Abstract
Jagged1 (JAG1) is one of 5 cell surface ligands that functions primarily in the highly conserved Notch signaling pathway. Notch signaling plays a critical role in cellular fate determination and is active throughout development and across many organ systems. The classic JAG1-NOTCH interaction leads to a cascade of proteolytic cleavages resulting in the NOTCH intracellular domain being transported into the nucleus where it functions to activate downstream transcription of target genes. JAG1 mutations have been associated with several disorders including the multisystem dominant disorder Alagille syndrome, and some cases of tetralogy of Fallot (although these may represent variable expressivity of Alagille syndrome). In addition, variations in JAG1 have been found to be associated with multiple types of cancer including breast cancer and adrenocortical carcinoma. Alagille syndrome, which primarily affects the liver, heart, skeleton, eye, face, kidney and vasculature is caused by loss of function mutations in JAG1, demonstrating that haploinsufficiency for JAG1 is disease causing, at least in these tissues. Expression and conditional gene knockout studies of JAG1 (Jag1) have correlated with tissue-specific disease phenotypes and have provided insight into both disease pathogenesis and human development.
Keywords: Jagged1, Notch signaling, Alagille Syndrome, Tetralogy of Fallot, Developmental disorder
History
What is now known as the NOTCH signaling pathway was first primitively observed and described during the early 1900’s in Drosophila melanogaster by Otto L. Mohr. Flies with small notches at the end of their wings were identified and crossed with wild-type flies and the notched phenotype was found to segregate in offspring. This observation demonstrated that the wing shape was genetically regulated and the locus responsible for this wing phenotype was termed the “Notch” locus (Mohr, 1919). Further work on Drosophila with NOTCH mutations revealed that Notch locus deficiencies also lead to abnormal embryonic development, with hypertrophy of the nervous system. In 1983, a portion of the Notch locus was cloned in Drosophila, paving the way to understand the structure of the gene and the protein (Artavanis-Tsakonas et al., 1983; Artavanis-Tsakonas et al., 1999). NOTCH signaling was first implicated in human disease in 1990 with the discovery of a patient with T-cell acute lymphoblastic leukemia who had a chromosomal translocation breaking TAN-1 (as it was then called), as the human homolog of the Drosophila Notch gene. (Ellisen et al., 1991). In 1995 the Weinmaster group cloned a mammalian (rat) Notch pathway ligand that was capable of activating NOTCH1. The ligand was named Jagged1 (because of its similarity to the Drosophila gene serrate) and initially, was shown to prevent muscle cell differentiation, and subsequent analyses extended these findings to other body tissues (Lindsell et al., 1995).
In 1997 the human homolog (JAG1) of the rat Jagged1 gene was cloned and mapped within a region of the short arm of chromosome 20, which coincided with the critical region for the autosomal dominant disorder, Alagille syndrome (ALGS) and immediately following this discovery, mutations within JAG1 were found to be associated with ALGS (Krantz et al., 1997; Li et al., 1997; Oda et al., 1997). Nine years later, some patients with ALGS were also identified who had mutations in the NOTCH2 receptor, although the percentage of patients with JAG1 mutations is much higher (94% have JAG1 mutations, and 1–2% have NOTCH2 mutations) (McDaniell et al., 2006; Kamath et al., 2012). Shimizu et al. demonstrated that JAG1 domains physically interact with NOTCH2 as well as other NOTCH receptors to activate Notch signaling (Shimizu et al., 2000). JAG1 was first implicated in cancer in 2005 with the discovery of a correlation between breast cancer survival and JAG1 expression levels (Reedijk et al., 2005). While Notch signaling was originally thought to be primarily involved in neurogenesis, it is now known that this pathway is implicated in the development of many tissues and organs, including the sensory regions of the inner ear, the heart, the kidney, the eye, the lung and other tissues (Adam et al., 1998; Kiernan et al., 2001; Cheng and Kopan, 2005; Penton et al., 2012)
