Abstract
The study of Drosophila Hox genes, located in the Antennapedia complex (ANT-C) and Bithorax complex (BX-C), have provided fundamental insights into mechanisms of how the segments of the animal body plan are specified. Notably, even though the analysis of the BX-C formally began over a century ago, surprises continue to emerge regarding its regulation and function. Even simply the gene content of the BX-C has been regularly revised in past years, especially with regard to non-coding RNAs (ncRNAs), including microRNAs. In this perspective, we review the history of studies of non-coding transcription in the BX-C, and highlight recent studies of its miRNAs that provide new insights into their tissue-specific roles in Hox gene regulation. In particular, we have demonstrated unexpected importance of endogenous BX-C miRNAs to restrict the spatial accumulation of Hox proteins and their TALE cofactors in the ventral nerve cord, and link this to aberrant neural differentiation and reproductive behavior. These findings open new directions on studying Hox miRNA function, and we speculate that further understanding of their roles in insect models may provide new leads for studying the enigmatic biological functions of analogous miRNAs located in vertebrate Hox clusters.
Introduction
microRNAs (miRNAs) are a particular class of short RNAs that are bound by Argonaute proteins, and act as antisense sequence guides to target genes for post-transcriptional repression (Bartel, 2009). Although several aspects of the ~22 nucleotide (nt) mature miRNA and target mRNA sequence can impact regulation, the dominant feature involves Watson-Crick pairing between nts 2–8 (the “seed”) of the miRNA and target (Brennecke et al., 2005; Doench and Sharp, 2004; Lai, 2002). Such minimal target recognition requirements in animals permit individual miRNAs to regulate many transcripts, and conserved animal miRNAs tend to have hundreds of conserved targets (Sun and Lai, 2013).
Beyond the inference that purifying selection acting on conserved target sites implies their functional constraint, transcriptomic (Lim et al., 2005) and proteomic (Baek et al., 2008; Selbach et al., 2008) studies confirm that individual miRNAs indeed confer detectable (although subtle) regulation of extensive target networks. On the other hand, the founding miRNAs, C. elegans lin-4 and let-7, were identified on the basis of characteristic mutant phenotypes that proved to be attributable to individual target genes (Ecsedi et al., 2015; Lee et al., 1993; Reinhart et al., 2000; Wightman et al., 1993). It remains an ongoing question whether modest regulation of panels of miRNA targets can directly explain the phenotypes ascribed to genetic manipulations of miRNAs. Moreover, most miRNA mutants generated in large-scale reverse genetic efforts tend to have modest phenotypes, if at all (Chen et al., 2014; Miska et al., 2007). The general paucity of miRNA mutant phenotypes, combined with the typically extensive catalogs of conserved miRNA target genes, together have made it challenging to define the contributions of miRNAs to in vivo biology (Lai, 2015).
The Hox genes comprise sets of genomically clustered, homeodomain-encoding, transcription factors that are well-known for establishing segmental identities along the anterior-posterior (A-P) axis of animal species (Pearson et al., 2005). Therefore, regulatory mechanisms that specify and maintain the appropriate A-P domains of Hox protein accumulation are central to normal development. Indeed, mutants of Hox genes were famously identified due to “homeotic” defects in which the identity of one body segment is transformed into another (Gehring and Hiromi, 1986). The discovery of multiple miRNAs encoded in invertebrate and vertebrate Hox clusters, and the finding that their predicted targets include some Hox genes, raised the possibility that they comprise a novel layer of regulatory complexity in the Hox hierarchy (Yekta et al., 2008). However, as is the case for most other miRNAs, recognition of their endogenous biological impact has been difficult to garner, and has led to the notion that Hox miRNAs confer fine-tuning, but are not intrinsically required for substantial aspects of Hox-mediated body patterning (Hornstein et al., 2005; Lemons et al., 2012). Nevertheless, recent studies shed light on the unexpectedly robust and non-redundant impact of Drosophila Hox miRNAs on Hox gene expression and organismal biology (Bender, 2008; Garaulet et al., 2014; Gummalla et al., 2012). Critical to these advances was the appreciation of novel locations of Hox miRNA expression and function (i.e., in the post-embryonic central nervous system), and the consideration of Hox gene function beyond body segmentation (i.e., to neural differentiation in reproductive circuits). These findings extend the regulatory complexity of the Hox hierarchy, and provide new vistas for probing Hox gene function. Although we focus this perspective on a particular set of Drosophila Hox miRNAs, the general principles gained from their analysis may potentially be applicable to other Hox miRNAs, not only in flies but also in vertebrates.
A look at the past: 30 years of non-coding transcription across the BX-C
A century ago, in 1915, Calvin Bridges isolated the first homeotic mutant of Drosophila melanogaster in Thomas Morgan’s laboratory at Columbia University. These exhibited transformation of the metathorax (T3) towards the mesothorax (T2), and given the much larger size of T2 relative to T3, the flies appeared to duplicate the entire thorax. Calvin named this mutant after the double-thorax phenotype: bithorax (Bridges and Morgan, 1923). Such mutants became the life work of the Nobel laureate Ed Lewis, who spent some 50 years generating and analyzing dozens of mutations that caused homeotic transformations in fly thoracic and abdominal segments (Figure 1A–C). He found that all these lesions mapped very close to each other in the third chromosome, and speculated that they might have originated by duplication and divergence during arthropod evolution. In the end, he categorized 9 classes of mutations, which he postulated to define a complex of 9 genes (the Bithorax complex, BX-C) that controlled the specification and development of the second thoracic segment, the abdomen, and the genital organs (Lewis, 1978). Similarly, the Antennapedia complex (ANT-C) encodes other homeotic functions that are required to specify head and anterior thoracic segments (Kaufman et al., 1990).
Figure 1.
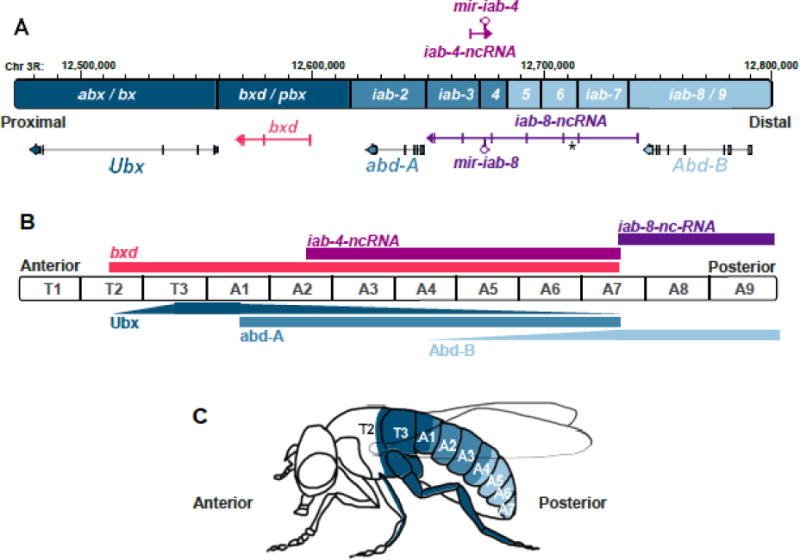
The Drosophila Bithorax complex (BX-C). (A) Physical and genetic organization of the BX-C. The regulatory regions that control the expression of the three Hox protein-coding genes (Ubx, abd-A and Abd-B) are shown in shades of blue and labeled on the schematic of the chromosome. For the infra-abdominal (iab) region, note that iab-4 through iab-6 are abbreviated as 4, 5, and 6. Transcription from the top strand of the BX-C (proximal to distal) is shown above the chromosome in A; transcription of the bottom strand is shown below. All of the transcripts of homeodomain-encoding genes derive from the bottom strand. Non-coding transcripts with cDNA evidence are shown in colored arrows except msa, which is similar to iab-8-ncRNA but differs in the transcription start site (marked with an asterisk). Note that other non-coding transcripts defined by in situ hybridization evidence are not shown in this diagram (see text for details). (B) Schematized domains of expression of the BX-C members along the anterior/posterior axis of an embryonic VNC. The ncRNAs are shown above the diagram, and the protein distributions are shown below. The figure represents a simplified view of the complex pattern of expression within each segment, thus no variations in the compartments are shown. The anterior limit of expression of the bxd transcript shown in the figure corresponds to bxd exon 5 described in Pease et al. (2013), not to the major bxd transcript referred in the same study. (C) Regions of the adult fly specified by each Hox protein-coding gene, inferred by mutations in either the coding region or the regulatory sequences of each transcription unit. No phenotype in the adult morphology has been identified for BX-C ncRNA mutants; however, as discussed in subsequent figures, the miRNAs affect VNC patterning and neural differentiation. T2-A7 stand for thoracic (T) or abdominal (A) segments.
