Abstract
We recently reported a specific mechanism that RAD54B, an important factor in homologous recombination, promotes genomic instability via the degradation of p53 protein in vitro. However, clinical significance of RAD54Bin colorectal cancer (CRC) remains unclear. Thus we analyzed RAD54B geneexpression in CRC patients. Using the training set (n = 123), the optimal cut-off value for stratification was determined, and validated in another cohort (n = 89). Kaplan–Meier plots showed that distant recurrence free survival was significantly lesser in high RAD54B expression group compared with that of low expression group in both training (P = 0.0013) and validation (P = 0.024) set. Multivariate analysis using Cox proportional-hazards model showed that high RAD54B expression was an independent predictor in both training (hazard ratio, 4.31; 95% CI, 1.53–13.1; P = 0.0060) and validation (hazard ratio, 3.63; 95% CI, 1.23–10.7; P = 0.021) set. In addition, a negative significant correlation between RAD54B and CDKN1A, a target gene of p53, was partially confirmed, suggesting that RAD54B functions via the degradation of p53 protein even in clinical samples. This study first demonstrated RAD54B expression has potential to serve as a novel prognostic biomarker, particularly for distant recurrence in CRC patients.
Keywords: RAD54B, colorectal cancer, homologous recombination, prognosis, distant recurrence
INTRODUCTION
Despite the recent progress in the therapeutic modalities, colorectal cancer (CRC) is a serious public health issue worldwide owing to its high incidence and cancer-related mortality [1]. In addition to TNM staging system, several biomarkers are currently applied to practical use for CRC patients. For example, UGT1A1 genetic polymorphism is associated with CPT-11 toxicity [2]. KRAS/NRAS mutations status predicts response to anti-EGFR antibody therapy [3, 4]. CRC with microsatellite instability has a better prognosis compared to CRC with intact mismatch repair [5]. Such biomarkers have improved clinical outcomes in CRC, however, the number of biomarkers still remains insufficient, and more studies exploring new biomarkers are required to establish further personalized therapeutic strategy.
Human RAD54B was first identified in 1999 as a homolog of RAD54. RAD54B belongs to SNF2/SWI2 superfamily, and biochemical studies have shown that RAD54B is involved in the homologous recombination (HR) [6-9]. HR is one of the most important DNA double-strand break repair pathway. It is generally accepted that imbalanced regulation of the HR system is associated with genomic instability, resulting in carcinogenesis and malignant tumor development [10-12].
Recently, our group found another mechanism different from the HR repair pathway in multiple cell line experiments, including the HCT116 colon cancer cell line. RAD54B is associated with cell cycle control and functions as a scaffold for p53 degradation through its direct interaction with the MDM2–MDMX ubiquitin–ligase complex, and constitutive upregulation of RAD54B activity promotes genomic instability [13]. Although one study concerning the effectiveness of capecitabine and concurrent radiation therapy for glioblastoma analyzed the expression profiles of eight genes including RAD54B and reported that high RAD54B expression was associated with a poor outcome [14], the clinical significance of RAD54B expression remains unknown particularly in CRC.
Therefore, we examined RAD54B expression in surgically resected CRC tissues by real-time PCR method and analyzed the correlation with various clinicopathological factors and patient's prognosis. In addition to RAD54B, we also analyzed RAD51 expression in the same cohort because RAD51 is another important factor involved in HR process [15-18] and several studies reported that RAD51 protein overexpression is associated with poor prognosis in various cancers [19-24]. Moreover, we measured CDKN1A (p21/WAF1) expression, a target gene of p53, and analyzed the association between RAD54B and CDKN1A expression to investigate the biological role of RAD54B in clinical samples. From a clinical perspective, this is the first study demonstrating the utility of RAD54B as a prognostic biomarker in CRC patients.
RESULTS
RAD54B expression was elevated in most CRC tissues
We first analyzed RAD54B and RAD51 expression in the training set (n = 123). Gene expression of RAD54B, RAD51 were quantified by real-time PCR as described in the Materials and Methods section. Table 1 shows the summary of RAD54B and RAD51 expression values in the training set. The median (inter-quartile range) RAD54B expression value was 2.60 (2.50–3.99), and RAD54B expression values were higher than 1.00 in 116 of the 123 samples (94.3%), indicating that RAD54B expression was elevated in most CRC tissues compared with corresponding normal mucosa. In contrast, RAD51 expression value [1.15 (0.80–1.56)] was not as elevated as RAD54B. The number of samples with values higher than 1.00 was 74 of the 123 samples (60.2%), which was significantly smaller than that of RAD54B (P = 0.0008).