Structure
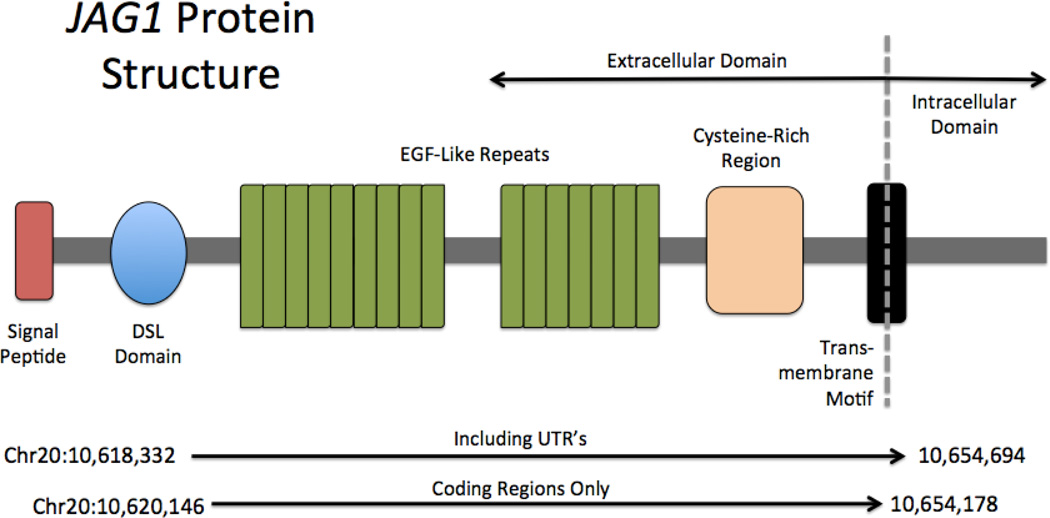
JAGGED1 (JAG1) is located on chromosome 20 at cytogenetic location 20p12.2 and genomic location (GRCh37) chr20:10,618,331–10,654,693. Encompassing 26 exons over 36kb, JAG1 produces a protein of 1218 amino acids (Oda et al., 1997). JAG1 is composed of a relatively small intracellular domain, a transmembrane domain and a larger extracellular component. The extracellular portion of the protein includes four motifs necessary for proper functioning of the protein, including a 21-amino acid signal peptide, an N-terminal region, a 40 amino-acid highly conserved DSL domain, (named for the Drosophila melanogaster and C.elegans ligands, delta, serrate and lag-2), followed by 16 epidermal growth factor-like (EGF-like) repeats, and a cysteine-rich region (Guarnaccia et al., 2004) (Figure 1). The signal peptide insures proper localization of the protein to the cell surface, and the DSL domain is required for binding of JAG1 to the NOTCH receptors. The EGF repeats (particularly EGF repeats 1 and 2) are important for increasing the affinity of JAG1 to the NOTCH receptors. Each EGF repeat contains 6 cysteine residues, which form disulfide bridges facilitating protein stabilization (Kopan and Ilagan, 2009; Chillakuri et al., 2012).
Figure 1.
Expression Profiles
JAG1 is widely expressed throughout mammalian development, across many tissues and developmental stages. Co-expression of JAG1 and Notch in the mammalian rat was first identified in developing nervous system. Expression was seen in neural progenitors forming both the central and peripheral nervous systems (Lindsell et al., 1995). Further expression studies by Lindsell et al. in the embryonic development of the rat showed broad expression of the gene in several body systems.
In humans, JAG1 expression varies throughout development and across tissues. In the adult, JAG1 is highly expressed in the heart, placenta, pancreas and prostate with lower levels found in lung, liver, kidney, thymus, testis and leucocytes (Jones et al., 2000; Gasperowicz and Otto, 2008). In the developing embryo, JAG1 expression is observed in the mesocardium, pulmonary artery, major arteries, distal cardiac outflow tract, metanephros, pancreas, branchial arches, around the major bronchial branches, portal vein, the neural tube optic vesicle, and the otocyst. (Jones et al., 2000). During development, JAG1 expression correlates with the organ systems involved in ALGS, although it is interesting to note that the highest levels are not necessarily found in organs that are affected in ALGS (Crosnier et al., 2000; Jones et al., 2000)
There is now ample evidence for the role of JAG1 in development of a number of organ systems. In the heart, in situ hybridization studies demonstrate JAG1 expression within vascular structures of the mammalian heart, correlating with the phenotypic cardiovascular problems found in ALGS (Loomes et al., 1999; High and Epstein, 2008). Several organ systems associated with ALGS including the kidney, vertebral column and the eye also show high expression levels of JAG1 during development through in situ hybridization studies on those tissues (Loomes et al., 1999).
