Abstract
MicroRNAs are differentially expressed in breast cancer cells and have been implicated in cancer formation, tumour invasion and metastasis. We investigated the miRNA expression profiles in the developing mammary gland. MiR-137 was expressed prominently in the developing mammary gland. When the miR-137 was over-expressed in the embryo, the mammary epithelium became thickened. Moreover, genes associated with mammary gland formation such as Tbx3 and Lef1 were not expressed. This suggests that miR-137 induces gland formation and invasion. When miR-137 was over-expressed in MDA-MB-231 cells, their ability to form tumours in adult mice was significantly reduced. These data support miR-137 decides epithelial cell behavior in the human breast cancer. It also suggests that miR-137 is a potential therapeutic target for amelioration of breast cancer progression.
Keywords: mammary gland development, breast cancer, miR-137, tumour suppressor, tachykinin-1
INTRODUCTION
Mammary gland development in mouse embryos is used as a model for breast cancer and can be divided into a series of stages [1-3]. In the initiation stage (in E10.5 mouse embryos), bilateral mammary lines are formed and epidermal cells of the mammary line become columnar and multilayered, defining the mammary ridge. At E11.5, five pairs of mammary placodes form at specific locations along the mammary line and these placodes then begin to invade the underlying mesenchyme (E12.5) to form a spherical epithelial bud enveloped by dense mammary mesenchyme and connected to the epidermis by a short stalk (E13.5) [4]. Subsequently, the epithelial bud grows and undergoes branching morphogenesis in female mice. Many genes expressed in the developing mouse mammary gland are also expressed in adult human breast cancer, including genes encoding molecules involved in signaling e.g. the estrogen receptor is expressed in the mesenchyme surrounding the invasive mammary epithelial bud [5] and various transcription factors [6-10]. For example, Tac1, the gene encoding Tachykinin-1, the precursor protein for neuroendocrine peptides, is expressed in the mammary mesenchyme during embryonic development [11-13] and the level of Tac1 expression in breast cancers is directly proportional to their aggressiveness [14, 15]. Another example is the GATA family zinc-finger transcription factor, GATA3, a crucial regulator of luminal differentiation during mammary gland development [16, 17] which is expressed within invasive breast carcinomas [18].
Alterations in several genes, including those encoding microRNAs (miRNAs), accumulate in cancer by a complex and multistep process [19]. MiRNAs are 19- to 25-nucleotide single-stranded RNA molecules that function as post-transcriptional regulators of target genes expression [20, 21]. Recent studies have reported that miRNAs may act as either tumour suppressors or oncogenes [22,23] and there is interest in identifying miRNAs associated with breast cancer as they could be novel therapeutic targets. Here we investigated which miRNAs are expressed in the early development of mouse mammary glands by comparing the 3rd mammary gland and the inter mammary gland region from E13.5 mouse embryos. We found that miRNA-137 (miR-137) is highly expressed in the 3rd mammary gland at E13.5 and over-expressing it in the flank region of developing mouse embryos perturbed invasion of the epithelial mammary bud.
Down-regulated miR-137 has been observed in various cancers such as colorectal cancer, gastric cancer, oral cancer, and squamous cell carcinoma of the head and neck [24-27]. A recent study also reported that miR-137 impairs proliferation and migration of cells of a breast cancer cell line by targeting expression of the nuclear receptor estrogen-related receptor alpha (ERRα) [28], suggesting that miR-137 may suppress the formation of breast cancer. To confirm this possibility in vivo, we over-expressed miR-137 in cells of a breast cancer cell line, then inoculated them into nude mice and found that their ability to form tumours was reduced.
The outcomes of this study reveal that miR-137 not only perturbs embryonic mammary gland development but also inhibits tumour formation by human breast cancer cells. These results bolster the idea that the embryonic mouse mammary gland is a good model for human breast cancer.
RESULTS
Microarray profiling of embryonic mouse mammary glands
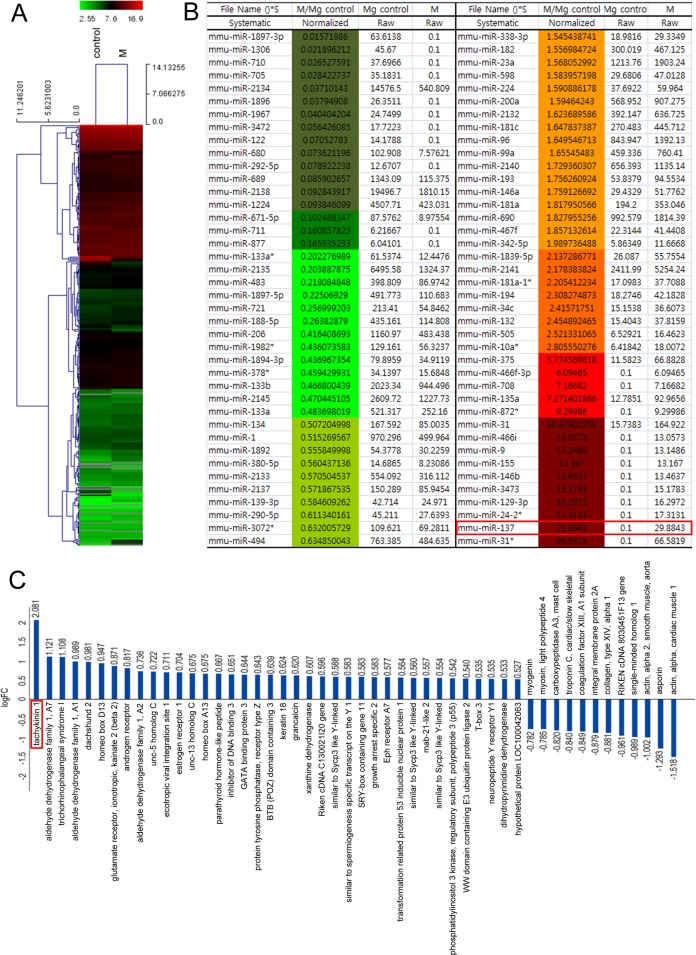
To identify miRNAs expressed in the embryonic mouse mammary gland during invagination of the epithelial bud, we used miRNA microarrays to compare the 3rd mammary gland and inter mammary gland flank region dissected from E13.5 mouse embryos (Supplementary Figure S1A-S1C). This analysis showed that expression of many miRNAs is correlated to mammary gland development (Figure 1A and 1B). Among these, the expression level of miR-137 was increased about 30 fold in the 3rd mammary gland compared to inter mammary gland region (red box in Figure 1B). This high level of expression in the 3rd mammary gland forming region was confirmed by carrying out in situ hybridization using a miR-137 locked nucleic acid (LNA) probe to detect miR-137 transcripts (Supplementary Figure S1D). We also carried out further microarray analysis using tissue dissected from mouse embryos in the same way to screen for protein-coding genes differentially expressed in the developing gland. As expected, we found many genes already known to be highly expressed in developing mouse gland at similar stages [5]. For example, ER1 and Gata3 were highly expressed in 3rd mammary gland compared to the intermammary gland region (Figure 1C). The gene that we found to be most highly expressed in the 3rd mammary gland at E13.5 was Tac1 (red boxes in Figure 1C).
