Abstract
Non-Communicable diseases (NCDs), including obesity, are emerging as the major health concern of the 21st century. Excess adiposity and related NCD metabolic disturbances have stimulated development of new lipid compartment measurement technologies to help us understand cellular energy exchange, to refine phenotypes, and to develop predictive markers of adverse clinical outcomes. Recent advances now allow for quantification of multiple intracellular lipid and adipose tissue compartments that can be evaluated across the human lifespan. With magnetic resonance methods leading the way, newer approaches will give molecular structural and metabolic information beyond the laboratory in real-world settings. The union between these new technologies and the growing NCD population is creating an exciting interface advancing our understanding of chronic disease mechanisms.
Keywords: obesity, adiposity, imaging, metabolism, body composition
Lipid Compartment Measurement Advances
A new view of body lipid compartments is emerging: once considered the main storage site for body lipids and a metabolically homogenous compartment, adipose tissue is increasingly recognized as comprised of many sub-compartments that have distinct anatomic and functional characteristics [1]. Non-adipose tissue lipids, that in the past were viewed primarily as essential components of cell membranes, are increasingly being recognized in the form of cytoplasmic organelles linked with pathological metabolic states [2]. This transformation in the science of lipid compartment biology coincides with the growing recognition that excess adiposity and its metabolic consequences are the pathological foundation for the emerging non-communicable disease epidemic [3].
One reason for the acceleration in the science of lipid compartment biology in both health and disease is the growing capability of safely and non-invasively quantifying body tissue lipids in living humans across the lifespan. Understanding cellular energy exchange, refining phenotypes in the search for causative genes, and developing predictive markers of adverse clinical outcomes are all activities enhanced by the emerging ability to quantify lipid compartments in vivo.
Our review provides a background on these multiple methods with a focus on selected recent technological advances that are filling important knowledge gaps. An expanded discussion of the more traditional methods can be found in earlier reports [1].
Emerging Methods Closing Knowledge Gaps
The most important development, one that continues to advance, was the introduction in the 1980s of magnetic resonance imaging (MRI) and related magnetic resonance spectroscopy (MRS) systems designed to accommodate human subjects [4]. From lipid droplets to whole body adipose tissue, and more recently brown adipose tissue (BAT), MRI and MRS are not only a technological tour-de-force, but studies are safe to conduct in a vast range of subjects differing in age, body size, and health status.
T1-Weighted Magnetic Resonance Imaging
T1-weighted imaging is based on the differential exponential growth or “relaxation” rates of fat and water proton signals as they recover after radiofrequency excitation. T1-weighted MRI is primarily used for white adipose tissue (WAT) quantification, as fat (i.e., triglycerides in lipid droplets) is characteristically bright in contrast to other lean tissues and organs due to its rapid T1 recovery rate [5]. The sharp contrast between adipose and water-containing lean tissues creates the visual basis that trained technicians exploit to “segment” each image, using partially automated software, into adipose and non-adipose tissue components. A single cross-sectional scan can be used to estimate a component’s area and multiple scans can be combined to calculate a component’s partial or total volume. As whole-body contiguous scan protocols involve collection of a large number of images and long analysis times, a frequent approach is to scan subjects at predefined intervals (e.g., 3–5 cm), with prediction errors a function of the component’s volume and slice gap [6]. Recent advances in both rapid data acquisition and image post-processing algorithms are increasingly providing the opportunity for automating the whole-body tissue segmentation process than can be achieved within a few minutes [5, 7–9]. The introduction and growing availability of large-bore MRI scanners now enables investigators to evaluate most subjects who are moderately obese with a high body mass index (BMI).
Since metabolically important visceral adipose tissue (VAT) is within the abdomen, a frequent approach is to only scan the trunk region. Approaches range from a single slice at a predefined abdominal level to the whole trunk that includes intrathoracic, intra-abdominal, and intra-pelvic cavities and associated adipose tissues [10]. The “VAT” reported in published studies thus depends on the specific scanning protocol. There are many VAT subcomponents within the trunk that potentially have distinct metabolic properties [11], and there are a growing number of studies designed to examine epicardial, perirenal, and other small adipose tissue compartments [12].
An example of this evolution is the changing approach used to quantify abdominal VAT in phenotyping and metabolic studies. Early single-slice computed tomography (CT) and MRI studies were usually obtained between the 4th and 5th lumbar vertebral level. That approach gained traction as L4–L5 is an easily identified landmark and usually has abundant VAT compared to other trunk regions. That largely empirical approach is now being replaced with studies showing improved representation of total VAT or stronger associations with cardiovascular and type 2 diabetes risk factors at other levels [5, 13–15]. Recent studies have identified the VAT image slice cross-sectional areas 5–10 cm above L4–L5 or at T12 (12th thoracic vertebra) to L1, L1–L2, L2–3, and L3–L4 as having the strongest associations with total VAT volume and cardiovascular disease risk in overweight and obese individuals [13, 14, 16–21]. New single slice VAT imaging protocols are thus favoring locations higher in the abdomen than the traditional L4–L5 level. Nevertheles, the field as yet lacks accepted “standard” approaches for acquiring VAT scans, segmenting images, and for training and certifying analysts.
Because of the unique properties of each fat depot and their differential contribution to metabolic derrangement in the face of obesity, seperation of pathophysiologically important mesenteric, omental, and extraperitoneal adipose tissue components during the segmentation process is important. However, this remains a concern with abdominal VAT analyses using T1-weighted MRI. Respiration, peristaltic activity, and irregular compartment shapes make it difficult for the image analyst to separate these three major portions of abdominal VAT. Single or multiple slice abdominal VAT analyses typically thus report only a total abdominal VAT value.
Subcutaneous adipose tissue (SAT) volume for the whole body can be estimated from a single slice at L5-S1 or 5 cm below L4–L5 [22, 23]. Compared to VAT, predictions of total SAT from single slice SAT images are less influenced by abdominal anatomic location. Although a single image slice often shows good associations with whole-body components in cross-sectional studies, these measures perform less well as VAT and SAT predictors during weight loss [23, 24]. This is a concern when studies are designed specifically to examine the relative magnitude of SAT or VAT changes with a specific weight loss intervention.
Chemical-Shift Based Magnetic Resonance
Chemical-shift based magnetic resonance methods represent a large group of methods that share in common underlying lipid measurement approaches [25–27]. These methods include single-voxel MRS, magnetic resonance spectroscopic imaging (MRSI), chemical-shift selective MRI, and chemical-shift encoded (CSE) MRI that are increasingly the mainstay for non-invasively quantifying tissue fat. All of these methods rely on the principle of chemical-shift, the fundamental basis of nuclear magnetic resonance (NMR). One common quantitative endpoint that has been validated in the literature is the fat-signal fraction, which is the ratio of the proton fat signal to the sum of the proton fat and bulk free water signals [28]. The fat-signal fraction is commonly used to measure lipid in ectopic locations and in organs [29–33]. However, the fat-signal fraction does not provide any information on tissue fatty acid composition (i.e., chain length, unsaturation) and it is not a requisite approach for measuring adipose tissue volume[34].
