Abstract
We studied if combination genetic signature potentially stratifies lower-grade gliomas better than histology by investigating 214 lower-grade gliomas for IDH1/2 and TERTp mutations, 1p/19q codeletion and EGFR amplification as to their impact on prognostication. Prognostic association of grading was independent of other prognostic variables including age, histological type, IDH1/2, 1p/19q and TERTp status. No single marker, including IDH1/2, superseded grading in prognostication, indicating grading was still a very important tool. Prognosis was most favorable in 31.7% of patients with IDH1/2 mutation and either 1p/19q codeletion or TERTp mutation (IDHmut-OT), intermediate in 45.8% of patients with IDH1/2 mutation only (IDHmut) and 16.9% of patients without any of the alterations (IDHwt), and poorest in 5.6% of patients with wild-type IDH1/2 and either TERTp mutation or EGFR amplification (IDHwt-ET). Our results suggested not all IDH1/2 wild-type lower-grade gliomas are aggressive and additional biomarkers are required to identify glioblastoma-equivalent tumors. Multivariate analysis revealed independent prognostic values of grading and genetic signature. Grade II IDHwt-ET gliomas exhibited shorter survival than IDH1/2 mutated grade III gliomas, suggesting combination genetic signature potentially superseded grading in prognostication. In summary, biomarker-based stratification is useful in the diagnosis and prognostication of lower-grade gliomas, and should be used together with grading.
Keywords: Pathology Section, glioma, IDH1/2, 1p/19q, TERT, EGFR
INTRODUCTION
Lower-grade glioma is a diversified group of infiltrative brain tumors comprising WHO grades II and III astrocytomas, oligodendrogliomas and mixed oligoastrocytomas. The tumors exhibit wide range of biological and clinical heterogeneity and remain as one of the major challenges in clinical practice in neuro-oncology. One of which is the interobserver variability arising from the morphology-based classification of diffuse gliomas which leads to suboptimal diagnostic reproducibility affecting both histological typing and grading [1–4]. Accurate differentiation of histological grade is crucial in clinical management of lower-grade gliomas not only for prognostic estimation but treatment guidance [5–9]. In most places, lower-grade gliomas are treated by maximal resection and high grade tumors need adjuvant therapy in most regimens [5, 9]. Such differentiation is of particular importance in surgical biopsy and yet there is major interobserver discrepancy [4]. Therefore, more objective and reproducible markers which can supersede histological grade are needed for clinical stratification of lower-grade gliomas. In order to improve the diagnostic and prognostic accuracy and facilitate therapeutic formulation, molecular markers will be supplemented into the coming WHO classification of lower-grade glioma so as to provide a more objective and precise tumor classification system [10]. Isocitrate dehydrogenase 1 (IDH1) mutation and chromosome 1p/19q codeletion represent the most important molecular markers with clinical implications in lower-grade gliomas. Both of the markers are strong favorable prognostic factors in lower-grade gliomas as shown in the literature [11–18], and their predictive values to PCV (procarbazine, CCNU and vincristine) chemotherapy in oligodendroglial tumors have also been recently revealed in clinical trials [11, 12, 17]. Promoter mutation of telomerase reverse transcriptase (TERTp) is another emerging molecular marker which can stratify lower-grade gliomas and glioblastomas into prognostic subgroups, particularly in combination with IDH1 mutation [19–24]. The activating mutation is associated with 1p/19q codeletion and oligodendroglial histology in the presence of IDH1 mutation and identifies a subset of aggressive astrocytic tumors lacking IDH1 mutation, highlighting its potential diagnostic and prognostic impact in lower-grade gliomas [19, 20, 23, 25, 26]. Incorporation of molecular markers into tumor classification not only provides additional prognostic information but potential influence on the histological grade of tumor diagnosis. For example, IDH1 mutation has been reported to exert a greater prognostic relevance in glioblastoma and anaplastic astrocytoma than histological grade, indicating that molecular markers might potentially supersede histological grade in tumor classification [27]. The influence of molecular markers in the prognostic value of histological grade in lower-grade gliomas, however, remains under-investigated. In this study, we aim to examine a panel of molecular markers including IDH1/2 mutation, 1p/19q codeletion, TERTp mutation and EGFR amplification as to their impact on the prognostic value of histological grade in a cohort of lower-grade gliomas. Our study reveals that combination genetic signature potentially supersedes histological grade in prognostic classification of lower-grade gliomas and has important clinical implication in refining the diagnostic and prognostic accuracy of grades II and III gliomas.
RESULTS
Cohort characteristics
Table 1 and Fig. 1 summarize the clinical and molecular data of the cohort. Seventy-eight cases from Prince of Wales Hospital (Hong Kong) and 136 cases from Huashan Hospital (Shanghai) formed the basis of the cohort. The cohort comprised 142 grade II diffuse gliomas which included 86 diffuse astrocytomas, 18 oligodendrogliomas and 38 oligoastrocytomas, as well as 72 grade III anaplastic gliomas which included 63 anaplastic astrocytomas, 6 anaplastic oligodendrogliomas and 3 anaplastic oligoastrocytomas. Male to female ratio of the cohort was 1:0.69. The mean and median ages were 41.9 and 41 years (range 20 to 79), respectively. Operation data was available in 199 of 214 (93%) patients and adjuvant treatment data was available in 188 of 214 (87.9%) patients. Total resection was performed in 83 of 137 (60.6%) grade II tumors and 29 of 62 (46.8%) grade III tumors (p = 0.07). Radiotherapy was given in 95 of 128 (74.2%) grade II tumors and 47 of 60 (78.3%) grade III tumors (p = 0.503). Chemotherapy was given in 83 of 128 (64.8%) grade II tumors and 34 of 60 (56.7%) grade III tumors (p = 0.281). Survival data was available in 214 patients. The median follow-up and median overall survival were 9.2 years and 8.4 years, respectively.
Table 1. Summary of clinical and molecular data of lower grade gliomas in the study.
| Grade II n = 142 | Grade III n = 72 | All tumors n = 214 | |
|---|---|---|---|
| Gender (Male /Female) | 79/63 | 48/24 | 127/87 |
| Age (Mean / median / range) | 40.4 / 39 (21–79) | 44.9 / 44.5 (20–70) | 41.9 / 41 (20–79) |
| Histological type | |||
| Astrocytomas | 86 (60.6%) | 63 (87.5%) | 149 (69.6%) |
| Oligodendrogliomas | 18 (12.7%) | 6 (8.3%) | 24 (11.2%) |
| Oligoastrocytomas | 38 (26.8%) | 3 (4.2%) | 41 (19.2%) |
| Operation | |||
| Total resection | 83 (58.5%) | 29 (40.3%) | 112 (52.3%) |
| Non-total resection | 54 (38%) | 33 (45.8%) | 87 (40.7%) |
| Not available | 5 (3.5%) | 10 (13.9%) | 15 (7%) |
| Adjuvant therapy | |||
| Radiotherapy only | 22 (15.5%) | 16 (22.2%) | 38 (17.8%) |
| Chemotherapy only | 10 (7%) | 3 (4.2%) | 13 (6.1%) |
| Both radiotherapy and chemotherapy | 73 (51.4%) | 31 (43.1%) | 104 (48.6%) |
| No | 23 (16.2%) | 10 (13.9%) | 33 (15.4%) |
| Not available | 14 (9.9%) | 12 (16.7%) | 26 (12.1%) |
| IDH1/2 | |||
| Mutant | 110 (77.5%) | 46 (63.9%) | 156 (72.9%) |
| Wild-type | 32 (22.5%) | 26 (36.1%) | 58 (27.1%) |
| 1p/19q | |||
| Codeleted | 35 (24.6%) | 3 (4.2%) | 38 (17.8%) |
| Non-codeleted | 107 (75.4%) | 69 (95.8%) | 176 (82.2%) |
| TERTp | |||
| Mutant | 41 (28.9%) | 20 (27.8%) | 61 (28.5%) |
| Wild-type | 101 (71.1%) | 52 (72.2%) | 153 (71.5%) |
| EGFR | |||
| Amplified | 6 (4.2%) | 3 (4.2%) | 9 (4.2%) |
| Non-amplified | 136 (95.8%) | 69 (95.8%) | 205 (95.8%) |
| Genetic signature | |||
| IDHmut-OT | 45 (31.7%) | 12 (16.7%) | 57 (26.6%) |
| IDHmut | 65 (45.8%) | 34 (47.2%) | 99 (46.3%) |
| IDHwt | 24 (16.9%) | 18 (25%) | 42 (19.6%) |
| IDHwt-ET | 8 (5.6%) | 8 (11.1%) | 16 (7.5%) |
n, number of case; IDHmut-OT, IDH1/2 mutated and either 1p/19q codeleted or TERTp mutated; IDHmut, IDH1/2 mutated only; IDHwt, IDH1/2 wild-type, TERTp wild-type and EGFR non-amplified; IDHwt-ET, IDH1/2 wild-type and either TERTp mutated or EGFR amplified.
