Abstract
DNA demethylation plays a central role during development and in adult physiology. Different mechanisms of active DNA demethylation have been established. For example, Growth Arrest and DNA Damage 45-(GADD45) and Ten-Eleven-Translocation (TET) proteins act in active DNA demethylation but their functional relationship is unresolved. Here we show that GADD45a physically interacts – and functionally cooperates with TET1 in methylcytosine (mC) processing. In reporter demethylation GADD45a requires endogenous TET1 and conversely TET1 requires GADD45a. On GADD45a target genes TET1 hyperinduces 5-hydroxymethylcytosine (hmC) in the presence of GADD45a, while 5-formyl-(fC) and 5-carboxylcytosine (caC) are reduced. Likewise, in global analysis GADD45a positively regulates TET1 mediated mC oxidation and enhances fC/caC removal. Our data suggest a dual function of GADD45a in oxidative DNA demethylation, to promote directly or indirectly TET1 activity and to enhance subsequent fC/caC removal.
Keywords: Gadd45, TET, DNA demethylation, LC–MS/MS, hmC
1. Introduction
DNA methylation at the C5 position of cytosine (mC) is a well-characterized epigenetic mark in higher eukaryotes (reviewed in Bird, 2002, Deaton and Bird, 2011, Hackett and Surani, 2013, Jones and Takai, 2001). mC typically confers transcriptional silencing within gene regulatory regions and propagates this silenced state to daughter cells. DNA methylation can be very dynamic e.g. during epigenetic reprogramming, early embryonic development and cellular differentiation (reviewed in Messerschmidt et al., 2014, Niehrs, 2009, Williams et al., 2012, Wu and Zhang, 2010). During these phases active DNA demethylation, the enzymatic removal of mC, is crucial to shape the epigenetic signature in order to activate key developmental genes (reviewed in Guo et al., 2011a, Messerschmidt et al., 2014, Niehrs and Schäfer, 2012, Pastor et al., 2013, Schäfer, 2013, Wu and Zhang, 2010). In animals, three main mechanisms of active DNA demethylation have been proposed: DNA demethylation (i) by nucleotide-excision repair (NER; Barreto et al., 2007), (ii) by base-excision repair (BER) upon mC deamination by AID (Activation Induced Deaminase; Cortellino et al., 2011, Morgan et al., 2004), and (iii) by mC oxidation mediated by the Ten-Eleven Translocation (TET) family enzymes followed by BER (Maiti and Drohat, 2011, Shen et al., 2013, Tahiliani et al., 2009).
A regulatory protein family in NER- and BER-based DNA demethylation is GADD45 (Growth Arrest and DNA Damage Protein 45a,-b,-g). GADD45 proteins are devoid of any obvious enzymatic activity and act as adapters between demethylation target genes and the DNA repair machinery. For example, GADD45a binds to distinct genomic loci via the H3K4me3 reader ING1b (Schäfer et al., 2013), the RNA polymerase cofactor TAF12 (Schmitz et al., 2009), or the lncRNA TARID (Arab et al., 2014) to recruit DNA repair enzymes such as the 3′-NER endonuclease XPG (Barreto et al., 2007, Le May et al., 2010, Schmitz et al., 2009), the BER enzyme Thymine DNA Glycosylase TDG (Arab et al., 2014, Cortellino et al., 2011, Li et al., 2015), and AID (Cortellino et al., 2011, Rai et al., 2008).
An important question is whether GADD45 also interacts with TET-mediated, oxidative DNA demethylation. TET dioxygenases iteratively oxidize the methyl group at C5 to yield 5-hydroxymethyl-(hmC) (Kriaucionis and Heintz, 2009, Tahiliani et al., 2009), 5-formyl-(fC) (Maiti and Drohat, 2011) and 5-carboxylcytosine (caC) (He et al., 2011, Maiti and Drohat, 2011). caC can be decarboxylated by bacterial and mammalian C5-DNA methyltransferases in vitro (Liutkeviciute et al., 2014). In vivo however, only TDG mediated excision of fC and caC has been shown to accomplish DNA demethylation. The resulting abasic site is processed by BER to incorporate unmethylated C (Cortellino et al., 2011, He et al., 2011, Maiti and Drohat, 2011). Recently it has been shown that GADD45a enhances TDG mediated removal of fC and caC (Li et al., 2015). Thus, TDG is a common component of both, TET- and GADD45 mediated DNA demethylation. Together with the finding that GADD45a and TDG are required for TET mediated demethylation of TCF21 (Arab et al., 2014) this raises the question, if Gadd45a may directly interact with TET enzymes.
Here we show that GADD45a and TET1 directly bind each other. Moreover, GADD45a positively regulates TET1 induced mC oxidation and the two proteins require each other for reporter demethylation. Furthermore, GADD45a reduces fC and caC levels, both gene-specifically as well as globally. Our data corroborate a close link between the GADD45a- and TET1-mediated DNA demethylation pathways.
2. Results
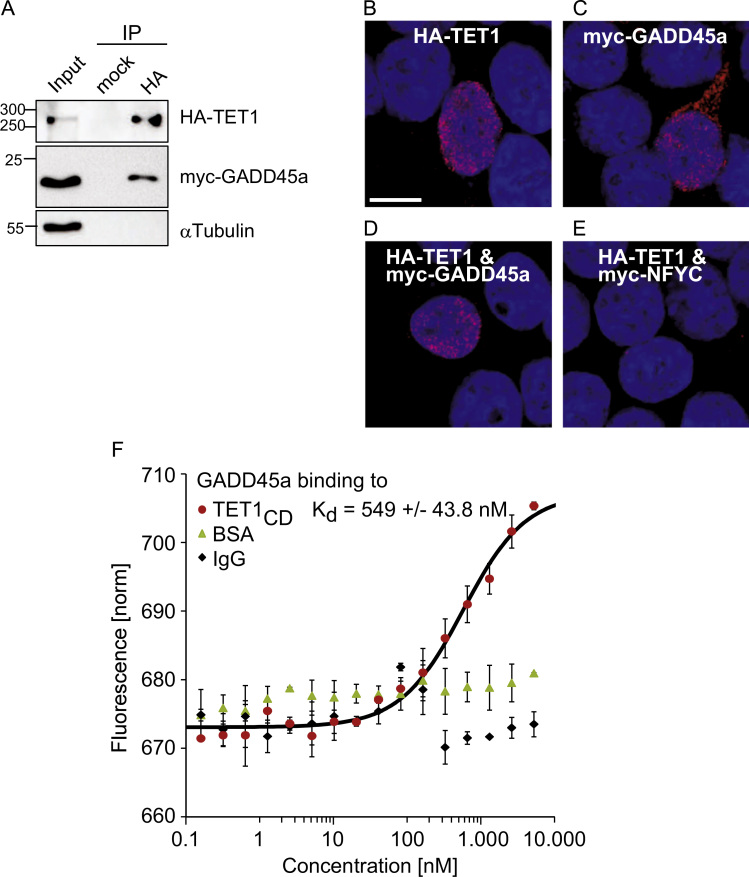
We first tested by three independent approaches if GADD45a- and TET-proteins physically interact. First, in co-immunoprecipitation (Co-IP) experiments using overexpressed tagged proteins, both full-length TET1 as well as TET-catalytic-domain-only (TET1CD) bound GADD45a (Fig. 1A; Supplement Fig. 1A). Second, we used in-situ Proximity Ligation Assay (PLA), in which protein–protein interactions are visualized as fluorescent speckles by rolling-circle amplification (Söderberg et al., 2006). PLA of HA-TET1 with itself (self-PLA) detected the protein expectedly in the nucleus (Fig. 1B; Tahiliani et al., 2009). Self-PLA of myc-GADD45a showed both cytoplasmic and nuclear staining, consistent with the reported bimodal GADD45a distribution (Fig. 1C; Fayolle et al., 2006). PLA between ectopic GADD45a and TET1 showed robust nuclear staining (Fig. 1D), while no signal was obtained for the control protein NFYC with TET1 (Fig. 1E). Third, to assess directness of this interaction, we performed Microscale Thermophoresis (MST) binding assays with recombinant proteins. GADD45a specifically bound TET1CD, but not control proteins (BSA, IgG; Fig. 1F). The apparent Kd was in the high nanomolar range (0.55 μM) and comparable to the Kd of GADD45a self-binding (0.57 μM; Supplement Fig. 1B). This apparently moderate affinity is likely an underestimate, since the active fraction of both recombinant proteins is probably less than 100%, notably for GADD45a after covalent modification with fluorescent dye. Taken together, we conclude that GADD45a and TET1 directly interact.