Table 1. RAD54B and RAD51 expression values in the training set (n = 123).
| Category | No. | RAD54B value | No. | RAD51 value | P value |
|---|---|---|---|---|---|
| All patients | 2.60 (2.50-3.99) | 1.15 (0.80-1.56) | |||
| Expression value <1.00 | 7 (5.69%) | 49 (39.8%) | 0.0008 | ||
| Expression value ≥1.00 | 116 (94.3%) | 74 (60.2%) | |||
RAD54B and RAD51 expression values and clinicopathological factors in the training set
We next analyzed the relationship between RAD54B or RAD51 expression and clinicopathological features in the training set. As shown in Table 2, no statistically significant correlations were found between RAD54B expression values and clinicopathological factors, such as sex, age, tumor location, tumor size, cell differentiation, lymphatic invasion, venous invasion, and preoperative CEA and CA19-9 levels. Similarly, no significant correlations were detected between RAD54B expression values and UICC stage (P = 0.20), T stage (P = 0.16), and N stage (P = 0.23). Likewise, there was no association between RAD51 expression values and clinicopathological factors including UICC stage (P = 0.078), T stage (P = 0.51), and N stage (P = 0.38), except that RAD51 expression values in lymphatic invasion-positive patients were significantly higher than those in negative patients (P = 0.027).
Table 2. Clinicopathological factors and RAD54B and RAD51 expression values in the training set (n = 123).
| Category | No (%) | RAD54B value | P value | RAD51 value | P value | |
|---|---|---|---|---|---|---|
| Sex | Male | 65 (52.8) | 2.46 (1.52-3.60) | 0.11 | 1.02 (0.71-1.50) | 0.65 |
| Female | 58 (47.2) | 2.78 (2.16-4.16) | 1.23 (0.84-1.62) | |||
| Age | <65 | 56 (45.5) | 2.72 (1.97-4.19) | 0.91 | 1.14 (0.81-1.56) | 0.97 |
| ≥65 | 67 (54.5) | 2.50 (2.10-3.65) | 1.15 (0.77-1.57) | |||
| Tumor location | Colon | 96 (78.0) | 2.69 (2.05-4.22) | 0.28 | 1.19 (0.79-1.58) | 0.34 |
| Rectum | 27 (22.0) | 2.42 (1.97-3.40) | 1.02 (0.80-1.48) | |||
| Tumor size (mm) | <50 | 66 (53.7) | 2.48 (2.05-3.24) | 0.12 | 1.17 (0.80-1.53) | 0.93 |
| ≥50 | 57 (46.3) | 2.88 (1.98-1.73) | 1.05 (0.78-1.58) | |||
| Cell differentiation | WD | 57 (46.3) | 2.48 (1.98-3.33) | 0.42 | 1.20 (0.79-1.56) | 0.68 |
| MD | 62 (50.4) | 2.62 (2.08-4.33) | 1.11 (0.78-1.57) | |||
| PD | 4 (3.3) | 4.47 (1.97-5.40) | 1.50 (0.82-2.74) | |||
| Lymphatic invasion | Positive | 47 (38.2) | 2.66 (1.57-4.57) | 0.92 | 1.35 (0.97-1.79) | 0.027 |
| Negative | 76 (61.8) | 2.56 (2.14-3.69) | 1.01 (0.79-1.48) | |||
| Venous invasion | Positive | 27 (22.0) | 2.58 (2.00-4.12) | 0.94 | 1.13 (0.80-1.57) | 0.97 |
| Negative | 96 (78.0) | 2.56 (2.05-3.41) | 1.18 (0.79-1.53) | |||
| Preoperative CEA | <5.0 | 61 (49.6) | 2.44 (1.87-4.11) | 0.67 | 1.17 (0.80-1.56 | 0.28 |
| ≥5.0 | 61 (49.6) | 2.72 (2.10-3.82) | 1.14 (0.79-1.56) | |||
| Preoperative CA19-9 | <37 | 99 (80.5) | 2.56 (1.98-3.78) | 0.59 | 1.10 (0.80-1.56) | 0.26 |
| ≥37 | 23 (18.7) | 2.97 (2.10-4.54) | 1.35 (0.69-1.93) | |||
| Tumor stage (UICC) | I | 13 (10.6) | 2.58 (2.13-3.04) | 0.20 | 1.09 (0.76-1.38) | 0.078 |
| II | 51(41.5) | 2.72 (223-4.45) | 1.18 (0.80-1.56) | |||
| III | 44 (35.8) | 2.42 (1.43-3.59) | 1.02 (0.73-1.53) | |||
| IV | 15 (12.2) | 2.98 (2.11-5.26) | 1.56 (1.13-2.67) | |||
| T stage | T1 | 5 (4.1) | 2.21 (1.05-5.31) | 0.16 | 0.68 (0.36-9.42) | 0.51 |
| T2 | 18 (14.6) | 2.76 (2.08-3.03) | 1.09 (0.87-1.40) | |||
| T3 | 60 (48.8) | 2.44 (1.62-3.94) | 1.13 (0.78-1.53) | |||
| T4 | 40 (32.5) | 2.87 (2.18-5.32) | 1.28 (0.81-1.79) | |||
| N stage | N0 | 67 (54.5) | 2.72 (2.21-3.87) | 0.23 | 1.15 (0.80-1.51) | 0.38 |
| N1 | 47 (38.2) | 2.46 (1.53-3.42) | 1.04 (0.78-2.28) | |||
| N2 | 9 (7.3) | 4.54 (1.77-4.00) | 1.39 (1.11-1.76) | |||
| Distant Recurrence in Stage I-III | Yes | 15 (12.2) | 3.87 (2.55-4.88) | 0.031 | 1.16 (0.81-1.79) | 0.69 |
| No | 93 (75.6) | 2.46 (1.92-3.24) | 1.07 (0.78-1.52) | |||
WD = well differentiated; MD = moderately differentiated; PD = poorly differentiated.
