Abstract
An elaborate metabolic response to fasting is orchestrated by the liver and is heavily reliant upon transcriptional regulation. In response to hormones (glucagon, glucocorticoids) many transcription factors (TFs) are activated and regulate various genes involved in metabolic pathways aimed at restoring homeostasis: gluconeogenesis, fatty acid oxidation, ketogenesis and amino acid shuttling. We summarize the recent discoveries regarding fasting-related TFs with an emphasis on genome-wide binding patterns. Collectively, the summarized findings reveal a large degree of co-operation between TFs during fasting which occurs at motif-rich DNA sites bound by a combination of TFs. These new findings implicate transcriptional and chromatin regulation as major determinants of the response to fasting and unravels the complex, multi-TF nature of this response.
Keywords: CREB, GR, PPARα, FoxO, FoxA, C/EBP, TR, p53, PLZF, NR4A, KLF15, CREBH, TFEB
The response to fasting – hormonal regulation of hepatic metabolism
Mammals are constantly met with varying nutrient availability and have developed integrated mechanisms to tune metabolism according to the excess or scarcity of energy sources. This systemic response is orchestrated mainly by the liver which responds to circulating endocrine cues. During fasting, blood glucose levels decrease and the pancreas secretes glucagon which initiates an elaborate set of responses in the liver aimed at restoring homeostasis. In the first few hours of fasting, glycogenolysis (i.e. glycogen breakdown, Glossary) is sufficient to supply extra-hepatic tissues with glucose. As fasting continues, gluconeogenesis (i.e. the de novo synthesis of glucose from non-carbohydrate substrates) assumes a more dominant role in producing glucose. In prolonged fasting, gluconeogenic precursors (mostly muscle-derived amino acids) are depleted and ketogenesis (i.e. the synthesis of ketone bodies from acetyl-CoA) becomes the main fuel-providing process in liver [1]. Both gluconeogenesis and ketogenesis are dependent upon extra-hepatic supply of substrates. Gluconeogenesis relies on muscle-derived amino acids and adipose tissue-derived glycerol as precursors. Also, gluconeogenesis has been suggested to be partially supported by fatty acid oxidation (FAO) for the supply of ATP and NADH, although this issue is controversial [2]. Excessive muscle protein catabolism to supply amino acids for gluconeogenesis might result in muscle wasting and thus the liver gradually shifts to ketogenesis as fasting persists. Ketogenesis is extremely reliant upon adipose tissue lipolysis to supply the liver with fatty acids (FA) which are oxidized to supply the ketogenic precursor acetyl CoA [3]. In sum, the core biochemical pathways during fasting rely on a constant stream of precursors from muscle and adipose tissue and are also dependent upon hepatic FAO (Figure 1).
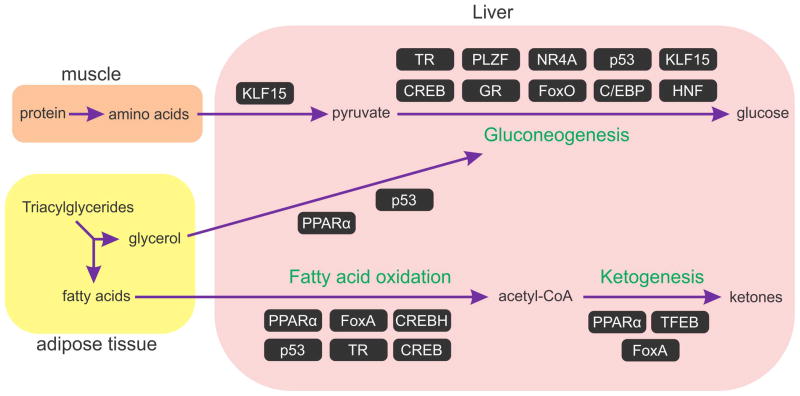
Figure 1. Transcription activators involved in the response to fasting.
The Figure depicts the major metabolic pathways activated during fasting and transcription factors involved in each of them. Gluconeogenesis precursors are supplied by muscle-derived amino acids and adipose tissue-derived glycerol. Shunting of these gluconeogenic precursors as well as gluconeogenesis itself are regulated transcriptionally. Ketogenesis mainly relies on fatty acid oxidation for its precursor – acetyl-CoA. Thus, TFs which regulate fatty acid oxidation often has an effect on ketogenesis. Accordingly, some of these TFs were directly implicated in ketogenesis. We determined the involvement of a TF in regulating a pathway based on evidence showing that the TF induces genes involved in the pathway and/or evidence showing that the metabolic output of the pathway is affected when the TF is inhibited or activated. (C/EBP - CCAAT enhancer binding protein, CREB - cAMP responsive element binding protein, CREBH - cAMP responsive element binding protein H, FoxA – forkhead box transcription factor class A, FoxO – forkhead box transcription factor class O, GR – glucocorticoid receptor, HNF –hepatocyte nuclear factor, KLF15 - kruppel-like Factor 15, NR4A – nuclear receptor subfamily 4 group A, PLZF - promyelocytic Leukemia Zinc Finger Protein, PPARα - peroxisome proliferator-activated receptor α, TFEB – transcription factor EB, TR – thyroid hormone receptor)
Upon feeding, ample glucose becomes available from dietary sources leading to an increase in blood glucose levels and insulin secretion from the pancreas leading to a rapid stop of glycogenolysis, gluconeogenesis and ketogenesis [4]. This strict and rapid regulation by insulin is brought about both by extra-hepatic actions (e.g. inhibition of glucagon secretion) and by direct effects of insulin on liver [5–7] (Figure 2). In the absence of insulin regulation, hepatic glucose production is uninhibited resulting in hyperglycemia. Such dysregulated glucose production occurs either in the lack of insulin secretion or in de-sensitization of metabolic organs to insulin (i.e. insulin resistance) and is a hallmark in the development of type 1 and 2 diabetes, respectively [5].
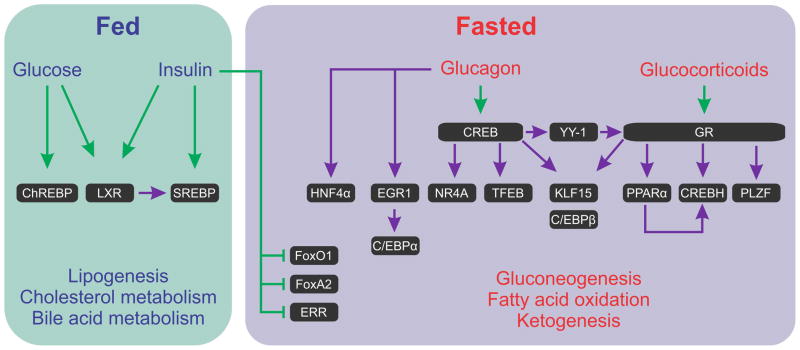
Figure 2. Transcriptional regulation in the fed vs. fasted states and transcription factor cascades.
The transition from a fed state to fasting results in increased glucagon/insulin ratio. This deactivates feeding-related TFs and alleviates the inhibitory effect of insulin on fasting-related TFs. In addition, glucagon and glucocorticoids activate CREB and GR, respectively. There is ample evidence to suggest that during fasting, TFs not only regulate ‘metabolic’ gene expression (i.e. regulating genes encoding enzymes involved in metabolic pathways) but also regulate the expression of other fasting-related TFs. Major initiators of these TF cascades are CREB and GR.
Green arrow indicates signal-dependent activation/inhibition of a TF not involving an increase in absolute TF level. Purple arrow indicates an increase in the level of the TF (in most cases this increased level was shown to be due to transcriptional induction of the gene encoding the TF). (C/EBP - CCAAT enhancer binding protein, ChREBP - Carbohydrate-responsive element-binding protein, CREB - cAMP responsive element binding protein, CREBH - cAMP responsive element binding protein H, EGR1 - early growth response 1, ERR - estrogen-related receptor, FoxA –forkhead box transcription factor class A, FoxO – forkhead box transcription factor class O, GR –glucocorticoid receptor, HNF4α - hepatocyte nuclear factor 4α, KLF15 - kruppel-like Factor 15, NR4A – nuclear receptor subfamily 4, group A, LXR - liver X nuclear receptor, PLZF -promyelocytic Leukemia Zinc Finger Protein, PPARα - peroxisome proliferator-activated receptor α, SREBP - sterol regulatory element binding transcription factor, TFEB – transcription factor EB, YY1 – ying yang 1)
Regulation of gene expression during fasting – the mainstay of transcriptional regulation
A major part of the livers’ response to fasting is achieved by eliciting a comprehensive transcriptional program. Gluconeogenesis, FAO and ketogenesis are partly dependent upon transcriptional regulation. The reliance of the hepatic response to fasting on transcriptional regulation has been documented for decades. Indeed, early studies delineating the regulation of Pck1, a gene encoding the gluconeogenic enzyme phosphoenolpyruvate carboxykinase (PEPCK) have become a mainstay to the field of signal-dependent transcriptional regulation. While certain sequence-specific transcription factors (TFs) have been frequently reported to regulate fasting-related transcription, recent advances in the field have greatly expanded the scope of TFs involved in this complex response. In the first part of the review (including Box 1), we will individually describe TFs involved in the hepatic response to fasting with an emphasis on recent advances and on the relatively unexplored role of chromatin regulation during fasting. In addition to newly-discovered fasting-related TFs, the advances in systems biology approaches, particularly genome-wide characterizations, have had a large impact on fasting research and are the focus of the second part of this review.