Figure 1. Clinical and molecular data of 214 lower-grade gliomas in the study.

1p/19q codeletion occurred exclusively in IDH1/2 mutated gliomas (p < 0.0001, Chi square test). EGFR amplification was mutually exclusive with IDH1/2 mutation (p < 0.0001, Fisher's exact test) and associated with TERTp mutation within the IDH1/2 wild-type gliomas (p = 0.002, Fisher's exact test). Tumors were assigned into genetic subgroups based on combinations of status of IDH1/2 mutation, 1p/19q codeletion, TERTp mutation and EGFR amplification. IDHmut-OT gliomas were defined as tumors harboring IDH1/2 mutation and either 1p/19q codeletion or TERTp mutation. IDHmut gliomas were defined as tumors harboring IDH1/2 mutation only and without 1p/19q codeletion nor TERTp mutation. IDHwt gliomas were defined as tumors harboring none of the molecular markers. IDHwt-ET gliomas were defined as IDH1/2 wild-type tumors with either TERTp mutation or EGFR amplification. The cases were sorted by overall survival within each genetic signature subgroup. OS, overall survival.
IDH1/2 mutation
IDH1/2 mutations were found in 156 of 214 (72.9%) of tumors examined, including 59 of 86 (68.6%) diffuse astrocytomas, 17 of 18 (94.4%) oligodendrogliomas, 34 of 38 (89.5%) oligoastrocytomas, 37 of 63 (58.7%) anaplastic astrocytomas, 6 of 6 (100%) anaplastic oligodendrogliomas and 3 of 3 (100%) anaplastic oligoastrocytomas. Regarding histological grade, IDH1/2 mutations were detected in 110 of 142 (77.5%) grade II tumors and 46 of 72 (63.9%) grade III tumors. Among the 156 IDH1/2 mutated tumors, IDH1 and IDH2 mutations were mutually exclusive and were observed in 148 (94.9%) and 8 (5.1%) cases, respectively. 147 of 148 (99.3%) IDH1 mutations were IDH1-R132H and 6 of 8 (75%) IDH2 mutations were IDH2-R172K. Other mutation subtype observed included one case of IDH1-R132C, one case of IDH2-R172M and one case of IDH2-R172G.
Chromosome 1p/19q codeletion
Chromosome 1p/19q codeletion was detectable in 38 of 214 (17.8%) of tumors examined, including 6 of 86 (7%) diffuse astrocytomas, 11 of 18 (61.1%) oligodendrogliomas, 18 of 38 (47.4%) oligoastrocytomas and 3 of 6 (50%) anaplastic oligodendrogliomas. Twenty-five percent of grade II tumors and 4.2% of grade III tumors harbored 1p/19q codeletion. 1p/19q codeletion was found exclusively in IDH1/2 mutated tumors (p < 0.0001). According to histological tumor type, 1p/19q codeletion was observed in 14 of 24 (58.3%) oligodendrogliomas, 18 of 41 (43.9%) oligoastrocytomas and 6 of 149 (4%) astrocytomas (p <0.00001) (Fig. 1).
TERT promoter mutation
TERTp mutation was identified in 61 of 214 (28.5%) tumors examined, including 41 of 142 (28.9%) grade II gliomas and 20 of 72 (27.8%) grade III gliomas. Among grade II tumors, 13 of 86 (15.1%) diffuse astrocytomas, 13 of 18 (72.2%) oligodendrogliomas and 15 of 38 (39.5%) oligoastrocytomas harbored TERTp mutation. Among grade III tumors, mutation was found in 12 of 63 (19%) anaplastic astrocytomas, 6 of 6 (100%) anaplastic oligodendrogliomas and 2 of 3 (66.7%) anaplastic oligoastrocytomas, respectively. Thirty-eight (62.3%) mutated tumors showed C228T mutation and 23 (37.7%) mutated tumors showed C250T mutation, of which the two mutation subtypes were mutually exclusive.
EGFR amplification
In all lower-grade gliomas examined for EGFR FISH analysis, EGFR amplification was found in 6 of 86 (7%) diffuse astrocytomas and 3 of 63 (4.8%) anaplastic astrocytomas, giving an overall amplification frequency of 4.2% (9/214). EGFR amplification was exclusively found in tumors with wild-type IDH1/2 (p < 0.0001) and showed co-occurring association with TERTp mutation in IDH1/2 wild-type tumor subset (p = 0.002).
Survival analysis (Individual variable)
We evaluated the survival data according to histological grade, patient age, histological type and various molecular markers by univariate analysis (Table 2, Fig. 2a to 2g). Patients with grade II gliomas had longer overall survival than grade III gliomas (p < 0.0001) (Fig. 2a). The median overall survival was 10.2 years for grade II tumors and 2.3 years for grade III tumors. Patients of 50 years or younger had longer overall survival than those above 50 years (p < 0.0001) (Fig. 2b). Stratifying the cohort according to histological type, tumors with oligodendroglial or oligoastrocytic histology showed longer overall survival than tumors with astrocytic histology (p = 0.0003) (Fig. 2c). Regarding molecular markers, patients with IDH1/2 mutation (p < 0.0001) and 1p/19q codeletion (p < 0.0001) had more favorable overall survival than those without the markers (Fig. 1d and 1e). Patients with TERTp mutated tumors showed trend of longer overall survival than those with TERTp wild-type tumors (Fig. 2f). Tumors with EGFR amplification exhibited very short overall survival comparing to tumors without the gene amplification (p < 0.0001) (Fig. 2g).
Table 2. Univariate analysis of clinical and molecular variables.
| n | HR | [95% CI] | Median OS (years) | p | |
|---|---|---|---|---|---|
| Histological grade | |||||
| Grade II | 142 | 1 | 10.2 | <0.0001 | |
| Grade III | 72 | 4.045 | [2.629–6.223] | 2.3 | |
| Age | |||||
| ≤50 years | 172 | 1 | 10 | <0.0001 | |
| >50 years | 42 | 2.43 | [1.549–3.812] | 2.3 | |
| Histological type | |||||
| Astrocytic | 149 | 2.266 | [1.445–3.555] | 5.5 | 0.0003 |
| Oligodendroglial / Oligoastrocytic | 65 | 1 | 11 | ||
| IDH1/2 | |||||
| Mutant | 156 | 0.431 | [0.286–0.649] | 10.1 | <0.0001 |
| Wild-type | 58 | 1 | 2.2 | ||
| 1p/19q codeletion | |||||
| Codeleted | 38 | 0.274 | [0.142–0.529] | 11.8 | <0.0001 |
| Non-codeleted | 176 | 1 | 5.6 | ||
| TERTp | |||||
| Mutant | 61 | 0.73 | [0.472–1.128] | 10.5 | 0.155 |
| Wild-type | 153 | 1 | 6.5 | ||
| EGFR | |||||
| Amplified | 9 | 9.082 | [4.373–18.863] | 1.1 | <0.0001 |
| Non-amplified | 205 | 1 | 9.9 | ||
| Genetic signature | |||||
| IDHmut-OT | 57 | 1 | 11.8 | <0.0001 | |
| IDHmut | 99 | 2.863 | [1.643–4.989] | 5.9 | |
| IDHwt | 42 | 2.883 | [1.498–5.547] | 5 | |
| IDHwt-ET | 16 | 17.65 | [8.603–36.208] | 0.6 |
n, number of case; HR, hazard ratio; 95% CI, 95% confidence interval; OS, overall survival; IDHmut-OT, IDH1/2 mutated and either 1p/19q codeleted or TERTp mutated; IDHmut, IDH1/2 mutated only; IDHwt, IDH1/2 wild-type, TERTp wild-type and EGFR non-amplified; IDHwt-ET, IDH1/2 wild-type and either TERTp mutated or EGFR amplified.
Figure 2. Kaplan-Meier survival analysis by histological grade, age, histological type, IDH1/2 mutation, 1p/19q codeletion, TERTp mutation and EGFR amplification.