The molecular identity of Hox genes followed the recovery of BX-C and ANT-C sequences, and was particularly informed by the discovery of colleagues in the Gehring and Kaufman groups of the homeobox. This 180 bp sequence encodes a DNA binding domain present in multiple coding regions of the ANT-C (McGinnis et al., 1984; Scott and Weiner, 1984), and is found three times within ~330kb comprising the entire BX-C (Regulski et al., 1985). Together with complementation tests involving recessive alleles performed by the Morata and Whittle groups (Sánchez-Herrero et al., 1985; Tiong et al., 1985), the molecular and genetic data supported a revised model of the BX-C. Instead of 9 genes, it was established that this region consists of three homeodomain-encoding genes, Ultrabithorax (Ubx), abdominal-A (abd-A) and Abdominal-B (Abd-B) (Figure 1A). The other homeotic mutant groups within the BX-C were inferred to affect cis-regulatory sequences contained in the large intergenic regions between the critical protein-coding homeotic loci.
Curiously, these intergenic regulatory regions proved to be extensively transcribed. The first evidence of this was provided by the Hogness group (Hogness et al., 1985; Lipshitz et al., 1987). Northern blotting and cDNA analysis identified two transcripts from the bithoraxoid (bxd) cis-regulatory region of the Ubx gene (Figure 1A). The bxd region is responsible for directing the expression of Ubx in the posterior half of the third thoracic segment (T3p) and the anterior half of the first abdominal segment (A1a). The bxd region is transcribed in early embryos and larval/pupal stages to yield polyadenylated transcripts of 1.1–1.3 kb and 0.8 kb. More recently, Bender’s group used embryo in situ hybridizations to provide evidence for 7 additional non-coding transcripts in the Ubx-bxd region, arising from both DNA strands (Pease et al., 2013).
Distal to Ubx, the BX-C encodes abd-A and Abd-B, which confer the appropriate metameric identity of the A2-A8 segments throughout the development of the fly (Figure 1A and 1C). The expression of abd-A and Abd-B is controlled by a titanic regulatory region that spans these genes (Celniker et al., 1990; Karch et al., 1990; Karch et al., 1985; Macias et al., 1990; Sánchez-Herrero, 1991). Ed Lewis catalogued the mutations in this region as “infra-abdominal” lesions (iab) (Figure 1A). In situ hybridizations using probes covering the entire iab region showed the majority of this genomic region was transcribed during embryogenesis (Sanchez-Herrero and Akam, 1989), in an analogous manner to the Ubx domain. These analyses did not distinguish the number or structure of the iab transcription units, but they defined three distinct expression domains along the anterior-posterior (A/P) axis of the embryo that corresponded with their relative position within the iab region. Such A/P-restricted patterns, in accord with relative 5′-3′ position along the genome (Figure 1A, B), are characteristic of the canonical BX-C Hox genes and referred to as colinearity (Lewis, 1978). In fact, the colinearity principle is a general feature not only of Hox genes in the Drosophila Antp-C, but also of Hox genes in a wide variety of other invertebrate and vertebrate species (Duboule, 2007).
Shortly thereafter, further study of the iab sequences that regulate abd-A expression in the embryo, revealed two novel transcripts arising from the opposite strand (thus, proximal to distal) of the protein-coding genes (distal to proximal), at the level of the iab-4 domain (Cumberledge et al., 1990) (Figure 1A). These apparently non-coding RNAs differ only in their 3′ termini, with two polyadenylation (polyA) sites separated by 304 bp, yielding stable transcripts of 1.7 and 2 kb. Termed accordingly iab-4 ncRNAs, their transcription was later demonstrated to originate from the iab-3 region (Bender and Hudson, 2000) and progress through iab-4 (Figure 1A), but their original naming has been preserved until now. More than a dozen years after their discovery, small RNA cloning by the Tuschl lab showed that a microRNA (miRNA) hairpin is encoded downstream of the initial polyA site that generates the short iab-4 isoform, yielding the mature species miR-iab-4-5p and miR-iab-4-3p (Aravin et al., 2003) (Figure 1A).
The discovery of a miRNA in the BX-C expanded the genic content of this homeotic cluster, and expanded the recognition of mir-10 located in the ANT-C, which is conserved in both sequence and position in vertebrate Hox clusters (Lagos-Quintana et al., 2001). Subsequent deep sequencing of Drosophila small RNAs revealed further surprises, an additional ANT-C miRNA (mir-993), and the existence of miRNAs produced from the antisense strand of the mir-iab-4 hairpin (Ruby et al., 2007). The latter were characterized in concurrent papers from our group and those of Kellis and Bender (Bender, 2008; Stark et al., 2008; Tyler et al., 2008), which demonstrate that sense and antisense transcription across the iab-4 locus produces hairpins that are independently processed by the miRNA biogenesis machinery into functional small RNAs. Although the initial nomenclature was “mir-iab-4AS”, the bottom strand miRNAs were subsequently renamed mir-iab-8 species (Figure 1A, see below). Interestingly, all four mature miRNAs from this locus (miR-iab-4-5p and 3p, miR-iab-8-5p and -3p) are highly conserved in insects, and the regulatory activity of all four possible mature miRNA products was detected in cultured cells and transgenic animals (Hui et al., 2013; Okamura et al., 2008). However, the dominant small RNAs from sense and antisense hairpins derive from the “5p” arms (Ruby et al., 2007). Since miRNAs function as concentration-dependent regulators, this may imply that miR-iab-4-5p and miR-iab-8-5p have greater biological impact than their respective “3p” species.
Although it is the same genomic sequence that produces the top and bottom strand miRNA hairpins, mir-iab-4 can be considered to exist more proximally on the chromosome to mir-iab-8, based on the relative locations of their promoters (Figure 1A, B). In accord with this, mir-iab-4 is transcribed in more anterior segments relative to mir-iab-8, and in fact their spatial domains abut each other but for the most part they are not co-expressed (Bender, 2008; Ronshaugen et al., 2005; Stark et al., 2008; Tyler et al., 2008) (Figure 1B). Therefore, BX-C miRNAs conform to the colinearity principle.
In fact, the notion of antisense transcription (from the bottom strand) across the iab-4 region of the BX-C was not a new one. After the initial in situ experiments by Sanchez-Herrero and Akam, Levine’s group detected transcription of this strand from an iab-8 internal promoter lying just downstream of the Abd-B transcription unit (Zhou et al., 1999). This putative iab-8 transcript is expressed in the A8 and A9 segments throughout embryonic development. Further in situ studies by the Drewell lab confirmed transcription from this strand in all the iab domains from iab-2 to iab-8, with different patterns of expression (Bae et al., 2002). During early embryonic development, the anterior limit of the expression domain of each transcript was colinear with its relative position of their DNA in the chromosome, in agreement with previous studies. But intriguingly, their patterns of expression in late embryos proved indistinguishable, and confined to only the A8–A9 segments of the ventral nerve cord (VNC, analogous to the spinal cord in vertebrates). The explanation of this discrepancy, although initially suggested by Bender’s lab (Bender and Fitzgerald, 2002) was not fully confirmed until later complementation experiments (Bender, 2008). These tests implied the existence of a massive iab-8 non-coding transcript (potentially >140 kb) spanning from the iab-8 promoter described by Levine and colleagues, and later corroborated in other studies (Enderle et al., 2011; Gummalla et al., 2012), to at least past the mir-iab-4 hairpin, thus generating miR-iab-8 species (Figure 1A). The previous observations of the coincident transcription patterns from distinct iab domains in late embryos (Bae et al., 2002) may likely be explained by the maintained transcription of a single iab-8 ncRNA through the entire iab region, once the transcriptional activity from all the other iab domains has ceased by mid-embryonic development.
Contemporary studies by the modENCODE project yielded higher-resolution molecular information on developmental and tissue-specific non-coding transcription within the BX-C (Graveley et al., 2011). While the boundaries of pri-mir-iab-4 transcripts are relatively well-delineated, the picture regarding iab-8 transcripts generated from the entire transcribed infra-abdominal region gained complexity. For example, novel spliced, male-specific isoforms of the long iab-8 ncRNA were identified from RNA-seq data and confirmed by cDNA evidence; the latter identified transcript initiates in the iab-6 region and terminates just upstream of abd-A (Graveley et al., 2011) (Figure 1A). This locus was named male specific abdominal (msa), based on its restriction to the male gonad. It is unclear whether msa exerts function as non-coding transcripts or potentially via short ORFs (Gummalla et al., 2014). In addition, msa necessarily transcribes mir-iab-8 (contained in its intron) and therefore may represent a source of this miRNA.
Despite the deep annotation of transcription from enormous datasets, recently expanded in the capstone modENCODE Drosophila transcriptome paper (Brown et al., 2014), it is important to bear in mind that highly cell-specific, temporal-specific, and/or short-lived transcripts may still elude recognition. For example, although a huge amount of embryonic mRNA-seq and total RNA-seq data now exist, for the most part, these were performed from whole embryo samples. Notably, earlier in situ studies detected transient expression of additional putative short iab RNAs in specific domains of the A/P axis other than A8 and A9 during early embryonic development (Bae et al., 2002; Sanchez-Herrero and Akam, 1989) (Pease et al., 2013), whose molecular identities remain undetermined. One can imagine that further sequencing of polyadenylated and rRNA-depleted RNAs from specific axial domains across development, may produce further insights.