Figure 1. Heat-map of 3rd mammary gland and inter mammary gland region miRNA expression illustrating cluster-analysis of the expression data.
A., B. The signal intensity converted to log2 is reflected in the color scale such that dark green, green and red show low, medium and strong miRNA expression, respectively. The dendrogram on the left shows hierarchical similarity. C. Microarray data sorting for protein-coding genes related to mammary gland development.
Relationship between miR-137 and mammary gland development
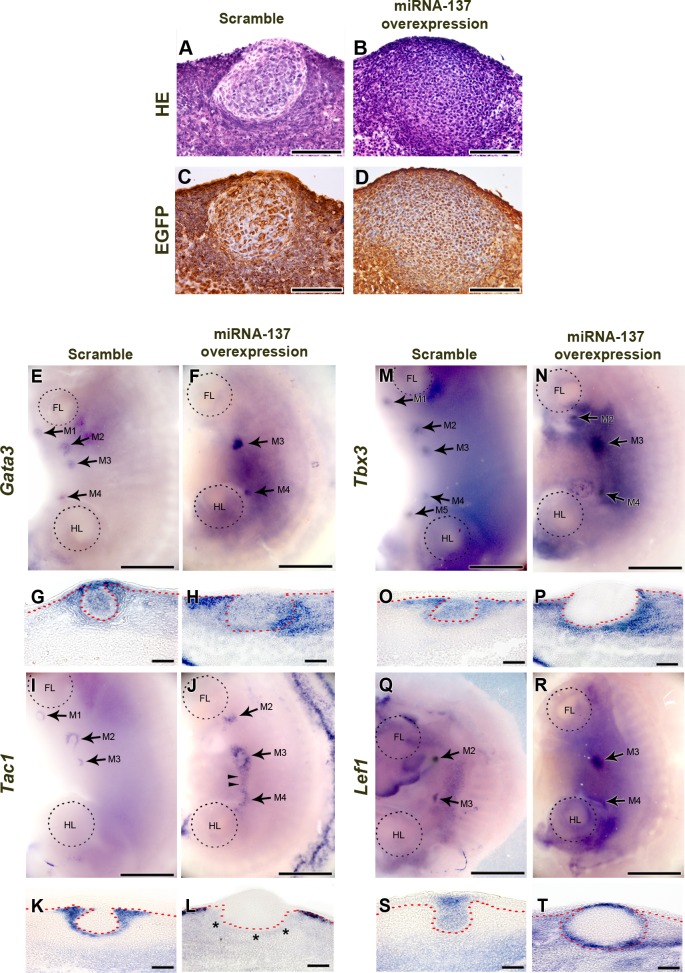
To investigate the function of miR-137 in mouse mammary gland development, we over-expressed miR-137 using a lentiviral system in the flank region of E11.0 mouse embryos and then cultured the flank for 72 h. As controls, we over-expressed scrambled miRNA. The lentiviral system also incorporated an EGFP cassette and successful over-expression of miR-137 or the control scrambled miRNA was confirmed by immunohistochemistry (IHC) using EGFP antibody and real-time quantitative polymerase chain reaction (RT-qPCR) (Figure 2C and 2D; Supplementary Figure S2A). In the controls, well-developed mammary buds were observed. With the over-expression of miR-137, thickened epithelium was observed in mammary placode region. However, this enlarged epithelium showed different cell morphology from normal mammary placode cells and they did not protrude into the underlying mesenchyme to give a spherical bud (compare Figure 2A and 2C with Figure 2B and 2D; see also for example Figure 2O versus 2P). To determine whether miRNA-137 inhibits or simulates expression of genes involved in mammary gland formation, we applied miR-137 to cultured mouse flanks and examined expression of Gata3, T-box transcription factor Tbx3, Tac1, and Lef1 (lymphoid enhancing factor 1) (Figure 2E-2T).
Figure 2. Function of miR-137 during mammary gland development.
A., B. HE staining of sections of mammary glands that developed after miR-137 over-expression, shows that normal invasion has not taken place although the placode has thickened. C., D. EGFP IHC indicates that scrambled miRNA and miR-137 have been successfully over-expressed. E.-H. Gata3 expression is disrupted by over-expressed miR-137. I.-L. MiR-137 over-expression alters Tac1 expression. (M-T) Expression patterns of the mammary gland markers, Tbx3 and Lef1, are changed after miR-137 treatment. Arrows, mammary bud. Arrowheads, ectopic Tac1 expression. Asterisk, disrupted Tac1 expression region ; FL, fore limb; HL, hind limb; Scale bar, A-D, G, H, K, L, O, P, S, T, 100 μm; E, F, I, J, M, N, Q, R, 1 mm.
Surface views of the embryos showed that the Gata3 expression pattern was ring shaped, in the mammary glands at E12.5. However, at E13.5, expression of Gata3 was seen in entire mammary gland region (compare Figure 2E and 2G with Supplementary Figure S3S and V). Gata3 expression pattern was altered by miR-137 over-expression. Mesenchymal expression of Gata3 was induced but epithelial expression was reduced by miR-137 over-expression (Figure 2F and 2H). Gata3 was expressed in the center of mammary epithelial, surface ectoderm and mammary mesenchyme just beneath epithelium of mammary bud in control (Figure 2G). However, Gata3 expression was lost its typical pattern in mammary epithelium and surface ectoderm after miR-137 over-expression (Figure 3H). Gata3 was observed in the flank mesenchyme besides the mammary bud (Figure 2F) extending both ventrally and dorsally after miR-137 over-expression (Figure 2H).