Fig. 1.
GADD45a directly binds TET1. (A) Western blot analysis of co-immunoprecipitation (Co-IP) experiments using protein lysates of HEK293T cells transfected with Flag-HA-TET1 and myc-GADD45a. α-Tubulin was used as specificity control. Input shows 5% of lysate used for IP. (B–E) In-situ analysis of TET1-GADD45a interaction by Proximity Ligation Assay (PLA) in HEK293T cells transiently transfected as indicated. Scale bar, 10 µm. (F) Microscale Thermophoresis binding assays of fluorescently labeled recombinant GADD45a to TET1CD or the negative controls BSA and IgG. Concentration of GADD45a was kept constant at 50 nM, while increasing concentrations of TET1CD, BSA and IgG were tested (0.1 nM to 5 µM). Each measurement point represents the mean of biological replicates (n=3) with error bars as±SD.
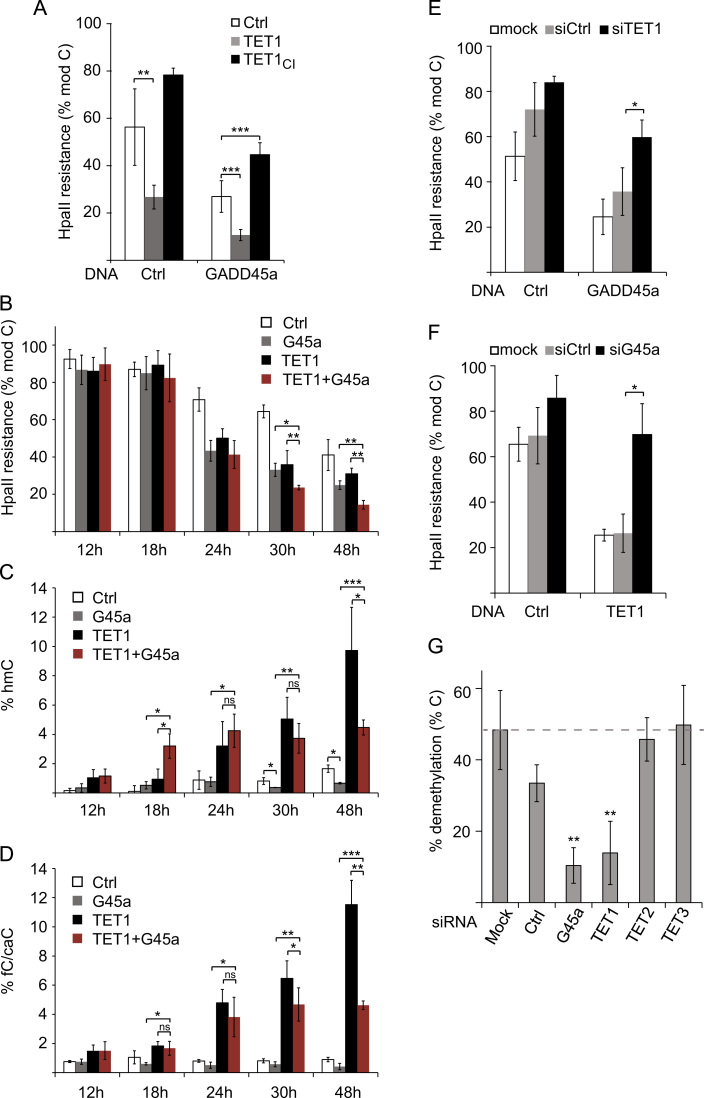
We next tested for functional cooperation of GADD45a and TET1 in reporter plasmid demethylation, an assay by which the demethylating activity of both proteins can be monitored (Barreto et al., 2007, Guo et al., 2011b, Schäfer et al., 2013, Schäfer et al., 2010). Expression of GADD45a and TET1 both reduced HpaII resistance of an in vitro methylated oct4TK-GFP reporter plasmid, i.e. they restored unmodified cytosine (Fig. 2A), as expected. Co-expression of GADD45a further enhanced TET1-mediated demethylation (Fig. 2A) without affecting TET1 protein levels (Supplement Fig. 2A). Notably, a catalytically inactive TET1 mutant (TET1CI) acted dominant negative and impaired GADD45a induced DNA demethylation (Fig. 2A), eventually competing with endogenous TET1 protein or TET1 interactors.
Fig. 2.