Three decades of work on non-coding transcripts across the regulatory regions of the BX-C provide extensive knowledge of the transcriptional status of specific cis-regulatory regions across developmental stages and certain tissues. During all this time, several groups have speculated about the possible functions of these transcripts. However, it has been difficult to separate potential effects of mutations, often involving chromosomal rearrangements or other complex aberrations, on the ncRNAs as intrinsic regulatory molecules from effects on transcriptional enhancers. Conversely, current knowledge points to the idea that it is not the sequence identity of such transcripts, but their transcription per se, that contributes to regulation of BX-C protein-coding genes. Earlier work established that transcription across a Polycomb-silenced region of the BX-C could lead to its activation (Bender and Fitzgerald, 2002; Hogga and Karch, 2002; Rank et al., 2002; Schmitt et al., 2005). On the other hand, non-coding transcription in the BX-C might interfere with that of downstream of protein-coding gene promoters, as proposed for the effect of bxd transcription on Ubx (Petruk et al., 2006), or iab-8 transcription on abd-A (Gummalla et al., 2012). Given that painstaking genetic efforts to precisely eliminate a major BX-C ncRNA (bxd) did not yield substantial regulatory or phenotypic defects (Pease et al., 2013), the possibility remains that many BX-C ncRNAs might represent products of transcriptional noise associated with enhancers. Since miRNAs are typically considered as trans-regulatory species, then, it becomes especially germane to ask whether these BX-C miRNAs indeed have any functional consequence to the regulation or function of the BX-C.
Homeotic function of BX-C miRNAs: conducting the conductors
Initial bioinformatic predictions of Hox-encoded miRNAs, not only in Drosophila but in vertebrates, included hints that they might directly regulate canonical Hox genes (Yekta et al., 2008). For example, analysis of genes bearing conserved miR-iab-4 seed matches identified Ubx as a candidate target (Grun et al., 2005; Stark et al., 2003). While other studies had shown the capacity for vertebrate Hox miRNAs to repress Hox homeobox genes in reporter and molecular assays (Yekta et al., 2004), experiments by the Levine and Lai groups provided the first evidence that manipulation of a Hox miRNA could induce a morphological homeotic phenotype in the animal. An endogenous function of Ubx is to suppress the thorax and wing developmental program within the developing haltere. Thus, Ubx mutants described by Lewis exhibit transformation of the haltere into a second thorax and extra pair of wings. Similarly, spatially-controlled misexpression of miR-iab-4 in imaginal disc haltere primordia resulted in larger, flatter halteres bearing characteristic rows of sensory organs on the anterior dorsoventral margin, evidence for a haltere-to-wing transformation (Ronshaugen et al., 2005).
The addition of mir-iab-8 to the BX-C landscape implied an interesting consideration, since its sequence is related to mir-iab-4. In particular, the dominant “5p” products of their respective hairpins have overlapping seed regions, albeit offset by two nucleotides (Ruby et al., 2007). Thus, while certain target sites might accommodate both miRNAs, their overall target networks are expected to be largely distinct. Interestingly, these attributes play out with respect to Hox targets. While the Ubx 3′ UTR contains multiple conserved pairings to miRNAs of both strands, only one of these is an optimal seed match site for miR-iab-4-5p whereas five of them are strong sites for miR-iab-8-5p. Even more to the extreme, the abd-A 3′ UTR contains six conserved, strong seed matches for miR-iab-8-5p whereas all of these sites represent weak pairings to miR-iab-4-5p (Stark et al., 2008; Tyler et al., 2008).
The relative strengths of these predicted miRNA-target pairings were borne out using in vivo assays. Clonal misexpression experiments showed that ectopic mir-iab-8 largely eliminates endogenous Ubx and/or abd-A proteins (normally found in the haltere and genital discs, respectively) whereas ectopic mir-iab-4 weakly suppresses Ubx and has no effect on abd-A accumulation (Tyler et al., 2008). These findings were confirmed using in vitro and in vivo sensor experiments, and most tellingly by the finding that ectopic mir-iab-8 generates much stronger haltere-to-wing transformations (i.e. Ubx phenocopies) than does mir-iab-4 (Stark et al., 2008; Tyler et al., 2008).
Thus, while the initial study of miR-iab-4 provided evidence that it suppresses one anteriorly-expressed Hox gene, miR-iab-8 actually is wired for massive targeting of multiple anteriorly-expressed Hox genes; neither miRNA contains binding sites within Abd-B, the most posteriorly-expressed Hox gene (Figure 2). Notably, miR-iab-8-5p even bears conserved seed matches within the Antp 3′ UTR. All of these relationships are analogous to the established transcriptional repression hierarchy by homeodomain-encoding Hox genes, whereby chromosomally distal Hox genes (which are normally expressed in posterior segments) can inactivate genes located more proximally (and are thus expressed more anteriorly) (Hafen et al., 1984; Harding et al., 1985; Struhl and White, 1985). Thus, the BX-C miRNAs conform to key principles established for Hox homeobox genes, that is, their spatial expression obeys the colinearity rule while their function appears to integrate into a posterior dominance hierarchy (Singh and Mishra, 2008; Yekta et al., 2008) (Figure 2).
Figure 2.
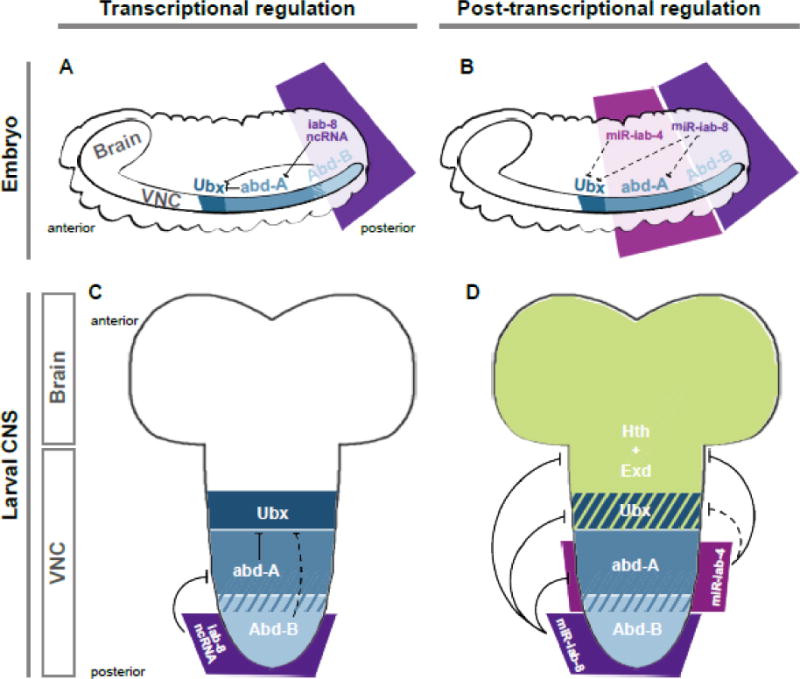
Interplay of regulatory strategies in the BX-C. Transcriptional (A, C) and post-transcriptional (B, D) regulation of the Hox protein-coding genes in the embryonic (A, B), and larval (C, D) VNC. The control of the Hox cofactors Hth and Exd by miRNAs is also shown in D. Note that for simplicity, the regulatory relationships of transcription factors with their genomic targets and of miRNAs with their RNA targets is indicated directly on the respective tissues. However, the regulation by TFs and miRNAs implicitly occurs within their cognate expression domains (i.e., Abd-B transcriptionally represses Ubx only within the terminal segments, and miR-iab-8 post-transcriptionally represses abd-A, Ubx and Hox cofactors only within the terminal segments). Note also that the patterns of distribution of the Hox proteins and cofactors are simplified, obviating overlapping domain of Ubx and abd-A. Exd is ubiquitously expressed, but is only functional/nuclear in the presence of Hth, which is segmentally patterned as shown. Solid lines indicate inhibition by miR-iab-4 or miR-iab-8 of a given target; dashed lines indicate subtle inhibition. The combined transcriptional and post-transcriptional inhibition in their corresponding domain of expression establishes the final distribution of homeotic factors in the VNC: preventing Hth and Exd protein from all abdominal segments, and Ubx and abd-A out from the most terminal segments (A8 onwards).
Nevertheless, while the phenotypes generated by ectopically expressed BX-C miRNAs were striking, they left open the question of the regulatory effects of the endogenous miRNAs. Bender addressed this by creating a precise deletion of the hairpin (referred to as Δmir), constituting a null allele for both mir-iab-4 and mir-iab-8 (Bender, 2008). A study of the spatial distribution of BX-C Hox proteins in Δmir embryos, revealed a mild derepression of Ubx and abd-A in A8–A9 in the VNC, which corresponds to the domain where the long iab-8 ncRNA which produced miR-iab-8 is expressed, where Ubx and Abd-A proteins do not normally accumulate. Notably, no misexpression was found in the embryonic epidermis (Bender, 2008; Gummalla et al., 2012). Alonso proposed an explanation to this differential regulation of Ubx expression based on binding site accessibility for miR-iab-4 and miR-iab-8: only long 3′ UTR isoforms of Ubx, which are restricted to the embryonic central nervous system (CNS), would carry enough binding sites to allow a substantial regulation of Ubx mRNA, compared to the smaller isoforms expressed also in the epidermis which would consequently escape to this mechanism (Thomsen et al., 2010). Notably, the principle of substantially extended neural 3′ UTRs has proven to be a broad feature of the Drosophila (Hilgers et al., 2011; Smibert et al., 2012) and mammalian (Miura et al., 2013) transcriptomes.