MRS and MRSI technology and applications
MRS and MRSI provide the most direct approach to discriminate tissue fat and water proton component signals in vivo [35]. These methods yield a detailed frequency spectrum of water and fat resonance peaks that provides an intuitive visualization of the presence and quantity (i.e., relative area under each spectral peak) of water and fat proton moieties within the interrogated voxel. Whereas water is a symmetrical molecule with two hydroxyl protons that give rise to a single resonance, protons in triglyceride molecules give rise to a group of resonances as shown in Figure 1 with MRS data from a 3.0 Tesla scan. The predominant resonance shown is the methylene (CH2) peak from the triglyceride carbon backbone.
Figure 1. Representative proton spectrum of safflower oil showing triglyceride resonances.
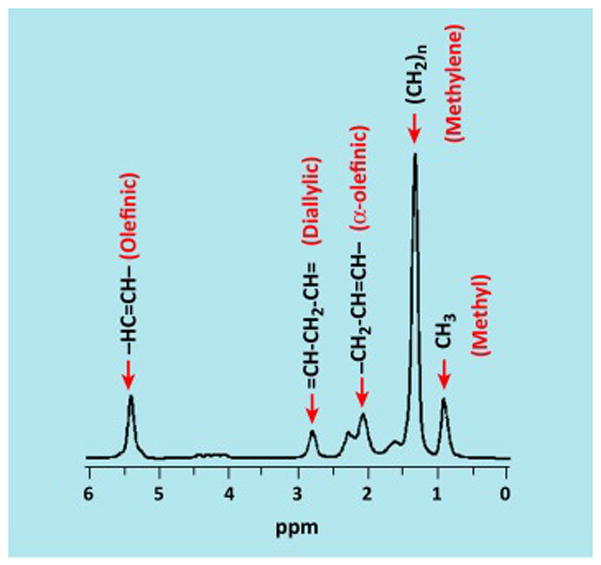
The localized safflower oil proton spectrum acquired at 3 Tesla that shows multiple triglyceride resonances. The predominant methylene peak, corresponding to hydrogens in the CH2 moiety, is located at 1.3 ppm. Additional proton resonances related to triglyceride’s carbon backbone are also visible [36]. Illustration courtesy of Jürgen Machann, University Hospital Tübingen, Germany.
Additionally, while MRS and MRSI techniques were used in the past to further quantify triglyceride properties such as carbon chain length, number of double bonds, and the degree of mono- and poly-unsaturation in vitro, advanced CSE algorithms are also emerging to estimate these molecular traits in living humans (Figure 2)[36]. The NMR spectrum shown in figure 2 demonstrates greater triglyceride fatty acid saturation in deep SAT and VAT compared to superficial and lower extremity SAT, in a healthy man. Adipose tissue triglyceride composition is the reference method for representing dietary fatty acid intake [37] and the potential to non-invasively analyze multiple sites extends the traditional SAT biopsy approach. Cardiovascular disease risk may be increased by a diet high in saturated triglyceride fatty acids [38] and an objective biomarker of dietary fat composition is an important advance over often-unreliable self-reported food records.
Figure 2. Representative MRS examples from a healthy man’s adipose tissue compartments.
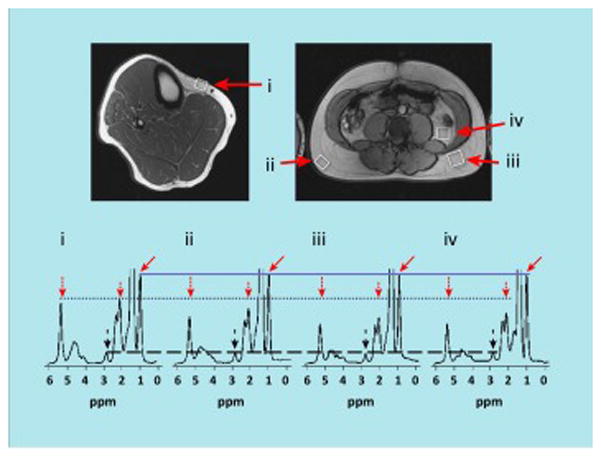
MRS examples obtained at 3 Tesla from various adipose tissue compartments of a healthy man: (i) lower leg subcutaneous, (ii) abdominal superficial subcutaneous, (iii) abdominal deep subcutaneous, and (iv) visceral. White boxes in top images show locations of the interrogating MRS voxel, corresponding to the enlarged spectra shown below. In each of the four spectra, the CH3 methyl resonance (solid arrows) serves as an internal reference (solid line) since each triglyceride molecule has a constant of 9 methyl protons. Marked differences in fatty acid composition between the adipose tissue compartments is evidenced by the varying amplitudes of the other triglyceride resonances (dotted line) near 2.0 ppm (short dotted arrows) and 5.3 ppm (long dotted arrows), namely the allylic and vinyl peaks, respectively. Little differences are seen in the diallylic peaks at 2.8 ppm (dashed lines and dashed arrows). Data suggests that triglycerides in (iii) deep subcutaneous and (iv) visceral adipose tissues are more saturated than (ii) superficial and (i) lower extremity subcutaneous adipose tissues. Illustration courtesy of Jürgen Machann, University Hospital Tübingen, Germany.
While 1.5 and 3 Tesla magnetic resonance systems are available in most clinical and research settings, higher field strength scanners are increasingly being used at specialized centers to conduct human imaging and spectroscopy studies. A recent study to understand the composition of adipose tissue and marrow fat in humans reported improved rapid analysis of triglyceride saturation level with 1H MRS at 7 Tesla [39].
Lipid droplets or “adiposomes” are the smallest recognized lipid compartments (20–40 nm to 100 μm diameter) that reside in the cytoplasm of many cell types. They are in close proximity to mitochondria [40], and serve as a link that provides a flow of triglyceride-derived free-fatty acids for energy production through beta-oxidation. The lipid droplet triglyceride core is surrounded by a phospholipid bilayer and membrane associated proteins [41]. These inducible organelles, in addition to serving as a storage site for energy-containing triglyceride, play an important role in cell signaling, the control of inflammatory mediator (e.g., eicosanoids) synthesis and secretion, and in the generation of free-fatty acid-derived intermediates (e.g., sphingolipid ceramides) that may play a key role in lipid-induced lipotoxicity, insulin resistance, and pancreatic β cell dysfunction [42]. Understanding the localization and composition of these organelles is thus important as accumulation of lipid droplets outside of adipocyte is often linked with adverse metabolic states [42].