Box 1. Transcription factor cascades during fasting.
The two major hormonal signals regulating fasting are glucagon and glucocorticoids. Through the direct activation of CREB and GR, these hormones also lead indirectly to increased activity of many other fasting-related TFs through induction of their encoding genes’ expression (we refer to these pathways as ‘TF cascades’).
Glucagon, through CREB, induces the expression of the Nr4a gene family, which enhance gluconeogenesis [76]. In addition to its role in gluconeogenesis, NUR77 (the product of Nr4a1) induces the expression of FGF21, a hormone responsible for a much broader, inter-organ response to fasting (Box 2) [77]. Another critical CREB-induced TF cascade involves transcription factor EB (TFEB) [78]. In the absence of TFEB, most of the PPARα target genes related to FAO and ketogenesis are not induced [79]. Thus, a chain of events is plausible in which in early fasting glucagon activates CREB, leading eventually to the activation of ketogenesis through the TFEB-PPARα cascade. In accordance with that concept, impaired glucagon signaling in the liver disrupts PPARα-dependent functions [80]. An additional cascade was recently described whereby CREB induces the Yin-Yang 1 (YY1) TF which in turn induces the expression of GR, thereby augmenting its glucogenic potential [81].
During fasting, both glucagon and glucocorticoids induce kruppel-like factor 15 (KLF15) [6, 82]. KLF15 is a unique fasting-related TF as it seems to support gluconeogenesis not only by inducing gluconeogenic genes [6, 82] but also by inducing genes responsible for the catabolism of amino acids shunted to the liver during fasting to serve as gluconeogenic precursors. Among these, the gene encoding alanine aminotransferase is chief. It is induced by KLF15 during fasting leading to increased supply of amino acid-derived gluconeogenic precursors, thus facilitating gluconeogenesis [83].
In addition to the collaboration with glucagon, glucocorticoids also induce the expression of promyelocytic leukemia zinc finger protein (PLZF), which induces gluconeogenic genes and elevate glucose production during fasting [84]. GR also induces the gene encoding PPARα [22]. Lastly, during fasting PPARα and GR both induce the expression of cAMP responsive element binding protein H (CREB-H) which induces both gluconeogenic genes expression [85, 86] and FGF21 [87–89].
These TF cascades (summarized in Figure 2) may help in propagating and fine-tuning the response to fasting. Moreover, although this avenue is under-explored, these TF cascades might be a driving force behind the temporal organization of fuel production during fasting.
The volume of research on the transcriptional regulation taking place during fasting is immense, with studies dating back decades. This review will only briefly summarize critical discoveries made during those decades with an emphasis on recent findings. Due to space limitations and to several recent excellent reviews, we will not cover co-factors and post-translational modifications of fasting-related TFs [8–10].
Transcription factors involved in the response to fasting
Below we provide a summary of the sequence-specific TFs involved in the response to fasting. These factors bind DNA at cis-acting regulatory elements (promoters and enhancers) either following a stimulus or constitutively to induce gene transcription. The TFs covered in this review and their roles in various fasting-related pathways are summarized in Figure 1.
cAMP responsive element binding protein (CREB)
The pancreatic hormone glucagon was isolated and reported to elevate blood glucose almost a century ago [11]. Since then, a series of studies have established the canonical pathway by which the glucogenic action of glucagon is brought about at the transcriptional level. Briefly, glucagon binds to its membrane receptor, activating adenylate cyclase which produces cAMP leading to the activation of protein kinase A (PKA). PKA phosphorylates CREB which, in turn induces a set of fasting-related genes [8]. The elaborate regulation imposed on the glucagon-PKA-CREB axis is mediated by post-translational modifications and co-activators and has been meticulously characterized [8]. Being the ‘first responder’ TF during fasting, CREB activates gluconeogenesis. Additionally, CREB regulates the rest of the fasting-related metabolic pathways either through direct induction of genes encoding metabolic enzymes, or by induction of genes encoding TFs which carry on and promote their own transcriptional signature (Box 1, Figure 2).
A major update to the classic CREB paradigm was recently introduced when two studies showed a role for histone modifications in mediating gene induction following fasting. These studies reported that fasting-related signals alter the chromatin state around CREB binding sites through histone modifications. The first study portrayed the role of the histone acetyl transferase KAT2B (a.k.a. PCAF) in CREB-dependent gluconeogenic gene induction [12]. Fasting increased a KAT2B-dependent histone mark associated with active transcription (H3K9Ac). KAT2B was recruited to gluconeogenic-related CREB binding sites upon glucagon treatment and this recruitment was important for efficient gene induction. The second study highlighted the importance of histone methylation by protein arginine methyltransferase 5 (PRMT5) [13]. PRMT5 increased the levels of di-methylated histone H3 arginine (H3R2me2) following glucagon. PRMT5 was recruited to CREB sites on gluconeogenic genes; downregulation of PRMT5 decreased circulating glucose levels and gluconeogenic gene expression. This study expands an earlier report of PRMT4, a similar methyltransferase which methylates histones at gluconeogenic loci thus augmenting transcription [14]. Collectively, the studies described above emphasize the central role of the chromatin environment and histone modifications in the execution of the glucagon-PKA-CREB pathway.
Glucocorticoid receptor (GR)
GR is a steroid hormone receptor that regulates transcription upon binding and stimulation with the stress-related hormones glucocorticoids. Although glucocorticoids are long-known to stimulate an increase in circulating glucose [15], it was only in the 1990’s that initial mechanisms behind this action were unraveled when glucocorticoids and GR were shown to induce the expression of Pck1 in the liver [16]. Hepatocyte-specific GR knock-out resulted in impaired ability to induce gluconeogenic genes and maintain normal blood glucose levels during fasting [17]. Moreover, a liver-selective GR antagonist was able to lower blood glucose levels [18]. These and other studies further established the glucogenic role of GR. While glucocorticoids are the major activator of GR, recent data might imply that GR activity is indirectly regulated by the predominant ketone body during fasting – β-hydroxybutyrate (β-OHB). Two well-established GR target genes, Lcn2 and Mt2, are within the top genes induced by β-OHB, which is presumed to regulate gene expression through inhibiting HDACs [19]. It is tempting to speculate that as fasting persists and β-OHB levels increase, GR transcriptional activity is augmented by a more promiscuous chromatin environment (i.e. hyper-acetylated histones) mediated by β-OHB. Similarly to CREB, GR also induces a set of genes encoding fasting-related TFs thereby contributing to fasting-induced transcription both directly and indirectly (Box 1, Figure 2)
Peroxisome proliferator-activated receptor α (PPARα)
The PPARα nuclear receptor binds a variety of FA and FA-derived compounds as ligands leading to transcriptional activation (following heterodimerization with retinoid X receptor – RXR). This wide ligand selection might be the basis for the relatively high activity of PPARα in the lack of an exogenous pharmacological agonist [20]. PPARα is involved in virtually every aspect of fasting by regulating genes, with direct roles in ketogenesis, gluconeogenesis, FAO and amino acid utilization as shown by gene knock-out models and transcriptomic studies [20]. Moreover, PPARα regulates fibroblast growth factor 21 (FGF21), a liver-produced hormone with systemic effects during fasting (Box 2). Additionally, PPARα has a substantial degree of co-operation with other fasting-related TFs. For example, PPARα is needed to exert the diabetogenic effect of glucocorticoids and GR [21]. This GR-PPARα relationship could be due to the fact that the Ppara gene is induced by GR [22] but might also indicate co-operation between the two TFs in gene regulation [23].
Box 2. Transcriptional regulation of FGF21.