We subsequently examined BX-C miRNA expression and activity in later stages of development, and showed that these are maintained for miR-iab-4 and miR-iab-8 in the larval posterior VNC, in the same spatial register as observed in late embryos (Garaulet et al., 2014). The larval VNC appears to be the relevant location of BX-C miRNA function, since in contrast to the subtle effects observed in the embryos, Ubx and Abd-A proteins are substantially derepressed in Δmir mutant VNCs (Garaulet et al., 2014) (Figure 3A). Moreover, this misexpression of Hox proteins is maintained through pupal stages to adult neurons (DLG and ECL, unpublished observations).
Figure 3.
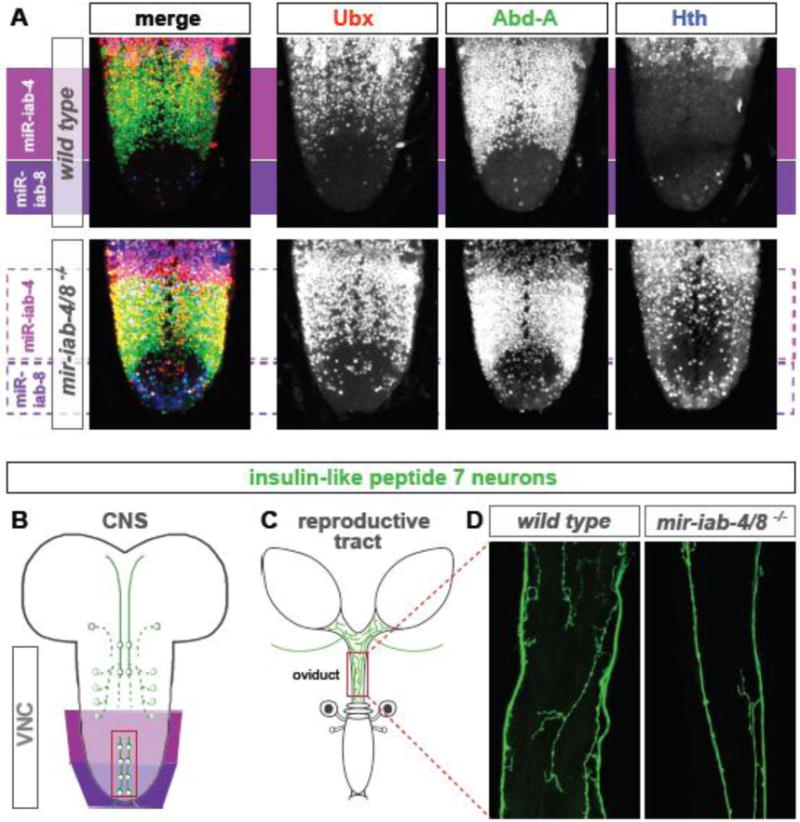
Larval expression of BX-C genes in the VNC and neural phenotypes in the mir-iab-4/8 mutant. (A) Distribution of Ubx, Abd-A, and Hth proteins in the larval VNC (region labeled as VNC in the CNS diagram in B). The domain of expression of miR-iab-4 and miR-iab-8 is shown in magenta and purple, respectively. The mir-iab-4/8−/− VNC exhibits elevation of Ubx protein within the miR-iab-4 domain, and misexpression of Ubx, Abd-A and Hth proteins within the miR-iab-8 domain. (B) The cell bodies of abdominal Ilp-7 neurons (red box) lie within the miRNA-expression domains in the larval VNC. (C) The adult derivatives of Ilp-7 neurons innervate the reproductive tract in females, especially the oviduct (red box). (D) Staining of Ilp-7 neuronal projections shows that their innervation of the oviduct is reduced in mir-iab-4/8−/− mutant relative to wildtype.
The degree of target derepression observed in Δmir VNC places it within a relatively rare realm of instances for miRNA targeting studied in the animal. Transcriptome and proteome studies provide the general view that the direct impact of miRNAs on the bulk of many thousands of documented targets is generally subtle (Flynt and Lai, 2008). On the other hand, mutants of BX-C miRNAs cause misexpression of Ubx and abd-A in some CNS cells to levels comparable to wildtype-expressing cells (Figure 3A). While the strength of targeting of these BX-C genes is exceptional, the “salt-and-pepper” derepression of the endogenous protein products does not mirror the relatively uniform repression of tub-GFP sensor transgenes linked to miR-iab-4 or miR-iab-8 sites detected within the cognate miRNA expression domains (Garaulet et al., 2014). This may suggest that the strong impact of miRNA targeting is amplified by mechanisms that control the transcriptional regulation of these Hox targets.
Another important conclusion from these studies is the detection of coexpressed Ubx and/or Abd-A proteins in posterior VNC cells that also accumulate Abd-B protein (Garaulet et al., 2014). This indicates a flagrant breakdown of posterior dominance, since according to this principle, Abd-B should not allow the expression of these anterior BX-C genes (Figure 2A, C). This suggests a more complex contest where miR-iab-8 downregulates Ubx and abd-A, not only as a “fail-safe” strategy, but indeed as a substantial mechanism. This notion builds on concepts from Karch’s group, who found that unlike in the embryonic epidermis, the loss of Abd-B protein in the VNC of embryos mutant for this posterior Hox gene, did not upregulate the immediately anterior Hox protein Abd-A in posterior segments (Gummalla et al., 2012). In contrast, the absence of the repression exerted by either miR-iab-8 or iab-8-ncRNA derepresses abd-A. This genuine, non-redundant role of miR-iab-8 to the established transcriptional hierarchy in the embryonic CNS, was later expanded by comparative analysis of the distribution of Ubx and Abd-A proteins in larval VNCs of wildtype, Δmir and Abd-B mutants. In contrast to the strong phenotype of Δmir mutants (Figure 3A), the absence of Abd-B only slightly de-represses Ubx and has no effect on Abd-A in larval VNCs (Garaulet et al., 2014). Thus, the posterior dominance dogma should now be reinterpreted according to the tissue: only in epidermal cells the control exerted by Hox protein-coding genes is enough to maintain their pattern of expression, while in the neural tissue, especially in post-embryonic contexts, post-transcriptional regulation is a major factor driving BX-C patterning in terminal segments (Figure 2).
Curiously, the control of homeotic activity by BX-C miRNAs extends beyond regulation of Hox genes. The homeodomain Hox cofactors of the TALE class encoded by homothorax (hth) and extradenticle (exd) (Peifer and Wieschaus, 1990; Pai et al., 1998; Rieckhof et al., 1997) are also targeted by miR-iab-4 and/or miR-iab-8 (Garaulet et al., 2014). hth 3′ UTRs contain four well-conserved binding sites for the different BX-C miRNAs. However, an unannotated isoform of the hth-RA 3′ UTR bears a 2.2 kb 3′ UTR extension that is specifically expressed in the CNS (Garaulet et al., 2014). This extension harbors many additional matches for various miR-iab-4/8 seeds, which collectively make hth more heavily targeted than most Drosophila genes studied in previous genomewide miRNA target scans (Ruby et al., 2007; Stark et al., 2005). Conversely, target predictions for exd were lost from earlier systematic assessments, due to a break within its 3′ UTR in genome alignments. When aligned manually, conserved seed matches for BX-C miRNAs were observed within exd 3′UTR. Once again, only some of them are shared by miR-iab-4 and miR-iab-8, and the majority of them are unique to one or the other, implying complex independent regulation of individual exd and hth isoforms by Hox miRNAs (Garaulet et al., 2014). Altogether, the fact that previous genomewide scans did not reveal the full extent of TALE cofactor targeting by BX-C miRNAs (due to insufficient 3′ UTR annotations or to problems in the genomewide alignments) are an important reminder that such prediction efforts are only a guide for the breadth of the miRNA target network, and are likely incomplete at present.
Hth and Exd (as are their vertebrate counterparts) are cofactors that increase binding affinity and specificity of Hox proteins (Mann et al., 2009; Moens and Selleri, 2006). Their spatial distribution in the larval CNS is restricted to the brain and thoracic segments, but their transcripts are almost absent from the abdominal neurons due to a negative control exerted by the BX-C proteins (Kurant et al., 1998). However, Δmir mutants exhibit massive misexpression of these proteins in the abdominal segments, expanding their pattern of expression to the most posterior end of larval VNC (Garaulet et al., 2014) (Figure 3A).
In sum, Ubx/abd-A/hth/exd not only generate functionally related protein complexes that direct segmental identity, they are all also functionally targeted by miR-iab-4 and miR-iab-8. This miRNA-mediated regulation is particularly manifest in the posterior abdominal region of the VNC, and adds to multiple transcriptional mechanisms (both protein and ncRNA-mediated) that restrict Hox gene expression in the posterior VNC (Figure 2). What then, are biological contexts that underlie the selection for such parallel, and apparently convergent, regulatory strategies?