GADD45a promotes TET1-mediated mC oxidation and reporter DNA demethylation. (A–G) Methylation analysis of in vitro methylated oct4TK-GFP reporter by methylation sensitive PCR. (A) HEK293T cells were transfected with oct4TK-GFP along with empty vector (Ctrl), GADD45a (G45a) or TET1 expression constructs as indicated. Plasmid DNA was recovered 48 h after transfection and subjected to HpaII restriction digest and qPCR. HpaII resistance reflecting the fraction of modified C is displayed. Ctrl, control; TET1CI, TET1 catalytically inactive mutant. Bar graphs represent the mean of biological triplicates (n=3) with error bars±SD. (B–D) Kinetics of mC and its oxidized derivatives during oct4TK-GFP reporter demethylation analyzed by modification-sensitive qPCR. Cells were treated as in (A) and plasmid DNA was recovered at indicated time points after transfection. mC and its oxidized derivatives were analyzed using qPCR following HpaII or MspI restriction digest on T4 β-glucosyltransferase (β-GT) treated or control treated plasmid DNA. In (B), % modified cytosine (mC+hmC+fC+caC) is displayed as % HpaII resistance. (C) and (D) display % hmC and % fC/caC determined by MspI resistance following β-GT treatment and MspI resistance in control treated DNA (without β-GT), respectively. Error bars indicate±SD (n=3). (E, F) HEK293T cells were treated as in (A), but pre-transfected with the indicated siRNAs 24 h before DNA transfection. HpaII resistance reflecting the fraction of modified C is displayed. (G) HEK293T cells were transfected with oct4TK-GFP without any effector protein with the indicated siRNAs, whereby “mock” represents transfection reagent only. oct4TK-GFP plasmid was recovered 72 h after siRNA and 48 h after oct4TK-GFP transfection. HpaII cleavage is displayed as % demethylation, reflecting formation of unmodified C. Bar graphs represent the mean of biological triplicates (n=3) with error bars±SD. p-Values: (*) p<0.05; (**) p<0.01; (***) p<0.001. Complete list of p-values in Supplementary material section.
TET1 carries out iterative oxidation of mC and we aimed to dissect the role of GADD45a in this progression in a time course experiment following transfection. Over the course of 48 h, mC was progressively replaced by unmodified C upon GADD45a or TET1 expression (Fig. 2B). Combination of TET1 with GADD45a again led to enhanced DNA demethylation, and this became apparent at 30 h and 48 h after transfection. To analyze the underlying mode for this enhanced demethylation we quantified hmC and fC/caC at the demethylated CpG of the reporter using qPCR following MspI restriction on T4 β-glucosyltransferase (β-GT) treated plasmid DNA (Ito et al., 2011, Kinney et al., 2011). TET1 mediated demethylation was accompanied by a rise in hmC and fC/caC levels, as expected (Fig. 2C and D). Interestingly, GADD45a co-expression enhanced hmC at early- and reduced it at late time points (Fig. 2C). The late-phase hmC reduction by GADD45a cannot be explained by impairment of TET1 activity, given the enhanced net demethylation upon GADD45a-TET1 combination (Fig. 2B). The bimodal kinetics rather suggests that GADD45a promotes not only TET1 activity to produce hmC, but also enhances its processivity to proceed with fC/caC formation, as TET1 tends to stop oxidation at the level of hmC (Lister et al., 2013, Tahiliani et al., 2009, Wu et al., 2014). fC/caC levels did not accumulate but were reduced by GADD45a at all late time points (Fig. 2D). This is consistent with the observation that TDG excision of fC/caC is also enhanced by GADD45a (Li et al., 2015). Hence, the data point to a dual role of GADD45a. First, it promotes the iterative oxidation of mC by TET1 and second, GADD45a enhances the TDG removal of fC/caC, thereby promoting DNA demethylation.
To further analyze the interaction of GADD45a and TET1, we performed loss of function experiments. GADD45a mediated DNA demethylation was strongly impaired by siRNA mediated knockdown of TET1 (Fig. 2E). Conversely, TET1 triggered demethylation was fully dependent on endogenous GADD45a (Fig. 2F), while TET1 protein levels remained unaffected (Supplement Fig. 2B). These results indicate that GADD45a and TET1 not only cooperate in reporter demethylation, but also require each other.
The previous experiments relied on ectopic expression of GADD45a and TET1. Therefore, we next analysed the endogenous proteins. We observed previously that the oct4TK-GFP reporter is demethylated cell-intrinsically upon transfection in HEK293T cells (Barreto et al., 2007, Schäfer et al., 2013, Schäfer et al., 2010; see also Fig. 2A and B). This endogenous demethylation is impaired by knockdown of either GADD45a or TET1, whereas siTET2 and siTET3 showed no effect (Fig. 2G; see Supplement Fig. 2C for knockdown). Hence, this endogenous reporter DNA demethylation requires both, GADD45a and TET1, but not TET2 or TET3.
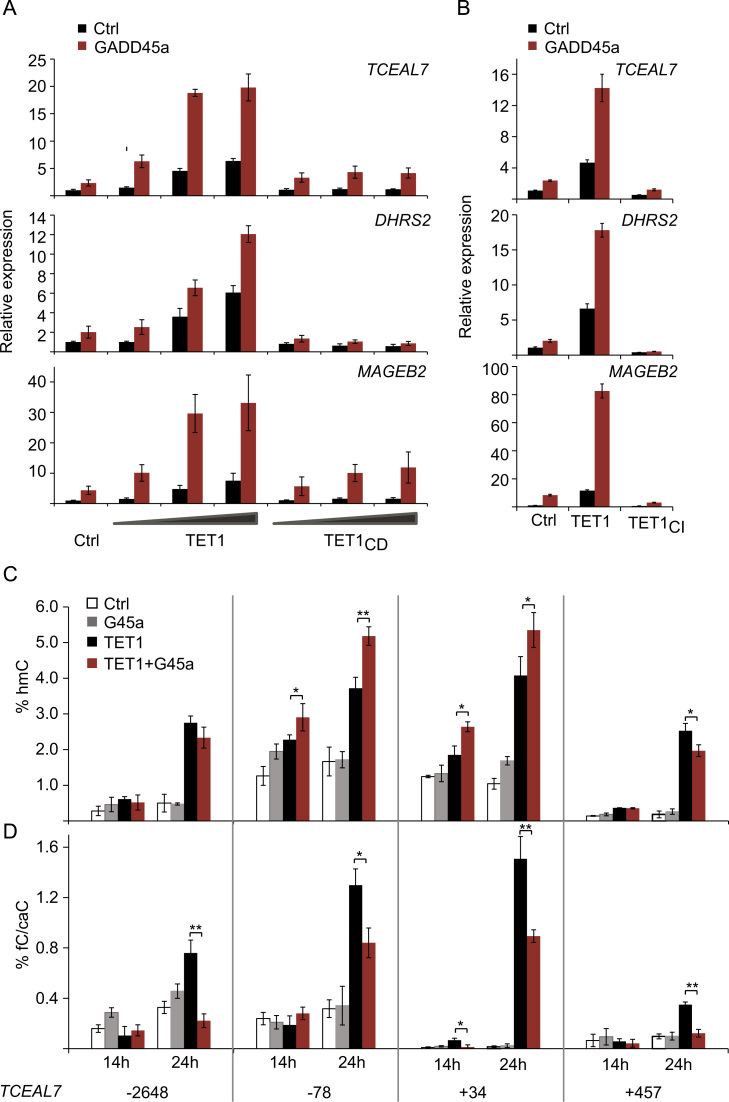
We extended our analysis from reporter genes to endogenous GADD45a target genes. Together with its cofactor, the histone reader ING1b, GADD45a activates a distinct set of methylation-silenced genes including TCEAL7, DHRS2 and MAGEB2 in HEK293T cells (Schäfer et al., 2013). These GADD45a targets were also induced by TET1 in a dose-dependent manner (Fig. 3A). GADD45a-TET1 co-expression led to synergistic gene activation (Fig. 3A), without affecting unrelated control gene expression (Supplement Fig. 3A) or TET1 protein levels (Supplement Fig. 3B). This synergy was specific for catalytically active, full-length TET1 as GADD45a neither cooperated with the TET1-catalytic domain-only (TET1CD) variant, nor the TET1CI mutant (Fig. 3A-B). The lack of TET1CD-GADD45a cooperation cannot be explained by the lack of protein interaction as GADD45a still binds TET1CD in MST-binding assays and Co-IP experiments (Fig. 1F, Supplement Fig. 1A). The results rather indicate a critical role of the TET1 N-terminus for transcriptional induction of GADD45a targets.