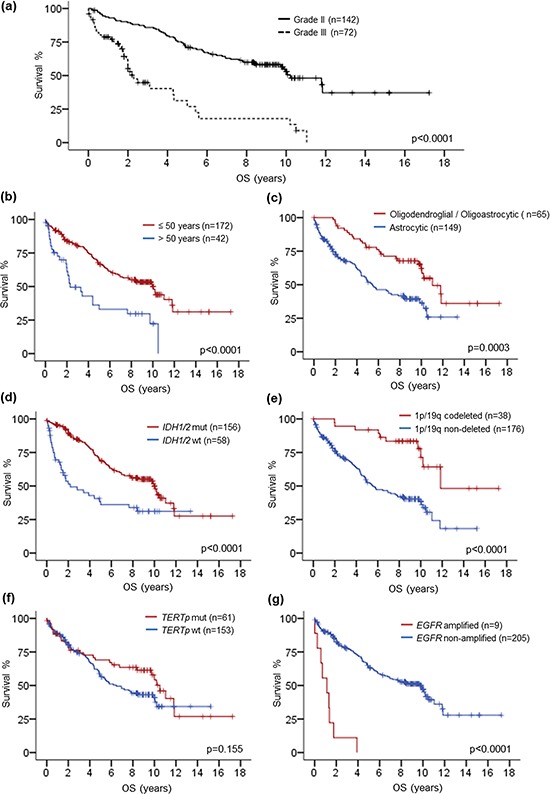
Longer overall survival was associated with grade II gliomas a. patients at 50 years or younger b. oligodendroglial or oligoastrocytic tumors c. IDH1/2 mutated gliomas d. 1p/19q codeleted gliomas e. and EGFR non-amplified gliomas g. TERTp mutated gliomas trended to longer overall survival f. OS, overall survival; mut, mutant; wt, wild-type.
Next, we investigated the influences of age, histological typing and molecular markers on the prognostic value of histological grade by co-evaluating histological grade and individual variables in survival analysis (Table 3, Fig. 3a to 3f). Histological grade could prognostically dichotomize patients of 50 years or younger (p < 0.0001) and the effect was marginal in patients above 50 years (p = 0.09) (Fig. 3a). Co-evaluating histological grade and type showed that the prognostic value of histological grade was independent of histological type (Fig. 3b). Grade III histology was associated with unfavorable prognosis in both astrocytic tumors (p < 0.0001) and oligodendroglial or oligoastrocytic tumors (p = 0.006). Prognostic stratification by histological grade in tumor groups defined by molecular markers was shown in Fig. 3c to 3f. Histological grade showed a good prognostic breakdown in IDH1/2 mutated gliomas (p < 0.0001), IDH1/2 wild-type gliomas (p < 0.0001), 1p/19q codeleted gliomas (p = 0.024), 1p/19q non-deleted gliomas (p < 0.0001), TERTp mutated gliomas (p < 0.0001) and TERTp wild-type gliomas (p < 0.0001), indicating that prognostic value of histological grade was independent of these three molecular markers. Co-evaluating histological grade and EGFR amplification status showed that grade could prognostically separate EGFR non-amplified lower-grade gliomas (p < 0.0001) but not EGFR amplified lower-grade gliomas (p < 0.811) (Fig. 3f). Though the number of EGFR amplified grade II gliomas was small (n = 6), it's worthwhile to note that this small subset of grade II tumors exhibited worse prognosis than EGFR non-amplified grade III gliomas (p = 0.006). This was consistent with the univariate analysis that the hazard ratio (HR) of amplified EGFR to non-amplified EGFR (HR = 9.082, 95% CI = 4.373–18.863) was greater than that of grade III to grade II histology (HR = 4.045, 95% CI = 2.629–6.223) (Table 2).
Table 3. Survival analysis of histological grade by clinical and molecular variables.
| Median OS (years) | p | ||
|---|---|---|---|
| Grade II | Grade III | ||
| Age | |||
| ≤50 years | 11.8 | 3.1 | <0.0001 |
| >50 years | 4.4 | 2.1 | 0.09 |
| Histological type | |||
| Astrocytic | 9.9 | 2.2 | <0.0001 |
| Oligodendroglial / Oligoastrocytic | 11.8 | 3.1 | 0.006 |
| IDH1/2 | |||
| Mutant | 10.2 | 4.3 | <0.0001 |
| Wild-type | 8.4 | 1.3 | <0.0001 |
| 1p/19q | |||
| Codeleted | NR | 2 | 0.024 |
| Non-codeleted | 8.5 | 2.3 | <0.0001 |
| TERTp | |||
| Mutant | 11.8 | 2 | <0.0001 |
| Wild-type | 9.9 | 2.3 | <0.0001 |
| EGFR | |||
| Amplified | 0.7 | 1.1 | 0.811 |
| Non-amplified | 11.8 | 2.5 | <0.0001 |
| Genetic signature | |||
| IDHmut-OT | NR | 10.2 | 0.004 |
| IDHmut | 7.8 | 4.3 | 0.0002 |
| IDHwt | NR | 1.5 | 0.0004 |
| IDHwt-ET | 1.3 | 0.4 | 0.04 |
OS, overall survival; NR, median survival not yet reached; IDHmut-OT, IDH1/2 mutated and either 1p/19q codeleted or TERTp mutated; IDHmut, IDH1/2 mutated only; IDHwt, IDH1/2 wild-type, TERTp wild-type and EGFR non-amplified; IDHwt-ET, IDH1/2 wild-type and either TERTp mutated or EGFR amplified.
Figure 3. Kaplan-Meier survival analysis by co-evaluating histological grade with clinical and molecular variables.
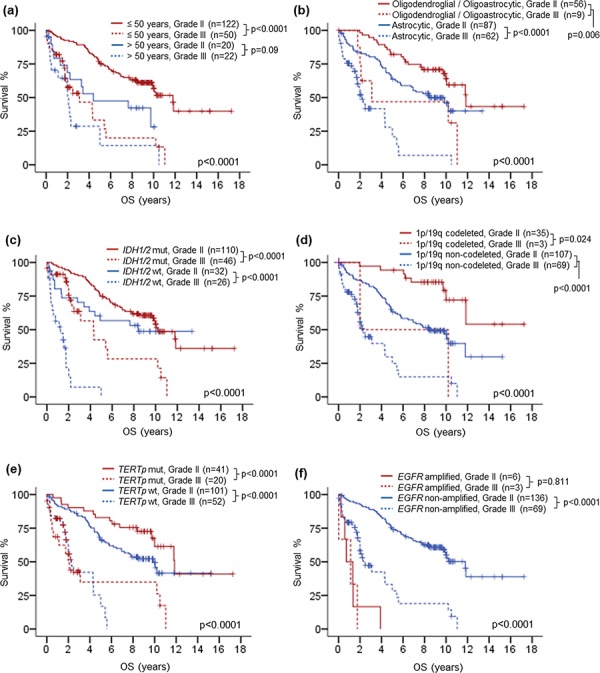
Histological grade showed prognostic relevance in patients of 50 years or younger and trend in patients above 50 years a. Prognostic relevance of histological grade was demonstrated in both astrocytic gliomas and oligodendroglial / oligoastrocytic gliomas b. Prognostic value of histological grade was independent of IDH1/2 mutation status c. 1p/19q codeletion status d. and TERTp mutation status e. No prognostic association was found for histological grade among EGFR amplified gliomas f. OS, overall survival; mut, mutant; wt, wild-type.
Survival analysis (Genetic signature)
To further interrogate the survival data of our cohort and compare the prognostic value of molecular markers and histological grade, we assigned each tumor into genetic subgroups defined by combination of the molecular markers. We defined tumors with IDH1/2 mutation and either 1p/19q codeletion or TERTp mutation as ‘IDHmut-OT’ gliomas, tumors with IDH1/2 mutation only as ‘IDHmut’ gliomas, tumors with wild-type IDH1/2 and either TERTp mutation or EGFR amplification as ‘IDHwt-ET’ gliomas, and tumors without any of the four molecular markers as ‘IDHwt’ gliomas. As shown in Fig. 4a, tumor subgroups defined by the genetic signature showed significant difference in overall survival across the whole cohort (p < 0.0001). Pair-wise comparison revealed that 57 IDHmut-OT gliomas showed longest overall survival compared to 99 IDHmut gliomas (p < 0.0001), 42 IDHwt gliomas (p = 0.002) and 16 IDHwt-ET gliomas (p < 0.0001). Comparing the survival between 99 IDHmut gliomas and 42 IDHwt gliomas, there was no statistical significance in overall survival between the two groups. Prognostic value of the genetic signature was further analyzed according to histological grade, patient age and tumor histology (Fig. 4b to 4g). The genetic signature showed statistical significance across both grade II (p < 0.0001) and grade III gliomas (p < 0.0001). In the 72 grade III gliomas, 12 IDHmut-OT tumors showed marginally favorable overall survival than 34 IDHmut tumors (p = 0.043), which in turn showed better survival than 18 IDHwt tumors (p = 0.002), followed by 8 IDHwt-ET tumors (p = 0.008) (Fig. 4c). In the 142 grade II gliomas, 45 IDHmut-OT tumors showed a trend of better prognosis than 24 IDHwt tumors in Kaplan-Meier's curve (p = 0.351), which in turn showed a trend of better prognosis than 65 IDHmut tumors (p = 0.097), followed by 8 IDHwt-ET tumors (p = 0.0001) (Fig. 4b). Analyzing according to patient age, the genetic signature showed a better stratification in patients of 50 years or younger (p < 0.0001) than in patients above 50 years (p = 0.0001) (Fig. 4d and 4e). The genetic signature was significant across 149 astrocytic gliomas (p < 0.0001) and marginally significant between 43 IDHmut-OT tumors and 17 IDHmut tumors with oligodendroglial or oligoastrocytic histology (p = 0.045) (Fig. 4f and 4g). In all subgroups of the cohort according to histological grade, age and astrocytic histology, tumors with IDHwt-ET signature showed poorest prognosis.