Hox transcripts, homeotic proteins, and sexual behavior
The majority of individual miRNAs in various species appear largely dispensable under normal laboratory conditions. Recent analysis of a genome-scale collection of Drosophila melanogaster miRNA knockouts showed that many exhibit quantitative defects in one or more settings, but on the other hand, nearly all were viable, fertile and lack obvious morphological malformations (Chen et al., 2014). Systematic analysis of C. elegans miRNA knockouts yielded a similar perspective of few and/or modest defects (Miska et al., 2007). Although Δmir mutants have no overt sign of homeotic transformations or other aberrant morphology, adult flies lacking BX-C miRNAs manifest a dramatic phenotype: both males and females are completely sterile (Bender, 2008). In his work, Bender conducted complementation tests in which he assayed fertility in trans-heterozygous animals carrying the Δmir allele over different breakpoints in the infra-abdominal region that likely truncate either the iab-4 or iab-8 ncRNAs (Bender, 2008). By these genetic tricks, he was able to generate flies that were null for only one or the other of the BX-C miRNAs. With this approach, he observed that flies lacking miR-iab-4 were fertile, while flies lacking miR-iab-8 recapitulate Δmir sterility. Since he assayed a variety of genomic breakpoints across the infra-abdominal region, his experiments provide genetic evidence that miR-iab-8 is indeed processed from an extremely long transcript, corroborating the previous notion of the iab-8 promoter downstream of the Abd-B transcription unit (Zhou et al., 1999).
Initial interrogation revealed that gamete production was apparently unaffected in both sexes. Nevertheless, females are unable to lay eggs, while males cannot bend their abdomen far enough to copulate (Bender, 2008). This suggested that the basis of such defects might involve different organs in the two sexes: the reproductive tract in females, and the abdominal muscles in males. However, the function of both is governed in part by a common tissue, namely the CNS. Indeed, it was only in the posterior VNC where an aberrant pattern of Hox proteins distribution was observed in the entire embryo, both in Bender’s paper for Ubx, and some years after by Karch’s group for Abd-A (Gummalla et al., 2012). Even though the misexpression of the two Hox proteins in embryos seemed too subtle to provoke such a profound disorder in adult flies, our studies supply missing links that support this notion (Garaulet et al., 2014). First, as discussed above, derepression of both Ubx and Abd-A proteins is broader in larval VNCs than reported in embryos, and an even more dramatic effect was observed in the misexpression of their cofactors, Hth and Exd (Figure 3A). Combining the Δmir background with heterozygous combinations of miR-iab-8 target genes revealed that female sterility was partially suppressed when reducing the dose of hth or Ubx, and more weakly, of abd-A (Garaulet et al., 2014). These results highlight two relevant conclusions. First, they establish a causal connection between the misexpression of target genes (observed only in the VNC) with the adult sterility observed in Δmir flies, which assigns a crucial role for the patterning of homeotic genes in an adult behavior. Second, they represent a notable situation of phenotypically critical, individual, miRNA target loci. As conserved miRNAs often have hundreds of conserved targets, miRNAs are often inferred to exert their function via cumulative, subtle effect across their target network (Lai, 2015; Smibert and Lai, 2010). In the case of BX-C miRNAs, the mere heterozygosity of hth was sufficient to restore fertility in nearly 60% of the adult females, illustrating an exceptionally high rescue of a miRNA mutant phenotype induced by mild reduction in a single target gene.
The lack of a substantial fertility rescue in abd-A or exd heterozygous flies does not rule out their contribution to Δmir sterility; nevertheless, it indicates that this defect is more sensitive to the dose of Ubx and hth. Notably, experiments from the Karch group show that flies lacking transcriptional interference of the iab-8 ncRNA over the abd-A promoter, but that retain the miRNA locus, are also sterile, and show acute derepression of abd-A in the posterior portion of the VNC during embryonic development (Gummalla et al., 2012). These observations indicate that apart from miR-iab-8 regulatory functions, the transcriptional interference on the abd-A promoter by the iab-8-ncRNA is crucial for the patterning of this Hox gene in the CNS (Figure 2). Together, the consequences of lacking either of these ncRNA-mediated functions emphasize the notion that misinterpretation of positional information conferred by the Hox activity in the posterior domain of the VNC can lead to abnormal adult sexual behavior.
A recent study by the Vosshall lab further supports this hypothesis by showing that the Abd-B protein is required during development for a normal female receptivity in adult virgin flies (Bussell et al., 2014). The reason for these sex-specific behavioral defects may originate in the fact that many of the implicated neurons, most of which are defined by the expression of the sex-determining genes doublesex and or fruitless, reside in the posterior VNC (Bussell et al., 2014; Feng et al., 2014), where Abd-B and miR-iab-8 are coexpressed. Therefore, it seems plausible that the misexpression of anterior Hox proteins and their cofactors observed within this region of the VNC of Δmir flies, could affect diverse aspects of female and male sexual behaviors beyond sterility.
At present, no specific functional or morphological defects of neurons are known to underlie the sexual receptivity phenotype of flies lacking Abd-B in the CNS (Bussell et al., 2014). However, discrete populations of neurons that reside in this domain project to the oviduct and are known to be critical for egg passage through the oviduct, including glutamatergic neurons that express insulin-like peptide 7 (Ilp7) (Yang et al., 2008) (Figure 3B,C) and octopaminergic neurons that express tyrosine decarboxylase 2 (Cole et al., 2005). In our study (Garaulet et al., 2014), we found that oviduct innervation and synaptic connectivity by Ilp7-expressing neurons was substantially compromised in Δmir flies (Figure 3D). Notably, this defect can be rescued by double heterozygosity of Ubx and abd-A, indicating a causal effect of the misexpression of these Hox genes in the neural development of motor neurons required for oviposition. This fact, although not the first, constitutes a notable failure of “phenotypic suppression” that is normally attributed to BX-C proteins. By this precept, when two Hox proteins are forcibly coexpressed in the same cell, the developmental program dictated by the posterior Hox factor predominates over the “anterior” Hox factor (Duboule and Morata, 1994; Gonzalez-Reyes and Morata, 1990). According to this notion, in the Abd-B region of the VNC of Δmir flies, derepression of Ubx or abd-A should not influence the development of the neurons. However, our evidence clearly illustrates this is not the case, since reduction of Ubx or abd-A can partially rescue Δmir phenotypes (Garaulet et al., 2014).
It is not yet known how Hox gene deregulation induces aberrant morphology of Ilp7 axons. However, work from Mann’s group describes how Hox gene functions in the fly thorax are required for specification of leg motoneuron morphologies (Baek et al., 2013). Whereas Hox activity is not required for the selection of the region of the leg to be targeted by Lin A motoneurons, it is determinant for the branching pattern of their axons within that region, as well as their dendritic arborizations within the VNC. Based on this, it is conceivable that aberrant Hox specification conferred to Ilp7 neurons in Δmir flies might result in defective axonal innervation observed in the oviduct of Δmir flies. Also, Ilp7 neurons differentiate from embryonic pioneer dMP2 neurons, which undergo apoptosis in anterior segments of the VNC but survive in the posterior region. This involves an anti-apoptotic function of Abd-B, which directly represses the activity of the cell death-promoting genes reaper and grim (Miguel-Aliaga and Thor, 2004; Miguel-Aliaga et al., 2008). It seems possible that the reduced innervation could be caused by a partial absence of Ilp7 neurons, because of an altered cell death/survival program induced by expression or coexpression of several homeotic proteins instead of Abd-B alone in the posterior region of the abdominal VNC. Nevertheless, no significant difference in the number of ventral Ilp7 neurons was found in the VNC of Δmir larvae suggesting that cell survival or cell death is not affected in this specific lineage of this mutant background (Garaulet et al., 2014).
Are Ilp7 neurons the solely responsible of Δmir infertility? Our results suggest that this is not the case. First, we were not able to rescue fecundity in mutant flies by knocking down any of the individual targets or overexpressing the mir-iab-8 in Ilp7 neurons. Second, unlike the results with BX-C proteins, heterozygosity of hth strongly rescues female sterility in Δmir mutants, but does not restore either the innervation nor the synapse defects of Ilp7 axons (Garaulet et al., 2014). These results imply that other neuronal lineages beyond Ilp7 neurons, that are relevant for fertility, may be affected by the loss of miR-iab-4 and miR-iab-8, at least by the derepression of hth. Further efforts are needed to identify these additional neurons that require miR-iab-4/8 activity for an accurate control of the reproductive organs in Drosophila females.