In cancer, high levels of JAG1 mRNA and protein have been shown to be a marker for poor overall outcome in breast cancer and possibly enhance adrenocortical carcinoma cell proliferation and tumor aggressiveness (Reedijk et al., 2005; Simon et al., 2012)
Role In Disease
Alagille Syndrome
ALGS, which is caused by mutations in JAG1 or more rarely NOTCH2, is an autosomal dominant, multi-system disorder. In the original, clinical description of ALGS, diagnosis was made by identification of 3 of the 5 classic features including liver disease (bile duct paucity and resulting cholestasis), cardiac disease (primarily pulmonic stenosis or intracardiac defects such as Tetralogy of Fallot), vertebral defects (most prominently butterfly vertebrae), eye findings (most commonly posterior embryotoxon), and characteristic facial features (triangular face, deep set eyes, and pointed chin). In addition to these classic features, it has been recognized that a high percentage of patients also have renal and vascular anomalies (Kamath et al., 2003; Turnpenny and Ellard, 2012). The spectrum of JAG1 mutations in ALGS includes full gene deletions, and other protein truncating mutations including nonsense, frameshift and splice site as well as missense mutations, suggesting that the clinical phenotype is caused by haploinsufficiency for JAG1 (Warthen et al., 2006). In some cases, missense mutations have been shown to be pathogenic because they are not properly trafficked to the cell membrane, which also supports haploinsufficiency as the disease mechanism for ALGS (Morrissette et al., 2001).
Phenotypic effects of a JAG1 mutation are highly penetrant, but with highly variable expressivity (Kamath et al., 2003). There is a high rate of de novo mutations, with approximately 60% of mutations in probands not found in either parent (Krantz et al., 1998; Crosnier et al., 2000; Warthen et al., 2006). There is no strong correlation between the type and location of JAG1 mutation and severity of the disease, suggesting other genomic modifiers beyond the known JAG1 mutation may be the cause of the variable expressivity that characterizes this disorder.
Tetralogy of Fallot and Pulmonary Stenosis
Tetralogy of Fallot (TOF) is a relatively common form of complex congenital heart disease that has highly heterogeneous etiology. Eldadah et al. 2001 detected a missense mutation in JAG1 (p.G274D) within a large family with multiple individuals with cardiac disease, most notably TOF (Eldadah et al., 2001). Though ALGS is caused by haploinsufficiency of this gene, no family member met the clinical criteria for the syndrome. In 2010 a larger cohort of individuals with TOF or pulmonic stenosis/peripheral pulmonary stenosis (PS/PPS) were screened for JAG1 mutations (Bauer et al., 2010). Within this cohort 2% of TOF and 4% of PS/PPS individuals had a JAG1 mutation. As JAG1 has been shown to be expressed in critical vascular structures of heart development, and the cardiac disease in these individuals is similar to that seen in ALGS patients, the mutations detected in this cohort are likely the cause of the cardiac disease (Loomes et al., 1999). Since these individuals do not have ALGS, but have cardiopulmonary involvement, there may be confounding genetic modifiers beyond the detectable JAG1 mutation.
Role in Cancer
The Notch signaling pathway and JAG1 in particular have been implicated in multiple forms of cancer and multiple aspects of cancer biology including tumor angiogenesis, neoplastic cell growth, cancer stem cell maintenance and the metastatic process (Li et al., 2014). One of the first demonstrations of the role of JAG1 in a human cancer was the recognition that JAG1 mRNA expression is upregulated in breast cancer and correlates with a poor overall breast cancer survival in a dose-dependent fashion (Reedijk et al., 2005; Dickson et al., 2007). JAG1 is strongly expressed by tumor associated blood vessels in both brain and ovarian cancer (Lu et al., 2007; Li et al., 2014) Upregulation of JAG1 in adrenocortical carcinoma has been shown to enhance proliferation of this aggressive cancer through the activation of NOTCH signaling in cells adjacent to the tumor (Simon et al., 2012). In addition to cancer of the breast, brain and ovaries, JAG1 has been implicated in cervical, colorectal, endometrial, gastric, head and neck, hepatocellular, lung, pancreatic, prostate, and kidney cancers. JAG1 has also been shown to play a role in multiple forms of leukemia and lymphoma. Targeting of JAG1 is currently thought to hold promise for cancer therapy on multiple fronts and work is in progress to target pathways associated with strong JAG1 expression profiles.