Figure 3. Tumour formation after SC inoculation of MDA-MB-231 cells over-expressing scrambled miRNA (controls) or over-expressing miR-137.
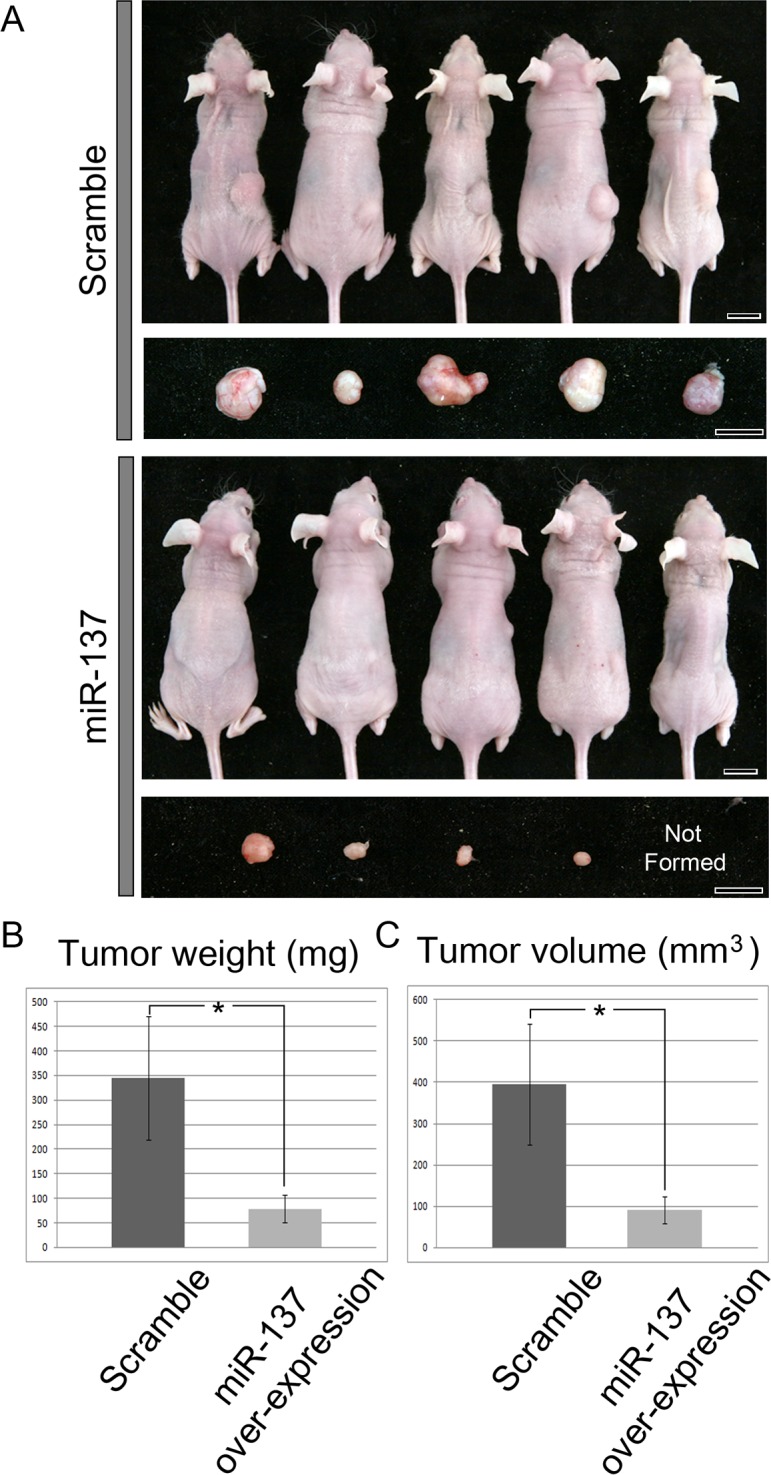
A. Cells over-expressing miR-137 form smaller tumours than cells over-expressing scrambled miRNA. B. Tumour weight is markedly reduced after miR-137 over-expression C. as is tumour volume compared to controls. Scale bar, 1 cm.
Tac1 is also known to be expressed only in mammary mesenchyme around in the developing mammary bud (Supplementary Figure S3G-S3L; ref 11-13) and this expression pattern was not changed by over-expression of scrambled miRNA (compare Figure 2I, 2K and Supplementary Figure S3G, S3J). Ectopic expression of Tac1 was detected between 3rd and 4th mammary gland after miR-137 over-expression (Figure 2I and 2J arrowheads). In transverse section showed that Tac1 was expressed in mesenchyme under surface ectoderm region but it was not expressed mesenchyme below mammary bud after miR-137 over-expression (Figure 2K and 2L asterisks).
Wnt signaling correlates expression of the transcription factor, Tbx3, which is essential for epithelial-mesenchymal interaction with Lef1 during early mammary gland development [30, 31]. In control, Tbx3 was expressed in mammary bud, mammary gland mesenchyme and dorsal part of flank in mouse embryo (Figure 2M and 2O). After miR-137 over-expression, Tbx3 expression pattern was similar as control in surface view of the dorsal part embryo flank (Figure 2N). However, transverse section showed that Tbx3 was completely obliterated in the mammary epithelium (Figure 2P). In addition, Tbx3 expression was induced in mammary mesenchyme and dermal mesenchyme region in dorsal part of flank (Figure 2P).
In the controls, expression of the Lef1 was observed in mammary epithelium from initiation (E10) to bud stage (E13) [31]. It was clearly observed in mammary epithelial buds in control (Figure 2Q and 2S). However, Lef1 expression was detected in broad region of mammary gland than control after miR-137 over-expression (Figure 2R). Moreover, Lef1 was not expressed in mammary epithelium except surface ectoderm and the margin of the enlarged mammary placodes between mammary epithelium and mesenchyme (Figure 2T). In addition, ectopic Lef1 expression was observed in mesenchyme around mammary gland after miR-137 over-exrpression (Figure 2T).
All these results indicate that over-expression of miRNA-137 suppresses development of the mammary gland in mouse embryos.