While the transcriptional regulation of the two major gluconeogenic genes Pck1 and G6pc was heavily studied, our understanding of most of the other fasting-induced genes’ regulatory TFs is only skin-deep (see Outstanding Questions Box). An emerging exception to that is the regulation of the gene encoding FGF21, a hormone secreted from the liver upon fasting which has drastic systemic effects on metabolism. The first glimpse into Fgf21 gene regulation was made with the observation that PPARα induces Fgf21 upon fasting and that FGF21 is responsible for some of the FAO-stimulating and ketogenic capabilities of PPARα [90, 91]. Following that initial observation, numerous studies showed that Fgf21 is induced by fasting-related TFs such as GR [92], NUR77 [77], CREBH [88, 89] as well as other TFs: aryl hydrocarbon receptor (AhR) [93], activating transcription factor 4 (ATF4) [94], RAR-related orphan receptor α (RORα) [95] and FXR [96]. Aspects relating to chromatin regulation are also starting to emerge with a study showing that sodium butyrate stimulates Fgf21 expression, presumably through inhibiting HDACs and keeping the chromatin environment around the Fgf21 gene accessible [97]. This is especially intriguing since butyrate is structurally similar to β-hydroxybutyrate which is increased during fasting. Thus, one might speculate that β-OHB propagates the transcriptional activity at the Fgf21 locus through chromatin relaxation in a manner similar to other fasting-induced genes [19]
Forkhead box proteins (FoxO1, FoxO3, FoxO4, FoxO6 and FoxA2)
Accumulated data has implicated all members of the class O forkhead box proteins in hepatic glucose production, with FoxO1 being the best characterized in that regard. FoxO1 is heavily regulated at the post-translational level, most notably by phosphorylation and acetylation (e.g. insulin significantly inhibits FoxO1 by phosphorylation-dependent cytoplasmic retention) [24]. A triple knock-out of FoxO1/3/4 led to the most pronounced impairment of glucose production compared to individual gene FoxO gene knock-outs [25–28]. Thus, there seems to be considerable redundancy between FoxO proteins in regulating gluconeogenesis. Remarkably, even FoxO6, which was long considered brain-specific, is now known to be expressed in liver and plays a role in hepatic gluconeogenesis [29, 30]. In addition to its role in enhancing gluconeogenesis directly, FoxO3 also mediates protein catabolism in muscle cells [31]. This might be a complementary mechanism to its role in increasing gluconeogenesis because gluconeogenesis largely relies on muscle-derived amino acids as precursors. An intriguing aspect pertaining to FoxO3’s role during fasting has emerged when the Foxo3a gene and some of FoxO3-induced genes were shown to be induced by β-OHB, which is the major metabolite produced at prolonged fasting [19].
Proteins belonging to class A of fox proteins, especially FoxA2, also play a role in the response to fasting [32]. Similarly to FoxO proteins, FoxA2 is inhibited by insulin-dependent phosphorylation [33]. However, while FoxO proteins mainly regulate gluconeogenesis, mouse models revealed the regulation of gluconeogenesis, FAO and ketogenesis by FoxA2 [33, 34].
CCAAT enhancer binding proteins (C/EBPs)
Early reports showed the involvement of the C/EBPα and C/EBPβ TFs in fasting-related metabolism and delineated the role of these two proteins in regulating gene expression in response to fasting. C/EBPs are often regarded as constitutive TFs and indeed, their signal-independent high expression and activity in liver supports that notion. However, evidence has accumulated to show that C/EBPβ expression and activity is increased upon the fasting-related signals glucocorticoids and glucagon. These classic roles for C/EBPs [35] served as the basis for what currently appears to be a broader role for C/EBPs during fasting. First, Cebpa, the gene encoding C/EBPα was found to be induced by glucagon through the early growth response protein 1 (EGR1) TF, leading to enhanced C/EBPα-dependent gene induction [36]. Second, reducing C/EBPα levels through chromatin regulation impairs gluconeogenesis. The histone de-methylase JHDM1A reduces the levels of an active transcription-associated histone mark (H3K36me2) at the Cebpa locus. This reduction leads to a decrease in C/EBPα expression, C/EBPα binding and in reduced gluconeogenic gene expression [37]. In another study, C/EBPα induced the gene encoding pyruvate carboxylase, a gluconeogenic enzyme, and regulated the response to fasting through an interaction with the fasting-induced co-activator SRC1 [38].
Hepatocyte Nuclear Factors (HNF1α, HNF1β and HNF4α)
The relevance of HNFs in metabolic disorders become apparent when mutations in three genes encoding HNFs (HNF1α, HNF1β and HNF4α) where found to result in maturity onset diabetes of the young (MODY) [39]. Although MODY genes usually affect pancreas functions, HNFs also have a clear role in hepatic functions and were shown to induce the expression of gluconeogenic genes [40]. HNFs are usually considered constitutive TFs which regulate transcription regardless of signal. Challenging these premises are evidence that the glucagon-PKA-cAMP pathway induces the expression of HNF4α [41, 42] and the fact that the ligand for HNF4α (a nuclear receptor) is still undefined. Recently, the expression of hepatic HNF1β was found to be regulated by microRNA-802 with an effect on glucose homeostasis and insulin sensitivity, further supporting the notion that HNF4α level is regulated to tune metabolism [43].
Thyroid hormone receptor (TR)
Thyroid hormone-dependent gene regulation is executed by TR, a nuclear receptor capable of regulating a wide array of genes following hetero-dimerization with RXR. In liver, the TRβ subtype is the predominant one and is responsible for many metabolic functions of thyroid hormone. Thyroid hormone enhances three critical pathways related to the response to fasting – gluconeogenesis, FAO and lipolysis (the latter taking place in adipose tissue) [44]. TR regulation of gluconeogenesis is mediated both through direct induction of gluconeogenic genes [45] and by enhancing alanine transport to the liver [46]. Cpt1a, a gene related to FAO, is regulated by TR via co-operation with C/EBP [47]. Remarkably and similarly to PPARα [48], TR also regulates FAO and ketogenesis by enhancing autophagy [49].
p53
p53 is a central tumor suppressor protein with vast gene regulatory capabilities affecting every aspect of cancer. p53 regulates primary metabolic pathways both as part of its anti-tumor roles and as a protein responsible for maintaining homeostasis [50]. Recently, three studies found a regulatory role for p53 during fasting. Two studies pointed to a glucogenic role of p53 in hepatocytes. p53 induced a set of gluconeogenic genes and genes regulating supply of gluconeogenesis precursors and primary hepatocytes lacking p53 showed impaired glucose production [51]. Furthermore, p53-deficient mice had lower fasting-glucose levels and impaired glucose production [52]. A putative mechanism by which p53 indirectly increases gluconeogenesis (and possibly ketogenesis) is by enhancing FAO [50, 53, 54]. Paradoxically, a third paper described an anti-glucogenic role for p53. The authors show a p53-dependent increase in SIRT6 levels, leading to FoxO1 deacetylation and cytoplasmic retention thus negating the glucogenic effect of FoxO1. In contrast to the first two studies, p53 knock-out mice had increased glucose production [55]. This puzzling discrepancy must be resolved by further research deciphering the signals which activate p53 during fasting. It is tempting to speculate that p53 activity is different in short term fasting compared to prolonged fasting and at each stage p53 plays either a glucogenic or an antiglucogenic role. A possible mediator of such a bipolar response of p53 may be SIRT1, which de-activates p53 in several circumstances [56]. SIRT1 is involved in a transcriptional switch occurring during fasting leading to a transition from a short-term to a long-term fasting transcriptional program [57]. Thus, SIRT1 may inhibit p53 in prolonged fasting whereas in short-term fasting p53 is free to elicit its glucogenic activities.
In sum, transcriptional regulation is at the heart of the mammalian response to fasting. The TFs covered in this part and in Box 1, along with other fasting-related TFs - retinoic acid receptor (RAR), RAR-related orphan receptor (ROR), testicular receptor 4 (TR4) and estrogen-related receptor (ERR) all contribute to this response in an elaborate program initiated by hormonal and metabolic factors (Figures 1 and 2).