Both MRS and MRSI have been widely used to evaluate lipid droplet biology through their detection of intracellular triglyceride[32]. These applications have identified lipid droplet triglycerides in the liver [31], heart [33], pancreas [30], skeletal muscles, myocardium, and more recently in BAT.
Chemical-shift selective MRI: in chemical-shift selective MRI, spectrally selective pulse sequences are applied to suppress either water or the predominant methylene fat signals prior to data acquisition. Subsequently, signals from the non-suppressed component are acquired for image formation. Thus, a water-suppressed image will exhibit fat-only signals, while conversely a fat-suppressed image will exhibit only tissue water signals. The presence of fat and subsequent fat-signal fraction measurements can be determined by acquiring a water-suppressed (i.e., fat-only) or a fat-suppressed (i.e., water-only) scan and comparing the results to a complementary non-suppressed scan (i.e., water and fat). Chemical-shift selective MRI techniques are commonly used in the quantification of lower-extremity skeletal muscle fat content. These approaches have also been explored as a means of quantifying SAT, VAT, and intermuscular adipose tissue volumes. As an adjunct to MRS, chemical-shift selective MRI protocols have been used to estimate hepatic fat content [5, 36, 43].
CSE MRI
CSE water-fat imaging, popularly known as Dixon MRI, is a class of techniques that has emerged over the past decade as a promising alternative approach to evaluating organ fat content with high spatial resolution [25]. Unlike chemical-shift selective MRI, CSE encoded MRI methods do not employ mechanisms to suppress water or fat signals. Rather, the combined water and fat signals are acquired using two or more echo times. Due to the difference in resonance frequencies between water and fat protons, chemical-shift effects are captured phase differences between water and fat signals via multiple echoes. Mathematical algorithms are subsequently used in data reconstruction to decompose the acquired multi-echo signals into separated water and fat components [44]. This procedure is performed on a voxel-wise basis and a fat-signal fraction map can then be computed from the estimated water and fat signals. CSE encoded MRI and quantitative fat-signal fraction imaging has validated applications for estimating whole-body composition and fat fractions in liver, pancreas, heart, bone marrow, and skeletal muscle [25].
One of the recently demonstrated advantages of CSE and its fat-signal fraction map is the technique’s ability to facilitate more robust automated segmentation post-processing routines for total-body adipose tissue quantification [45, 46](Figure 3). A critical question in this rapidly emerging area is how well these automated methods compare to the currently accepted semi-automated segmentation T1-weighted imaging approach reviewed in an earlier section and these studies are now underway[5, 46, 47]. Additionally, another automated option for quantifying the metabolically-relevant VAT compartment was recently added to dual-energy X-ray absorptiometry (DXA) software [48, 49]. Predicted VAT values are derived by this DXA approach from geometric models of the abdominal region and were validated against corresponding information gathered from CT scans in cross-sectional studies. It remains unknown how well DXA VAT estimates predict longitudinal changes in VAT with interventions. Carver et al. [50] evaluated DXA VAT in subjects with severe obesity and found a between-measurement coefficient of variation and root-mean standard deviation of 8.8% and 0.3 kg, respectively. In addition to segmentation accuracy and precision, a question also remains how well these automated VAT estimates correlate with clinical measures and health outcomes relative to other available phenotyping methods.
Figure 3. Fat-fraction abdominal adipose tissue images acquired from a woman with obesity.
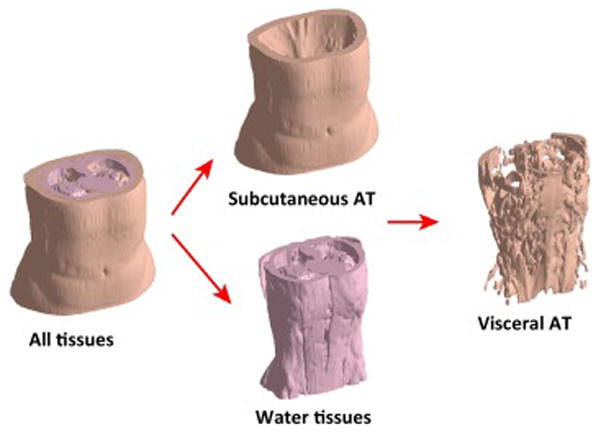
Abdominal fat-fraction image from the dome of the liver to the femoral heads of a woman with obesity showing the total volumetric image and the three component images (water-containing lean tissues, SAT, and VAT+intermuscular adipose tissue) that were segmented using automated software [45]. The scan was completed using an investigational parallel MRI accelerated IDEAL sequence on a GE 3.0 Tesla MR750 scanner (GE Healthcare, Waukesha, WI) and a 32-coil torso array (Neo Coil, Pewaukee, WI). Illustration courtesy of Charles Mckenzie, Western University, London, Ontario, Canada.
In the past five years, advanced reconstruction algorithms have been formulated that not only compute the separated water and fat signals on a voxel-wise basis, but additionally provide estimates of triglyceride characteristics [51]. These advanced algorithms can now estimate triglyceride characteristics in WAT depots including the number of double bonds, methylene-interrupted double bonds, and fatty acid chain length. The number of double bonds present can be used to calculate the proportion of mono- vs. poly-unsaturated fatty acids [51]. Tissue triglyceride saturation provides an important biomarker of dietary fat composition, and the capability of non-invasive measurement advances the often-impractical traditional SAT biopsy approach [52].
Going Beyond Traditional Adipose Tissue Compartment Measurements
The classic view held that total body non-membrane lipids, largely as adipose tissue triglycerides, are confined to two main compartments, one located in the subcutaneous space and the other found in the visceral cavity. Throughout our review we describe the measurement of non-membrane lipids across many organs and tissues, transcending the limited boundaries of the traditional adipose tissue compartments. Perhaps the best example of the renaissance in evaluating non-membrane lipids in vivo is the wide scope of lipids now recognized in skeletal muscles.
The largest of these compartments are the intermuscular adipose tissue component found between skeletal muscle groups and the intramuscular adipose tissue embedded within individual skeletal muscles [10]. The volume of these components, now extensively linked with metabolic and physiologic processes [53, 54], can be quantified mainly by the well-established T1-weighted and Dixon MRI methods reviewed earlier.
At the subcellular level, the metabolically important intramyocellular lipids, primarily triglycerides within lipid droplets, can be separated from extramyocellular lipids present within nearby adipocytes by also now well-established MRS and MRSI methods [55, 56].