Mouse Models
In mice, homozygous Jag1 null mutations were shown to be lethal during the embryonic stage due to cranial hemorrhaging (Xue et al., 1999). The heterozygous phenotype has proven to be more difficult to understand, and initial studies demonstrated that mice heterozygous for Jag1 mutations (with the mutation on a mixed genetic background) show only an eye phenotype Interestingly, subsequent studies of pure C57BL/6 Jag1 heterozygous mice do demonstrate the bile duct anomalies seen in ALGS (Xue et al., 1999; Thakurdas et al., 2015) Mice with haploinsufficiency for both Jag1 and Notch2 have been shown to exhibit classic features of ALGS (McCright et al., 2002). Mice heterozygous for mutations in Notch2 and Jag1 have a renal phenotype, including glomerular vascularization defects (McCright et al., 2001). Furthermore, glomerular defects as well as elevated blood urea nitrogen levels are present in mice heterozygous for the Jag1 null allele and a Notch2 hypomorphic allele (McCright et al., 2002). This kidney phenotype correlates with the renal involvement identified in patients with ALGS (Kamath et al., 2013).
Jag1 conditional knockout models have given insight into the organ specific effects caused by variations in the gene. Loomes et al. 2007 demonstrated that while liver-specific Jag1 knockout mice showed normal bile-duct maturation, crossing them with mice carrying a Jag1 null allele produced bile duct proliferation (Loomes et al., 2007). A Jag1 deletion in the portal vein mesenchyme (PVM) has been shown to disrupt normal bile duct development resulting in paucity (Hofmann et al., 2010). These studies highlight Jag1’s critical role in normal bile duct development and how its expression in the PVM signals to Notch2 within the ductal plate cells. An endothelial-specific knockout of Jag1 results in embryonic lethality resulting from severely impaired vascular smooth muscle and cardiovascular development. Additionally, the phenotype of the endothelial specific knockout was indistinguishable from the phenotype of global Jag1 knockouts, underlining the importance of the vascular smooth muscle phenotype (High et al. 2008). Further work demonstrated that when Jag1 is deleted in the endothelium, disruption of the endothelial-to-mesenchymal transition occurs resulting in valve calcification and congenital heart defects similar to those present in ALGS (Hofmann et al., 2012). Deletion of Jag1 in the cranial neural crest (CNC) cells cause midface anomalies as well as other craniofacial growth problems correlating with the classic inverted triangular facial features present in individuals with ALGS (Humphreys et al., 2012).
Highlights.
Jagged1 is a cell surface ligand that functions in the Notch signaling pathway.
JAG1 mutations are responsible for Alagille syndrome and other disorders.
Expression and mouse models of Jag1 correlate with disease phenotypes.