The morphological and behavioral consequences of miR-iab-4 and miR-iab-8 on the regulation of Hox proteins and cofactors highlight the importance of homeotic proteins in neural function and development. Apart from the Hox requirements described for leg motoneurons (Baek et al., 2013), many other roles for homeotic proteins in the nervous system have been documented (Philippidou and Dasen, 2013; Rogulja-Ortmann and Technau, 2008). For example, BX-C genes have been shown to be necessary to confer the segmental specificity of several neuroblast progenitors (Prokop et al., 1998; Rogulja-Ortmann et al., 2008), and the cell fate of some embryonic VNC abdominal neuroblasts (Berger et al., 2005; Birkholz et al., 2013). They can also regulate the number of neurons in a certain segment by regulating cell proliferation (Prokop et al., 1998; Tsuji et al., 2008), or programmed cell death (Bello et al., 2003; Rogulja-Ortmann et al., 2008; Suska et al., 2011). Taken together, the ample evidence of crucial roles for the BX-C Hox proteins in diverse aspects of neural development suggests that the misexpression of homeotic targets observed in Δmir flies, could very likely preclude in the correct development and functions of many populations of neurons beyond Ilp7 cells, within the posterior region of the VNC.
Conclusions and Perspectives
A century of work to decipher the Bithorax complex has led to hundreds of studies, which arguably rank the BX-C amongst the highest in the Drosophila genome in terms of ratio of publications per base pair. As a consequence, we have extensive knowledge of their components, the mechanisms underlying their regulation, and the functions of the proteins that they encode shaping the body of the fruit fly. Nevertheless, few studies have focused on the relevance of BX-C functions in larval or adult Drosophila behavior. As required for the specification, survival, number, and branching patterns of several lineages of neurons during development, the activity of Hox genes must be relevant for any potential behavior controlled by neuronal circuits affected by these processes: larval and adult locomotion, sexual courtship and intestinal physiology are some plausible examples. In addition to these, given the wide complexity of neuronal connections throughout several segments of the VNC and their intercommunication with the brain, the impact of Hox activity could affect diverse behaviors in unforeseen manners. With the implementation of physiological and quantitative methods to accurately monitor and probe Drosophila behaviors, we anticipate that new insights regarding adult Hox functions to emerge in the coming years.
Beyond the study of insects, given that myriad principles of Drosophila Hox gene organization, regulation and function have proven to be broadly conserved across animals, it will be interesting to see how the knowledge BX-C ncRNAs may further inform vertebrate studies. The existence of extensive non-coding transcription across the BX-C has been recapitulated in vertebrate Hox clusters (Gupta et al., 2010; Rinn et al., 2007), and includes multiple miRNAs (Yekta et al., 2008). Notably, vertebrate mir-196 loci are located in the posterior Hox complex regions, analogous to mir-iab-4/8, and substantially repress several more anteriorly-located Hox genes (Mansfield et al., 2004; Yekta et al., 2004). Although it was proposed that these have limited “fine-tuning” effects that are secondary to transcriptional regulation of murine Hox genes (Hornstein et al., 2005), injection of miR-196 morpholinos in zebrafish and chick caused axial defects and altered expression of Hox genes (He et al., 2011; McGlinn et al., 2009). It will be interesting to learn if chromosomal deletion of mir-196 genes, perhaps involving compound mutants of the multiple mir-196 loci, recapitulates any homeotic defects. Based on our studies in flies, we speculate that it may be important to go beyond gross axial patterning, and search for potential tissue-restricted and/or behavioral phenotypes. For example, there is evidence that regulation of Hox genes by miR-196 in the mammalian neural tube is involved in specifying appropriate motoneuron subtypes (Asli and Kessel, 2010).
In all systems, a more detailed understanding of how non-Hox targets mediate Hox miRNA function is warranted. While our work implies, but does not yet prove, a scenario whereby derepressed BX-C and TALE cofactors might drive mutant phenotypes as protein complexes (Garaulet et al., 2014), TALE factors certainly have Hox-independent functions (Cerda-Esteban and Spagnoli, 2014). It remains to be seen if de-repressed Hth, Exd, Ubx and Abd-A work together, or independently, or both, to mediate the observed physiological defects. In the zebrafish system, miR-196 was proposed to mediate developmental patterning at least in part by targeting the retinoic acid receptor rarab, whose activity itself can impact Hox gene expression (He et al., 2011). In mammalian systems, a variety of non-Hox targets of miR-196 have been proposed in disease and cancer (Brest et al., 2011; Yang et al., 2014). We advocate that precision engineering of Hox miRNA target sites in individual target genes, as recently demonstrated (Bassett et al., 2014; Ecsedi et al., 2015), will be critical for future elucidation of the impact of Hox miRNAs in developmental, physiology, and disease.
Highlights.
We provide a synthesis of the history of non-coding transcription and fundamental regulatory mechanisms in the Drosophila Bithorax-Complex (BX-C).
BX-C miRNAs post-transcriptionally regulate BX-C protein-coding genes and their cofactors hth and exd in the posterior abdominal segments of the VNC in Drosophila.
miRNA regulation of Hox patterning in the VNC is required for female fertility in Drosophila.
Perspectives on future directions for studying BX-C miRNA neural biology.
Acknowledgments
We thank Ernesto Sanchez-Herrero for critical reading and discussion. Work in E.C.L.’s group was supported by the Burroughs Wellcome Fund and the NIH (R01-NS074037 and R01-NS083833).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aravin A, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Asli NS, Kessel M. Spatiotemporally restricted regulation of generic motor neuron programs by miR-196-mediated repression of Hoxb8. Dev Biol. 2010;344:857–68. doi: 10.1016/j.ydbio.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Bae E, Calhoun VC, Levine M, Lewis EB, Drewell RA. Characterization of the intergenic RNA profile at abdominal-A and Abdominal-B in the Drosophila bithorax complex. Proc Natl Acad Sci U S A. 2002;99:16847–52. doi: 10.1073/pnas.222671299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek M, Enriquez J, Mann RS. Dual role for Hox genes and Hox co-factors in conferring leg motoneuron survival and identity in Drosophila. Development. 2013;140:2027–38. doi: 10.1242/dev.090902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AR, Azzam G, Wheatley L, Tibbit C, Rajakumar T, McGowan S, Stanger N, Ewels PA, Taylor S, Ponting CP, Liu JL, Sauka-Spengler T, Fulga TA. Understanding functional miRNA-target interactions in vivo by site-specific genome engineering. Nat Commun. 2014;5:4640. doi: 10.1038/ncomms5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello BC, Hirth F, Gould AP. A pulse of the Drosophila Hox protein Abdominal-A schedules the end of neural proliferation via neuroblast apoptosis. Neuron. 2003;37:209–19. doi: 10.1016/s0896-6273(02)01181-9. [DOI] [PubMed] [Google Scholar]
- Bender W. MicroRNAs in the Drosophila bithorax complex. Genes Dev. 2008;22:14–9. doi: 10.1101/gad.1614208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W, Fitzgerald DP. Transcription activates repressed domains in the Drosophila bithorax complex. Development. 2002;129:4923–30. doi: 10.1242/dev.129.21.4923. [DOI] [PubMed] [Google Scholar]
- Bender W, Hudson A. P element homing to the Drosophila bithorax complex. Development. 2000;127:3981–92. doi: 10.1242/dev.127.18.3981. [DOI] [PubMed] [Google Scholar]
- Berger C, Pallavi SK, Prasad M, Shashidhara LS, Technau GM. Cyclin E acts under the control of Hox-genes as a cell fate determinant in the developing central nervous system. Cell Cycle. 2005;4:422–5. doi: 10.4161/cc.4.3.1524. [DOI] [PubMed] [Google Scholar]
- Birkholz O, Vef O, Rogulja-Ortmann A, Berger C, Technau GM. Abdominal-B and caudal inhibit the formation of specific neuroblasts in the Drosophila tail region. Development. 2013;140:3552–64. doi: 10.1242/dev.096099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of MicroRNA-Target Recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brest P, Lapaquette P, Souidi M, Lebrigand K, Cesaro A, Vouret-Craviari V, Mari B, Barbry P, Mosnier JF, Hebuterne X, Harel-Bellan A, Mograbi B, Darfeuille-Michaud A, Hofman P. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nat Genet. 2011;43:242–5. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- Bridges C, Morgan TH. The third-chromosome group of mutant characters of Drosophila melanogaster. Publ Carnegie Inst. 1923;327:1–251. [Google Scholar]