A single instrument, whole-body MRI coupled with MRS, can thus separate and quantify the volume of adipocyte groupings found within the skeletal muscle compartment, and the amounts of triglyceride lipids within and outside of skeletal muscle fibers. These lipid components are central to the pathogenesis and adverse outcomes associated with cardiovascular diseases, cancer, diabetes, and other chronic diseases that share in common variable contributions of excess adiposity, insulin resistance, impaired mitochondrial function, systemic inflammation, and oxidative stress [3, 53, 54].
Linking Metabolic Activity to Compartment Size
Measurements of lipid compartments is extending beyond simply deriving an estimate of the amount of a particular substrate present in vivo. Now investigators are equally interested in directly measuring the metabolic activity of lipid-containing tissues, notably thermogenic BAT. The study of BAT in both animals and humans has progressed rapidly over the past decade [57], [58]. Thermogenesis in BAT and so-called beige (brite) fat cells may play a role in the pathogenesis of excess adiposity and also provides a target for pharmacologic interventions [58, 59]. Central to the study of human BAT is an array of radiological imaging techniques that can characterize the tissue non-invasively in vivo, namely MRI, positron emission tomography (PET), and CT [60, 61].
The characteristic mixed content of water and fat in BAT versus triglyceride-rich WAT directly translates to differences in X-ray attenuation (measured in Hounsfield units, HU) in CT and quantitative fat fraction measurements in MRI. On a calibrated HU scale in CT where pure water is at 0 HU and pure fat is at -100 HU, the greater density of BAT generally leads to the tissue occupying a more positive, albeit still negative, HU range than WAT. In proton-based MRI, spectroscopy, water- and fat-suppressed techniques, and more recently advanced CSE water-fat strategies have all been successfully employed to consistently show a lower fat content and higher water content in supraclavicular and interscapular BAT than subcutaneous WAT [5].
The strategy of delivering an exogenous chemical tracer into biochemical pathways as a means of visualizing BAT in vivo with imaging has been explored by multiple investigators. While 18F-fluorodeoxyglucose (FDG) has been the hallmark metabolic probe for PET, more BAT-specific 18F variants using fatty acid substrates [62, 63], 18F tracers that specifically target the mitochondria [64], as well as 11C agents that target the intermediate products within the Krebs’ cycle [63], have been developed. Analogous to 11C probes in PET, preliminary results using hyperpolarized 13C MRI for visualizing BAT metabolic activity have also been reported in mice [65]. In contrast, WAT does not typically exhibit any uptake of the tracers. The use of T2*-weighted blood-oxygen-level-dependent (BOLD) MRI [66] and hyperpolarized 129Xe gas MRI [67] have also been employed to measure BAT’s metabolic activity and temperature in vivo.
Quantifying BAT metabolic activity while the subject rests quietly inside a PET, CT, or MRI scanner creates conditions that are not representative of those experienced during daily life. In the future, next-generation infrared sensors [68] could fill major knowledge gaps in the real-world dynamics of BAT activation, including circadian fluctuations, changes due to exercise and food intake, and effects of various environmental conditions such as ambient temperature. These gaps raise critical questions about the real-world impact that BAT activating pharmacological agents will have on energy expenditure. The reason for the knowledge gaps is that chronic, ecological surveillance of BAT activity level, using worn or implanted sensors, is an unmet technological challenge, and thus almost all prior measurements have taken place in a laboratory setting. Skin temperature measurements from microwave radiometry or conventional infrared sensors have been proposed for ecological assessment, but influences of blood flow and non-fat tissue temperatures confound measurements [69, 70] [71–73]. Novel time-resolved Fourier transform infrared (FTIR) sensing techniques, meanwhile, provide much richer information about tissue composition including the path length of light, absorption, scattering, and concentrations of tissue oxygen and hemoglobin [74, 75]. One group has investigated FTIR spectroscopy quantification of BAT activation, with promising correlations with FDG PET measurements in humans [76]. Further technological advances in infrared sensing have enabled rapid, diffraction-limited spatial resolution imaging [77, 78] and an ability to probe length scales that are on par with sub-cellular structures such mitochondria, vasculature, and sympathetic nerves that are conspicuous within BAT. Thus, infrared sensing is poised to continue to contribute to the ecological measurement of BAT activity moving forward.
Extending Techniques to Early Life Measurements
Several decades ago our knowledge of lipid compartments, notably body fat, was mainly limited to adolescents and adults. The term “fat” in this context refers to neutral lipids, mainly triglycerides, as distinct from total lipids or adipose tissue [79]. Methods for quantifying adiposity in infants and children were complex, costly, and in some cases exposed subjects to high radiation doses [80]. Because of advances taking place over the past decade, in some cases within the past few years, several body lipid compartments can now be measured in the fetus, infants, and young children. A growing literature implicates intrauterine environmental events, maternal diet, and early feeding practices on having long term metabolic health and clinical outcomes [81]. These advances are allowing investigators to quantify and track changes in selected body lipid compartments across the full human lifespan, opening up new opportunities to link early life events with long term health outcomes.
Two recently introduced and validated systems are designed to measure infant total body fat without radiation exposure, neonatal air displacement plethysmographyi (ADP) [82] and quantitative nuclear magnetic resonanceii (QMR)[83]. The ADP approach is based on the classic two-compartment model that assumes body fat in the form on neutral lipids has a density of 0.900 g/cc and fat-free mass has a density of 1.00 g/cc [84]. The proportion of body weight as fat can be calculated using the model and the subject’s measured body density as derived by the ADP system.
Another rapidly developing field involves a broad area largely encompassed by “photogrammetry” methods that capture body shape and dimensions from surface mapping technologies ranging from 3D whole-body laser systems [85] to cellular telephone cameras [86, 87]. All of these approaches share in common the derivation of body volumes, linear dimensions, and circumferences that are used to predict whole body and regional fat mass. As with ADP and QMR, these approaches can potentially be used to make quantitative morphological measurements and to predict various fat compartments without radiation exposure early in life [88]. In utero approaches are also being developed using 3D ultrasound at 33 weeks of gestation to predict birth weight and neonatal body composition using ADP as the reference [89].
DXA systems can also be used to evaluate total and regional fat mass from birth onwards [82, 90]. Photon attenuation characteristics are used by DXA software algorithms to separate evaluated pixels into fat, lean soft tissue, and bone mineral. While DXA is now often used as a reference method in cross-sectional studies of infant adiposity, repeated scanning is limited by radiation exposure. Magnetic resonance imaging has recently been used to quantify SAT and VAT during the second trimester of pregnancy [91]. Neonatal BAT can also now be quantified using water-separated MRI [92] and intrahepatocellular lipid can be measured with MRS [93].