Figure 4. Kaplan-Meier survival analysis by genetic signature and co-evaluating genetic signature with clinical variables.
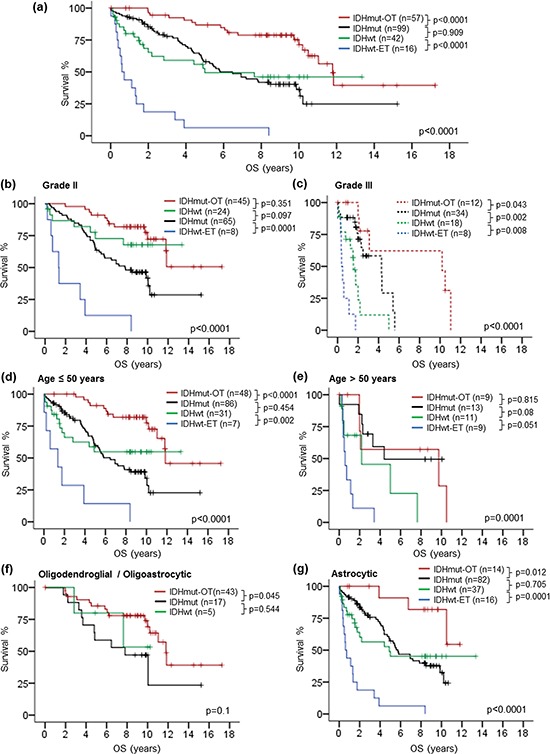
The genetic signature showed prognostic relevance across the whole cohort a. The genetic signature was associated with overall survival in grade II gliomas b. and grade III gliomas c. Prognostic association of the genetic signature was demonstrated in patients at or younger than 50 years d. as well as patients above 50 years e. The genetic signature exhibited prognostic relevance in astrocytic gliomas g. and trend in oligodendroglial / oligoastrocytic gliomas f. OS, overall survival; mut, mutant; wt, wild-type.
Prognostic value of histological grade in each of the genetic signature subgroups was also evaluated in order to investigate the influence of the genetic signature on prognostic value of histological grade (Fig. 5a to 5d). In all genetic signature subgroups, patients with grade II tumors had longer overall survival than those with grade III tumors, suggesting that histological grade was independent with the genetic signature in prognostication. Importantly, 8 grade II IDHwt-ET gliomas showed significantly shorter overall survival than 12 grade III IDHmut-OT gliomas (p = 0.005) and trend of shorter overall survival than 34 grade III IDHmut gliomas (p = 0.207) (Fig. 5e), suggesting that the genetic signature potentially had a more dominant prognostic value than histological grade in prognostication of lower-grade gliomas.
Figure 5. Kaplan-Meier survival analysis of histological grade in each of the genetic signature subgroup.
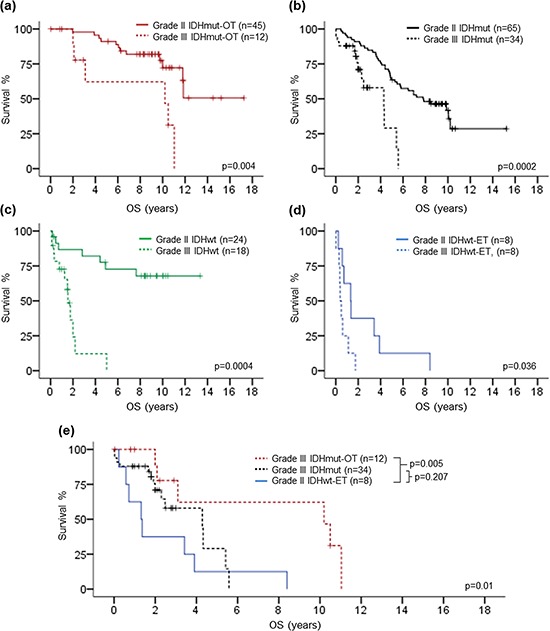
Histological grade showed prognostic association in IDHmut-OT gliomas a. IDHmut gliomas b. IDHwt gliomas c. as well as IDHwt-ET gliomas. Grade II IDHwt-ET gliomas demonstrated shorter overall survival than grade III IDHmut-OT gliomas and trend of shorter overall survival than grade III IDHmut gliomas e. OS, overall survival.
Multivariate analysis was performed by Cox proportional hazards model to evaluate the independent prognostic value of histological grade and genetic signature (Table 4). In the multivariate model adjusting for patient age, tumor histology and operation, both histological grade (p < 0.0001) and genetic signature (p < 0.0001) were independent prognostic factor for overall survival. The hazard ratio of grade III histology to grade II histology was 3.753 (95% CI = 2.214–6.363). For genetic signature, hazard ratio was greatest for IDHwt-ET tumors to IDHmut-OT tumors (HR = 19.491, 95% CI = 7.605–49.955, p < 0.0001), followed by IDHmut tumors to IDHmut-OT tumors (HR = 3.417, 95% CI = 1.704–6.852, p = 0.0005) and IDHwt tumors to IDHmut-OT tumors (HR = 2.958, 95% CI = 1.309–6.681, p = 0.009).
Table 4. Multivariate analysis of overall survival.
| HR | [95% CI] | p | |
|---|---|---|---|
| Age | 1.027 | [1.005–1.050] | 0.015 |
| Histological type | 0.639 | ||
| Astrocytic | 1 | ||
| Oligodendroglial / Oligoastrocytic | 0.869 | [0.485–1.560] | |
| Operation | <0.0001 | ||
| Total resection | 0.382 | [0.248–0.589] | |
| Non-total resection | 1 | ||
| Histological grade | <0.0001 | ||
| Grade II | 1 | ||
| Grade III | 3.753 | [2.214–6.363] | |
| Genetic signature | <0.0001 | ||
| IDHmut-OT | 1 | ||
| IDHmut | 3.417 | [1.704–6.852] | 0.0005 |
| IDHwt | 2.958 | [1.309–6.681] | 0.009 |
| IDHwt-ET | 19.491 | [7.605–49.955] | <0.0001 |
HR, hazard ratio; 95% CI, 95% confidence interval; IDHmut-OT, IDH1/2 mutated and either 1p/19q codeleted or TERTp mutated; IDHmut, IDH1/2 mutated only; IDHwt, IDH1/2 wild-type, TERTp wild-type and EGFR non-amplified; IDHwt-ET, IDH1/2 wild-type and either TERTp mutated or EGFR amplified.
DISCUSSION
Our study evaluated the influence of IDH1/2 mutation, 1p/19q codeletion, TERTp mutation and EGFR amplification in the prognostic value of histological grade in a cohort of 214 cases of lower-grade gliomas. Co-evaluation of histological grade and molecular markers demonstrated the prognostic association of grading in lower-grade gliomas irrespective of the status of IDH1/2, 1p/19q and TERTp. Notably, no single molecular marker, including IDH1/2 mutation, superseded histological grading in prognostication. IDH1 mutation was found to have more dominant prognostic effect than histological grade in anaplastic astrocytomas and glioblastomas [27]. Our study provided complementary results as to the prognostic role of IDH1 mutation relative to histological grade in lower-grade gliomas. A prognostic genetic signature was further constructed based on the biomarkers. Most favorable prognosis was observed in IDHmut-OT gliomas (median OS = 11.8 years), followed by IDHmut gliomas (median OS = 5.9 years) and IDHwt gliomas (median OS = 5 years), and IDHwt-ET gliomas showed the poorest prognosis (median OS = 0.6 year). Multivariate analysis further demonstrated the independent prognostic value of histological grading and genetic signature in lower-grade gliomas. Although histological grade exhibited prognostic association in each of the genetic subgroups, the overall survival of grade II IDHwt-ET gliomas was shorter than grade III gliomas harboring IDH1/2 mutation, signifying combination genetic signature could potentially supersede histological grade in prognostication of lower-grade gliomas.