Fig. 3.
Synergistic target gene activation by GADD45a-TET1 is accompanied by increase in hmC and reduction of fC/caC in promoter CpGs. (A, B) TCEAL7, DHRS2, MAGEB2 expression in HEK293T cells upon transfection of empty vector (Ctrl, control), GADD45a (G45a) alone or with increasing doses of TET1, catalytic domain only (TET1CD), or catalytically inactive TET1 (TETCI) as indicated. Relative expression was monitored by qPCR. Bar graphs represent the mean of n=4 (A) or n=3 (B) experiments with error bars as±SD. (C, D) Kinetics of hmC (C) and fC/caC (D) level changes in the TCEAL7 locus upon GADD45a and TET expression. HEK293T cells were transfected with empty vector (Ctrl) or GADD45a (G45a) or TET1 as indicated. Genomic DNA was harvested 14 h or 24 h after transfection. hmC and fC/caC were analyzed at positions −2648, −78, +34 and +457 relative to the transcription start site (TSS). Analysis was by modification-sensitive qPCR following MspI restriction on T4 β-glucosyltransferase (β-GT) treated or control treated plasmid DNA. % of MspI resistance following β-GT treatment is displayed as % hmC. % of MspI resistance in control treated DNA (−β-GT) is displayed as % fC/caC. Bar graphs represent the mean of biological triplicates (n=3) with error bars as±SD. p-Values: (*) p<0.05; (**) p<0.01; (***) p<0.001. Complete list of p-values (A–D) in supplementary material section.
TET proteins can regulate transcription not only by active DNA demethylation but also by recruitment of the O-linked N-acetylglucosamine transferase (OGT) (Chen et al., 2013, Shi et al., 2013). However, treatment even with a high dose of the OGT inhibitor alloxan (Konrad et al., 2002) did not affect target gene activation by TET1 and GADD45a (Supplement Fig. 3C), making an involvement of O-GlcNAcylation in this process unlikely. In contrast, the dioxygenase activity of TET1 was required for synergistic gene activation (Fig. 3B). Therefore, we assayed the formation of hmC and fC/caC within four MspI sites in the TCEAL7 locus (Fig. 3C and D) where GADD45a is binding (Supplement Fig. 4A and Schäfer et al., 2013). After 24 h of TET1 expression, hmC formed at all CpGs analyzed (Fig. 3C). Interestingly, hmC at promoter proximal sites (−78 and +34 relative to the transcription start site) was further enhanced by GADD45a+TET1 and appeared precociously after 14 h. This result supports the model that GADD45a facilitates CpG oxidation by TET1. TET1 induced fC/caC predominantly at promoter proximal sites, which was once again reduced by GADD45a (Fig. 3D), paralleling the findings in reporter analysis (Fig. 2D). These results support that synergistic target gene activation by active TET1 and GADD45a is accompanied by enhanced hmC formation in proximal CpGs and overall reduced fC/caC levels in the TCEAL7 locus. However, while the relative changes of induced hmC and fC/caC at the CpG sites analyzed were significant, their absolute levels did not exceed 5% and 1.6%, respectively. This suggests that the strong gene activation stems from only a small subfraction of the cells responding. Consistent with this interpretation, synergistic TCEAL7 gene activation and hmC production by TET1 and GADD45a were not accompanied by significant TCEAL7 promoter demethylation, as assayed by HpaII resistance, even at 48 h after transfection (Supplement Fig. 3D and data not shown).
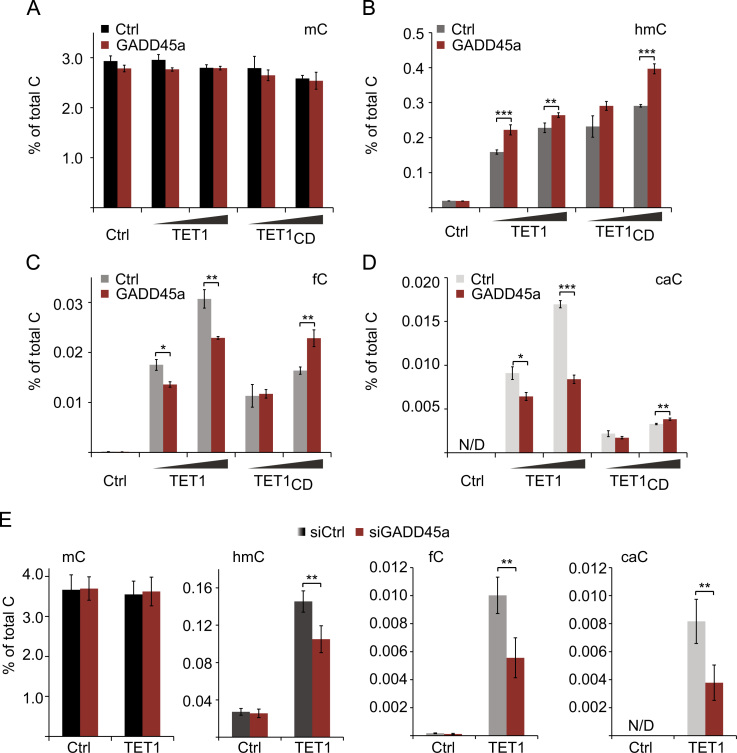
Since the results pointed to a dual role of GADD45a-promoting TET1 activity and removing fC/caC-we determined global levels of oxidized mC derivatives by quantitative SID-LC-MS/MS (Stable-Isotope Dilution Liquid Chromatography tandem Mass Spectrometry) (Liu et al., 2013, Pfaffeneder et al., 2014, Tsuji et al., 2014). Full-length TET1 or TET1CD strongly induced hmC, fC, and caC in HEK293T cells (Fig. 4A–D), as expected. Combination of GADD45a with TET1 led to a mild increase in hmC (Fig. 4B), whereas GADD45a alone had no effect (Supplement Fig. 4B). GADD45a co-expression with TET1 reduced fC and caC levels (Fig. 4C and D), consistent with the reporter- (Fig. 2C) and gene-specific analyses (Fig. 3D), as well as a recent report (Li et al., 2015). Notably, this fC/caC reduction was restricted to combined expression of GADD45a with TET1 full-length protein, but not TET1CD (Fig. 4C and D). These results again support the notion that the increase in hmC and the decrease of fC/caC are two distinct functions of GADD45a, the former requiring the TET1 C-terminal catalytic domain, the latter the TET1 N-terminus.