Taken collectively, these advances fill a large void in the ability of investigators to quantify body lipid compartments during the early phase of life, thus opening new research directions and potentially creating clinical diagnostic and treatment opportunities. All of these methods, ADP, QMR, DXA, photogrammetry, ultrasound, and MRI all are also well established approaches for evaluating body composition in older children, adolescents and adults.
CONCLUDING REMARKS AND FUTURE PERSPECTIVES
Once the exclusive domain of bench scientists working with animal models or in vitro, the emerging capability of measuring lipid compartments ranging in size from microscopic to the whole human body across the lifespan is creating a wealth of new research questions opportunities for clinical investigators (see outstanding questions). Refined older and new technologies are going beyond quantifying compartment size to probe lipid structure, adipocyte type, metabolic flux rates, and BAT heat production rates in real-world settings. Will these real-world dynamic measurements supplant the largely structural lipid compartment measurements now the main focus of research efforts? Can uniform standards be developed for measuring lipid compartments across vendors, platforms, and protocols? These advances are timely as a fundamental understanding of lipid compartment biology in living humans is increasingly recognized as key to facilitating advances that promote healthy aging and reducing non-communicable chronic disease risk.
Outstanding Questions.
What are the limits of magnetic resonance-based quantification of adipose tissue structure? A high priority is to establish if it is feasible using widely-available magnetic resonance equipment to discriminate between metabolically distinct mesenteric, omental, and extraperitoneal adipose tissue components; quantify adipocyte and lipid droplet size; and estimate adipose tissue macrophage infiltration, fibrosis, and triglyceride concentrations. What are the compelling clinical applications of such techniques?
What are the limits of magnetic resonance-based quantification of adipose tissue dynamics? An important research direction is to determine if everyday quantification of triglyceride metabolism, metabolic fluxes, and mitochondrial function that capture brown and beige (brite) adipocyte metabolic states is a reality. If so, will these methods render adipose tissue structural measurements irrelevant?
Will everyday non-invasive in utero measurement of fat content in organs and structures in the fetus become a reality? We need to establish if magnetic resonance or ultrasound techniques are viable options for this application and what properties they will be able to quantify. What are the compelling applications in pre-natal care for such techniques?
What distinct roles will next-generation X-ray, magnetic resonance, and infrared sensing technologies play in quantifying adipose tissue structure and dynamics?
Will phase contrast X-ray, FTIR, or ultra-high-field magnetic resonance enable higher-fidelity, finer-grained, or ecological quantification of adipose tissue properties?
Trends.
Not long ago the main measureable lipid compartment was total body fat, mainly triglyceride, in adults.
Advances now enable investigators to quantify lipid compartments in vivo that range in size from microscopic intracellular lipid-containing organelles found in a wide range of tissues, to those that account for a large fraction of body weight, across the human lifespan.
This emerging area of lipid compartment measurement in vivo is providing new insights into the pathophysiology of multiple disease states, notably non-communicable chronic diseases.
Future developments promise to go beyond measurements of lipid compartment size, to lipid structural and dynamic estimates, with metabolic insights gained by moving from restricted laboratory settings to real-world conditions.
Acknowledgments
We thank Robin Post for critically reading the manuscript.
The authors and their close relatives and their professional associates have no financial interests in the study outcome nor do they serve as an officer, director, member, owner, trustee, or employee of an organization with a financial interest in the outcome or as an expert witness, advisor, consultant, or public advocate on behalf of an organization with a financial interest in the study outcome.
Funding Sources: HHH acknowledges support from Philips Healthcare for biomedical imaging related projects. OC has received support from the Pennington Biomedical Research Foundation.
Abbreviations
- BAT
brown adipose tissue
- BOLD
blood-oxygen-level-dependent
- BMI
body mass index
- CSE
chemical-shift encoded
- CT
computed tomography
- DXA
dual-energy X-ray absorptiometry
- FTIR
Fourier Transform Infrared spectrometry
- HU
Hounsfield units
- MRI
magnetic resonance imaging
- MRS
magnetic resonance spectroscopy
- MRSI
magnetic resonance spectroscopic imaging
- PET
positron emission tomography
- VAT
visceral adipose tissue
- WAT
white adipose tissue
- SAT
subcutaneous adipose tissue
- NMR
nuclear magnetic resonance
- SGI
stepped-grating interferometry
Footnotes
ADP; COSMED USA, Concord, CA
QMR; EchoMRI Corp., Houston, TX
Authors’ contributions to manuscript: SBH, HHH, WS, and OC all contributed equally to the preparation and editing of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Heymsfield SB, et al. Handbook of obesity. CRC Press, Taylor & Francis Group; 2014. Measurement of Total Adiposity, Regional Fat Depots, and Ectopic Fat. [Google Scholar]
- 2.Szendroedi J, Roden M. Ectopic lipids and organ function. Current opinion in lipidology. 2009;20:50–56. doi: 10.1097/mol.0b013e328321b3a8. [DOI] [PubMed] [Google Scholar]
- 3.Kuh D, et al. A life course approach to chronic disease epidemiology. Oxford University Press; 2004. http://dx.doi.org/10.1093/acprof:oso/9780198578154.001.0001. [Google Scholar]
- 4.Heymsfield SB, et al. Skeletal muscle mass and quality: evolution of modern measurement concepts in the context of sarcopenia. The Proceedings of the Nutrition Society. 2015:1–12. doi: 10.1017/S0029665115000129. [DOI] [PubMed] [Google Scholar]
- 5.Hu HH, et al. Segmentation and Quantification of Adipose Tissue by Computed Tomography and Magnetic Resonance Imaging. Magnetic Resonance Materials in Physics, Biology and Medicine. 2015 In Press. [Google Scholar]
- 6.Shen W, et al. Between-slice intervals in quantification of adipose tissue and muscle in children. Int J Pediatr Obes. 2011;6:149–156. doi: 10.3109/17477166.2010.486833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orgiu S, et al. Automatic muscle and fat segmentation in the thigh from T1-Weighted MRI. J Magn Reson Imaging. 2015 doi: 10.1002/jmri.25031. [DOI] [PubMed] [Google Scholar]