In vivo tumour formation assay after miR-137 over-expression
Recent work has shown that miR-137 impairs proliferation and migration of MDA-MB-231 human breast cancer cells in culture [28]. In order to investigate the relationship between miR-137 and breast tumour growth in vivo, MDA-MB-231 cells over-expressing miR-137 were inoculated subcutaneously (SC) in nude mice (n = 34) and formation of tumours were monitored 7 weeks later. Over-expression of miR-137 in the cells was confirmed by RT-qPCR (Supplementary Figure S2B). As a control, MDA-MB-231 cells over-expressing scrambled miRNA were inoculated (n = 16). The tumours formed by miR-137 over-expressing cells were noticeably smaller than those formed by cells transfected with the scrambled miRNA (Figure 3A). In fact, in 3 of the mice injected with MDA-MB-231 cells over-expressing miR-137 no tumours formed at all. In order to quantitate these results, we measured tumour weight and volume. Both weight and volume of tumours developing from miR-137 over-expressing cells were significantly reduced compared with tumours developing from cells over-expressing scrambled miRNA (Figure 3B and 3C). Thus, these results showed that miR-137 can suppress tumour formation in vivo.
Expression pattern of angiogenesis and tumour suppressor markers after miR-137 overexpression
To explore the mechanisms involved in suppressing tumour formation when miR-137 is over-expressed in MDA-MB-231 cells, we performed IHC to monitor cell proliferation, angiogenesis and expression of tumour suppressors. Cell proliferation was reduced in the tumours formed by miR-137 over-expressing cells. A large number of Ki67-positive proliferating cells were observed in tumours formed by cells expressing the scrambled miRNA compared to tumours formed by cells over-expressing miR-137 (Supplementary Figure S4A-S4D). The number of Ki67-positive cells was counted in scramble and miR-137 over-expression group. The numbers of Ki67-positive cells were 38.57 cells/100 × 100 μm and 14.14 cells/100 × 100 μm in scramble miRNA and miR-137 over-expressing group, respectively (Supplementary Figure S4E).
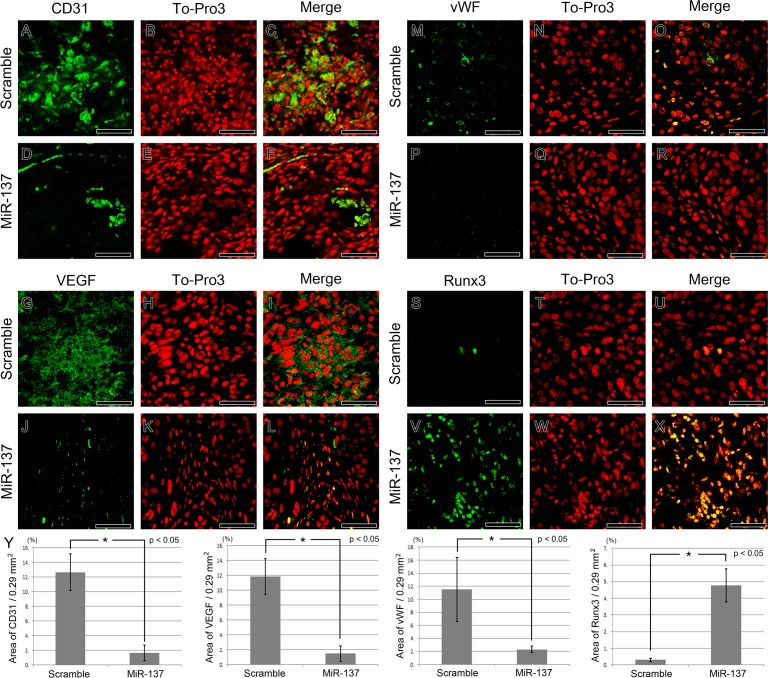
In addition, we found that angiogenesis was markedly reduced after miR-137 over-expression. Staining for the sinusoidal endothelial cell marker CD31 indicated that there were a smaller number of blood vessels in the tumours made by miR-137 over-expressing MDA-MB-231 cells (Figure 4A-4F) and furthermore the expression of vasculogenesis markers, VEGF and vWF were dramatically reduced (Figure 4G-4L; Figure 4M-4R). On the other hand, expression of known tumour suppressors, Runx3 and p53, was increased in the tumours formed by miR-137 over-expressing cells (Figure 4S-4X; Figure 5A-5C). The area of CD31, VEGF, vWF and Runx3 expression in the scramble miRNA and miR-137 over-expression group was measured (n = 7 for each). The CD31, VEGF and vWF positive area was reduced in the miR-137 over-expression group than in those treated with scramble miRNA treated group. Runx3 expression region was increased in miR-137 treated group compared to control group (Figure 4Y).
Figure 4. IHC of vasculogenesis and tumour suppressor markers after miR-137 over-expression.
A.-F. Sinusoidal endothelial cell marker CD31 is reduced after miR-137 over-expression. G.-L. Over-expressed miR-137 inhibits VEGF expression. M.-R. vWF expression is decreased in breast cancer after over-expression of miR-137 in MDA-MB-231. S.-X. Tumour suppressor Runx3 expression is substantially increased after miR-137 treatment. Y. The area of CD31, VEGF, vWF and Runx3 expression in the scramble miRNA and miR-137 over-expression group (n = 7 for each). IHC staining positive areas Scale bar, 100 μm.
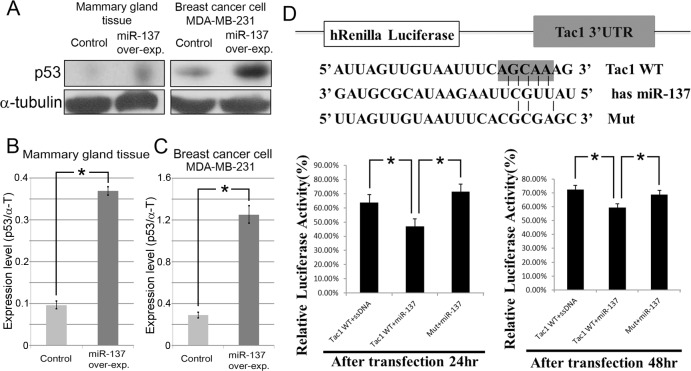
Figure 5. MiR-137 induces p53 expression in mammary gland tissue and breast cancer.
A.-C. Expression level of p53 is increased in mammary gland tissue and MDA-MB-231 cells after over-expressing miR-137. D. A luciferase assay result indicating that Tac1 is a direct target of miR-137.