Genomic approaches in fasting research
Sequencing technologies developed in recent years allowed for a genome-wide characterization of TF occupancy on DNA (i.e. cistrome). The ability to define the entire binding site repertoire of TFs led to a paradigm shift in the field of transcriptional regulation. It became evident that while promoter-proximal regulatory elements (ranging a few hundred base pairs from the transcriptional start site) are important for gene regulation, distal enhancers also play a critical role in it [58]. In addition, genome-wide maps of histone and DNA modifications were generated, showing an extra layer of regulation on gene expression. Lastly, the physical accessibility of DNA to TF binding has been assessed and shown to correlate to enhancer activity [59]. Thus, a combination of cistromes, genome-wide chromatin modifications maps and global chromatin accessibility characterizations can be useful in defining enhancers. Enhancers involved in the response to fasting are beginning to be identified as studies define TF cistromes involved in that response. The case of GR exemplifies how genome-wide technologies promote our understanding of the response to fasting. Until recently, GR binding to chromatin and subsequent gene induction was only described for a handful of fasting-related genes. The first step in defining the broad role of GR during fasting was characterization of hepatic GR binding at promoter-proximal regions using ChIP-chip following a combination of fasting and a synthetic glucocorticoid (dexamethasone - dex) [60]. A more recent report describes the whole GR cistrome following dex treatment using ChIP-seq, revealing 11K GR binding sites throughout the genome of liver cells [61]. Although the functional link between these sites and fasting-related genes was not specifically examined in the study, GR did show binding at the Pck1 locus. The ChIP-seq technology not only led to a better estimation of the number of binding events of fasting-related TFs, it also provided mechanistic insights to the mode of action of two of them –CREB and TR. Before the genomic area, CREB was thought to bind DNA only upon hormone stimulation. However, a genome-wide analysis of hepatic CREB binding (by ChIP-seq) surprisingly showed virtually no changes in the CREB cistrome in the transition from a fed to a fasted state [62]. This finding is in accordance with a previous ChIP-chip experiment mapping CREB and phospho-CREB in primary hepatocytes [63]. Thus, under current data, the conclusion is drawn that neither CREB recruitment to chromatin nor its phosphorylation state is indicative for gene induction; rather, phosphorylation-dependent recruitment of co-activators [8, 63] seems to be a critical step.
The reciprocal conceptual shift was recently made in the case of TR. TR was classically considered to bind DNA prior to hormone stimulation thereby repressing gene transcription. Only upon hormone stimulation was TR thought to induce genes through recruitment of cofactors and chromatin modulating proteins. Two recent papers profiling the hepatic TR cistrome challenge that view, showing considerable increase in TR binding upon hormone treatment (for endogenous TRβ, 2K sites following ligand were found compared to 0.8K sites in the unliganded state) [64, 65].
Another major discovery made possible by ChIP-seq relates to the co-operation between fasting-related TFs. A study mapping the C/EBPβ cistrome found 25K binding sites in the genome, further attesting to C/EBPβ’s extensive role in liver physiology which extends beyond the regulation of a few genes [61]. Interestingly, mapping the GR cistrome in the presence or absence of C/EBPβ revealed that C/EBPβ assisted the loading of GR to many sites in the genome by increasing chromatin accessibility, suggesting another layer of regulatory complexity between these two fasting-related TFs. This observation proposes a mechanism not relying on protein-protein interactions for the classic ‘accessory factor’ model for TFs whereby C/EBPs enhance the gene-inducing capacity of other fasting-related TFs [35]. Further evidence for an assisted loading model for GR in the liver comes from studies showing that optimal GR transcriptional activity and the diabetogenic potential of glucocorticoids is reliant on liver X receptor β (LXRβ) and PPARα [21, 66].
Similarly to GR and CREB, PPARα is also activated by an upstream ligand. However, in contrast to GR and CREB, which are activated by bona fide hormones that increase at well-defined physiological situations, PPARα is activated by a range of lipid-derived compounds [20]. This complicates the assessment of an ‘activated’ versus an ‘un-activated’ cistrome for PPARα. Attempts at defining the PPARα cistrome began with a first ChIP-chip experiment [67]. More recently, three studies reported ChIP-seq of PPARα in mouse liver [48, 68] and in human hepatocytes [69]. In mouse liver, PPARα’s cistrome (16K-20K sites, depending on the study) showed extensive overlap with both LXR [68] and FXR [48] cistromes. In both cases the shared binding sites of these TFs promotes mostly an antagonistic relationship between PPARα and LXR/FXR. This is not surprising giving the fact that LXR/FXR primarily promote feeding-related pathways in the liver (LXR increases lipogenesis while FXR inhibits gluconeogenesis and autophagy) [48, 70]. The PPARα cistrome also helped in obtaining a mechanistic insight regarding PPARα-dependent autophagy during fasting [48].
In the case of FoxO1, the cistrome following fasting was mapped using ChIP-seq (but not compared to the fed state) [71]. In compliance with its role during fasting, FoxO1 was bound next to fasting-induced genes. The FoxO1 cistrome was relatively small (0.4K sites). This might imply a restricted transcriptional repertoire of FoxO1 during fasting but might also reflect the extensive redundancy between FoxO family members.
A very different situation is observed in the case of FoxA2 which was observed to bind thousands of sites in the genome. Its cistrome is very sexually dimorphic (11K binding sites in male mice and 17K sites in female mice) [72]. Unfortunately, comparing FoxA2 binding in the fed vs. the fasted states is not available; thus it is currently unclearhow the FoxA2 cistrome will change following fasting, and whether there will be a sexual dimorphism phenotype in this subset of fasting-altered sites. Nonetheless, in agreement with the role of FoxA2 during fasting, genes around FoxA2 binding sites are functionally related to amino acid, FA and ketone metabolism [73].
Towards defining ‘fasting enhancers’
Perhaps the most far-reaching discovery made possible by genome-wide studies is that the binding of TFs is not randomly distributed throughout the genome. Rather, TFs tend to bind at ‘hot-spots’, i.e. sites on chromatin which are enriched with many TF binding motifs, are more accessible to TF binding and are enriched with certain histone modifications. These three characteristics are usually used to define these hot-spots as enhancers [58]
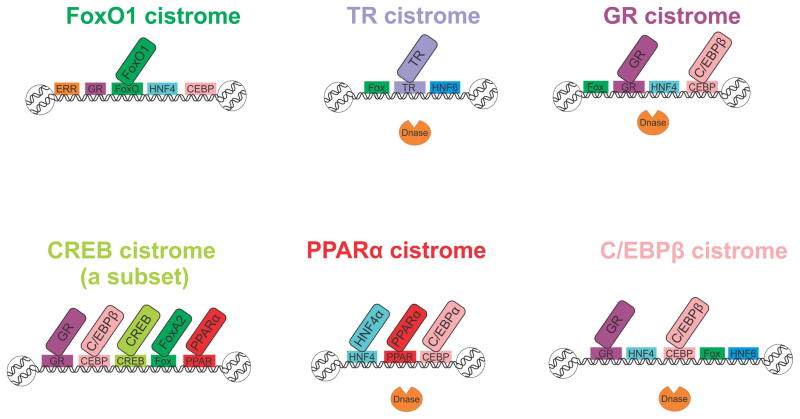
Indeed, it soon became apparent that the cistromes of fasting-related TFs are speckled with binding motifs for other factors involved in the same response. The regions surrounding GR binding sites are enriched with CEBP, HNF4 and Fox motifs whereas the C/EBPβ cistrome is enriched with HNF4, HNF6, Fox and GR motifs [61]. In accordance, FoxO1 binding sites are flanked by GR, CEBP, ERR and HNF4 motifs [71]. The TR cistrome is also surrounded by the fasting-related motifs HNF6 and Fox [65]. The abundance of the HNF4 motif is not surprising due to its established role in fasting and its relatively extended cistrome (20K sites) [74]. In addition to mere motif presence, these hot-spots are actually bound by multiple fasting-related TFs as was exemplified in two cases. A subset of the CREB cistrome was shown to be enriched for binding of GR, FoxA2, C/EBPβ and PPARα [62]. However, this enrichment was only examined at CREB binding sites proximally associated with genes and not on the entire CREB cistrome. It is now established that distal enhancers also play crucial roles in gene expression. Therefore, examining the entire CREB cistrome might reveal a different, more precise picture. In the case of PPARα, such a genome-wide analysis has been reported; the PPARα cistrome overlapped with the HNF4α and C/EBPα cistromes [68]. Moreover, the cistromes of PPARα, GR and TR reside in DNase-I accessible regions, further attesting to its regulatory role and possible function as enhancers [61, 65, 68]. It seems that these PPARα hot-spots are somewhat ‘fasting-specific’ as binding of the feeding-related TF sterol regulatory element-binding protein (SREBP) at those regions is not enriched [68].
In sum, we are beginning to gain an understanding of the genome-wide ‘fasting enhancers’ repertoire through the overlap between fasting-related TF cistromes (Figure 3). The results summarized above show that fasting-related TFs bind nearby to each other and assist each other’s loading onto DNA. Most evidence lead to a mechanism not relying on tethering together of TFs but rather indirect facilitation of binding by enhancing DNA accessibility [61, 68, 75].
Figure 3. fasting-related TF cistromes are clustered together in specific, motif-rich hot-spots.