Fig. 4.
GADD45a enhances hmC formation by TET1 and reduces global fC and caC levels. (A–E) SID-LC-MS/MS analysis of mC and its oxidized derivatives in HEK293T cells transiently transfected with empty vector (Ctrl), GADD45a (G45a), and increasing doses of TET1 or its catalytic domain only (TET1CD) (A–D). For (E), 24 h prior to DNA transfection, cells were transfected with control or GADD45a specific siRNA and harvested 48 h after DNA transfection. Bar graphs represent the mean of biological replicates (for A–D n=3, for E n=4) with error bars as±SD. p-Values: (*) p<0.05; (**) p<0.01; (***) p<0.001. Complete list of p-values (A–E) in Supplementary material section. N/D, no data.
If GADD45a stimulates TET1 activity, its knockdown should impair TET1 mediated mC oxidation. Therefore, we combined TET1 expression with siGADD45a. Depletion of GADD45a reduced hmC formation by TET1 (Fig. 4E) without affecting TET1 protein level (Supplement Fig. 2B). The concomitant fC/caC reduction in this context may likely be a consequence of the hmC reduction. Taken together, our data support a model whereby GADD45a positively regulates TET1 activity and enhances fC/caC removal.
3. Discussion
A major unanswered question in DNA demethylation is the relationship of the different pathways proposed. Our results provide the first direct evidence that GADD45a and TET1 mediated DNA demethylation are connected and interdependent. The results support a dual role of GADD45a in TET1 mediated DNA demethylation and we propose that this reflects GADD45a binding and modulation of the two key enzymes in oxidative DNA demethylation, TET1 and TDG. First, GADD45a binds TET1 and positively regulates its activity, thereby enhancing mC oxidation. Elucidating the mechanism of TET1 activation by GADD45a is a highly interesting question to be addressed in future studies, and it may involve e.g. more effective TET1 recruitment to target CpGs, TET1 conformational change, or recruitment of cofactors conferred by GADD45a binding. Second, GADD45a reduces fC and caC levels and we propose that this occurs via TDG: fC/caC removal by the glycosylase TDG is well-established and GADD45a directly binds TDG (Arab et al., 2014, Cortellino et al., 2011, Li et al., 2015). Importantly, a recent study demonstrated that GADD45a promotes processing of fC/caC by TDG by an unknown mechanism (Li et al., 2015). This study also showed that knockout of both Gadd45a and Gadd45b from mouse ES cells leads to hypermethylation of specific genomic loci, many of which are also targets of TDG. Hence, GADD45a may function as a bridging protein between TET1 and TDG, thereby physically coupling mC oxidation with repair.
Although the data suggest that GADD45a boosts TET1 mediated DNA demethylation, global mC levels were not affected. This is expected since the bulk of mC arises from constitutively methylated repetitive elements, which mask gene-specific effects of GADD45a (reviewed in Niehrs and Schäfer, 2012, Schäfer, 2013). Importantly, the functional cooperation between TET1 and GADD45a may explain the previously observed co-requirement of Gadd45 and Tet1 for demethylation of a number of genes, including Bdnf IX, Fgf-1b in mouse brain (Guo et al., 2011b, Ma et al., 2009), TCF21 in cancer cells (Arab et al., 2014), and Oct4- and Nanog-reporters in embryonic stem (ES) cells (Sabag et al., 2014).
TET1 has been implicated in both, gene activation and gene repression. In ES cells, loss of Tet1 leads to gene up- and downregulation (Williams et al., 2012, Wu et al., 2011, Xu et al., 2011). Similarly, in HEK293T cells, TET1 represses and activates a number of genes, however mostly independent of TET1 catalytic activity (Jin et al., 2014). For gene activation with GADD45a the TET1 dioxygenase activity is required (Fig. 3A and B). This supports that GADD45a can engage TET1 for epigenetic gene activation, e.g. by targeting TET1 to specific loci, bridging TDG, or by recruiting cofactors. The fact that transcriptional activation as well as enhanced fC/caC removal are dependent on the TET1 N-terminus (Fig. 4B) suggests that fC/caC removal might be crucial for transcriptional activation. This is in line with the observation that a single fC or caC stalls RNA polymerase II in vitro (Kellinger et al., 2012).
The link between GADD45a and TET1 raises new questions. What is the mechanism by which GADD45a stimulates TET1 activity and fC/caC removal by TDG? Does GADD45a regulate the pattern of hmC, fC and caC deployment during development? Does combined knockout of Gadd45a and Tet1 affect development in mice? These questions and the greater physiological significance of the GADD45a-TET1 interaction can now be addressed using e.g. double-mutant mice.
4. Material and methods
4.1. Tissue culture, transfection, siRNA treatment
HEK293T cells were grown at 37 °C in 10% CO2 in Dulbecco's Modified Eagle's Medium (DMEM), 10% fetal calf serum, 2 mM l-Glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin. Plasmid DNA was transfected using X-tremeGENE 9 at a DNA:X-tremeGENE ratio of 1:3. 40 nM stealth siGADD45a (Invitrogen) or Dharmacon Smart pools were transfected using Lipofectamine 2000 (Invitrogen) or Dharmafect (Dharmacon) 24 h prior to DNA transfection. For OGT inhibition, 10 mM alloxan was added after transfection and refreshed after 24 h, prior to harvesting after 48 h.
4.2. Expression and reporter constructs and antibodies
Flag-HA-TET1 was from Addgene (ID 49792; FH-TET1-pEF). The catalytically inactive human TET1H1672Y/D1674A mutation has been described (Guo et al., 2011b). The point mutation was introduced in the HxD motif using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies) using the following primers: c5519t_a5526c antisense, 5′-ttattcatgttgtgaatggccctgtagggatgagcacagaagtc-3′ c5519t_a5526c sense; 5′-gacttctgtgctcatccctacagggccattcacaacatgaataa-3′. Flag-TET1 catalytic domain (Flag-TET1CD, aa1416-2136) was PCR amplified from Flag-HA-TET1 and subcloned into pCS2-2xFlag expression construct. Oct4TK-GFP reporter, human pHA-GADD45a and mouse myc-Tet2 expression plasmids have been described (Barreto et al., 2007, Ko et al., 2010), respectively. Human GADD45a and NFYC coding sequences were PCR amplified and cloned into pCS2+-myc to express N-terminally myc-tagged fusion proteins. Mouse anti-alpha-Tubulin, anti-flag M2, and anti-myc 9E10 were from Sigma Aldrich, rabbit anti-GADD45a from Santa-Cruz, rabbit anti-HA and IgG control from Abcam.