Moreover, the cells in the tumours formed from miR-137 over-expressing cells induced the epithelial marker E-cadherin expression (Supplementary Figure S5A-S5F) but mesenchymal marker, Vimentin expression was reduced compare to control (Supplementary Figure S5G-S5L).
MiR-137 binding to Tac1 was examined by measuring luciferase activity with a dual-luciferase assay vector containing cloned 3′UTR of Tac1 transcripts (Figure 5D). Wild-type (WT) Tac1 reporters showed an inhibition of Renilla luciferase activity compared with the mutant reporter (Mut), with mismatched sequences inserted into the seed sequences of the predicted miR-137 binding site (Figure 5D). Luciferase assay results indicate that Tac1, which confers a poor prognosis in breast cancer [14, 15], was the directly correlated to miR-137 (Figure 5D).
DISCUSSION
From screens of tissue from mouse embryos for miRNAs differentially expressed in the developing mammary gland, we identified miR-137 was highly expressed. We also identified, via screening of the same tissues, that many protein-coding genes were already known to be expressed in the developing mammary gland and in breast tumours. Upon over-expression of miR-137 in mouse embryo flank during organ culture, the mammary epithelium thickened but it did not express several key genes known to be required for gland development and it also failed to invaginate into the underlying mesenchyme. MiR-137 is also highly expressed in an epithelial cell line derived from the adult human breast. When we over-expressed miR-137 in this breast cancer cell line, tumour formation in vivo was suppressed.
We identified miR-137 as a miRNA highly expressed in the developing mouse mammary gland at a stage when the mammary placode has invaginated the underlying mesenchyme to form a spherical bud. Our data and that of others [5] reveal that around this stage in development, many genes encoding proteins known to be associated with breast cancer are expressed in the gland. Thus, the molecular analysis of the mouse embryonic mammary gland is a good strategy for identifying genes that are potentially involved in breast cancer and any miRNAs identified from the mammary gland are good candidates for miRNAs associated with breast cancer.
ER1, GATA3 and Trps1 are among the genes that are highly expressed in the E13.5 mouse mammary gland and have also been implicated in human breast cancer [5, 16-18]. These genes were also identified by microarray analysis of E12.5 mammary glands. In addition, our analysis identified genes encoding proteins involved in retinoic acid signaling which appears to be involved in gland initiation [32]. Some genes identified are expressed in the mesenchyme and some in the epithelial bud and it is well-established that there are complex reciprocal signaling correlations between these tissues during normal development.
Our functional analysis suggests that miR-137 is involved in modulating early stages of mammary gland formation in the embryo especially invasion. When miR-137 is over-expressed, mammary placode formed and thickened but failed to invade the underlying mesenchyme and did not express either Tbx3 or Lef1 in mammary epithelium. Tbx3 mutations are found in Ulnar mammary syndrome in human patients, which is characterized by mammary gland hypoplasia [33, 34]. In Tbx3 knockout (KO) mouse embryos, mammary placode do not form and expression of Lef1 is absent [34]. Lef1 expression is one of the earliest molecular markers of placode formation and is also maintained in the epithelial cells during invasion of the epithelial bud. Thus, the failure of the mammary placode invagination by loss of Tbx3 and Lef1 after miR-137 over-expression suggests that miR-137 correlated the early stages of mammary gland development. Tac1 and Gata3 genes whose expression is also altered by over-expression of miR-137 are also detected at early bud stages. Moreover, luciferase assay results indicate that Tac1, which confers a poor prognosis in breast cancer [14, 15], was the direct target of miR-137. These data suggest that miR-137 is a potential therapeutic target for breast cancer by controlling of Tac1 expression.
In breast cancer cells, miR-137 has been shown to target expression of ERRα [28]. It is not clear whether ERRα could be the target of miR-137 in the developing mouse mammary gland. Interestingly, ER1 is expressed in the mesenchyme surrounding the mammary bud at E12.5 [5] and is a target of miR-206, which is also expressed in the mesenchyme at E11.5 and E12.5 (earlier than miR-137) but not at E13.5 [35]. When miR-206 was over-expressed in the same flank culture system used here, mammary placode development was very severely retarded; the placode formed but did not thicken [35]. This is consistent with miR-206 having a function at a slightly earlier stage in mammary gland development than miR-137.
Our results also showed that over-expression of miR-137 reduced tumour formation in vivo. Interestingly, we observed that cell proliferation of subcutaneously inoculated breast cancer cells was reduced when miR-137 was over-expressed. Moreover, epithelial-mesenchymal transition (EMT) was inhibited in the tumours formed from miR-137 over-expressing MDA-MB-231 cells. These results indicate that miR-137 have multiple tumour suppress function including prevention of the cell proliferation, epithelial invasion and EMT.
Our focus on miR-137 initially arose from screening for genes differentially expressed in embryonic mammary glands and this shows how the study of the developing embryonic mammary gland can provide new perspectives on breast cancer.
MATERIALS AND METHODS
All experiments were performed according to the guidelines of the Intramural Animal Use and Care Committee of Yonsei University College of Dentistry.
Animals
Adult Institute of Cancer Research Research; Caesarian Derived-1 (ICR; CD-1) mice were housed in a temperature-controlled room (22°C) under artificial illumination (lights on from 0500 to 1700 hours) and 55% relative humidity. The mice had access to food and water ad libitum. Embryos were obtained from time-mated pregnant mice. E0 was designated as the day a vaginal plug was confirmed. Embryos at developmental stages E11.0, E12.5 and E13.5 were used in this study. To study tumour formation, subcutaneous (SC) inoculation of 2.0×106 MDA-MB-231 cells was performed in the BALB/c nude mice (Orientbio, Korea). No immunosuppressive medication was used. Tumour size was measured seven weeks after SC inoculation. The host nude mice were sacrificed, and tumours were dissected for analysis.