- Brown JB, Boley N, Eisman R, May G, Stoiber M, Duff M, Booth B, Park S, Suzuki A, Wan K, Yu C, Zhang C, Carlson J, Cherbas L, Eads B, Miller C, Mockaitis K, Roberts J, Davis C, Frise E, Hammonds A, Olson S, Shenker S, Sturgill D, Andrews J, Wen J, Robinson G, Hernandez J, Bickel PJ, Carninci P, Cherbas P, Gingeras T, Hoskins R, Kaufman TC, Lai EC, Oliver B, Graveley BR, Celniker SE. Diversity and dynamics of the Drosophila transcriptome. Nature. 2014;512:393–399. doi: 10.1038/nature12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussell JJ, Yapici N, Zhang SX, Dickson BJ, Vosshall LB. Abdominal-B neurons control Drosophila virgin female receptivity. Curr Biol. 2014;24:1584–95. doi: 10.1016/j.cub.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celniker SE, Sharma S, Keelan DJ, Lewis EB. The molecular genetics of the bithorax complex of Drosophila: cis-regulation in the Abdominal-B domain. EMBO J. 1990;9:4277–86. doi: 10.1002/j.1460-2075.1990.tb07876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerda-Esteban N, Spagnoli FM. Glimpse into Hox and tale regulation of cell differentiation and reprogramming. Dev Dyn. 2014;243:76–87. doi: 10.1002/dvdy.24075. [DOI] [PubMed] [Google Scholar]
- Chen YW, Song S, Weng R, Verma P, Kugler JM, Buescher M, Rouam S, Cohen SM. Systematic Study of Drosophila MicroRNA Functions Using a Collection of Targeted Knockout Mutations. Dev Cell. 2014;31:784–800. doi: 10.1016/j.devcel.2014.11.029. [DOI] [PubMed] [Google Scholar]
- Cole SH, Carney GE, McClung CA, Willard SS, Taylor BJ, Hirsh J. Two functional but noncomplementing Drosophila tyrosine decarboxylase genes: distinct roles for neural tyramine and octopamine in female fertility. J Biol Chem. 2005;280:14948–55. doi: 10.1074/jbc.M414197200. [DOI] [PubMed] [Google Scholar]
- Cumberledge S, Zaratzian A, Sakonju S. Characterization of two RNAs transcribed from the cis-regulatory region of the abd-A domain within the Drosophila bithorax complex. Proc Natl Acad Sci U S A. 1990;87:3259–63. doi: 10.1073/pnas.87.9.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–11. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboule D. The rise and fall of Hox gene clusters. Development. 2007;134:2549–60. doi: 10.1242/dev.001065. [DOI] [PubMed] [Google Scholar]
- Duboule D, Morata G. Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet. 1994;10:358–64. doi: 10.1016/0168-9525(94)90132-5. [DOI] [PubMed] [Google Scholar]
- Ecsedi M, Rausch M, Grosshans H. The let-7 microRNA Directs Vulval Development through a Single Target. Dev Cell. 2015;32:335–44. doi: 10.1016/j.devcel.2014.12.018. [DOI] [PubMed] [Google Scholar]
- Enderle D, Beisel C, Stadler MB, Gerstung M, Athri P, Paro R. Polycomb preferentially targets stalled promoters of coding and noncoding transcripts. Genome Res. 2011;21:216–26. doi: 10.1101/gr.114348.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng K, Palfreyman MT, Hasemeyer M, Talsma A, Dickson BJ. Ascending SAG neurons control sexual receptivity of Drosophila females. Neuron. 2014;83:135–48. doi: 10.1016/j.neuron.2014.05.017. [DOI] [PubMed] [Google Scholar]
- Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9:831–42. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garaulet DL, Castellanos MC, Bejarano F, Sanfilippo P, Tyler DM, Allan DW, Sanchez-Herrero E, Lai EC. Homeotic Function of Drosophila Bithorax-Complex miRNAs Mediates Fertility by Restricting Multiple Hox Genes and TALE Cofactors in the CNS. Dev Cell. 2014 doi: 10.1016/j.devcel.2014.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring WJ, Hiromi Y. Homeotic genes and the homeobox. Annu Rev Genet. 1986;20:147–73. doi: 10.1146/annurev.ge.20.120186.001051. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Reyes A, Morata G. The developmental effect of overexpressing a Ubx product in Drosophila embryos is dependent on its interactions with other homeotic products. Cell. 1990;61:515–22. doi: 10.1016/0092-8674(90)90533-k. [DOI] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, Brown JB, Cherbas L, Davis CA, Dobin A, Li R, Lin W, Malone JH, Mattiuzzo NR, Miller D, Sturgill D, Tuch BB, Zaleski C, Zhang D, Blanchette M, Dudoit S, Eads B, Green RE, Hammonds A, Jiang L, Kapranov P, Langton L, Perrimon N, Sandler JE, Wan KH, Willingham A, Zhang Y, Zou Y, Andrews J, Bickel PJ, Brenner SE, Brent MR, Cherbas P, Gingeras TR, Hoskins RA, Kaufman TC, Oliver B, Celniker SE. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun D, Wang YL, Langenberger D, Gunsalus KC, Rajewsky N. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comp Biol. 2005;1:0051–0063. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gummalla M, Galetti S, Maeda RK, Karch F. Hox gene regulation in the central nervous system of Drosophila. Front Cell Neurosci. 2014;8:96. doi: 10.3389/fncel.2014.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gummalla M, Maeda RK, Castro Alvarez JJ, Gyurkovics H, Singari S, Edwards KA, Karch F, Bender W. abd-A Regulation by the iab-8 Noncoding RNA. PLoS Genet. 2012;8:e1002720. doi: 10.1371/journal.pgen.1002720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafen E, Levine M, Gehring WJ. Regulation of Antennapedia transcript distribution by the bithorax complex in Drosophila. Nature. 1984;307:287–9. doi: 10.1038/307287a0. [DOI] [PubMed] [Google Scholar]
- Harding K, Wedeen C, McGinnis W, Levine M. Spatially regulated expression of homeotic genes in Drosophila. Science. 1985;229:1236–42. doi: 10.1126/science.3898362. [DOI] [PubMed] [Google Scholar]
- He X, Yan YL, Eberhart JK, Herpin A, Wagner TU, Schartl M, Postlethwait JH. miR-196 regulates axial patterning and pectoral appendage initiation. Dev Biol. 2011;357:463–77. doi: 10.1016/j.ydbio.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V, Perry MW, Hendrix D, Stark A, Levine M, Haley B. Neural-specific elongation of 3′ UTRs during Drosophila development. Proc Natl Acad Sci U S A. 2011;108:15864–9. doi: 10.1073/pnas.1112672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogga I, Karch F. Transcription through the iab-7 cis-regulatory domain of the bithorax complex interferes with maintenance of Polycomb-mediated silencing. Development. 2002;129:4915–22. doi: 10.1242/dev.129.21.4915. [DOI] [PubMed] [Google Scholar]
- Hogness DS, Lipshitz HD, Beachy PA, Peattie DA, Saint RB, Goldschmidt-Clermont M, Harte PJ, Gavis ER, Helfand SL. Regulation and products of the Ubx domain of the bithorax complex. Cold Spring Harb Symp Quant Biol. 1985;50:181–94. doi: 10.1101/sqb.1985.050.01.024. [DOI] [PubMed] [Google Scholar]
- Hornstein E, Mansfield JH, Yekta S, Hu JK, Harfe BD, McManus MT, Baskerville S, Bartel DP, Tabin CJ. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–4. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- Hui JH, Marco A, Hunt S, Melling J, Griffiths-Jones S, Ronshaugen M. Structure, evolution and function of the bi-directionally transcribed iab-4/iab-8 microRNA locus in arthropods. Nucleic Acids Res. 2013;41:3352–61. doi: 10.1093/nar/gks1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch F, Bender W, Weiffenbach B. abdA expression in Drosophila embryos. Genes Dev. 1990;4:1573–87. doi: 10.1101/gad.4.9.1573. [DOI] [PubMed] [Google Scholar]
- Karch F, Weiffenbach B, Peifer M, Bender W, Duncan I, Celniker S, Crosby M, Lewis EB. The abdominal region of the bithorax complex. Cell. 1985;43:81–96. doi: 10.1016/0092-8674(85)90014-5. [DOI] [PubMed] [Google Scholar]
- Kaufman TC, Seeger MA, Olsen G. Molecular and genetic organization of the antennapedia gene complex of Drosophila melanogaster. Adv Genet. 1990;27:309–62. doi: 10.1016/s0065-2660(08)60029-2. [DOI] [PubMed] [Google Scholar]
- Kurant E, Pai CY, Sharf R, Halachmi N, Sun YH, Salzberg A. Dorsotonals/homothorax, the Drosophila homologue of meis1, interacts with extradenticle in patterning of the embryonic PNS. Development. 1998;125:1037–48. doi: 10.1242/dev.125.6.1037. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lai EC. microRNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–4. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- Lai EC. Two decades of miRNA biology: lessons and challenges. RNA. 2015;21:675–7. doi: 10.1261/rna.051193.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lemons D, Pare A, McGinnis W. Three Drosophila Hox complex microRNAs do not have major effects on expression of evolutionarily conserved Hox gene targets during embryogenesis. PLoS ONE. 2012;7:e31365. doi: 10.1371/journal.pone.0031365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–70. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lipshitz HD, Peattie DA, Hogness DS. Novel transcripts from the Ultrabithorax domain of the bithorax complex. Genes Dev. 1987;1:307–22. doi: 10.1101/gad.1.3.307. [DOI] [PubMed] [Google Scholar]
- Macias A, Casanova J, Morata G. Expression and regulation of the abd-A gene of Drosophila. Development. 1990;110:1197–207. doi: 10.1242/dev.110.4.1197. [DOI] [PubMed] [Google Scholar]