Acknowledgments
This review and the corresponding Gene Wiki article are written as part of the Cardiac Gene Wiki Review series--a series resulting from a collaboration between the journal GENE, the Gene Wiki Initiative, and the BD2K initiative. The Cardiac Gene Wiki Initiative is supported by National Institutes of Health (GM089820 and GM114833). Additional support for Gene Wiki Reviews is provided by Elsevier, the publisher of GENE. The authors would also like to acknowledge grant R01DK081702 (NBS) and to thank the Children’s Hospital of Philadelphia for their continued support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The corresponding Gene Wiki entry for this review can be found here: https://en.wikipedia.org/wiki/JAG1
References
- Adam J, Myat A, Le Roux I, Eddison M, Henrique D, Ish-Horowicz D, Lewis J. Cell fate choices and the expression of Notch, Delta and Serrate homologues in the chick inner ear: parallels with Drosophila sense-organ development. Development. 1998;125:4645–4654. doi: 10.1242/dev.125.23.4645. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Muskavitch M, Yedvobnick B. Molecular cloning of Notch, a locus affecting neurogenesis in Drosophila melanogaster. Proceedings of the National Academy of Sciences. 1983;80:1977–1981. doi: 10.1073/pnas.80.7.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch Signaling: Cell Fate Control and Signal Integration in Development. Science. 1999;30:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Bauer RC, Laney AO, Smith R, Gerfen J, Morrissette JJ, Woyciechowski S, Garbarini J, Loomes KM, Krantz ID, Urban Z, Gelb BD, Goldmuntz E, Spinner NB. Jagged1 (JAG1) mutations in patients with tetralogy of Fallot or pulmonic stenosis. Hum Mutat. 2010;31:594–601. doi: 10.1002/humu.21231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H-T, Kopan R. The role of Notch signaling in specification of podocyte and proximal tubules within the developing mouse kidney. Kidney international. 2005;68:1951–1952. doi: 10.1111/j.1523-1755.2005.00627.x. [DOI] [PubMed] [Google Scholar]
- Chillakuri CR, Sheppard D, Lea SM, Handford PA. Notch receptor–ligand binding and activation: Insights from molecular studies. Seminars in cell & developmental biology. 2012;23:421–428. doi: 10.1016/j.semcdb.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosnier C, Attie-Bitach T, Encha-Razavi F, Audollent S, Soudy F, Hadchouel M, Meunier-Rotival M, Vekemans M. JAGGED1 gene expression during human embryogenesis elucidates the wide phenotypic spectrum of Alagille syndrome. Hepatology. 2000;32:574–581. doi: 10.1053/jhep.2000.16600. [DOI] [PubMed] [Google Scholar]
- Dickson BC, Mulligan AM, Zhang H, Lockwood G, O'Malley FP, Egan SE, Reedijk M. High-level JAG1 mRNA and protein predict poor outcome in breast cancer. Modern Pathology. 2007;20:685–693. doi: 10.1038/modpathol.3800785. [DOI] [PubMed] [Google Scholar]
- Eldadah ZA, Hamosh A, Biery NJ, Montgomery RA, Duke M, Elkins R, Dietz HC. Familial Tetralogy of Fallot caused by mutation in the jagged1 gene. Hum Mol Genet. 2001;10:163–169. doi: 10.1093/hmg/10.2.163. [DOI] [PubMed] [Google Scholar]
- Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, Sklar J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- Gasperowicz M, Otto F. The notch signalling pathway in the development of the mouse placenta. Placenta. 2008;29:651–659. doi: 10.1016/j.placenta.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Guarnaccia C, Pintar A, Pongor S. Exon 6 of human Jagged-1 encodes an autonomously folding unit. FEBS Letters. 2004;574:156–160. doi: 10.1016/j.febslet.2004.08.022. [DOI] [PubMed] [Google Scholar]
- High FA, Epstein JA. The multifaceted role of Notch in cardiac development and disease. Nat Rev Genet. 2008;9:49–61. doi: 10.1038/nrg2279. [DOI] [PubMed] [Google Scholar]