MiRNA microarray analysis
For mammary gland miRNAs, the synthesis of target miRNA probes and hybridization were performed using Agilent's miRNA Labelling Reagent and Hybridization kit (Agilent Technology, USA) according to the manufacturer's instructions. Briefly, 100 ng of total RNA from each region (the 3rd mammary gland and the inter-mammary gland region) at E13.5 was dephosphorylated with ∼15 Units of calf intestine alkaline phosphatase (CIP), followed by RNA denaturation with ∼40% DMSO and a 10 min incubation at 100 °C. Dephosphorylated RNA was ligated with pCp-Cy3 mononucleotide and purified with MicroBioSpin 6 columns (Bio-Rad, USA). After purification, labelled samples were resuspended with Gene Expression blocking Reagent and Hi-RPM Hybridization buffer, followed by boiling for 5 min at 100 °C and 5 min chilled on ice. Finally, denatured labelled probes were pipetted onto the assembled Agilent miRNA Microarray (15K) and hybridized for 20 hours at 55 °C with rotation at 20 rpm in an Agilent Hybridization oven (Agilent Technology, USA). The hybridized microarrays were washed as the manufacturer's washing protocol (Agilent Technology, USA). Location of array raw data; GEO (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE66546)
Data acquisition and analysis
The hybridized images were scanned using Agilent's DNA microarray scanner and quantified with Feature Extraction Software (Agilent Technology, Palo Alto, CA). All data normalization and selection of fold-changed genes were performed using GeneSpringGX 7.3 (Agilent Technology, USA). The averages of normalized ratios were calculated by dividing the average of normalized signal channel intensity by the average of normalized control channel intensity. Functional annotation of genes was performed according to the Gene OntologyTM Consortium (http://www.geneontology.org/index.shtml) by GeneSpringGX 7.3.
Lentivirus production by transfecting 293T cells
We used miExpressTM precursor miRNA expression clone (GeneCopoeiaTM, USA) for miR-137 over-expression in mouse embryo flank and MDA-MB-231 cell. This clone is pEZX-MR04 vector system which carries EGFP and CMV promoter. The pEZX-MR04 vector confers resistance to puromycin (5-15 μg/ml) in transduced or transfected cells. The minimum antibiotic concentration to use is the lowest concentration that kills 100% of the cells in 3-6 days from the start of antibiotic selection. Finally we used 10 μg/ml purimycin for transfection of lentivirus production. Lentiviral production was carried out by transfecting human embryonic kidney 293T cells with lentivital and packaging vectors. For viral tittering, 5 × 104 293T cells were seed into 24-well culture plate with DMEM (10% FBS, 1% pen-strep) and counted the EGFP expressing cells or colonies of cells. EGFP positive colonies X 125 (dilution factor) X 40 = 2.76 × 105 TU/mL. For each 100 mm dish, helper plasmid pCD-NL/BH*ΔΔΔ 3 μg, envelope pLTR-G 300 ng, target miR-137 expressing lentivirus DNA 3 μg were mixed in 500 μl transfection reagent FuGENE HD (Roche). This mixture was incubated at room temperature for 20 min, and then added to 293T cells. The medium was changed after 4 h into culture medium containing 30% FBS. The resulting supernatant was collected 48 h, filtered through syringe driven filter Unit 0.45μg (Millipore, Cat No. SLHV 033 RS) and concentrated by Amicon Ultra-15 Centrifugal Filter Devices (Millipore, Cat No. UFC910024). Virus medium was used for in vitro organ and breast cancer cell culture.
Over-expression of miR-137 in in vitro organ culture and breast cancer cells
During cell culture (MDA-MB-231), miR-137 expressing lentiviral vector was transduced into cells. Concentrated miR-137 expressing lentivirus was added 1% (v/v) in culture medium containing polybrene (Santa Cruz Biotechnology). ICR mouse embryos were isolated at E11.0 and placed in culture medium (BGJb; Sigma, USA). The culture medium was supplemented with 20 μg/mL (v/v) ascorbic acid (Sigma, USA) and 1% (v/v) penicillin/streptomycin. Individual embryos were dissected into left and right halves using fine tungsten needles to bisect the neural tube. The left flank was designated the experimental tissue, and the right acted as control. Each flank was placed onto filter membranes (Track-etch, 1.0 μm pore; Whatman Nuclepore) that were supported on stainless steel grids in sterile culture dishes. Tissues were cultured in the air–medium interface at 37 °C and 5% CO2 for 12 h and 48 h by using a slight modification of the culture method reported by Trowell [29]. MDA-MB-231 human breast cancer cells were cultured in MEM (Lonza) supplemented with 10% (v/v) fetal bovine serum (Gibco), 100 unit/ml penicillin and 100 ug/ml streptomycin. Cells were grown to confluence at 37 °C in a humidified atmosphere containing 5% CO2 in air.
In situ hybridization
Cultured specimens were fixed overnight in 4% paraformaldehyde in phosphate buffered saline (PBS). For in situ hybridization, the specimens were treated with 20 μg/mL proteinase K for 5 min at room temperature [30]. Antisense RNA probes were labeled with digoxigenin (Roche). MiR-137 locked nucleic acid (LNA) modified probe was synthesized in Exiqon. After in situ hybridization, the specimens were cryosectioned at a thickness of 12 μm. At least 20 specimens were examined for each stage.
Histology and IHC
Samples were fixed in 4% paraformaldehyde in phosphate buffered saline (PBS) and then embedded in paraffin using standard procedures. Serial paraffin sections (4-μm thickness) were prepared, and individual slides were stained with haematoxylin and eosin. Antigen retrieval was achieved by citrate buffer, pH 6.0. After antigen retrieval, immunohistochemical analyses were performed using the DakoCytomation Envision System (using horseradish peroxidase with diaminobenzidine enhancer) (Dako, CA, USA) according to the manufacturer's instructions. The following primary antibodies were used for immunostaining or immunofluorescence: Ki67 (Thermo Scientific, USA), EGFP (Novus, USA), CD31 (BD Pharmingen, USA), vWF (Millipore, USA), VEGF (Santa Cruz Biotechnology, Inc., USA), Runx3 (Abcam, USA), E-cadherin (R&D systems, USA), and Vimentin (Thermo Scientific, USA). Alexa Fluor-conjugated secondary antibodies (Invitrogen, Carlsbad, CA, USA) were used for immunofluorescent staining. The stained sections were examined with a Leica MD5500D and a Zeiss LSM700.
Quantification of Ki67-positive cells
Ten slides that contained five sections each were used to determine the number of total cells and Ki67-positive cells, and 15 sections were chosen at random from the 10 slides. The number of cells was counted in an area of 100 × 100 μm. After counting the proliferating cells, we corrected the counting results using Abercrombie's method [36].
The equations used is
P is the average number of nuclear points per section, A the crude count of number of nuclei seen in the section, M the thickness of the section and L is the average length of the nuclei. Data were expressed as the mean ± S.D.