4.3. DNA methylation analysis and T4 β-glucosyltransferase (β-GT) treatment
Methylation-sensitive PCR (MS-PCR) was performed as described (Schäfer et al., 2010). Briefly, HEK293T cells were transfected with in vitro methylated oct4TK-GFP reporter and/or effector expression plasmids. Plasmid DNA or genomic DNA (gnDNA) was recovered 48 h after transfection or at indicated time points using DNeasy Blood & Tissue kit (Qiagen). Plasmid DNA was split into two parts and digested with either methylation-sensitive HpaII restriction enzyme or PvuII (unrelated control enzyme to fragment the genomic DNA) for 3 h. Via qPCR and the Roche Absolute quantification software tool we determined the arbitrary concentrations, taking into account the PCR-efficiency of each individual primer obtained by standard-curve measurements. HpaII resistance in % was determined from the ratio of HpaII digested versus control digested DNA and represents the fraction of modified cytosine residues (mC+hmC+fC+caC). T4 β-glucosyltransferase (β-GT) treatment was performed according to manufacturer's instructions (NEB EpiMark Kit). Briefly, reporter plasmid or gnDNA was split into two parts and incubated with or without β-GT for 16 h at 37 °C. Both fractions were again split and digested with PvuII (control) or MspI and analyzed by qPCR. Primer sequences are listed in Supplementary information. MspI restriction is blocked by fC and caC (Ito et al., 2011). Hence, the ratio of MspI digested vs PvuII digested DNA reflects the combined fraction of fC/caC. β-GT treatment specifically glucosylates hmC, resulting in MspI resistance (Kinney et al., 2011). The ratio of MspI digested vs PvuII digested DNA (both +β-GT) thus reflects the combined fraction of hmC/fC/caC. Substraction of fC/caC values (−β-GT) from MspI resistant fraction (+β-GT) results in % hmC.
4.4. Co-immunoprecipitation (Co-IP)
48 h after transient TET1 and GADD45a transfection, HEK293T cells were lysed (150 mM NaCl, 20 mM Tris pH 7.5, 2 mM EDTA, 10% glycerol, and cOmplete Mini Protease Inhibitor Cocktail (Roche)) and needle-syringe-homogenized. Following pre-clearing of the lysate by centrifugation and incubation with Agarose G beads (Roche) for 1 h at 4 °C, lysates were incubated on a rotating wheel over night at 4 °C with specific antibody or IgG as control. 2 h after addition of the beads and incubation at 4 °C on a rotating wheel, beads were washed 3 times in lysis buffer and protein was eluted with 4× Laemmli buffer containing ß-Mercaptoethanol. For Western blot analysis, 5% of input was compared to 1/3 of the Co-IP eluate.
4.5. Proximity Ligation Assay (PLA)
For visualization of protein–protein interaction in situ, Proximity Ligation Assay (PLA) was performed according to manufacturer's instructions (Duolink using PLA Technology, Sigma). Briefly, 48 h after transient HA-TET1 and/or myc-GADD45a and myc-NFYC (control protein) transfection, HEK293T cells were fixed in 100% MeOH (Sigma) for 10 min at −20 °C, permeabilized with 0.5% TritonX100 (Sigma) for 10 min, blocked and incubated with primary antibodies anti-HA (Abcam) and anti-myc (Sigma) (both diluted 1:1000) overnight at 4 °C. Then, cells were incubated with respective secondary antibodies (Duolink In Situ PLA Probes Anti-Mouse and Anti-Rabbit both PLUS and MINUS, SIGMA) followed by a ligation and subsequent amplification step to visualize protein interaction by red fluorescence signals (Duolink In Situ Detection Reagents Red, Sigma). To visualize individual proteins by Self-PLA, the respective primary antibody was combined with two secondary antibodies (PLA probes both PLUS and MINUS). Nuclei were counterstained by DAPI-containing mounting medium (Duolink In Situ Mounting Medium with DAPI, Sigma) and analysis was performed using a TCS SP5 confocal microscope (Leica) and 63× oil immersion objective lens. The speckles are likely limited to a fraction of cells due to incomplete transfection efficiency.
4.6. Microscale Thermophoresis
For binding studies, recombinant GADD45a (purified from E. coli), recombinant mouse Tet1 (catalytic domain, Active Motif), BSA (Sigma) or IgG (Abcam) were used. GADD45a was labeled with the MonolithTM NT.115 Protein Labeling Kit RED-NHS according to manufacturer's recommendations. Concentration of labeled GADD45a was kept constant at 50 nM and mixed with increasing concentrations of Tet1, BSA and IgG (0.1 nM to 5 µM) in a sixteen step 1:1 dilution series in MST buffer (50 mM Tris–HCl pH 7.6, 150 mM NaCl, 10 mM MgCl2 and 0.05% Tween-20). After 15 min of incubation at room temperature, samples were transferred into hydrophilic capillaries and measurements were conducted using a NanoTemper Monolith NT.115 instrument with 60% MST power and 60% LED power. Laser-On time was 30 s, Laser-Off time 5 s. The sigmoidal binding curve fitting was performed according to the law of mass action using NTAnalysis software provided by NanoTemper technologies. No fitting was obtained for GADD45a binding to the negative controls BSA and IgG.
4.7. RT-qPCR
Total RNA was isolated using Qiagen RNeasy mini kit with on-column DNase digest (Qiagen). First strand cDNA was generated using SuperScriptII reverse transcriptase (Invitrogen). Real-Time PCR was performed in technical duplicates using Roche LightCycler480 probes master and primers (see Supplementary information for primer sequences) in combination with predesigned mono-color hydrolysis probes of the Roche Universal probe library (UPL) or SYBR green. For quantification, the Roche LC480 quantification software module was used. All values were normalized to the level of the housekeeping gene Gapdh. Mean values shown were calculated using the mean value of the qPCR replicates from independent experiments (n≥3).
4.8. Protein expression and purification
E. coli BL21-CodonPlus (DE3)-RIL (Stratagene) was transformed with expression constructs encoding C-terminally hexahistidin tagged human GADD45a (pET24b-hGADD45a) and N-terminally hexahistidin-SUMO-tagged Naegleria gruberi Tet1 (pXC1010) (Hashimoto et al., 2014). Cultures were grown at 37 °C to an OD600 of 0.6 and induced with 1 mM isopropyl β-d-thiogalactopyranoside (IPTG, Thermo Scientific). Protein expression was performed at 16 °C overnight. Pelleted cells were resuspended in lysis buffer (25 mM HEPES/KOH pH 7.6, 0.5 M NaCl, 40 mM Imidazol) and lysed by passage through a Constant Systems LTD cell disrupter (1.8 kbar, constant run). Cell debris were pelleted by centrifugation at 38,400g for 30 min and supernatant subjected to a Ni2+-charged chelating sepharose Fast Flow column (GE Healthcare). Bound proteins were eluted by a stepwise gradient of imidazol (60–500 mM) in lysis buffer. Fractions containing pure proteins were concentrated by Vivaspin ultrafiltration devices (GE Healthare). Concentrations of proteins were determined by Bradford assay (Bio-Rad) using BSA as standard.