- 8.Wang D, et al. Fully automatic and nonparametric quantification of adipose tissue in fat-water separation MR imaging. Med Biol Eng Comput. 2015 doi: 10.1007/s11517-015-1347-y. [DOI] [PubMed] [Google Scholar]
- 9.Wurslin C, et al. Topography mapping of whole body adipose tissue using A fully automated and standardized procedure. J Magn Reson Imaging. 2010;31:430–439. doi: 10.1002/jmri.22036. [DOI] [PubMed] [Google Scholar]
- 10.Shen W, et al. Adipose tissue quantification by imaging methods: a proposed classification. Obesity research. 2003;11:5–16. doi: 10.1038/oby.2003.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mathieu P, et al. Ectopic visceral fat: a clinical and molecular perspective on the cardiometabolic risk. Reviews in endocrine & metabolic disorders. 2014;15:289–298. doi: 10.1007/s11154-014-9299-3. [DOI] [PubMed] [Google Scholar]
- 12.Gustafson B, Smith U. Regulation of white adipogenesis and its relation to ectopic fat accumulation and cardiovascular risk. Atherosclerosis. 2015;241:27–35. doi: 10.1016/j.atherosclerosis.2015.04.812. [DOI] [PubMed] [Google Scholar]
- 13.Kuk JL, et al. Does measurement site for visceral and abdominal subcutaneous adipose tissue alter associations with the metabolic syndrome? Diabetes Care. 2006;29:679–684. doi: 10.2337/diacare.29.03.06.dc05-1500. [DOI] [PubMed] [Google Scholar]
- 14.Shen W, et al. Visceral adipose tissue: relationships between single slice areas at different locations and obesity-related health risks. Int J Obes (Lond) 2007;31:763–769. doi: 10.1038/sj.ijo.0803474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwenzer NF, et al. Quantitative analysis of adipose tissue in single transverse slices for estimation of volumes of relevant fat tissue compartments: a study in a large cohort of subjects at risk for type 2 diabetes by MRI with comparison to anthropometric data. Investigative radiology. 2010;45:788–794. doi: 10.1097/RLI.0b013e3181f10fe1. [DOI] [PubMed] [Google Scholar]
- 16.Schaudinn A, et al. Predictive accuracy of single- and multi-slice MRI for the estimation of total visceral adipose tissue in overweight to severely obese patients. NMR in biomedicine. 2015;28:583–590. doi: 10.1002/nbm.3286. [DOI] [PubMed] [Google Scholar]
- 17.Demerath EW, et al. Approximation of total visceral adipose tissue with a single magnetic resonance image. The American journal of clinical nutrition. 2007;85:362–368. doi: 10.1093/ajcn/85.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen W, et al. Visceral adipose tissue: relations between single-slice areas and total volume. The American journal of clinical nutrition. 2004;80:271–278. doi: 10.1093/ajcn/80.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abate N, et al. Prediction of total subcutaneous abdominal, intraperitoneal, and retroperitoneal adipose tissue masses in men by a single axial magnetic resonance imaging slice. The American journal of clinical nutrition. 1997;65:403–408. doi: 10.1093/ajcn/65.2.403. [DOI] [PubMed] [Google Scholar]
- 20.Irlbeck T, et al. Association between single-slice measurements of visceral and abdominal subcutaneous adipose tissue with volumetric measurements: the Framingham Heart Study. Int J Obes (Lond) 2010;34:781–787. doi: 10.1038/ijo.2009.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maislin G, et al. Single slice vs. volumetric MR assessment of visceral adipose tissue: reliability and validity among the overweight and obese. Obesity. 2012;20:2124–2132. doi: 10.1038/oby.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen W, et al. Total body skeletal muscle and adipose tissue volumes: estimation from a single abdominal cross-sectional image. J Appl Physiol. 2004;97:2333–2338. doi: 10.1152/japplphysiol.00744.2004. [DOI] [PubMed] [Google Scholar]
- 23.Schweitzer L, et al. What is the best reference site for a single MRI slice to assess whole-body skeletal muscle and adipose tissue volumes in healthy adults? The American journal of clinical nutrition. 2015;102:58–65. doi: 10.3945/ajcn.115.111203. [DOI] [PubMed] [Google Scholar]
- 24.Shen W, et al. A Single mri Slice Does Not Accurately Predict Visceral and Subcutaneous Adipose Tissue Changes During Weight Loss. Obesity. 2012;20:2458–2463. doi: 10.1038/oby.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eggers H, Bornert P. Chemical shift encoding-based water-fat separation methods. J Magn Reson Imaging. 2014;40:251–268. doi: 10.1002/jmri.24568. [DOI] [PubMed] [Google Scholar]
- 26.Hu HH, Kan HE. Quantitative proton MR techniques for measuring fat. NMR in biomedicine. 2013;26:1609–1629. doi: 10.1002/nbm.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma J. Dixon techniques for water and fat imaging. J Magn Reson Imaging. 2008;28:543–558. doi: 10.1002/jmri.21492. [DOI] [PubMed] [Google Scholar]
- 28.Reeder SB, et al. Quantitative Assessment of Liver Fat with Magnetic Resonance Imaging and Spectroscopy. J Magn Reson Imaging. 2011:34. doi: 10.1002/jmri.22580. spcone. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischer MA, et al. Quantification of muscle fat in patients with low back pain: comparison of multi-echo MR imaging with single-voxel MR spectroscopy. Radiology. 2013;266:555–563. doi: 10.1148/radiol.12120399. [DOI] [PubMed] [Google Scholar]
- 30.Lingvay I, et al. Noninvasive quantification of pancreatic fat in humans. J Clin Endocrinol Metab. 2009;94:4070–4076. doi: 10.1210/jc.2009-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meisamy S, et al. Quantification of hepatic steatosis with T1-independent, T2-corrected MR imaging with spectral modeling of fat: blinded comparison with MR spectroscopy. Radiology. 2011;258:767–775. doi: 10.1148/radiol.10100708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas EL, et al. Whole body fat: content and distribution. Prog Nucl Magn Reson Spectrosc. 2013;73:56–80. doi: 10.1016/j.pnmrs.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 33.Venkatesh BA, et al. MR proton spectroscopy for myocardial lipid deposition quantification: a quantitative comparison between 1.5T and 3T. J Magn Reson Imaging. 2012;36:1222–1230. doi: 10.1002/jmri.23761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu HH, et al. Assessment of abdominal adipose tissue and organ fat content by magnetic resonance imaging. Obesity reviews: an official journal of the International Association for the Study of Obesity. 2011;12:e504–515. doi: 10.1111/j.1467-789X.2010.00824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cassidy FH, et al. Fatty liver disease: MR imaging techniques for the detection and quantification of liver steatosis. Radiographics: a review publication of the Radiological Society of North America, Inc. 2009;29:231–260. doi: 10.1148/rg.291075123. [DOI] [PubMed] [Google Scholar]