When examining the motifs enriched in a TF cistrome and comparing different cistromes, the observation wass made that fasting-related TFs tend to bind in the same regions. Horizontal rectangles represent a binding motif sequence found to be enriched in a given cistrome. Diagonal rectangles represent a TF with increased binding at a given cistrome.(C/EBP - CCAAT enhancer binding protein, CREB - cAMP responsive element binding protein, ERR - estrogen-related receptor, FoxA – forkhead box transcription factor class A, FoxO –forkhead box transcription factor class O, GR – glucocorticoid receptor, HNF - hepatocyte nuclear factor, PPARα - peroxisome proliferator-activated receptor α, TR – thyroid hormone receptor)
Concluding remarks
The transcriptional response to food deprivation is a complex, multi-stage one. Although this area has been heavily studied for decades, recent technologies and conceptual breakthroughs were recently introduced, reshaping this field. The binding pattern of fasting-related TFs is now recognized to be extensive (in terms of number of binding sites) and has provided mechanistic insights regarding TF mode of action. We now realize that transcriptional regulation during fasting parallels in its complexity to the multitude of biochemical processes taking place upon fasting.
However, a complete understanding of gene expression changes, TF binding patterns and linkage to metabolic output is far from being resolved. Moreover, while the studies described here helped to provide a genome-wide perspective of fasting-related TF binding in liver, the experiments were mostly done in either unstimulated or agonist-stimulated situations. The field is still desperately lacking experiments examining TF cistromes in the fed vs. the fasted state. Finally, while defining cistromes is a valuable tool, it is an intrinsically biased approach, as it only detects sites bound by a specific TF in a specific context. In contrast, assessing the genome-wide pattern of chromatin accessibility and histone modifications is a more general approach to defining enhancers, and should be implemented in fasting research (see Outstanding Questions Box). It is becoming clear that understanding these regulatory mechanisms will shed light on many metabolic disorders such as diabetes.
Outstanding Questions Box.
-
What is the cistrome of fasting-related TFs following fasting?
Although most of the cistromes of fasting-related TFs have been defined, those cistromes were not compared in the physiological fed vs. fasted states (except CREB). This would help in defining what subset of a given cistrome is directly related to fasting.
-
What chromatin transitions occur during fasting?
TF access to binding sites in chromatin is crucial for proper gene regulation. Genome-wide assays to define open chromatin are now available and would provide insight as to how chromatin accessibility is altered following fasting, resulting in changes in transcription.
-
What are the ‘fasting-enhancers’?
In a process so dependent on TFs such as fasting, we need a genome-wide description of fasting-related DNA regulatory elements. This can only be achieved by characterizing TF cistromes following fasting together with a genome-wide assessment of changes in histone modifications and chromatin accessibility following fasting.
-
What is the complete transcriptional response to fasting?
Fasting research is biased towards examining gene regulation of gluconeogenesis. Moreover, within gluconeogenesis the two most heavily studied genes are Pck1 and G6pc. During fasting, a myriad of metabolic reactions occur as part of many metabolic pathways. In the current genomic era, we now have the tools to look at the complete transcriptional response to fasting.
Trends box.
Many transcription factors (TFs) regulate the various metabolic pathways needed to restore homeostasis upon fasting.
Recent advances in genome biology have established a genome-wide characterization of TF binding profiles, showing an extensive, genome-wide transcriptional response to fasting and revealing mechanistic insights regarding TF mode of action.
There is a large degree of co-operation between TFs during fasting which occurs at motif-rich sites on DNA bound by several fasting-related TFs. Much of this collaboration between factors is achieved through localized opening of restrictive chromatin structures.
Fasting initiates transcription cascades wherein TFs regulate the expression of genes encoding other TFs that augment and advance the response to fasting.
Glossary
- Glycogenolysis
catabolism of the polysaccharide glycogen to glucose-6 phosphate which can enter intra-cellular metabolic pathways or be de-phosphorylated to glucose and exit the cell.
- Gluconeogenesis
production of glucose from three- and four-carbon precursors, mainly amino acids, lactate and glycerol. It is essentially the reverse process to glycolysis.
- Ketogenesis
ketone bodies are produced mostly from fatty acid-derived acetyl-CoA during fasting. The three ketone bodies are β-hydroxybutyrate, aceto-acetate and acetone.
- Sequence-specific transcription factors (TFs)
TFs which bind specific sequences in cis-acting DNA regulatory elements (enhancers and promoters). Following binding, these TFs recruit chromatin modifying machinery and transcriptional machinery leading to gene expression.
- Cistrome
the genome-wide binding patterns of a transcription factor to cis-acting DNA regulatory elements (enhancers and promoters)
- Chip-seq and Chip-chip
Chromatin immunoprecipitation followed by massive parallel sequencing (ChIP-seq) is the current technique for defining cistromes. A TF (or a histone modification) is immunoprecipitated along with bound DNA which is then fragmentized, isolated and sequenced. An earlier version of defining TF binding sites in a high throughput manner is ChIP-chip in which the isolated DNA is hybridized to a pre-defined set of DNA fragments. ChIP-seq provides a genome-wide profile of TF binding whereas ChIP-chip is more biased as it covers only selected regions, usually promotes.
- Histone modifications and gene regulation
DNA is wound around the basic unit of chromatin organization – nucleosomes, which are composed of histone proteins. The chemical modification of histone tails (mainly methylation and acetylation) at various positions affects gene expression. Mainly, histone acetyl transferases (HATs, e.g. PCAF, p300 and CREB-binding protein – CBP) are recruited by sequence-specific TFs leading to local histone acetylation, relaxation (‘opening’) of the chromatin environment which enables the activity of more TFs and the basal transcriptional machinery. In contrast, histone de-acetylases (HDACs, e.g. sirtuins) remove acetyl groups from histones leading to compaction (‘closing’) of chromatin and repression of gene expression.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cahill GF., Jr Fuel metabolism in starvation. Annual review of nutrition. 2006;26:1–22. doi: 10.1146/annurev.nutr.26.061505.111258. [DOI] [PubMed] [Google Scholar]
- 2.Derks TG, van Dijk TH, Grefhorst A, Rake JP, Smit GP, Kuipers F, Reijngoud DJ. Inhibition of mitochondrial fatty acid oxidation in vivo only slightly suppresses gluconeogenesis but enhances clearance of glucose in mice. Hepatology. 2008;47:1032–1042. doi: 10.1002/hep.22101. [DOI] [PubMed] [Google Scholar]
- 3.Newman JC, Verdin E. Ketone bodies as signaling metabolites. Trends in endocrinology and metabolism: TEM. 2014;25:42–52. doi: 10.1016/j.tem.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rizza RA. Pathogenesis of fasting and postprandial hyperglycemia in type 2 diabetes: implications for therapy. Diabetes. 2010;59:2697–2707. doi: 10.2337/db10-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saltiel AR. New perspectives into the molecular pathogenesis and treatment of type 2 diabetes. Cell. 2001;104:517–529. doi: 10.1016/s0092-8674(01)00239-2. [DOI] [PubMed] [Google Scholar]
- 6.Teshigawara K, Ogawa W, Mori T, Matsuki Y, Watanabe E, Hiramatsu R, Inoue H, Miyake K, Sakaue H, Kasuga M. Role of Kruppel-like factor 15 in PEPCK gene expression in the liver. Biochemical and biophysical research communications. 2005;327:920–926. doi: 10.1016/j.bbrc.2004.12.096. [DOI] [PubMed] [Google Scholar]