Prognostic stratification of diffuse gliomas by combinations of molecular markers was demonstrated by other research groups but the relative prognostic relevance between histological grading and marker combinations has not been intensively studied. Jiao and colleagues examined a cohort of grades II to IV diffuse gliomas for IDH, ATRX, TP53, CIC and FUBP1 mutations as well as 1p/19q codeletion and classified the tumors into ‘I-CF’ (IDH1/2 mutation with either 1p/19q loss, CIC mutation or FUBP1 mutation), ‘I-A’ (IDH1/2 mutation and ATRX mutation) and ‘I-X’ gliomas (neither I-CF nor I-A). I-CF group was associated with most favorable prognosis and I-X group was associated with poorest prognosis, with I-A group being in between. This I-CF/I-A/I-X genetic signature exhibited a better prognostic classification than histopathology in their cohort of grade III gliomas [34]. In another study, Wiestler and colleagues defined ‘molecular astrocytoma’, ‘molecular oligodendroglioma’ and ‘molecular glioblastoma’ by combinations of status of IDH1/2, 1p/19q and ATRX immunohistochemical expression and demonstrated that loss of ATRX expression refined classification of anaplastic gliomas by identifying a subgroup of IDH mutated anaplastic astrocytomas with better prognosis [35]. Mur and colleagues analyzed IDH1/2 mutation, 1p/19q codeletion and ATRX expression and categorized the tumors into ‘I-CD’ (IDH1/2 mutation, 1p/19q codeletion and ATRX positive expression), ‘I-A’ (IDH1/2 mutation, absence of ATRX expression and intact 1p/19q), ‘I’ (IDH1/2 mutation, intact 1p/19q and positive ATRX expression) and ‘NA’ (wild-type IDH1/2, intact 1p/19q, and positive ATRX expression) molecular groups. They showed that such molecular classification defined four prognostically distinct glioma groups irrespective of tumor diagnosis and grade [36]. On the other hand, combination of TERTp mutation with IDH1/2 mutation was also shown to refine prognostic stratification of diffuse gliomas by several studies [19, 20, 23]. In these studies, four distinct prognostic groups were outlined by the two markers with IDH1/2 mutated / TERTp mutated tumors having the most favorable survival, followed by IDH1/2 mutated / TERTp wild-type tumors and IDH1/2 wild-type / TERTp wild-type tumors, and IDH1/2 wild-type / TERTp mutated tumors exhibiting the worst survival [19, 20, 23]. Our current study combined IDH1/2 mutated tumors with either 1p/19q codeletion or TERTp mutation as IDHmut-OT gliomas since both of the markers were closely associated with oligodendroglial differentiation with the presence of IDH1/2 mutation [19, 20, 23, 25, 26]. This combination allowed us to identify ‘molecular oligodendrogliomas’ with favorable prognosis, of which 9.4% of astrocytomas, 87.5% of oligodendrogliomas and 53.7% of oligoastrocytomas harbored the IDHmut-OT genetic signature. On the other end, IDH1/2 wild-type tumors with either TERTp mutation or EGFR amplification were defined as IDHwt-ET gliomas. Wild-type IDH1/2 and mutated TERTp were identified in up to 80% of glioblastomas and was associated with poor prognosis in patients with the grade IV tumors [20, 22, 24]. A recent study by Labussiere and colleagues also demonstrated that combined analysis of TERTp, EGFR and IDH1/2 defined distinct prognostic classes in glioblastomas, with the absence of both EGFR amplification and TERTp mutation associating with longer survival in patients with glioblastomas [22]. The prognostic value of IDHwt-ET genetic signature in lower-grade gliomas in our cohort was consistent with these studies. Patients with IDHwt-ET gliomas exhibited a similarly poor outcome (median OS = 0.6 year) compared with other lower-grade gliomas, suggesting that these patients may require more follow-up and treatment. Importantly, grade II IDHwt-ET gliomas had poorer prognosis than grade III IDHmut-OT gliomas and grade III IDHmut gliomas, indicating that IDHwt-ET genetic signature potentially supersedes histological grade in prognostication of lower-grade gliomas. Evaluation of EGFR amplification and TERTp mutation in a background of wild-type IDH1/2 in lower-grade gliomas could help to identify aggressive tumors with seemingly low grade histology, facilitating a more precise prognostic estimation. Nevertheless, histological grade showed prognostic relevance in each genetic signature subgroup, suggesting its irreplaceable role in this biomarker-based stratification.
In our cohort, IDHmut gliomas and IDHwt gliomas demonstrated intermediate prognosis and there was no significant prognostic difference between these two groups. While IDH1/2 mutation showed a wide prognostic split across the whole cohort (Fig. 1d), the lack of prognostic difference between IDHmut gliomas and IDHwt gliomas could be partially due to the elimination of the favorable prognostic effect of IDHmut-OT gliomas from the IDH1/2 mutated tumors as well as the unfavorable prognostic effect of IDHwt-ET gliomas from the IDH1/2 wild-type tumors, therefore bringing the two survival curves overlapping with each other (Fig. 4a). Alternatively, our data suggested that there existed an under-characterized subset of IDH1/2 wild-type lower-grade gliomas with comparable survival to IDH1/2 mutated gliomas lacking 1p/19q codeletion and TERTp mutation. The data further suggested that not all IDH1/2 wild-type lower grade gliomas are uniformly aggressive and additional markers including TERTp mutation and EGFR amplification should be examined in the clinical management of lower-grade gliomas with wild-type IDH1/2. This was corroborated by two recent studies by Weller et al. and Olar et al. [37, 38]. Weller and colleagues conducted comprehensive genomic and transcriptomic profiling in grades II and III gliomas to determine the prognostic utility of molecular profiling in the tumor entity. The authors demonstrated the existence of a genomic group of IDH1/2 wild-type lower-grade gliomas (Group IV) lacking glioblastoma-like genomic aberrations such as 7q gains and 10q losses. This group of tumors had intermediate prognosis as IDH1/2 mutated tumors lacking 1p/19q codeletion (Group II and III), indicating that merely wild-type IDH1/2 status may not be invariably associated with poor clinical outcome and additional glioblastoma-like molecular alterations are required to prognosticate a particularly unfavorable survival [38]. Olar and colleagues investigated a large cohort of lower-grade gliomas to determine the prognostic role of histological grade and mitotic index following stratification by IDH1/2 mutation. The authors showed that combining IDH1/2 mutation, 1p/19q codeletion and mitotic index (4/1000 tumor cells as cut-off) could stratify lower-grade gliomas into four prognostic groups, with 1p/19q codeletion identified tumors with better prognosis in IDH1/2 mutated group and mitotic index >4 identified tumors with poorer prognosis in IDH1/2 wild-type group [37]. Notably, there was a portion of IDH1/2 wild-type / mitotic index = 0–4 tumors showing comparable survival with IDH1/2 wild type / 1p/19q non-codeleted tumors. The results provided further evidence that additional investigations should be carried out in lower-grade gliomas with wild-type IDH1/2 for better prognostication.
The prognosis of IDHwt-ET lower-grade gliomas was poor and comparable to glioblastomas. Among the 16 IDHwt-ET gliomas, nine cases harbored EGFR amplification as demonstrated by FISH (Supplementary Fig. S1 & Table S1). The definitions of EGFR amplification varied in different cancer types and even within diffuse gliomas, the cut-off criteria varied in different studies [31, 39–47]. This was partially because the pattern of EGFR gain or amplification could not be seen normally in non-neoplastic brain tissues and therefore the cut-off could not be determined by statistical methods using mean plus three standard deviations as in deletion detection [31, 44, 48, 49]. In this study, all EGFR amplified cases demonstrated a pattern of clusters of tumor cells with high level amplification (Supplementary Fig. S1). Presence of EGFR amplification and involvement of deep midline structures (including corpus callosum, thalamus, basal ganglia, ventricles and midbrain) in four cases suggested the possibility that these tumors are under-sampled high-grade gliomas [43, 50]. Nevertheless, examination of the biomarkers represented a satisfactory way of removing sampling issues in these aggressive tumors. We also reviewed the pre-operative radiological reports still available in our hospital system in four available cases and contrast enhancement was demonstrated in one anaplastic astrocytoma while others presented as hypodense lesions. Although we could not identify any concrete evidence within the available retrospective clinical data that the EGFR amplified tumors involved more than two cerebral lobes with extraordinary diffuse infiltration, i.e. gliomatosis cerebri [51, 52], the possibility that gliomatosis cerebri presented with low grade histology but harboring high grade molecular aberrations could not be excluded. Notably, EGFR amplification was not detected in two large gliomatosis cerebri series [53, 54], though 7q gain as detected by comparative genomic hybridization was found to be a poor prognostic factor in gliomatosis cerebri [55].