- High FA, Lu MM, Pear WS, Loomes KM, Kaestner KH, Epstein JA. Endothelial expression of the Notch ligand Jagged1 is required for vascular smooth muscle development. Proc. Natl. Acad. Sci. 2008;105:1955–1959. doi: 10.1073/pnas.0709663105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann JJ, Briot A, Enciso J, Zovein AC, Ren S, Zhang ZW, Radtke F, Simons M, Wang Y, Iruela-Arispe ML. Endothelial deletion of murine Jag1 leads to valve calcification and congenital heart defects associated with Alagille syndrome. Development. 2012;139:4449–4460. doi: 10.1242/dev.084871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann JJ, Zovein AC, Koh H, Radtke F, Weinmaster G, Iruela-Arispe ML. Jagged1 in the portal vein mesenchyme regulates intrahepatic bile duct development: insights into Alagille syndrome. Development. 2010;137:4061–4072. doi: 10.1242/dev.052118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys R, Zheng W, Prince LS, Qu X, Brown C, Loomes K, Huppert SS, Baldwin S, Goudy S. Cranial neural crest ablation of Jagged1 recapitulates the craniofacial phenotype of Alagille syndrome patients. Hum. Mol. Genet. 2012;6:1374–1383. doi: 10.1093/hmg/ddr575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EA, Clement-Jones M, Wilson DI. JAGGED1 expression in human embryos: correlation with the Alagille syndrome phenotype. Journal of Medical Genetics. 2000;37:658–662. doi: 10.1136/jmg.37.9.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath BM, Bason L, Piccoli DA, Krantz ID, Spinner NB. Consequences of JAG1 mutations. J Med Genet. 2003;40:891–895. doi: 10.1136/jmg.40.12.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath BM, Bauer RC, Loomes KM, Chao G, Gerfen J, Hutchinson A, Hardikar W, Hirschfield G, Jara P, Krantz ID, Lapunzina P, Leonard L, Ling S, Ng VL, Hoang PL, Piccoli DA, Spinner NB. NOTCH2 mutations in Alagille syndrome. J Med Genet. 2012;49:138–144. doi: 10.1136/jmedgenet-2011-100544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath BM, Spinner NB, Rosenblum ND. Renal involvement and the role of Notch signalling in Alagille syndrome. Nature Reviews Nephrology. 2013;9:409–418. doi: 10.1038/nrneph.2013.102. [DOI] [PubMed] [Google Scholar]
- Kiernan AE, Ahituv N, Fuchs H, Balling R, Avraham KB, Steel KP, Hrabé de Angelis M. The Notch ligand Jagged1 is required for inner ear sensory development. PNAS. 2001;98:3873–3878. doi: 10.1073/pnas.071496998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Ilagan MXG. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz ID, Colliton RP, Genin A, Rand EB, Li L, Piccoli DA, Spinner NB. Spectrum and frequency of jagged1 (JAG1) mutations in Alagille syndrome patients and their families. Am J Hum Genet. 1998;62:1361–1369. doi: 10.1086/301875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz ID, Piccoli DA, Spinner NB. Alagille syndrome. J Med Genet. 1997;34:152–157. doi: 10.1136/jmg.34.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Masiero M, Banham AH, Harris AL. The notch ligand JAGGED1 as a target for anti-tumor therapy. Frontiers in oncology. 2014;4:1–13. doi: 10.3389/fonc.2014.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Krantz ID, Deng Y, Genin A, Banta AB, Collins CC, Qi M, Trask BJ, Kuo WL, Cochran J, Costa T, Pierpont ME, Rand EB, Piccoli DA, Hood L, Spinner NB. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat. 1997;16:243–251. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- Lindsell CE, Shawber CJ, Boulter J, Weinmaster G. Jagged: A mammalian ligand that activates notch1. Cell. 1995;80:909–917. doi: 10.1016/0092-8674(95)90294-5. [DOI] [PubMed] [Google Scholar]
- Loomes KM, Russo P, Ryan M, Nelson A, Underkoffler L, Glover C, Fu H, Gridley T, Kaestner KH, Oakey RJ. Bile duct proliferation in liver-specific Jag1 conditional knockout mice: effects of gene dosage. Hepatology. 2007;45:323–330. doi: 10.1002/hep.21460. [DOI] [PubMed] [Google Scholar]
- Loomes KM, Underkoffler LA, Morabito J, Gottlieb S, Piccoli DA, Spinner NB, Baldwin HS, Oakey RJ. The expression of Jagged1 in the developing mammalian heart correlates with cardiovascular disease in Alagille syndrome. Hum. Mol. Genet. 1999;8:2443–2449. doi: 10.1093/hmg/8.13.2443. [DOI] [PubMed] [Google Scholar]