- 7.Unger RH, Roth MG. A new biology of diabetes revealed by leptin. Cell metabolism. 2015;21:15–20. doi: 10.1016/j.cmet.2014.10.011. [DOI] [PubMed] [Google Scholar]
- 8.Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nature reviews. Molecular cell biology. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chalkiadaki A, Guarente L. Sirtuins mediate mammalian metabolic responses to nutrient availability. Nature reviews. Endocrinology. 2012;8:287–296. doi: 10.1038/nrendo.2011.225. [DOI] [PubMed] [Google Scholar]
- 10.Mihaylova MM, Shaw RJ. Metabolic reprogramming by class I and II histone deacetylases. Trends in endocrinology and metabolism: TEM. 2013;24:48–57. doi: 10.1016/j.tem.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimball CP, Murlin JR. AQUEOUS EXTRACTS OF PANCREAS: III. SOME PRECIPITATION REACTIONS OF INSULIN. Journal of Biological Chemistry. 1923;58:337–346. [Google Scholar]
- 12.Ravnskjaer K, Hogan MF, Lackey D, Tora L, Dent SY, Olefsky J, Montminy M. Glucagon regulates gluconeogenesis through KAT2B- and WDR5-mediated epigenetic effects. The Journal of clinical investigation. 2013;123:4318–4328. doi: 10.1172/JCI69035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai WW, Niessen S, Goebel N, Yates JR, 3rd, Guccione E, Montminy M. PRMT5 modulates the metabolic response to fasting signals. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:8870–8875. doi: 10.1073/pnas.1304602110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krones-Herzig A, Mesaros A, Metzger D, Ziegler A, Lemke U, Bruning JC, Herzig S. Signal-dependent control of gluconeogenic key enzyme genes through coactivator-associated arginine methyltransferase 1. The Journal of biological chemistry. 2006;281:3025–3029. doi: 10.1074/jbc.M509770200. [DOI] [PubMed] [Google Scholar]
- 15.Britton SW, Silvette H. Some Observations on the Cortico-Adrenal Hormone. Science. 1931;73:373–374. doi: 10.1126/science.73.1892.373. [DOI] [PubMed] [Google Scholar]
- 16.Patel R, Williams-Dautovich J, Cummins CL. Minireview: new molecular mediators of glucocorticoid receptor activity in metabolic tissues. Molecular endocrinology. 2014;28:999–1011. doi: 10.1210/me.2014-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Opherk C, Tronche F, Kellendonk C, Kohlmuller D, Schulze A, Schmid W, Schutz G. Inactivation of the glucocorticoid receptor in hepatocytes leads to fasting hypoglycemia and ameliorates hyperglycemia in streptozotocin-induced diabetes mellitus. Molecular endocrinology. 2004;18:1346–1353. doi: 10.1210/me.2003-0283. [DOI] [PubMed] [Google Scholar]
- 18.Zinker B, Mika A, Nguyen P, Wilcox D, Ohman L, von Geldern TW, Opgenorth T, Jacobson P. Liver-selective glucocorticoid receptor antagonism decreases glucose production and increases glucose disposal, ameliorating insulin resistance. Metabolism: clinical and experimental. 2007;56:380–387. doi: 10.1016/j.metabol.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 19.Shimazu T, Hirschey MD, Newman J, He W, Shirakawa K, Le Moan N, Grueter CA, Lim H, Saunders LR, Stevens RD, Newgard CB, Farese RV, Jr, de Cabo R, Ulrich S, Akassoglou K, Verdin E. Suppression of oxidative stress by beta-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science. 2013;339:211–214. doi: 10.1126/science.1227166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kersten S. Integrated physiology and systems biology of PPARalpha. Mol Metab. 2014;3:354–371. doi: 10.1016/j.molmet.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernal-Mizrachi C, Weng S, Feng C, Finck BN, Knutsen RH, Leone TC, Coleman T, Mecham RP, Kelly DP, Semenkovich CF. Dexamethasone induction of hypertension and diabetes is PPAR-alpha dependent in LDL receptor-null mice. Nat Med. 2003;9:1069–1075. doi: 10.1038/nm898. [DOI] [PubMed] [Google Scholar]
- 22.Steineger HH, Sorensen HN, Tugwood JD, Skrede S, Spydevold O, Gautvik KM. Dexamethasone and insulin demonstrate marked and opposite regulation of the steady-state mRNA level of the peroxisomal proliferator-activated receptor (PPAR) in hepatic cells. Hormonal modulation of fatty-acid-induced transcription. Eur J Biochem. 1994;225:967–974. doi: 10.1111/j.1432-1033.1994.0967b.x. [DOI] [PubMed] [Google Scholar]
- 23.Lee HY, Gao X, Barrasa MI, Li H, Elmes RR, Peters LL, Lodish HF. PPAR-alpha and glucocorticoid receptor synergize to promote erythroid progenitor self-renewal. Nature. 2015 doi: 10.1038/nature14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tikhanovich I, Cox J, Weinman SA. Forkhead box class O transcription factors in liver function and disease. J Gastroenterol Hepatol. 2013;28(Suppl 1):125–131. doi: 10.1111/jgh.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haeusler RA, Kaestner KH, Accili D. FoxOs function synergistically to promote glucose production. The Journal of biological chemistry. 2010;285:35245–35248. doi: 10.1074/jbc.C110.175851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang K, Li L, Qi Y, Zhu X, Gan B, DePinho RA, Averitt T, Guo S. Hepatic suppression of Foxo1 and Foxo3 causes hypoglycemia and hyperlipidemia in mice. Endocrinology. 2012;153:631–646. doi: 10.1210/en.2011-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiong X, Tao R, DePinho RA, Dong XC. Deletion of hepatic FoxO1/3/4 genes in mice significantly impacts on glucose metabolism through downregulation of gluconeogenesis and upregulation of glycolysis. PloS one. 2013;8:e74340. doi: 10.1371/journal.pone.0074340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haeusler RA, Hartil K, Vaitheesvaran B, Arrieta-Cruz I, Knight CM, Cook JR, Kammoun HL, Febbraio MA, Gutierrez-Juarez R, Kurland IJ, Accili D. Integrated control of hepatic lipogenesis versus glucose production requires FoxO transcription factors. Nature communications. 2014;5:5190. doi: 10.1038/ncomms6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim DH, Perdomo G, Zhang T, Slusher S, Lee S, Phillips BE, Fan Y, Giannoukakis N, Gramignoli R, Strom S, Ringquist S, Dong HH. FoxO6 integrates insulin signaling with gluconeogenesis in the liver. Diabetes. 2011;60:2763–2774. doi: 10.2337/db11-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calabuig-Navarro V, Yamauchi J, Lee S, Zhang T, Liu YZ, Sadlek K, Goudriet GM, Piganelli JD, Jiang CL, Miller R, Lowe M, Harashima H, Dong HH. FoxO6 Depletion Attenuates Hepatic Gluconeogenesis and Protects Against Fat-Induced Glucose Disorder in Mice. The Journal of biological chemistry. 2015 doi: 10.1074/jbc.M115.650994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell metabolism. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 32.Kaestner KH. The FoxA factors in organogenesis and differentiation. Current opinion in genetics & development. 2010;20:527–532. doi: 10.1016/j.gde.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolfrum C, Asilmaz E, Luca E, Friedman JM, Stoffel M. Foxa2 regulates lipid metabolism and ketogenesis in the liver during fasting and in diabetes. Nature. 2004;432:1027–1032. doi: 10.1038/nature03047. [DOI] [PubMed] [Google Scholar]
- 34.Zhang L, Rubins NE, Ahima RS, Greenbaum LE, Kaestner KH. Foxa2 integrates the transcriptional response of the hepatocyte to fasting. Cell metabolism. 2005;2:141–148. doi: 10.1016/j.cmet.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 35.Roesler WJ. The role of C/EBP in nutrient and hormonal regulation of gene expression. Annual review of nutrition. 2001;21:141–165. doi: 10.1146/annurev.nutr.21.1.141. [DOI] [PubMed] [Google Scholar]
- 36.Shen N, Jiang S, Lu JM, Yu X, Lai SS, Zhang JZ, Zhang JL, Tao WW, Wang XX, Xu N, Xue B, Li CJ. The constitutive activation of Egr-1/C/EBPa mediates the development of type 2 diabetes mellitus by enhancing hepatic gluconeogenesis. The American journal of pathology. 2015;185:513–523. doi: 10.1016/j.ajpath.2014.10.016. [DOI] [PubMed] [Google Scholar]
- 37.Pan D, Mao C, Zou T, Yao AY, Cooper MP, Boyartchuk V, Wang YX. The histone demethylase Jhdm1a regulates hepatic gluconeogenesis. PLoS genetics. 2012;8:e1002761. doi: 10.1371/journal.pgen.1002761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Louet JF, Chopra AR, Sagen JV, An J, York B, Tannour-Louet M, Saha PK, Stevens RD, Wenner BR, Ilkayeva OR, Bain JR, Zhou S, DeMayo F, Xu J, Newgard CB, O’Malley BW. The coactivator SRC-1 is an essential coordinator of hepatic glucose production. Cell metabolism. 2010;12:606–618. doi: 10.1016/j.cmet.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anik A, Catli G, Abaci A, Bober E. Maturity-onset diabetes of the young (MODY): an update. J Pediatr Endocrinol Metab. 2015;28:251–263. doi: 10.1515/jpem-2014-0384. [DOI] [PubMed] [Google Scholar]