In summary, our study demonstrates the independent prognostic values of histological grading and genetic signature based on IDH1/2 mutation, 1p/19q codeletion, TERTp mutation and EGFR amplification. The biomarker-based stratification provides complementary prognostic information potentially superseding histology and refines the diagnostic and prognostic classification of lower-grade gliomas.
MATERIALS AND METHODS
Patients, tissue samples and clinico-pathological data
A total of 214 cases of lower-grade gliomas diagnosed between 1990 and 2012 with formalin-fixed paraffin embedded tissue available were retrieved from the tissue archive of Department of Anatomical and Cellular Pathology, Prince of Wales Hospital (Hong Kong) and Department of Neurosurgery, Huashan Hospital (Shanghai). All tumor sections were stained with haematoxylin and eosin (H&E) and reviewed carefully by two senior neuropathologists (H.K. Ng and Y. Wang). Histologic classification and grading are according to the World Health Organization (WHO) 2007 classification [28]. For diagnosis of Grade III astrocytoma, the criteria were cellularity, atypia and mitosis. For diagnosis of Grade III oligodendroglioma, the criteria are also cellularity, atypia and mitosis, and/or with microvascular proliferation or necrosis. Necrosis and endothelial proliferation are carefully looked for and those cases are excluded. Patient demographics and clinical follow-up data were retrieved from the respective institutional medical record systems. The cohort was overlapped with previous studies [19, 29]. The study was approved by the New Territories East Cluster-Chinese University of Hong Kong Ethics Committee and Shanghai Huashan Hospital Ethics Committee.
Fluorescence in situ hybridization for chromosome 1p/19q codeletion and EGFR amplification
Chromosome 1p and 19q codeletion and EGFR amplification were evaluated by fluorescence in situ hybridization [19, 29, 30]. The loci examined for 1p and 19q were 1p36.3 (RP11–62M23 labeled red) / 1q25.3-q31.1 (RP11–162L13 labeled green) and 19q13.3 (CTD-2571L23 labeled red)/19p12 (RP11–420K14 labeled green), respectively. Locus-specific probes for chromosome 1 and 19 were generated from bacterial artificial chromosome clones using nick translation with the presence of Spectrum Orange deoxyuridine triphosphate (dUTP) or Spectrum Green deoxyuridine triphosphate (dUTP). The labelled probes were then mixed with cot-1 DNA (Life Technologies) in Hybrisol VI solution (Appligene Oncor, Graffenstaden, France). Vysis EGFR/CEP7 FISH Probe Kit (Abbott Laboratories, Illinois, USA) was used to examine EGFR and the loci interrogated were 7p12 (EGFR) / 7p11.1-q11.1 (CEP7). Four-μm thick formalin-fixed, paraffin embedded tissue sections were deparaffinized by xylene and treated with 1 M sodium thiocyanate at 80°C for 10 minutes, followed with tissue digestion by pepsin solution at 37°C for 20 to 30 minutes. Sections were then rinsed in milli-Q water and dehydrated. The FISH probes were applied to the digested tissue sections and denature. The sections were incubated at 37°C overnight for 16 hours. After overnight hybridization, sections were washed in 1.5 M urea in 0.1X saline sodium citrate at 48°C for 30 minutes, followed by 2X saline sodium citrate at 48°C for 5 minutes. Sections were then counterstained with Vectashield mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories) and evaluated under a Zeiss Axioplan fluorescence microscope (Carl Zeiss Microscopy LLC, NY, USA). Fluorescent signals in at least 100 non-overlapping nuclei were evaluated. 1p loss or 19q loss were defined as more than 50% of counted nuclei showing target (red) to reference (green) signal ratio of 1:2. EGFR amplification was defined as more than 5% tumor cells showing target (red) to reference (green) signal >2 [31].
Mutational analysis for IDH1, IDH2 and TERT promoter
Mutations of IDH1, IDH2 and TERTp were detected by direct sequencing as described previously [19, 29, 32, 33]. In brief, tissues from representative area with tumor content of at least 70% were scrapped off from deparaffinized sections and treated in 10 mM Tris-HCl buffer (pH8.5) with proteinase K at a final concentration of 2 μg/μl at 55°C for 2 to 18 hours and then 98°C for 10 minutes. The cell lysate was centrifuged and supernatant was collected and used for subsequent PCR amplification. Primer pairs spanning the mutation hotspots of IDH1 (codon 132), IDH2 (codon 172) and TERTp [chr5, 1,295,228 (C228T) and 1,295,250 (C250T)] were used for PCR amplification and primer sequences were shown in Supplementary Table S2. For PCR of IDH1 and IDH2, DNA amplification was conducted in a 10-μl reaction volume containing 1μl of cell lysate, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2.5 mM MgCl2, 0.2 mM of each deoxyribonucleoside triphosphate, 0.4 μM of each primer and 0.2U of AmpliTaq Gold DNA polymerase (Life Technologies Corporation, Hong Kong), and was incubated at 95°C for 10 minutes, followed by 40–45 cycles of 95°C for 20 seconds, 60°C for 20 seconds and 72°C for 30 seconds, and a final extension of 72°C for 1 minute. For amplification of TERTp, PCR was performed in 10μl reaction mixture containing 1μl of cell lysate, 0.3 mM of each dNTP, 2.5 mM MgCl2, 0.3 μM of each primer and 0.2U of KAPA HiFi HotStart DNA Polymerase (Kapa Biosystem Inc., Wilmington, USA). The reaction mixture was incubated at 95°C for 5 minutes, followed by 40–45 cycles of 98°C for 20 seconds, 68°C for 15 seconds and 72°C for 30 seconds, and a final extension of 72°C for 1 minute. PCR products were then treated with exonuclease I and alkaline phosphatase (Takara, Japan) at 37°C for 15 minutes and then at 80°C for 15 minutes. Sequencing was performed using BigDye Terminator Cycle Sequencing kit v1.1 (Life Technologies). The products were resolved in Genetic Analyzer 3130xl and analyzed by Sequencing Analysis software.
Statistical analysis
Statistical analysis was conducted using IBM SPSS Statistics 20 (IBM Corporation, NY, USA). Association between molecular markers and clinical parameters were examined by Chi square test or Fisher's exact test, whichever was appropriate. Overall survival (OS) was defined as the time between diagnosis and death or last follow-up. Survival curves were constructed by Kaplan-Meier method and survival distribution between groups was compared by Log-rank test. Multivariate analysis was performed by Cox proportional hazards model. P-value of <0.05 (two sided) was considered statistically significant.
SUPPLEMENTARY FIGURE AND TABLES
ACKNOWLEDGMENTS AND FUNDING
This study was supported by the National Science Foundation of China (grant no. 81172412) and Health and Medical Research Fund of Hong Kong (grant no. 02133146).