- Lu C, Bonome T, Li Y, Kamat AA, Han LY, Schmandt R, Coleman RL, Gershenson DM, Jaffe RB, Birrer MJ. Gene alterations identified by expression profiling in tumor-associated endothelial cells from invasive ovarian carcinoma. Cancer research. 2007;67:1757–1768. doi: 10.1158/0008-5472.CAN-06-3700. [DOI] [PubMed] [Google Scholar]
- McCright B, Gao X, Shen L, Lozier J, Lan Y, Maguire M, Herzlinger D, Weinmaster G, Jiang R, Gridley T. Defects in development of the kidney, heart and eye vasculature in mice homozygous for a hypomorphic Notch2 mutation. Development. 2001;128:491–502. doi: 10.1242/dev.128.4.491. [DOI] [PubMed] [Google Scholar]
- McCright B, Lozier J, Gridley T. A mouse model of Alagille syndrome: Notch2 as a genetic modifier of Jag1 haploinsufficiency. Development. 2002;129:1075–1082. doi: 10.1242/dev.129.4.1075. [DOI] [PubMed] [Google Scholar]
- McDaniell R, Warthen DM, Sanchez-Lara PA, Pai A, Krantz ID, Piccoli DA, Spinner NB. NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway. Am J Hum Genet. 2006;79:169–173. doi: 10.1086/505332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr OL. Character changes caused by mutation of an entire region of a chromosome in Drosophila. Genetics. 1919;4:275. doi: 10.1093/genetics/4.3.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissette JD, Colliton RP, Spinner NB. Defective intracellular transport and processing of JAG1 missense mutations in Alagille syndrome. Hum Mol Genet. 2001;10:405–413. doi: 10.1093/hmg/10.4.405. [DOI] [PubMed] [Google Scholar]
- Oda T, Elkahloun AG, Pike BL, Okajima K, Krantz ID, Genin A, Piccoli DA, Meltzer PS, Spinner NB, Collins FS, Chandrasekharappa SC. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat. Genet. 1997;16:235–242. doi: 10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- Penton AL, Leonard LD, Spinner NB. Notch signaling in human development and disease. Semin. Cell Dev. Biol. 2012;23:450–457. doi: 10.1016/j.semcdb.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reedijk M, Odorcic S, Chang L, Zhang H, Miller N, McCready DR, Lockwood G, Egan SE. High-level Coexpression of JAG1 and NOTCH1 Is Observed in Human Breast Cancer and Is Associated with Poor Overall Survival. Cancer Res. 2005;65:8530–8537. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- Shimizu K, Chiba S, Saito T, Kumano K, Hirai H. Physical Interaction of Delta1, Jagged1, and Jagged2 with Notch1 and Notch3 Receptors. Biochemical and Biophysical Research Communications. 2000;276:385–389. doi: 10.1006/bbrc.2000.3469. [DOI] [PubMed] [Google Scholar]
- Simon DP, Giordano TJ, Hammer GD. Upregulated JAG1 Enhances Cell Proliferation in Adrenocortical Carcinoma. Clin Cancer Res. 2012;18:2452–2464. doi: 10.1158/1078-0432.CCR-11-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakurdas SM, Lopez MF, Kakuda S, Fernandez-Valdivia R, Zarrin-Khameh N, Haltiwanger RS, Jafar-Nejad H. Jagged1 heterozygosity in mice results in a congenital cholangiopathy which is reversed by concomitant deletion of one copy of Poglut1 (Rumi) Hepatology. 2015 doi: 10.1002/hep.28024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnpenny PD, Ellard S. Alagille syndrome: pathogenesis, diagnosis and management. European Journal of Human Genetics. 2012;20:251–257. doi: 10.1038/ejhg.2011.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warthen DM, Moore EC, Kamath BM, Morrissette JJ, Sanchez-Lara PA, Piccoli DA, Krantz ID, Spinner NB. Jagged1 (JAG1) mutations in Alagille syndrome: increasing the mutation detection rate. Hum Mutat. 2006;27:436–443. doi: 10.1002/humu.20310. [DOI] [PubMed] [Google Scholar]
- Xue Y, Gao X, Lindsell CE, Norton CR, Chang B, Hicks C, Gendron-Maguire M, Rand EB, Weinmaster G, Gridley T. Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum Mol Genet. 1999;8:723–730. doi: 10.1093/hmg/8.5.723. [DOI] [PubMed] [Google Scholar]