- 40.Jitrapakdee S. Transcription factors and coactivators controlling nutrient and hormonal regulation of hepatic gluconeogenesis. The international journal of biochemistry & cell biology. 2012;44:33–45. doi: 10.1016/j.biocel.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 41.Viollet B, Kahn A, Raymondjean M. Protein kinase A-dependent phosphorylation modulates DNA-binding activity of hepatocyte nuclear factor 4. Molecular and cellular biology. 1997;17:4208–4219. doi: 10.1128/mcb.17.8.4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dankel SN, Hoang T, Flageng MH, Sagen JV, Mellgren G. cAMP-mediated regulation of HNF-4alpha depends on the level of coactivator PGC-1alpha. Biochimica et biophysica acta. 2010;1803:1013–1019. doi: 10.1016/j.bbamcr.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 43.Kornfeld JW, Baitzel C, Konner AC, Nicholls HT, Vogt MC, Herrmanns K, Scheja L, Haumaitre C, Wolf AM, Knippschild U, Seibler J, Cereghini S, Heeren J, Stoffel M, Bruning JC. Obesity-induced overexpression of miR-802 impairs glucose metabolism through silencing of Hnf1b. Nature. 2013;494:111–115. doi: 10.1038/nature11793. [DOI] [PubMed] [Google Scholar]
- 44.Mullur R, Liu YY, Brent GA. Thyroid hormone regulation of metabolism. Physiological reviews. 2014;94:355–382. doi: 10.1152/physrev.00030.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Ma K, Song S, Elam MB, Cook GA, Park EA. Peroxisomal proliferator-activated receptor-gamma coactivator-1 alpha (PGC-1 alpha) enhances the thyroid hormone induction of carnitine palmitoyltransferase I (CPT-I alpha) The Journal of biological chemistry. 2004;279:53963–53971. doi: 10.1074/jbc.M406028200. [DOI] [PubMed] [Google Scholar]
- 46.Singh SP, Snyder AK. Effect of thyrotoxicosis on gluconeogenesis from alanine in the perfused rat liver. Endocrinology. 1978;102:182–187. doi: 10.1210/endo-102-1-182. [DOI] [PubMed] [Google Scholar]
- 47.Jackson-Hayes L, Song S, Lavrentyev EN, Jansen MS, Hillgartner FB, Tian L, Wood PA, Cook GA, Park EA. A thyroid hormone response unit formed between the promoter and first intron of the carnitine palmitoyltransferase-Ialpha gene mediates the liver-specific induction by thyroid hormone. The Journal of biological chemistry. 2003;278:7964–7972. doi: 10.1074/jbc.M211062200. [DOI] [PubMed] [Google Scholar]
- 48.Lee JM, Wagner M, Xiao R, Kim KH, Feng D, Lazar MA, Moore DD. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014;516:112–115. doi: 10.1038/nature13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sinha RA, You SH, Zhou J, Siddique MM, Bay BH, Zhu X, Privalsky ML, Cheng SY, Stevens RD, Summers SA, Newgard CB, Lazar MA, Yen PM. Thyroid hormone stimulates hepatic lipid catabolism via activation of autophagy. The Journal of clinical investigation. 2012;122:2428–2438. doi: 10.1172/JCI60580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goldstein I, Rotter V. Regulation of lipid metabolism by p53 - fighting two villains with one sword. Trends in endocrinology and metabolism: TEM. 2012;23:567–575. doi: 10.1016/j.tem.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 51.Goldstein I, Yizhak K, Madar S, Goldfinger N, Ruppin E, Rotter V. p53 promotes the expression of gluconeogenesis-related genes and enhances hepatic glucose production. Cancer & metabolism. 2013;1:9. doi: 10.1186/2049-3002-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang SJ, Yu G, Jiang L, Li T, Lin Q, Tang Y, Gu W. p53-Dependent regulation of metabolic function through transcriptional activation of pantothenate kinase-1 gene. Cell cycle. 2013;12:753–761. doi: 10.4161/cc.23597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goldstein I, Ezra O, Rivlin N, Molchadsky A, Madar S, Goldfinger N, Rotter V. p53, a novel regulator of lipid metabolism pathways. Journal of hepatology. 2012;56:656–662. doi: 10.1016/j.jhep.2011.08.022. [DOI] [PubMed] [Google Scholar]
- 54.Liu Y, He Y, Jin A, Tikunov AP, Zhou L, Tollini LA, Leslie P, Kim TH, Li LO, Coleman RA, Gu Z, Chen YQ, Macdonald JM, Graves LM, Zhang Y. Ribosomal protein-Mdm2-p53 pathway coordinates nutrient stress with lipid metabolism by regulating MCD and promoting fatty acid oxidation. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E2414–2422. doi: 10.1073/pnas.1315605111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang P, Tu B, Wang H, Cao Z, Tang M, Zhang C, Gu B, Li Z, Wang L, Yang Y, Zhao Y, Wang H, Luo J, Deng CX, Gao B, Roeder RG, Zhu WG. Tumor suppressor p53 cooperates with SIRT6 to regulate gluconeogenesis by promoting FoxO1 nuclear exclusion. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:10684–10689. doi: 10.1073/pnas.1411026111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yi J, Luo J. SIRT1 and p53, effect on cancer, senescence and beyond. Biochimica et biophysica acta. 2010;1804:1684–1689. doi: 10.1016/j.bbapap.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, Milne J, Meyers DJ, Cole P, Yates J, 3rd, Olefsky J, Guarente L, Montminy M. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456:269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 59.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 60.Phuc Le P, Friedman JR, Schug J, Brestelli JE, Parker JB, Bochkis IM, Kaestner KH. Glucocorticoid receptor-dependent gene regulatory networks. PLoS genetics. 2005;1:e16. doi: 10.1371/journal.pgen.0010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grontved L, John S, Baek S, Liu Y, Buckley JR, Vinson C, Aguilera G, Hager GL. C/EBP maintains chromatin accessibility in liver and facilitates glucocorticoid receptor recruitment to steroid response elements. The EMBO journal. 2013;32:1568–1583. doi: 10.1038/emboj.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Everett LJ, Le Lay J, Lukovac S, Bernstein D, Steger DJ, Lazar MA, Kaestner KH. Integrative genomic analysis of CREB defines a critical role for transcription factor networks in mediating the fed/fasted switch in liver. BMC genomics. 2013;14:337. doi: 10.1186/1471-2164-14-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, Chen H, Jenner R, Herbolsheimer E, Jacobsen E, Kadam S, Ecker JR, Emerson B, Hogenesch JB, Unterman T, Young RA, Montminy M. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramadoss P, Abraham BJ, Tsai L, Zhou Y, Costa-e-Sousa RH, Ye F, Bilban M, Zhao K, Hollenberg AN. Novel mechanism of positive versus negative regulation by thyroid hormone receptor beta1 (TRbeta1) identified by genome-wide profiling of binding sites in mouse liver. The Journal of biological chemistry. 2014;289:1313–1328. doi: 10.1074/jbc.M113.521450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grontved L, Waterfall JJ, Kim DW, Baek S, Sung MH, Zhao L, Park JW, Nielsen R, Walker RL, Zhu YJ, Meltzer PS, Hager GL, Cheng SY. Transcriptional activation by the thyroid hormone receptor through ligand-dependent receptor recruitment and chromatin remodelling. Nature communications. 2015;6:7048. doi: 10.1038/ncomms8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Patel R, Patel M, Tsai R, Lin V, Bookout AL, Zhang Y, Magomedova L, Li T, Chan JF, Budd C, Mangelsdorf DJ, Cummins CL. LXRbeta is required for glucocorticoid-induced hyperglycemia and hepatosteatosis in mice. The Journal of clinical investigation. 2011;121:431–441. doi: 10.1172/JCI41681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van der Meer DL, Degenhardt T, Vaisanen S, de Groot PJ, Heinaniemi M, de Vries SC, Muller M, Carlberg C, Kersten S. Profiling of promoter occupancy by PPARalpha in human hepatoma cells via ChIP-chip analysis. Nucleic acids research. 2010;38:2839–2850. doi: 10.1093/nar/gkq012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boergesen M, Pedersen TA, Gross B, van Heeringen SJ, Hagenbeek D, Bindesboll C, Caron S, Lalloyer F, Steffensen KR, Nebb HI, Gustafsson JA, Stunnenberg HG, Staels B, Mandrup S. Genome-wide profiling of liver X receptor, retinoid X receptor, and peroxisome proliferator-activated receptor alpha in mouse liver reveals extensive sharing of binding sites. Molecular and cellular biology. 