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Aldape K, Simmons ML, Davis RL, Miike R, Wiencke J, Barger G, Lee M, Chen P, Wrensch M. Discrepancies in diagnoses of neuroepithelial neoplasms: the San Francisco Bay Area Adult Glioma Study. Cancer. 2000;88:2342–2349. [PubMed] [Google Scholar]
- 2.Brat DJ, Prayson RA, Ryken TC, Olson JJ. Diagnosis of malignant glioma: role of neuropathology. J Neurooncol. 2008;89:287–311. doi: 10.1007/s11060-008-9618-1. [DOI] [PubMed] [Google Scholar]
- 3.Kros JM, Gorlia T, Kouwenhoven MC, Zheng PP, Collins VP, Figarella-Branger D, Giangaspero F, Giannini C, Mokhtari K, Mørk SJ, Paetau A, Reifenberger G, van den Bent MJ. Panel review of anaplastic oligodendroglioma from European Organization For Research and Treatment of Cancer Trial 26951: assessment of consensus in diagnosis, influence of 1p/19q loss, and correlations with outcome. J Neuropathol Exp Neurol. 2007;66:545–551. doi: 10.1097/01.jnen.0000263869.84188.72. [DOI] [PubMed] [Google Scholar]
- 4.van den Bent MJ. Interobserver variation of the histopathological diagnosis in clinical trials on glioma: a clinician's perspective. Acta Neuropathol. 2010;120:297–304. doi: 10.1007/s00401-010-0725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourne TD, Schiff D. Update on molecular findings, management and outcome in low-grade gliomas. Nat Rev Neurol. 2010;6:695–701. doi: 10.1038/nrneurol.2010.159. [DOI] [PubMed] [Google Scholar]
- 6.Huse JT, Phillips HS, Brennan CW. Molecular subclassification of diffuse gliomas: seeing order in the chaos. Glia. 2011;59:1190–1199. doi: 10.1002/glia.21165. [DOI] [PubMed] [Google Scholar]
- 7.Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J Neuropathol Exp Neurol. 2005;64:479–489. doi: 10.1093/jnen/64.6.479. [DOI] [PubMed] [Google Scholar]
- 8.Okamoto Y, Di Patre PL, Burkhard C, Horstmann S, Jourde B, Fahey M, Schüler D, Probst-Hensch NM, Yasargil MG, Yonekawa Y, Lütolf UM, Kleihues P, Ohgaki H. Population-based study on incidence, survival rates, and genetic alterations of low-grade diffuse astrocytomas and oligodendrogliomas. Acta Neuropathol. 2004;108:49–56. doi: 10.1007/s00401-004-0861-z. [DOI] [PubMed] [Google Scholar]
- 9.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 10.Louis DN, Perry A, Burger P, Ellison DW, Reifenberger G, von Deimling A, Aldape K, Brat D, Collins VP, Eberhart C, Figarella-Branger D, Fuller GN, Giangaspero F, et al. International Society of Neuropathology-Haarlem Consensus Guidelines, for Nervous System Tumor Classification and Grading. Brain Pathol. 2014;24:429–435. doi: 10.1111/bpa.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cairncross G, Wang M, Shaw E, Jenkins R, Brachman D, Buckner J, Fink K, Souhami L, Laperriere N, Curran W, Mehta M. Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long-term results of RTOG 9402. J Clin Oncol. 2013;31:337–343. doi: 10.1200/JCO.2012.43.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cairncross JG, Wang M, Jenkins RB, Shaw EG, Giannini C, Brachman DG, Buckner JC, Fink KL, Souhami L, Laperriere NJ, Huse JT, Mehta MP, Curran WJ., Jr Benefit from procarbazine, lomustine, and vincristine in oligodendroglial tumors is associated with mutation of IDH. J Clin Oncol. 2014;32:783–790. doi: 10.1200/JCO.2013.49.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartmann C, Hentschel B, Tatagiba M, Schramm J, Schnell O, Seidel C, Stein R, Reifenberger G, Pietsch T, von Deimling A, Loeffler M. Molecular markers in low-grade gliomas: predictive or prognostic? Clin Cancer Res. 2011;17:4588–4599. doi: 10.1158/1078-0432.CCR-10-3194. [DOI] [PubMed] [Google Scholar]
- 14.Jenkins RB, Blair H, Ballman KV, Giannini C, Arusell RM, Law M, Flynn H, Passe S, Felten S, Brown PD, Shaw EG, Buckner JC. A t(1,19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res. 2006;66:9852–9861. doi: 10.1158/0008-5472.CAN-06-1796. [DOI] [PubMed] [Google Scholar]
- 15.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith JS, Perry A, Borell TJ, Lee HK, O'Fallon J, Hosek SM, Kimmel D, Yates A, Burger PC, Scheithauer BW, Jenkins RB. Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J Clin Oncol. 2000;18:636–645. doi: 10.1200/JCO.2000.18.3.636. [DOI] [PubMed] [Google Scholar]
- 17.van den Bent MJ, Brandes AA, Taphoorn MJ, Kros JM, Kouwenhoven MC, Delattre JY, Bernsen HJ, Frenay M, Tijssen CC, Grisold W, Sipos L, Enting RH, French PJ, et al. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol. 2013;31:344–350. doi: 10.1200/JCO.2012.43.2229. [DOI] [PubMed] [Google Scholar]
- 18.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan AK, Yao Y, Zhang Z, Chung NY, Liu JS, Li KK, Shi Z, Chan DT, Poon WS, Zhou L, Ng HK. TERT promoter mutations contribute to subset prognostication of lower-grade gliomas. Mod Pathol. 2015;28:177–186. doi: 10.1038/modpathol.2014.94. [DOI] [PubMed] [Google Scholar]
- 20.Killela PJ, Pirozzi CJ, Healy P, Reitman ZJ, Lipp E, Rasheed BA, Yang R, Diplas BH, Wang Z, Greer PK, Zhu H, Wang CY, Carpenter AB, et al. Mutations in IDH1, IDH2, and in the TERT promoter define clinically distinct subgroups of adult malignant gliomas. Oncotarget. 2014;5:1515–1525. doi: 10.18632/oncotarget.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, Jr, Friedman AH, Friedman H, Gallia GL, Giovanella BC, Grollman AP, He TC, He Y, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Labussière M, Boisselier B, Mokhtari K, Di Stefano AL, Rahimian A, Rossetto M, Ciccarino P, Saulnier O, Paterra R, Marie Y, Finocchiaro G, Sanson M. Combined analysis of TERT, EGFR, and IDH status defines distinct prognostic glioblastoma classes. Neurology. 2014;83:1200–1206. doi: 10.1212/WNL.0000000000000814. [DOI] [PubMed] [Google Scholar]
- 23.Labussière M, Di Stefano AL, Gleize V, Boisselier B, Giry M, Mangesius S, Bruno A, Paterra R, Marie Y, Rahimian A, Finocchiaro G, Houlston RS, Hoang-Xuan K, et al. TERT promoter mutations in gliomas, genetic associations and clinico-pathological correlations. Br J Cancer. 2014;111:2024–2032. doi: 10.1038/bjc.2014.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nonoguchi N, Ohta T, Oh JE, Kim YH, Kleihues P, Ohgaki H. TERT promoter mutations in primary and secondary glioblastomas. Acta Neuropathol. 2013;126:931–937. doi: 10.1007/s00401-013-1163-0. [DOI] [PubMed] [Google Scholar]
- 25.Arita H, Narita Y, Fukushima S, Tateishi K, Matsushita Y, Yoshida A, Miyakita Y, Ohno M, Collins VP, Kawahara N, Shibui S, Ichimura K. Upregulating mutations in the TERT promoter commonly occur in adult malignant gliomas and are strongly associated with total 1p19q loss. Acta Neuropathol. 2013;126:267–276. doi: 10.1007/s00401-013-1141-6. [DOI] [PubMed] [Google Scholar]
- 26.Koelsche C, Sahm F, Capper D, Reuss D, Sturm D, Jones DT, Kool M, Northcott PA, Wiestler B, Böhmer K, Meyer J, Mawrin C, Hartmann C. Distribution of TERT promoter mutations in pediatric and adult tumors of the nervous system. Acta Neuropathol. 2013;126:907–915. doi: 10.1007/s00401-013-1195-5. [DOI] [PubMed] [Google Scholar]
- 27.Hartmann C, Hentschel B, Wick W, Capper D, Felsberg J, Simon M, Westphal M, Schackert G, Meyermann R, Pietsch T, Reifenberger G, Weller M, Loeffler M, et al. Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta Neuropathol. 2010;120:707–718. doi: 10.1007/s00401-010-0781-z. [DOI] [PubMed] [Google Scholar]
- 28.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. WHO Classification of Tumours of the Central Nervous System. International Agency for Research on Cancer; Lyon: 2007. pp. 14–67. [Google Scholar]
- 29.Chan AK, Pang JC, Chung NY, Li KK, Poon WS, Chan DT, Shi Z, Chen L, Zhou L, Ng HK. Loss of CIC and FUBP1 expressions are potential markers of shorter time to recurrence in oligodendroglial tumors. Mod Pathol. 2014;27:332–342. doi: 10.1038/modpathol.2013.165. [DOI] [PubMed] [Google Scholar]