Evaluation and statistics of IHC staining
Pictures were taken by the confocal microscope (Carl Zeiss LSM700, Germany). The positive pixels of images were counted by the software Leica Application (Leica Microsystems, Germany). All parameters of the image acquisition were kept the same to allow comparison. Data were expressed as mean ± SD. The mean expression levels were compared between the experimental and control groups using ANOVA (SPSS 10.0), with a probability value of P < 0.05 (Tukey's HSD test) being considered statistically significant.
Luciferase assay
For the luciferase assay, the full length 3′UTRs of Tac1 genes (from 686nt to 1230nt of mRNA) were amplified from E13.5 mouse genomic DNA by PCR using sense and antisense primers carrying an XhoI restriction site and a NotI restriction site, respectively. The 3′UTR of the Tac1 gene was then cloned downstream of the Renilla luciferase gene in the psiCHECKTM-2 vector (Promega) between the XhoI and NotI sites. A mutant Tac1 3′UTR was synthesized using the QuikChange Lightning Multi Site-Directed Mutagenesis Kit (Agilent Technologies). Primer sequences used for cloning and mutagenesis are reported in Supplementary Figure S6. Cos-7 cells, a fibroblast-like cell line derived from monkey kidney tissue, were cultured in RPMI 1640 Medium (Gibco) supplemented with 10% (v/v) fetal bovine serum (Gibco) and 100X GlutaMaxTM-I (Gibco), without penicillin and streptomycin antibiotics. After cells grew to 80-90% confluence, wild type (psiCHECKTM-2 Tac1) or mutant (Mut) reporter vectors were co-transfected into Cos-7 cells with or without a miR-137 expressing lentiviral vector using FuGENE®H (Promega) and salmon sperm DNA (ssDNA, Sigma). A scrambled miRNA was used as control. Renilla and Firefly luciferase activities were measured 24 h and 48 h after transfection. Firefly and Renilla luciferase activities were measured using the dual-luciferase reporter system (Promega) on a GLOMAXTM 20/20 Luminometer (Promega). The data generated were expressed as relative ratios by normalizing Renilla luciferase readings to Firefly luciferase readings. All transfection experiments were performed in triplicate.
Western blotting analyses
Mammary gland tissue and MDA-MB-231 cells treated with the scrambled miRNA and miR-137-expressing lentiviral vectors underwent lysis by sonication (Nextadvance) in radio-immunoprecipitation assay (RIPA) buffer (50 nM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100). Anti-p53 (Santa Cruz Biotechnology, Inc., USA) antibody was used. Horseradish peroxidase conjugated secondary antibodies (Santa Cruz Biotechnology, Inc., USA) were used, and the protein bands were visualized by enhanced chemiluminescence (Amersham Biosciences, USA). α-tubulin expression served as an internal control.
SUPPLEMENTARY MATERIAL FIGURES
ACKNOWLEDGMENTS AND FUNDING
This research was supported by the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (2012M3A9B4028738). This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI14C3266).
Footnotes
CONFLICTS OF INTEREST
There is no conflict of interest.
REFERENCES
- 1.Robinson GW, Karpf AB, Kratochwil K. Regulation of mammary gland development by tissue interaction. J Mammary Gland Biol Neoplasia. 1999;4:9–19. doi: 10.1023/a:1018748418447. [DOI] [PubMed] [Google Scholar]
- 2.Sakakura T, Kusano I, Kusakabe M, Inaguma Y, Nishizuka Y. Biology of mammary fat pad in fetal mouse: capacity to support development of various fetal epithelia in vivo. Development. 1987;100:421–430. doi: 10.1242/dev.100.3.421. [DOI] [PubMed] [Google Scholar]
- 3.Veltmaat JM, Mailleux AA, Thiery JP, Bellusci S. Mouse embryonic mammogenesis as a model for the molecular regulation of pattern formation. Differentiation. 2003;71:1–17. doi: 10.1046/j.1432-0436.2003.700601.x. [DOI] [PubMed] [Google Scholar]
- 4.Hens JR, Wysolmerski JJ. Key stages of mammary gland development: molecular mechanisms involved in the formation of the embryonic mammary gland. Breast Cancer Res. 2005;7:220–224. doi: 10.1186/bcr1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wansbury O, Mackay A, Kogata N, Mitsopoulos C, Kendrick H, Davidson K, Ruhrberg C, Reis-Filho JS, Smalley MJ, Zvelebil M, Howard BA. Transcriptome analysis of embryonic mammary cells reveals insights into mammary lineage establishment. Breast Cancer Res. 2011;13:R79. doi: 10.1186/bcr2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siegel PM, Muller WJ. Transcription factor regulatory networks in mammary epithelial development and tumorigenesis. Oncogene. 2010;29:2753–2759. doi: 10.1038/onc.2010.43. [DOI] [PubMed] [Google Scholar]
- 7.Asselin-Labat ML, Sutherland KD, Vaillant F, Gyorki DE, Wu D, Holroyd S, Breslin K, Ward T, Shi W, Bath ML, Deb S, Fox SB, Smyth GK, et al. Gata-3 negatively regulates the tumor-initiating capacity of mammary luminal progenitor cells and targets the putative tumor suppressor caspase-14. Mol Cell Biol. 2011;31:4609–4622. doi: 10.1128/MCB.05766-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127:1041–1055. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pensa S, Watson CJ, Poli V. Stat3 and the inflammation/acute phase response in involution and breast cancer. J Mammary Gland Biol Neoplasia. 2009;14:121–129. doi: 10.1007/s10911-009-9124-x. [DOI] [PubMed] [Google Scholar]
- 10.Ferrari A, Petterino C, Ratto A, Campanella C, Wurth R, Thellung S, Vito G, Barbieri F, Florio T. CXCR4 expression in feline mammary carcinoma cells: evidence of a proliferative role for the SDF-1/CXCR4 axis. BMC Vet Res. 2012;8:27. doi: 10.1186/1746-6148-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weil M, Itin A, Keshet E. A role for mesenchyme-derived tachykinins in tooth and mammary gland morphogenesis. Development. 1995;121:2419–2428. doi: 10.1242/dev.121.8.2419. [DOI] [PubMed] [Google Scholar]