- 36.Machann J, et al. Fraction of unsaturated fatty acids in visceral adipose tissue (VAT) is lower in subjects with high total VAT volume - a combined 1 H MRS and volumetric MRI study in male subjects. NMR in biomedicine. 2013;26:232–236. doi: 10.1002/nbm.2849. [DOI] [PubMed] [Google Scholar]
- 37.Hodson L, et al. Fatty acid composition of adipose tissue and blood in humans and its use as a biomarker of dietary intake. Progress in lipid research. 2008;47:348–380. doi: 10.1016/j.plipres.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 38.Schwab U, et al. Effect of the amount and type of dietary fat on cardiometabolic risk factors and risk of developing type 2 diabetes, cardiovascular diseases, and cancer: a systematic review. Food & nutrition research. 2014:58. doi: 10.3402/fnr.v58.25145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ren J, et al. Composition of adipose tissue and marrow fat in humans by 1H NMR at 7 Tesla. Journal of lipid research. 2008;49:2055–2062. doi: 10.1194/jlr.D800010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Welte MA. Expanding roles for lipid droplets. Current biology: CB. 2015;25:R470–481. doi: 10.1016/j.cub.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wingfield HL, et al. Body composition assessment in overweight women: validation of air displacement plethysmography. Clinical physiology and functional imaging. 2014;34:72–76. doi: 10.1111/cpf.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gross DA, Silver DL. Cytosolic lipid droplets: from mechanisms of fat storage to disease. Critical reviews in biochemistry and molecular biology. 2014;49:304–326. doi: 10.3109/10409238.2014.931337. [DOI] [PubMed] [Google Scholar]
- 43.Pokharel SS, et al. Current MR imaging lipid detection techniques for diagnosis of lesions in the abdomen and pelvis. Radiographics: a review publication of the Radiological Society of North America, Inc. 2013;33:681–702. doi: 10.1148/rg.333125068. [DOI] [PubMed] [Google Scholar]
- 44.Hernando D, et al. Chemical shift-based water/fat separation: a comparison of signal models. Magnetic resonance in medicine. 2010;64:811–822. doi: 10.1002/mrm.22455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Addeman BT, et al. Validation of volumetric and single-slice MRI adipose analysis using a novel fully automated segmentation method. J Magn Reson Imaging. 2015;41:233–241. doi: 10.1002/jmri.24526. [DOI] [PubMed] [Google Scholar]
- 46.Karlsson A, et al. Automatic and quantitative assessment of regional muscle volume by multi-atlas segmentation using whole-body water-fat MRI. J Magn Reson Imaging. 2015;41:1558–1569. doi: 10.1002/jmri.24726. [DOI] [PubMed] [Google Scholar]
- 47.Alabousi A, et al. Evaluation of adipose tissue volume quantification with IDEAL fat-water separation. J Magn Reson Imaging. 2011;34:474–479. doi: 10.1002/jmri.22603. [DOI] [PubMed] [Google Scholar]
- 48.Kaul S, et al. Dual-energy X-ray absorptiometry for quantification of visceral fat. Obesity. 2012;20:1313–1318. doi: 10.1038/oby.2011.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Micklesfield LK, et al. Dual-energy X-ray performs as well as clinical computed tomography for the measurement of visceral fat. Obesity. 2012;20:1109–1114. doi: 10.1038/oby.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carver TE, et al. Precision of the iDXA for visceral adipose tissue measurement in severely obese patients. Med Sci Sports Exerc. 2014;46:1462–1465. doi: 10.1249/MSS.0000000000000238. [DOI] [PubMed] [Google Scholar]
- 51.Peterson P, Mansson S. Simultaneous quantification of fat content and fatty acid composition using MR imaging. Magnetic resonance in medicine. 2013;69:688–697. doi: 10.1002/mrm.24297. [DOI] [PubMed] [Google Scholar]
- 52.Hudgins LC, et al. Correlation of isomeric fatty acids in human adipose tissue with clinical risk factors for cardiovascular disease. The American journal of clinical nutrition. 1991;53:474–482. doi: 10.1093/ajcn/53.2.474. [DOI] [PubMed] [Google Scholar]
- 53.Hausman GJ, et al. Intermuscular and intramuscular adipose tissues: Bad vs. good adipose tissues. Adipocyte. 2014;3:242–255. doi: 10.4161/adip.28546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vettor R, et al. The origin of intermuscular adipose tissue and its pathophysiological implications. Am J Physiol Endocrinol Metab. 2009;297:E987–998. doi: 10.1152/ajpendo.00229.2009. [DOI] [PubMed] [Google Scholar]
- 55.Boesch C, et al. In vivo determination of intra-myocellular lipids in human muscle by means of localized 1H-MR-spectroscopy. Magnetic resonance in medicine. 1997;37:484–493. doi: 10.1002/mrm.1910370403. [DOI] [PubMed] [Google Scholar]
- 56.Schick F, et al. Comparison of localized proton NMR signals of skeletal muscle and fat tissue in vivo: two lipid compartments in muscle tissue. Magnetic resonance in medicine. 1993;29:158–167. doi: 10.1002/mrm.1910290203. [DOI] [PubMed] [Google Scholar]
- 57.Betz MJ, Enerback S. Human Brown Adipose Tissue: What We Have Learned So Far. Diabetes. 2015;64:2352–2360. doi: 10.2337/db15-0146. [DOI] [PubMed] [Google Scholar]
- 58.Cypess AM, et al. Brown fat in humans: consensus points and experimental guidelines. Cell metabolism. 2014;20:408–415. doi: 10.1016/j.cmet.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Claussnitzer M, et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. The New England journal of medicine. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bauwens M, et al. Molecular imaging of brown adipose tissue in health and disease. European journal of nuclear medicine and molecular imaging. 2014;41:776–791. doi: 10.1007/s00259-013-2611-8. [DOI] [PubMed] [Google Scholar]
- 61.Borga M, et al. Brown adipose tissue in humans: detection and functional analysis using PET (positron emission tomography), MRI (magnetic resonance imaging), and DECT (dual energy computed tomography) Methods in enzymology. 2014;537:141–159. doi: 10.1016/B978-0-12-411619-1.00008-2. [DOI] [PubMed] [Google Scholar]
- 62.Karmi A, et al. Increased brain fatty acid uptake in metabolic syndrome. Diabetes. 2010;59:2171–2177. doi: 10.2337/db09-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ouellet V, et al. Brown adipose tissue oxidative metabolism contributes to energy expenditure during acute cold exposure in humans. The Journal of clinical investigation. 2012;122:545–552. doi: 10.1172/JCI60433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Madar I, et al. 18F-fluorobenzyl triphenyl phosphonium: a noninvasive sensor of brown adipose tissue thermogenesis. Journal of nuclear medicine: official publication, Society of Nuclear Medicine. 2011;52:808–814. doi: 10.2967/jnumed.110.084657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lau AZ, et al. Noninvasive identification and assessment of functional brown adipose tissue in rodents using hyperpolarized (1)(3)C imaging. Int J Obes (Lond) 2014;38:126–131. doi: 10.1038/ijo.2013.58. [DOI] [PubMed] [Google Scholar]