4.9. Stable-isotope dilution based Liquid Chromatography tandem Mass Spectrometry (LC–MS/MS)
The genomic DNA (gnDNA) of HEK293T cells was isolated using DNeasy blood & tissue kit (Qiagen), ethanol precipitated and dissolved in ddH2O. 1 µg of gnDNA was degraded with nuclease P1 (Roche Diagnostics, Mannheim, Germany), snake venom phosphodiesterase (Worthington, Lakewood, USA) and alkaline phosphatase (Fermentas, St. Leon-Roth, Germany). All oligonucleotides as well as dC and 5mdC nucleosides were from Sigma Aldrich (Germany). Modified cytidine nucleosides, hmdC, fdC, cadC were purchased from Berry & Associates, Inc. (Dexter, MI). 15N3-dCTP, 15N3-dC, 15N5-dG were from Silantes, GmbH (Munich, Germany). 2H3-5mdC was from TRC, Inc (Toronto, Canada). 15N3-5hmdC, 15N3-5fdC, 15N3-5cadC, were self-synthesized by a series of in vitro reactions (see Supplementary information). All solutions were prepared using Millipore quality water (Barnstead GenPure xCAD Plus, Thermo Scientific). Quantitative LC–MS/MS analysis of the nucleosides was performed using an Agilent 1290 UHPLC system equipped with ReproSil 100 C18 column (3 µm, 4.6×150 mm2, Jasco GmbH, Groß-Umstadt, Germany) and an Agilent 6490 triple quadrupole mass spectrometer, coupled with the stable isotope dilution technique, which allows for accurate quantification (Kellner et al., 2015, Liu et al., 2013, Pfaffeneder et al., 2014, Tsuji et al., 2014). The details of the procedure are described in Supplementary information.
4.10. Statistical analyses
Data are expressed as mean values±standard deviation (SD) of biological replicates (n≥3). Two-tailed, unpaired Student's t-test was used to calculate the level of significance. A p-value<0.05 was considered significant. *, p<0.05; **, p<0.01; ***, p<0.001. Full listing of p-values of the individual experiments is provided in Supplementary information.
Competing interests
The authors declare that they have no competing interests.
Acknowledgments
We thank X. Cheng and A. Rao for reagents, S. Melcea for generation of the TET1H1672Y/D1674A mutant, and Stefanie Kellner for assistance with LC/MS–MS analyses. Support by the IMB Core Facility Microscopy is gratefully acknowledged. This work was supported by an ERC senior investigator grant to C.N. (“DNA Demethylase"). M.U.M was supported by Natural Sciences and Engineering Research Council of Canada Postdoctoral Fellowship (NSERC-PDF 403829-2011).
Footnotes
Supplementary data associated with this article can be found in the online version at 10.1016/j.diff.2015.10.003.
Appendix A. Supplementary material
Supplementary material
References
- Arab K., Park Y.J., Lindroth A.M., Schäfer A., Oakes C., Weichenhan D., Lukanova A., Lundin E., Risch A., Meister M., Dienemann H., Dyckhoff G., Herold-Mende C., Grummt I., Niehrs C., Plass C. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol. Cell. 2014;55:604–614. doi: 10.1016/j.molcel.2014.06.031. [DOI] [PubMed] [Google Scholar]
- Barreto G., Schäfer A., Marhold J., Stach D., Swaminathan S.K., Handa V., Doderlein G., Maltry N., Wu W., Lyko F., Niehrs C. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Chen Q., Chen Y., Bian C., Fujiki R., Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013;493:561–564. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortellino S., Xu J., Sannai M., Moore R., Caretti E., Cigliano A., Le Coz M., Devarajan K., Wessels A., Soprano D., Abramowitz L.K., Bartolomei M.S., Rambow F., Bassi M.R., Bruno T., Fanciulli M., Renner C., Klein-Szanto A.J., Matsumoto Y., Kobi D., Davidson I., Alberti C., Larue L., Bellacosa A. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton A.M., Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayolle C., Pourchet J., Cohen A., Pedeux R., Puisieux A., Caron de Fromentel C., Dore J.F., Voeltzel T. UVB-induced G2 arrest of human melanocytes involves Cdc2 sequestration by Gadd45a in nuclear speckles. Cell Cycle. 2006;5:1859–1864. doi: 10.4161/cc.5.16.3119. [DOI] [PubMed] [Google Scholar]
- Guo J.U., Su Y., Zhong C., Ming G.L., Song H. Emerging roles of TET proteins and 5-hydroxymethylcytosines in active DNA demethylation and beyond. Cell Cycle. 2011;10:2662–2668. doi: 10.4161/cc.10.16.17093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.U., Su Y., Zhong C., Ming G.L., Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett J.A., Surani M.A. DNA methylation dynamics during the mammalian life cycle. Philos. Trans. R. Soc. Lond. Ser. B: Biol. Sci. 2013;368:20110328. doi: 10.1098/rstb.2011.0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H., Pais J.E., Zhang X., Saleh L., Fu Z.Q., Dai N., Correa I.R., Jr., Zheng Y., Cheng X. Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA. Nature. 2014;506:391–395. doi: 10.1038/nature12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., Sun Y., Li X., Dai Q., Song C.X., Zhang K., He C., Xu G.L. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C., Lu Y., Jelinek J., Liang S., Estecio M.R., Barton M.C., Issa J.P. TET1 is a maintenance DNA demethylase that prevents methylation spreading in differentiated cells. Nucleic Acids Res. 2014;42:6956–6971. doi: 10.1093/nar/gku372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P.A., Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- Kellinger M.W., Song C.X., Chong J., Lu X.Y., He C., Wang D. 5-formylcytosine and 5-carboxylcytosine reduce the rate and substrate specificity of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 2012;19:831–833. doi: 10.1038/nsmb.2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner S., Ochel A., Thuring K., Spenkuch F., Neumann J., Sharma S., Entian K.D., Schneider D., Helm M. Absolute and relative quantification of RNA modifications via biosynthetic isotopomers. Nucleic Acids Res. 2015;42:e142. doi: 10.1093/nar/gku733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney S.M., Chin H.G., Vaisvila R., Bitinaite J., Zheng Y., Esteve P.O., Feng S., Stroud H., Jacobsen S.E., Pradhan S. Tissue-specific distribution and dynamic changes of 5-hydroxymethylcytosine in mammalian genomes. J. Biol. Chem. 2011;286:24685–24693. doi: 10.1074/jbc.M110.217083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M., Huang Y., Jankowska A.M., Pape U.J., Tahiliani M., Bandukwala H.S., An J., Lamperti E.D., Koh K.P., Ganetzky R., Liu X.S., Aravind L., Agarwal S., Maciejewski J.P., Rao A. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad R.J., Zhang F., Hale J.E., Knierman M.D., Becker G.W., Kudlow J.E. Alloxan is an inhibitor of the enzyme O-linked N-acetylglucosamine transferase. Biochem. Biophys. Res. Commun. 2002;293:207–212. doi: 10.1016/S0006-291X(02)00200-0. [DOI] [PubMed] [Google Scholar]