- Mann RS, Lelli KM, Joshi R. Hox specificity unique roles for cofactors and collaborators. Curr Top Dev Biol. 2009;88:63–101. doi: 10.1016/S0070-2153(09)88003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield JH, Harfe BD, Nissen R, Obenauer J, Srineel J, Chaudhuri A, Farzan-Kashani R, Zuker M, Pasquinelli AE, Ruvkun G, Sharp PA, Tabin CJ, McManus MT. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet. 2004;36:1079–83. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- McGinnis W, Levine MS, Hafen E, Kuroiwa A, Gehring WJ. A conserved DNA sequence in homoeotic genes of the Drosophila Antennapedia and bithorax complexes. Nature. 1984;308:428–33. doi: 10.1038/308428a0. [DOI] [PubMed] [Google Scholar]
- McGlinn E, Yekta S, Mansfield JH, Soutschek J, Bartel DP, Tabin CJ. In ovo application of antagomiRs indicates a role for miR-196 in patterning the chick axial skeleton through Hox gene regulation. Proc Natl Acad Sci U S A. 2009;106:18610–5. doi: 10.1073/pnas.0910374106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miguel-Aliaga I, Thor S. Segment-specific prevention of pioneer neuron apoptosis by cell-autonomous, postmitotic Hox gene activity. Development. 2004;131:6093–105. doi: 10.1242/dev.01521. [DOI] [PubMed] [Google Scholar]
- Miguel-Aliaga I, Thor S, Gould AP. Postmitotic specification of Drosophila insulinergic neurons from pioneer neurons. PLoS Biol. 2008;6:e58. doi: 10.1371/journal.pbio.0060058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, Bartel DP, Ambros VR, Horvitz HR. Most Caenorhabditis elegans microRNAs Are Individually Not Essential for Development or Viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura P, Shenker S, Andreu-Agullo C, Westholm JO, Lai EC. Widespread and extensive lengthening of 3′ UTRs in the mammalian brain. Genome Res. 2013;23:812–25. doi: 10.1101/gr.146886.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens CB, Selleri L. Hox cofactors in vertebrate development. Dev Biol. 2006;291:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol. 2008;15:354–63. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat Rev Genet. 2005;6:893–904. doi: 10.1038/nrg1726. [DOI] [PubMed] [Google Scholar]
- Pease B, Borges AC, Bender W. Noncoding RNAs of the Ultrabithorax domain of the Drosophila bithorax complex. Genetics. 2013;195:1253–64. doi: 10.1534/genetics.113.155036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell. 2006;127:1209–21. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippidou P, Dasen JS. Hox genes: choreographers in neural development, architects of circuit organization. Neuron. 2013;80:12–34. doi: 10.1016/j.neuron.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokop A, Bray S, Harrison E, Technau GM. Homeotic regulation of segment-specific differences in neuroblast numbers and proliferation in the Drosophila central nervous system. Mech Dev. 1998;74:99–110. doi: 10.1016/s0925-4773(98)00068-9. [DOI] [PubMed] [Google Scholar]
- Rank G, Prestel M, Paro R. Transcription through intergenic chromosomal memory elements of the Drosophila bithorax complex correlates with an epigenetic switch. Mol Cell Biol. 2002;22:8026–34. doi: 10.1128/MCB.22.22.8026-8034.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regulski M, Harding K, Kostriken R, Karch F, Levine M, McGinnis W. Homeo box genes of the Antennapedia and bithorax complexes of Drosophila. Cell. 1985;43:71–80. doi: 10.1016/0092-8674(85)90013-3. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack F, Basson M, Pasquinelli A, Bettinger J, Rougvie A, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogulja-Ortmann A, Renner S, Technau GM. Antagonistic roles for Ultrabithorax and Antennapedia in regulating segment-specific apoptosis of differentiated motoneurons in the Drosophila embryonic central nervous system. Development. 2008;135:3435–45. doi: 10.1242/dev.023986. [DOI] [PubMed] [Google Scholar]
- Rogulja-Ortmann A, Technau GM. Multiple roles for Hox genes in segment-specific shaping of CNS lineages. Fly (Austin) 2008;2:316–9. doi: 10.4161/fly.7464. [DOI] [PubMed] [Google Scholar]
- Ronshaugen M, Biemar F, Piel J, Levine M, Lai EC. The Drosophila microRNA iab-4 causes a dominant homeotic transformation of halteres to wings. Genes Dev. 2005;19:2947–52. doi: 10.1101/gad.1372505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Herrero E. Control of the expression of the bithorax complex genes abdominal-A and abdominal-B by cis-regulatory regions in Drosophila embryos. Development. 1991;111:437–49. doi: 10.1242/dev.111.2.437. [DOI] [PubMed] [Google Scholar]
- Sanchez-Herrero E, Akam M. Spatially ordered transcription of regulatory DNA in the bithorax complex of Drosophila. Development. 1989;107:321–9. doi: 10.1242/dev.107.2.321. [DOI] [PubMed] [Google Scholar]
- Sánchez-Herrero E, Vernos I, Marco R, Morata G. Genetic organization of Drosophila bithorax complex. Nature. 1985;313:108–13. doi: 10.1038/313108a0. [DOI] [PubMed] [Google Scholar]
- Schmitt S, Prestel M, Paro R. Intergenic transcription through a polycomb group response element counteracts silencing. Genes Dev. 2005;19:697–708. doi: 10.1101/gad.326205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MP, Weiner AJ. Structural relationships among genes that control development: sequence homology between the Antennapedia, Ultrabithorax, and fushi tarazu loci of Drosophila. Proc Natl Acad Sci U S A. 1984;81:4115–9. doi: 10.1073/pnas.81.13.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Singh NP, Mishra RK. A double-edged sword to force posterior dominance of Hox genes. Bioessays. 2008;30:1058–61. doi: 10.1002/bies.20847. [DOI] [PubMed] [Google Scholar]
- Smibert P, Lai EC. A view from Drosophila: multiple biological functions for individual microRNAs. Semin Cell Dev Biol. 2010;21:745–53. doi: 10.1016/j.semcdb.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P, Miura P, Westholm JO, Shenker S, May G, Duff MO, Zhang D, Eads B, Carlson J, Brown JB, Eisman RC, Andrews J, Kaufman TC, Cherbas P, Celniker SE, Graveley BR, Lai EC. Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Reports. 2012;1:277–289. doi: 10.1016/j.celrep.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–46. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila MicroRNA Targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Bushati N, Jan CH, Kheradpour P, Hodges E, Brennecke J, Bartel DP, Cohen SM, Kellis M. A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes Dev. 2008;22:8–13. doi: 10.1101/gad.1613108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G, White RA. Regulation of the Ultrabithorax gene of Drosophila by other bithorax complex genes. Cell. 1985;43:507–19. doi: 10.1016/0092-8674(85)90180-1. [DOI] [PubMed] [Google Scholar]
- Sun K, Lai EC. Adult-specific functions of animal microRNAs. Nat Rev Genet. 2013;14:535–48. doi: 10.1038/nrg3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suska A, Miguel-Aliaga I, Thor S. Segment-specific generation of Drosophila Capability neuropeptide neurons by multi-faceted Hox cues. Dev Biol. 2011;353:72–80. doi: 10.1016/j.ydbio.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen S, Azzam G, Kaschula R, Williams LS, Alonso CR. Developmental RNA processing of 3′UTRs in Hox mRNAs as a context-dependent mechanism modulating visibility to microRNAs. Development. 2010;137:2951–60. doi: 10.1242/dev.047324. [DOI] [PubMed] [Google Scholar]
- Tiong S, Bone LM, Whittle JR. Recessive lethal mutations within the bithorax-complex in Drosophila. Mol Gen Genet. 1985;200:335–42. doi: 10.1007/BF00425445. [DOI] [PubMed] [Google Scholar]
- Tsuji T, Hasegawa E, Isshiki T. Neuroblast entry into quiescence is regulated intrinsically by the combined action of spatial Hox proteins and temporal identity factors. Development. 2008;135:3859–69. doi: 10.1242/dev.025189. [DOI] [PubMed] [Google Scholar]
- Tyler DM, Okamura K, Chung WJ, Hagen JW, Berezikov E, Hannon GJ, Lai EC. Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes Dev. 2008;22:26–36. doi: 10.1101/gad.1615208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Yang CH, Belawat P, Hafen E, Jan LY, Jan YN. Drosophila egg-laying site selection as a system to study simple decision-making processes. Science. 2008;319:1679–83. doi: 10.1126/science.1151842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Han D, Chen X, Zhang D, Wang L, Shi C, Zhang W, Li C, Chen X, Liu H, Zhang D, Kang J, Peng F, Liu Z, Qi J, Gao X, Ai J, Shi C, Zhao S. MiR-196a exerts its oncogenic effect in glioblastoma multiforme by inhibition of IkappaBalpha both in vitro and in vivo. Neuro Oncol. 2014;16:652–61. doi: 10.1093/neuonc/not307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–6. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- Yekta S, Tabin CJ, Bartel DP. MicroRNAs in the Hox network: an apparent link to posterior prevalence. Nat Rev Genet. 2008;9:789–96. doi: 10.1038/nrg2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Ashe H, Burks C, Levine M. Characterization of the transvection mediating region of the abdominal-B locus in Drosophila. Development. 1999;126:3057–65. doi: 10.1242/dev.126.14.3057. [DOI] [PubMed] [Google Scholar]