- 30.Dong Z, Pang JS, Ng MH, Poon WS, Zhou L, Ng HK. Identification of two contiguous minimally deleted regions on chromosome 1p36.31-p36.32 in oligodendroglial tumours. Br J Cancer. 2004;91:1105–1111. doi: 10.1038/sj.bjc.6602093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horbinski C, Miller CR, Perry A. Gone FISHing: clinical lessons learned in brain tumor molecular diagnostics over the last decade. Brain Pathol. 2011;21:57–73. doi: 10.1111/j.1750-3639.2010.00453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yao Y, Chan AK, Qin ZY, Chen LC, Zhang X, Pang JC, Li HM, Wang Y, Mao Y, Ng HK, Zhou LF. Mutation analysis of IDH1 in paired gliomas revealed IDH1 mutation was not associated with malignant progression but predicted longer survival. PLoS One. 2013;8:e67421. doi: 10.1371/journal.pone.0067421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang ZY, Chan AK, Ng HK, Ding XJ, Li YX, Shi ZF, Zhu W, Zhong P, Wang Y, Mao Y, Yao Y, Zhou LF. Surgically treated incidentally discovered low-grade gliomas are mostly IDH mutated and 1p19q co-deleted with favorable prognosis. Int J Clin Exp Pathol. 2014;7:8627–8636. [PMC free article] [PubMed] [Google Scholar]
- 34.Jiao Y, Killela PJ, Reitman ZJ, Rasheed AB, Heaphy CM, de Wilde RF, Rodriguez FJ, Rosemberg S, Oba-Shinjo SM, Nagahashi Marie SK, Bettegowda C, Agrawal N, Lipp E, et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3:709–722. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiestler B, Capper D, Holland-Letz T, Korshunov A, von Deimling A, Pfister SM, Platten M, Weller M, Wick W. ATRX loss refines the classification of anaplastic gliomas and identifies a subgroup of IDH mutant astrocytic tumors with better prognosis. Acta Neuropathol. 2013;126:443–451. doi: 10.1007/s00401-013-1156-z. [DOI] [PubMed] [Google Scholar]
- 36.Mur P, Mollejo M, Hernández-Iglesias T, de Lope ÁR, Castresana JS, García JF, Fiaño C, Ribalta T, Rey JA, Meléndez B. Molecular classification defines 4 prognostically distinct glioma groups irrespective of diagnosis and grade. J Neuropathol Exp Neurol. 2015;74:241–249. doi: 10.1097/NEN.0000000000000167. [DOI] [PubMed] [Google Scholar]
- 37.Olar A, Wani KM, Alfaro-Munoz KD, Heathcock LE, van Thuijl HF, Gilbert MR, Armstrong TS, Sulman EP, Cahill DP, Vera-Bolanos E, Yuan Y, Reijneveld JC, Ylstra B, et al. IDH mutation status and role of WHO grade and mitotic index in overall survival in grade II-III diffuse gliomas. Acta Neuropathol. 2015;129:585–596. doi: 10.1007/s00401-015-1398-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weller M, Weber RG, Willscher E, Riehmer V, Hentschel B, Kreuz M, Felsberg J, Beyer U, Löffler-Wirth H, Kaulich K, Steinbach JP, Hartmann C, Gramatzki D, et al. Molecular classification of diffuse cerebral WHO grade II/III gliomas using genome- and transcriptome-wide profiling improves stratification of prognostically distinct patient groups. Acta Neuropathol. 2015;129:679–93. doi: 10.1007/s00401-015-1409-0. [DOI] [PubMed] [Google Scholar]
- 39.Nakazawa K, Dobashi Y, Suzuki S, Fujii H, Takeda Y, Ooi A. Amplification and overexpression of c-erbB-2, epidermal growth factor receptor, and c-met in biliary tract cancers. J Pathol. 2005;206:356–65. doi: 10.1002/path.1779. [DOI] [PubMed] [Google Scholar]
- 40.Reis-Filho JS, Pinheiro C, Lambros MB, Milanezi F, Carvalho S, Savage K, Simpson PT, Jones C, Swift S, Mackay A, Reis RM, Hornick JL, Pereira EM, et al. EGFR amplification and lack of activating mutations in metaplastic breast carcinomas. J Pathol. 2006;209:445–53. doi: 10.1002/path.2004. [DOI] [PubMed] [Google Scholar]
- 41.Lee HJ, Seo AN, Kim EJ, Jang MH, Kim YJ, Kim JH, Kim SW, Ryu HS, Park IA, Im SA, Gong G, Jung KH, Kim HJ, Park SY. Prognostic and predictive values of EGFR overexpression and EGFR copy number alteration in HER2-positive breast cancer. Br J Cancer. 2015;112:103–11. doi: 10.1038/bjc.2014.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stahl P, Seeschaaf C, Lebok P, Kutup A, Bockhorn M, Izbicki JR, Bokemeyer C, Simon R, Sauter G, Marx AH. Heterogeneity of amplification of HER2, EGFR, CCND1 and MYC in gastric cancer. BMC Gastroenterol. 2015;15:7. doi: 10.1186/s12876-015-0231-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith JS, Tachibana I, Passe SM, Huntley BK, Borell TJ, Iturria N, O'Fallon JR, Schaefer PL, Scheithauer BW, James CD, Buckner JC, Jenkins RB. PTEN mutation, EGFR amplification, and outcome in patients with anaplastic astrocytoma and glioblastoma multiforme. J Natl Cancer Inst. 2001;93:1246–56. doi: 10.1093/jnci/93.16.1246. [DOI] [PubMed] [Google Scholar]
- 44.Korshunov A, Sycheva R, Golanov A. Molecular stratification of diagnostically challenging high-grade gliomas composed of small cells: the utility of fluorescence in situ hybridization. Clin Cancer Res. 2004;10:7820–6. doi: 10.1158/1078-0432.CCR-04-0710. [DOI] [PubMed] [Google Scholar]
- 45.Pollack IF, Hamilton RL, James CD, Finkelstein SD, Burnham J, Yates AJ, Holmes EJ, Zhou T, Finlay JL, Children's Oncology Group Rarity of PTEN deletions and EGFR amplification in malignant gliomas of childhood: results from the Children's Cancer Group 945 cohort. J Neurosurg. 2006;105:418–24. doi: 10.3171/ped.2006.105.5.418. [DOI] [PubMed] [Google Scholar]
- 46.Faulkner C, Palmer A, Williams H, Wragg C, Haynes HR, White P, DeSouza RM, Williams M, Hopkins K, Kurian KM. EGFR, and EGFRvIII analysis in glioblastoma as therapeutic biomarkers. Br J Neurosurg. 2014 Aug 20; doi: 10.3109/02688697.2014.950631. [DOI] [PubMed] [Google Scholar]
- 47.Xie D, Zeng YX, Wang HJ, Tai LS, Wen JM, Tao Y, Ma NF, Hu L, Sham JS, Guan XY. Amplification and overexpression of epidermal growth factor receptor gene in glioblastomas of Chinese patients correlates with patient's age but not with tumor's clinicopathological pathway. Acta Neuropathol. 2005;110:481–9. doi: 10.1007/s00401-005-1072-y. [DOI] [PubMed] [Google Scholar]
- 48.Scheie D, Andresen PA, Cvancarova M, Bø AS, Helseth E, Skullerud K, Beiske K. Fluorescence in situ hybridization (FISH) on touch preparations: a reliable method for detecting loss of heterozygosity at 1p and 19q in oligodendroglial tumors. Am J Surg Pathol. 2006;30:828–37. doi: 10.1097/01.pas.0000213250.44822.2e. [DOI] [PubMed] [Google Scholar]
- 49.Woehrer A, Sander P, Haberler C, Kern S, Maier H, Preusser M, Hartmann C, Kros JM, Hainfellner JA. FISH-based detection of 1p 19q codeletion in oligodendroglial tumors: procedures and protocols for neuropathological practice - a publication under the auspices of the Research Committee of the European Confederation of Neuropathological Societies (Euro-CNS) Clin Neuropathol. 2011;30:47–55. doi: 10.5414/npp30047. [DOI] [PubMed] [Google Scholar]
- 50.Horbinski C. Practical molecular diagnostics in neuropathology: making a tough job a little easier. Semin Diagn Pathol. 2010;27:105–13. doi: 10.1053/j.semdp.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 51.Fuller GN, Kros JM. Gliomatosis cerebri. In: Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, editors. WHO Classification of Tumours of the Central Nervous System. Lyon: IARC; 2007. pp. 50–52. [Google Scholar]
- 52.Jennings MT, Frenchman M, Shehab T, Johnson MD, Creasy J, LaPorte K, Dettbarn WD. Gliomatosis cerebri presenting as intractable epilepsy during early childhood. J Child Neurol. 1995;10:37–45. doi: 10.1177/088307389501000111. [DOI] [PubMed] [Google Scholar]
- 53.Herrlinger U, Felsberg J, Küker W, Bornemann A, Plasswilm L, Knobbe CB, Strik H, Wick W, Meyermann R, Dichgans J, Bamberg M, Reifenberger G, Weller M. Gliomatosis cerebri: molecular pathology and clinical course. Ann Neurol. 2002;52:390–9. doi: 10.1002/ana.10297. [DOI] [PubMed] [Google Scholar]
- 54.Mawrin C, Kirches E, Schneider-Stock R, Boltze C, Vorwerk CK, von Mawrin A, Kirches E, Schneider-Stock R, Boltze C, Vorwerk CK, von Mawrin A, Kirches E, Schneider-Stock R, et al. Alterations of cell cycle regulators in gliomatosis cerebri. J Neurooncol. 2005;72:115–22. doi: 10.1007/s11060-004-2061-z. [DOI] [PubMed] [Google Scholar]
- 55.Ware ML, Hirose Y, Scheithauer BW, Yeh RF, Mayo MC, Smith JS, Chang S, Cha S, Tihan T, Feuerstein BG. Genetic aberrations in gliomatosis cerebri. Neurosurgery. 2007;60:150–8. doi: 10.1227/01.NEU.0000249203.73849.5D. discussion 158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