- 12.Severini C, Improta G, Falconieri-Erspamer G, Salvadori S, Erspamer V. The tachykinin peptide family. Pharmacol Rev. 2002;54:285–322. doi: 10.1124/pr.54.2.285. [DOI] [PubMed] [Google Scholar]
- 13.David S, Kan T, Cheng Y, Agarwal R, Jin Z, Mori Y. Aberrant silencing of the endocrine peptide gene tachykinin-1 in gastric cancer. Biochem Biophys Res Commun. 2009;378:605–609. doi: 10.1016/j.bbrc.2008.11.078. [DOI] [PubMed] [Google Scholar]
- 14.Ellsworth RE, Seebach J, Field LA, Heckman C, Kane J, Hooke JA, Love B, Shriver CD. A gene expression signature that defines breast cancer metastases. Clin Exp Metastasis. 2009;26:205–213. doi: 10.1007/s10585-008-9232-9. [DOI] [PubMed] [Google Scholar]
- 15.Reddy BY, Greco SJ, Patel PS, Trzaska KA, Rameshwar P. RE-1-silencing transcription factor shows tumor-suppressor functions and negatively regulates the oncogenic TAC1 in breast cancer cells. Proc Natl Acad Sci USA. 2009;106:4408–4413. doi: 10.1073/pnas.0809130106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG, van der Wees J, Lindeman GJ, Visvader JE. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- 17.Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci. USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cimino-Mathews A, Subhawong AP, Illei PB, Sharma R, Halushka MK, Vang R, Fetting JH, Park BH, Argani P. GATA3 expression in breast carcinoma: utility in triple-negative, sarcomatoid, and metastatic carcinomas. Hum Pathol. 2013;44:1341–1349. doi: 10.1016/j.humpath.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Visone R, Croce CM. MiRNAs and cancer. Am J Pathol. 2009;174:1131–1138. doi: 10.2353/ajpath.2009.080794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mouw JK, Yui Y, Damiano L, Bainer RO, Lakins JN, Acerbi I, Ou G, Wijekoon AC, Levental KR, Gilbert PM, Hwang ES, Chen YY, Weaver VM. Tissue mechanics modulate microRNA-dependent PTEN expression to regulate malignant progression. Nat Med. 2014;20:360–367. doi: 10.1038/nm.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miska EA. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev. 2005;15:563–568. doi: 10.1016/j.gde.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Zhang N, Wang X, Huo Q, Sun M, Cai C, Liu Z, Hu G, Yang Q. MicroRNA-30a suppresses breast tumor growth and metastasis by targeting metadherin. Oncogene. 2014;33:3119–3128. doi: 10.1038/onc.2013.286. [DOI] [PubMed] [Google Scholar]
- 23.Dyrskjøt L, Ostenfeld MS, Bramsen JB, Silahtaroglu AN, Lamy P, Ramanathan R, Fristrup N, Jensen JL, Andersen CL, Zieger K, Kauppinen S, Ulhøi BP, Kjems J, et al. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009;69:4851–4860. doi: 10.1158/0008-5472.CAN-08-4043. [DOI] [PubMed] [Google Scholar]
- 24.Balaguer F, Link A, Lozano JJ, Cuatrecasas M, Nagasaka T, Boland CR, Goel A. Epigenetic silencing of miR-137 is an early event in colorectal carcinogenesis. Cancer Res. 2010;70:6609–6618. doi: 10.1158/0008-5472.CAN-10-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- 26.Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, Vandenberg SR, Ginzinger DG, James CD, Costello JF, Bergers G, Weiss WA, Alvarez-Buylla A, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Q, Chen X, Zhang M, Fan Q, Luo S, Cao X. miR-137 is frequently down-regulated in gastric cancer and is a negative regulator of Cdc42. Dig Dis Sci. 2011;56:2009–2016. doi: 10.1007/s10620-010-1536-3. [DOI] [PubMed] [Google Scholar]
- 28.Zhao Y, Li Y, Lou G, Zhao L, Xu Z, Zhang Y, He F. MiR-137 targets estrogen-related receptor alpha and impairs the proliferative and migratory capacity of breast cancer cells. PLoS One. 2012;7:e39102. doi: 10.1371/journal.pone.0039102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trowell OA. The culture of mature organs in a synthetic medium. Exp Cell Res. 1959;16:118–147. doi: 10.1016/0014-4827(59)90201-0. [DOI] [PubMed] [Google Scholar]
- 30.Cho KW, Kim JY, Song SJ, Farrell E, Eblaghie MC, Kim HJ, Tickle C, Jung HS. Molecular interactions between Tbx3 and Bmp4 and a model for dorsoventral positioning of mammary gland development. Proc Natl Acad Sci USA. 2006;103:16788–16793. doi: 10.1073/pnas.0604645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boras-Granic K, Chang H, Grosschedl R, Hamel PA. Lef1 is required for the transition of Wnt signaling from mesenchymal to epithelial cells in the mouse embryonic mammary gland. Dev Biol. 2006;295:219–231. doi: 10.1016/j.ydbio.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 32.Cho KW, Kwon HJ, Shin JO, Lee JM, Cho SW, Tickle C, Jung HS. Retinoic acid signaling and the initiation of mammary gland development. Dev Biol. 2012;365:259–266. doi: 10.1016/j.ydbio.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 33.Bamshad M, Lin RC, Law DJ, Watkins WC, Krakowiak PA, Moore ME, Franceschini P, Lala R, Holmes LB, Gebuhr TC, Bruneau BG, Schinzel A, Seidman JG, et al. Mutations in human TBX3 alter limb, apocrine and genital development in ulnar-mammary syndrome. Nat Genet. 1997;16:311–315. doi: 10.1038/ng0797-311. [DOI] [PubMed] [Google Scholar]
- 34.Davenport TG, Jerome-Majewska LA, Papaioannou VE. Mammary gland, limb and yolk sac defects in mice lacking Tbx3, the gene mutated in human ulnar mammary syndrome. Development. 2003;130:2263–2273. doi: 10.1242/dev.00431. [DOI] [PubMed] [Google Scholar]
- 35.Lee MJ, Yoon KS, Cho KW, Kim KS, Jung HS. Expression of miR-206 during the initiation of mammary gland development. Cell Tissue Res. 2013;353:425–433. doi: 10.1007/s00441-013-1653-3. [DOI] [PubMed] [Google Scholar]
- 36.Abercrombie M. Estimation of nuclear population from microtome sections. Anat Rec. 1946;94:239–247. doi: 10.1002/ar.1090940210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.