- Kriaucionis S., Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le May N., Mota-Fernandes D., Velez-Cruz R., Iltis I., Biard D., Egly J.M. NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Mol. Cell. 2010;38:54–66. doi: 10.1016/j.molcel.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Li Z., Gu T.P., Weber A.R., Shen J.Z., Li B.Z., Xie Z.G., Yin R., Guo F., Liu X., Tang F., Wang H., Schär P., Xu G.L. Gadd45a promotes DNA demethylation through TDG. Nucleic Acids Res. 2015;43:3986–3997. doi: 10.1093/nar/gkv283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R., Mukamel E.A., Nery J.R., Urich M., Puddifoot C.A., Johnson N.D., Lucero J., Huang Y., Dwork A.J., Schultz M.D., Yu M., Tonti-Filippini J., Heyn H., Hu S., Wu J.C., Rao A., Esteller M., He C., Haghighi F.G., Sejnowski T.J., Behrens M.M., Ecker J.R. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Wang J., Su Y., Guerrero C., Zeng Y., Mitra D., Brooks P.J., Fisher D.E., Song H., Wang Y. Quantitative assessment of Tet-induced oxidation products of 5-methylcytosine in cellular and tissue DNA. Nucleic Acids Res. 2013;41:6421–6429. doi: 10.1093/nar/gkt360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liutkeviciute Z., Kriukiene E., Licyte J., Rudyte M., Urbanaviciute G., Klimasauskas S. Direct decarboxylation of 5-carboxylcytosine by DNA C5-methyltransferases. J. Am. Chem. Soc. 2014;136:5884–5887. doi: 10.1021/ja5019223. [DOI] [PubMed] [Google Scholar]
- Ma D.K., Jang M.H., Guo J.U., Kitabatake Y., Chang M.L., Pow-Anpongkul N., Flavell R.A., Lu B., Ming G.L., Song H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti A., Drohat A.C. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt D.M., Knowles B.B., Solter D. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 2014;28:812–828. doi: 10.1101/gad.234294.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan H.D., Dean W., Coker H.A., Reik W., Petersen-Mahrt S.K. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J. Biol. Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- Niehrs C. Active DNA demethylation and DNA repair. Differentiation. 2009;77:1–11. doi: 10.1016/j.diff.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Niehrs C., Schäfer A. Active DNA demethylation by Gadd45 and DNA repair. Trends Cell Biol. 2012;22:220–227. doi: 10.1016/j.tcb.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Pastor W.A., Aravind L., Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffeneder T., Spada F., Wagner M., Brandmayr C., Laube S.K., Eisen D., Truss M., Steinbacher J., Hackner B., Kotljarova O., Schuermann D., Michalakis S., Kosmatchev O., Schiesser S., Steigenberger B., Raddaoui N., Kashiwazaki G., Muller U., Spruijt C.G., Vermeulen M., Leonhardt H., Schär P., Muller M., Carell T. Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat. Chem. Biol. 2014;10:574–581. doi: 10.1038/nchembio.1532. [DOI] [PubMed] [Google Scholar]
- Rai K., Huggins I.J., James S.R., Karpf A.R., Jones D.A., Cairns B.R. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabag O., Zamir A., Keshet I., Hecht M., Ludwig G., Tabib A., Moss J., Cedar H. Establishment of methylation patterns in ES cells. Nat. Struct. Mol. Biol. 2014;21:110–112. doi: 10.1038/nsmb.2734. [DOI] [PubMed] [Google Scholar]
- Schäfer A. Gadd45 proteins: key players of repair-mediated DNA demethylation. Adv. Exp. Med. Biol. 2013;793:35–50. doi: 10.1007/978-1-4614-8289-5_3. [DOI] [PubMed] [Google Scholar]
- Schäfer A., Karaulanov E., Stapf U., Döderlein G., Niehrs C. Ing1 functions in DNA demethylation by directing Gadd45a to H3K4me3. Genes Dev. 2013;27:261–273. doi: 10.1101/gad.186916.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer A., Schomacher L., Barreto G., Döderlein G., Niehrs C. Gemcitabine functions epigenetically by inhibiting repair mediated DNA demethylation. PLoS One. 2010;5:e14060. doi: 10.1371/journal.pone.0014060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz K.M., Schmitt N., Hoffmann-Rohrer U., Schäfer A., Grummt I., Mayer C. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Mol. Cell. 2009;33:344–353. doi: 10.1016/j.molcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Shen L., Wu H., Diep D., Yamaguchi S., D'Alessio A.C., Fung H.L., Zhang K., Zhang Y. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153:692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F.T., Kim H., Lu W., He Q., Liu D., Goodell M.A., Wan M., Songyang Z. Ten-eleven translocation 1 (Tet1) is regulated by O-linked N-acetylglucosamine transferase (Ogt) for target gene repression in mouse embryonic stem cells. J. Biol. Chem. 2013;288:20776–20784. doi: 10.1074/jbc.M113.460386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderberg O., Gullberg M., Jarvius M., Ridderstrale K., Leuchowius K.J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.G., Landegren U. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji M., Matsunaga H., Jinno D., Tsukamoto H., Suzuki N., Tomioka Y. A validated quantitative liquid chromatography-tandem quadrupole mass spectrometry method for monitoring isotopologues to evaluate global modified cytosine ratios in genomic DNA. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2014;953–954:38–47. doi: 10.1016/j.jchromb.2014.01.050. [DOI] [PubMed] [Google Scholar]
- Williams K., Christensen J., Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2012;13:28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., D'Alessio A.C., Ito S., Xia K., Wang Z., Cui K., Zhao K., Sun Y.E., Zhang Y. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Wu X., Shen L., Zhang Y. Single-base resolution analysis of active DNA demethylation using methylase-assisted bisulfite sequencing. Nat. Biotechnol. 2014;32:1231–1240. doi: 10.1038/nbt.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.C., Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Wu F., Tan L., Kong L., Xiong L., Deng J., Barbera A.J., Zheng L., Zhang H., Huang S., Min J., Nicholson T., Chen T., Xu G., Shi Y., Zhang K., Shi Y.G. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol. Cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material