- 66.Khanna A, Branca RT. Detecting brown adipose tissue activity with BOLD MRI in mice. Magnetic resonance in medicine. 2012;68:1285–1290. doi: 10.1002/mrm.24118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Branca RT, et al. Detection of brown adipose tissue and thermogenic activity in mice by hyperpolarized xenon MRI. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:18001–18006. doi: 10.1073/pnas.1403697111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nirengi S, et al. Human brown adipose tissue assessed by simple, noninvasive near-Infrared time-resolved spectroscopy. Obesity. 2015;23:973–980. doi: 10.1002/oby.21012. [DOI] [PubMed] [Google Scholar]
- 69.Rodrigues DB, et al. Numerical 3D modeling of heat transfer in human tissues for microwave radiometry monitoring of brown fat metabolism. 2013:85840S-85840S–85812. doi: 10.1117/12.2004931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Symonds ME, et al. Thermal imaging to assess age-related changes of skin temperature within the supraclavicular region co-locating with brown adipose tissue in healthy children. The Journal of pediatrics. 2012;161:892–898. doi: 10.1016/j.jpeds.2012.04.056. [DOI] [PubMed] [Google Scholar]
- 71.Boon MR, et al. Supraclavicular skin temperature as a measure of 18F-FDG uptake by BAT in human subjects. PloS one. 2014;9:e98822. doi: 10.1371/journal.pone.0098822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Crane JD, et al. A standardized infrared imaging technique that specifically detects UCP1-mediated thermogenesis in vivo. Molecular metabolism. 2014;3:490–494. doi: 10.1016/j.molmet.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jang C, et al. Infrared thermography in the detection of brown adipose tissue in humans. Physiological reports. 2014:2. doi: 10.14814/phy2.12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hamaoka T, et al. The use of muscle near-infrared spectroscopy in sport, health and medical sciences: recent developments. Philosophical transactions Series A, Mathematical, physical, and engineering sciences. 2011;369:4591–4604. doi: 10.1098/rsta.2011.0298. [DOI] [PubMed] [Google Scholar]
- 75.Hamaoka T, et al. Near-infrared spectroscopy/imaging for monitoring muscle oxygenation and oxidative metabolism in healthy and diseased humans. Journal of biomedical optics. 2007;12:062105. doi: 10.1117/1.2805437. [DOI] [PubMed] [Google Scholar]
- 76.Nirengi S, et al. Human brown adipose tissue assessed by simple, noninvasive near-infrared time-resolved spectroscopy. Obesity. 2015;23:973–980. doi: 10.1002/oby.21012. [DOI] [PubMed] [Google Scholar]
- 77.Kastyak-Ibrahim MZ, et al. Biochemical label-free tissue imaging with subcellular-resolution synchrotron FTIR with focal plane array detector. NeuroImage. 2012;60:376–383. doi: 10.1016/j.neuroimage.2011.11.069. [DOI] [PubMed] [Google Scholar]
- 78.Nasse MJ, et al. High-resolution Fourier-transform infrared chemical imaging with multiple synchrotron beams. Nature methods. 2011;8:413–416. doi: 10.1038/nmeth.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Comizio R, et al. Total body lipid and triglyceride response to energy deficit: relevance to body composition models. The American journal of physiology. 1998;274:E860–866. doi: 10.1152/ajpendo.1998.274.5.E860. [DOI] [PubMed] [Google Scholar]
- 80.Ellis KJ. Human body composition: in vivo methods. Physiological reviews. 2000;80:649–680. doi: 10.1152/physrev.2000.80.2.649. [DOI] [PubMed] [Google Scholar]
- 81.Gali Ramamoorthy T, et al. Developmental programming of hypothalamic neuronal circuits: impact on energy balance control. Frontiers in neuroscience. 2015;9:126. doi: 10.3389/fnins.2015.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fields DA, et al. Body composition at 6 months of life: comparison of air displacement plethysmography and dual-energy X-ray absorptiometry. Obesity. 2012;20:2302–2306. doi: 10.1038/oby.2012.102. [DOI] [PubMed] [Google Scholar]
- 83.Andres A, et al. Quantitative nuclear magnetic resonance to measure fat mass in infants and children. Obesity. 2011;19:2089–2095. doi: 10.1038/oby.2011.215. [DOI] [PubMed] [Google Scholar]
- 84.Heymsfield SB, et al. Multi-component molecular-level body composition reference methods: evolving concepts and future directions. Obesity reviews: an official journal of the International Association for the Study of Obesity. 2015;16:282–294. doi: 10.1111/obr.12261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Soileau L, et al. Automated Anthropometric Phenotyping with Novel Kinect-Based Three-Dimensional Imaging Method: Comparison to a Reference Laser Imaging System. European Journal of Clinical Nutrition. 2015 doi: 10.1038/ejcn.2015.132. In press. [DOI] [PubMed] [Google Scholar]
- 86.Pradhan L, et al. Feature Extraction from 2D Images for Body Composition Analysis. In Review. [Google Scholar]
- 87.Xie B, et al. Accurate body composition measures from whole-body silhouettes. Medical physics. 2015;42:4668. doi: 10.1118/1.4926557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li Z, et al. Anthropometric body measurements based on multi-view stereo image reconstruction. Conference proceedings: … Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. Annual Conference; 2013; 2013. pp. 366–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.O’Connor C, et al. Birth weight and neonatal adiposity prediction using fractional limb volume obtained with 3D ultrasound. Fetal diagnosis and therapy. 2014;36:44–48. doi: 10.1159/000360417. [DOI] [PubMed] [Google Scholar]
- 90.Demerath EW, Fields DA. Body composition assessment in the infant. American journal of human biology: the official journal of the Human Biology Council. 2014;26:291–304. doi: 10.1002/ajhb.22500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Anblagan D, et al. Measurement of fetal fat in utero in normal and diabetic pregnancies using magnetic resonance imaging. Ultrasound in obstetrics & gynecology: the official journal of the International Society of Ultrasound in Obstetrics and Gynecology. 2013;42:335–340. doi: 10.1002/uog.12382. [DOI] [PubMed] [Google Scholar]
- 92.Rasmussen JM, et al. Brown adipose tissue quantification in human neonates using water-fat separated MRI. PloS one. 2013;8:e77907. doi: 10.1371/journal.pone.0077907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Brumbaugh DE, et al. Intrahepatic fat is increased in the neonatal offspring of obese women with gestational diabetes. The Journal of pediatrics. 2013;162:930–936. e931. doi: 10.1016/j.jpeds.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]