2012;32:852–867. doi: 10.1128/MCB.06175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McMullen PD, Bhattacharya S, Woods CG, Sun B, Yarborough K, Ross SM, Miller ME, McBride MT, LeCluyse EL, Clewell RA, Andersen ME. A map of the PPARalpha transcription regulatory network for primary human hepatocytes. Chem Biol Interact. 2014;209:14–24. doi: 10.1016/j.cbi.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 70.Calkin AC, Tontonoz P. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nature reviews. Molecular cell biology. 2012;13:213–224. doi: 10.1038/nrm3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shin DJ, Joshi P, Hong SH, Mosure K, Shin DG, Osborne TF. Genome-wide analysis of FoxO1 binding in hepatic chromatin: potential involvement of FoxO1 in linking retinoid signaling to hepatic gluconeogenesis. Nucleic acids research. 2012;40:11499–11509. doi: 10.1093/nar/gks932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Z, Tuteja G, Schug J, Kaestner KH. Foxa1 and Foxa2 are essential for sexual dimorphism in liver cancer. Cell. 2012;148:72–83. doi: 10.1016/j.cell.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Johnson ME, Schug J, Wells AD, Kaestner KH, Grant SF. Genome-wide analyses of ChIP-Seq derived FOXA2 DNA occupancy in liver points to genetic networks underpinning multiple complex traits. The Journal of clinical endocrinology and metabolism. 2014;99:E1580–1585. doi: 10.1210/jc.2013-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, Talianidis I, Flicek P, Odom DT. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Voss TC, Schiltz RL, Sung MH, Yen PM, Stamatoyannopoulos JA, Biddie SC, Johnson TA, Miranda TB, John S, Hager GL. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011;146:544–554. doi: 10.1016/j.cell.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pei L, Waki H, Vaitheesvaran B, Wilpitz DC, Kurland IJ, Tontonoz P. NR4A orphan nuclear receptors are transcriptional regulators of hepatic glucose metabolism. Nature medicine. 2006;12:1048–1055. doi: 10.1038/nm1471. [DOI] [PubMed] [Google Scholar]
- 77.Min AK, Bae KH, Jung YA, Choi YK, Kim MJ, Kim JH, Jeon JH, Kim JG, Lee IK, Park KG. Orphan nuclear receptor Nur77 mediates fasting-induced hepatic fibroblast growth factor 21 expression. Endocrinology. 2014;155:2924–2931. doi: 10.1210/en.2013-1758. [DOI] [PubMed] [Google Scholar]
- 78.Seok S, Fu T, Choi SE, Li Y, Zhu R, Kumar S, Sun X, Yoon G, Kang Y, Zhong W, Ma J, Kemper B, Kemper JK. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature. 2014;516:108–111. doi: 10.1038/nature13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Settembre C, De Cegli R, Mansueto G, Saha PK, Vetrini F, Visvikis O, Huynh T, Carissimo A, Palmer D, Klisch TJ, Wollenberg AC, Di Bernardo D, Chan L, Irazoqui JE, Ballabio A. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nature cell biology. 2013;15:647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Longuet C, Sinclair EM, Maida A, Baggio LL, Maziarz M, Charron MJ, Drucker DJ. The glucagon receptor is required for the adaptive metabolic response to fasting. Cell metabolism. 2008;8:359–371. doi: 10.1016/j.cmet.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu Y, Xiong X, Wang X, Zhang Z, Li J, Shi G, Yang J, Zhang H, Ning G, Li X. Yin Yang 1 promotes hepatic gluconeogenesis through upregulation of glucocorticoid receptor. Diabetes. 2013;62:1064–1073. doi: 10.2337/db12-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Takashima M, Ogawa W, Hayashi K, Inoue H, Kinoshita S, Okamoto Y, Sakaue H, Wataoka Y, Emi A, Senga Y, Matsuki Y, Watanabe E, Hiramatsu R, Kasuga M. Role of KLF15 in regulation of hepatic gluconeogenesis and metformin action. Diabetes. 2010;59:1608–1615. doi: 10.2337/db09-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gray S, Wang B, Orihuela Y, Hong EG, Fisch S, Haldar S, Cline GW, Kim JK, Peroni OD, Kahn BB, Jain MK. Regulation of gluconeogenesis by Kruppel-like factor 15. Cell metabolism. 2007;5:305–312. doi: 10.1016/j.cmet.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen S, Qian J, Shi X, Gao T, Liang T, Liu C. Control of hepatic gluconeogenesis by the promyelocytic leukemia zinc finger protein. Molecular endocrinology. 2014;28:1987–1998. doi: 10.1210/me.2014-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Danno H, Ishii KA, Nakagawa Y, Mikami M, Yamamoto T, Yabe S, Furusawa M, Kumadaki S, Watanabe K, Shimizu H, Matsuzaka T, Kobayashi K, Takahashi A, Yatoh S, Suzuki H, Yamada N, Shimano H. The liver-enriched transcription factor CREBH is nutritionally regulated and activated by fatty acids and PPARalpha. Biochemical and biophysical research communications. 2010;391:1222–1227. doi: 10.1016/j.bbrc.2009.12.046. [DOI] [PubMed] [Google Scholar]
- 86.Lee MW, Chanda D, Yang J, Oh H, Kim SS, Yoon YS, Hong S, Park KG, Lee IK, Choi CS, Hanson RW, Choi HS, Koo SH. Regulation of hepatic gluconeogenesis by an ER-bound transcription factor, CREBH. Cell metabolism. 2010;11:331–339. doi: 10.1016/j.cmet.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 87.Bae KH, Min AK, Kim JG, Lee IK, Park KG. Alpha lipoic acid induces hepatic fibroblast growth factor 21 expression via up-regulation of CREBH. Biochemical and biophysical research communications. 2014;455:212–217. doi: 10.1016/j.bbrc.2014.10.147. [DOI] [PubMed] [Google Scholar]
- 88.Kim H, Mendez R, Zheng Z, Chang L, Cai J, Zhang R, Zhang K. Liver-enriched transcription factor CREBH interacts with peroxisome proliferator-activated receptor alpha to regulate metabolic hormone FGF21. Endocrinology. 2014;155:769–782. doi: 10.1210/en.2013-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nakagawa Y, Satoh A, Yabe S, Furusawa M, Tokushige N, Tezuka H, Mikami M, Iwata W, Shingyouchi A, Matsuzaka T, Kiwata S, Fujimoto Y, Shimizu H, Danno H, Yamamoto T, Ishii K, Karasawa T, Takeuchi Y, Iwasaki H, Shimada M, Kawakami Y, Urayama O, Sone H, Takekoshi K, Kobayashi K, Yatoh S, Takahashi A, Yahagi N, Suzuki H, Yamada N, Shimano H. Hepatic CREB3L3 controls whole-body energy homeostasis and improves obesity and diabetes. Endocrinology. 2014;155:4706–4719. doi: 10.1210/en.2014-1113. [DOI] [PubMed] [Google Scholar]
- 90.Badman MK, Pissios P, Kennedy AR, Koukos G, Flier JS, Maratos-Flier E. Hepatic fibroblast growth factor 21 is regulated by PPARalpha and is a key mediator of hepatic lipid metabolism in ketotic states. Cell metabolism. 2007;5:426–437. doi: 10.1016/j.cmet.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 91.Inagaki T, Dutchak P, Zhao G, Ding X, Gautron L, Parameswara V, Li Y, Goetz R, Mohammadi M, Esser V, Elmquist JK, Gerard RD, Burgess SC, Hammer RE, Mangelsdorf DJ, Kliewer SA. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell metabolism. 2007;5:415–425. doi: 10.1016/j.cmet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 92.Patel R, Bookout AL, Magomedova L, Owen BM, Consiglio GP, Shimizu M, Zhang Y, Mangelsdorf DJ, Kliewer SA, Cummins CL. Glucocorticoids regulate the metabolic hormone FGF21 in a feed-forward loop. Mol Endocrinol. 2015;29:213–223. doi: 10.1210/me.2014-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cheng X, Vispute SG, Liu J, Cheng C, Kharitonenkov A, Klaassen CD. Fibroblast growth factor (Fgf) 21 is a novel target gene of the aryl hydrocarbon receptor (AhR) Toxicol Appl Pharmacol. 2014;278:65–71. doi: 10.1016/j.taap.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.De Sousa-Coelho AL, Marrero PF, Haro D. Activating transcription factor 4-dependent induction of FGF21 during amino acid deprivation. The Biochemical journal. 2012;443:165–171. doi: 10.1042/BJ20111748. [DOI] [PubMed] [Google Scholar]
- 95.Wang Y, Solt LA, Burris TP. Regulation of FGF21 expression and secretion by retinoic acid receptor-related orphan receptor alpha. The Journal of biological chemistry. 2010;285:15668–15673. doi: 10.1074/jbc.M110.102160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cyphert HA, Ge X, Kohan AB, Salati LM, Zhang Y, Hillgartner FB. Activation of the farnesoid X receptor induces hepatic expression and secretion of fibroblast growth factor 21. J Biol Chem. 2012;287:25123–25138. doi: 10.1074/jbc.M112.375907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li H, Gao Z, Zhang J, Ye X, Xu A, Ye J, Jia W. Sodium butyrate stimulates expression of fibroblast growth factor 21 in liver by inhibition of histone deacetylase 3. Diabetes. 2012;61:797–806. doi: 10.2337/db11-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]