sanchez A, et al. DICER inactivation identifies pancreatic beta-cell “disallowed” genes targeted by microRNAs. Mol Endocrinol. 2015;29:1067–1079. doi: 10.1210/me.2015-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mudhasani R, et al. Dicer is required for the formation of white but not brown adipose tissue. J Cell Physiol. 2011;226:1399–1406. doi: 10.1002/jcp.22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chendrimada TP, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xie M, et al. Versatile microRNA biogenesis in animals and their viruses. RNA Biol. 2014;11:673–681. doi: 10.4161/rna.28985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang JS, et al. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell. 2011;43:892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Babiarz JE, et al. Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xie M, et al. Mammalian 5′-capped microRNA precursors that generate a single microRNA. Cell. 2013;155:1568–1580. doi: 10.1016/j.cell.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cheloufi S, et al. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cifuentes D, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ansari KI, et al. Glucose-based regulation of miR-451/AMPK signaling depends on the OCT1 transcription factor. Cell Rep. 2015;11:902–909. doi: 10.1016/j.celrep.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]