RAD54B and RAD51 expression and patient prognosis in the training set
Among the 108 stage I–III CRC patients (excluding stage IV patients) in the training set, 15 patients developed distant recurrence during the median follow-up period of 50.7 (40.9–59.9) months (Table 2). RAD54B expression values in patients developing distant recurrence were significantly higher than those in patients without distant recurrence [3.87 (2.55–4.88) vs. 2.46 1.92–3.24); P = 0.031; Table 2], suggesting that high RAD54B expression is associated with inferior prognostic outcome, specifically by causing postoperative distant metastasis. In contrast, RAD51 expression values were not significantly different between patients with and without distant recurrence [1.16 (0.81–1.79) vs. 1.07 (0.78–1.52); P = 0.69; Table 2]. According to these findings, we selected DRFS as the primary endpoint to further evaluate the prognostic ability of RAD54B.
Clinicopathological features of RAD54B high and low groups in the training set
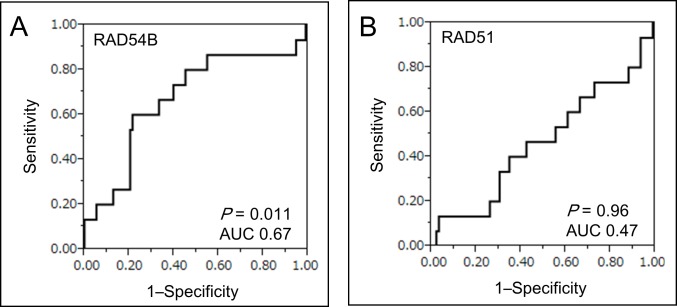
To evaluate the prognostic ability of RAD54B on DRFS, the optimal cut-off value of RAD54B expression for segregating DRFS was determined by receiver operating characteristic curve (ROC) analysis (Figure 1A). With the optimal cut-off value of 3.63, 108 stage I–III CRC patients in the training set were divided into two groups: low group (n = 79) and high group (n = 29). In contrast, we could not determine the optimal cut-off value of RAD51 expression in the training set due to a low area under curve (AUC) score (Figure 1B).
Figure 1. ROC curve analysis of RAD54B and RAD51 expression.
A. ROC curve analysis using RAD54B expression values for distinguishing patients developing distant recurrence in the training set. The estimated optimal cut-off value of RAD54B expression was 3.63 (AUC 0.67, P = 0.011). B. ROC curve analysis using RAD51 expression values for distinguishing patients developing distant recurrence in the training set (AUC 0.47, P = 0.96).
Table 3 demonstrates a comparison of clinicopathological features of the RAD54B high and low groups in the training set (n = 108). Although the RAD54B high group had more advanced T stage (P = 0.027), factors including sex, age, tumor location, tumor size, cell differentiation, lymphatic invasion, venous invasion, preoperative tumor markers, UICC stage, and N stage were similar in both groups. Twenty-four patients (30.4%) in the RAD54B low group and nine patients (31.0%) in the high group received postoperative adjuvant chemotherapy consisting of 5-fluorouracil (5-FU)-based regimens (P = 0.95). During the median follow-up period of 50.7 (40.9–59.9) months, six patients (7.6%) in the RAD54B low group (n = 79) and nine patients (31.0%) in the high group (n = 29) developed distant recurrence (P = 0.0018).
Table 3. Comparison of clinicopathological factors between RAD54B high and low groups.
| Category | Training set (n = 108) | P value | Validation set (n = 71) | P value | |||
|---|---|---|---|---|---|---|---|
| RAD54B Group | RAD54B Group | ||||||
| LOW (n = 79) | High (n = 29) | Low (n = 53) | High (n =18) | ||||
| No (%) | No (%) | No (%) | No (%) | ||||
| Sex | Male | 44 (55.7) | 13 (44.8) | 0.32 | 33 (62.3) | 10 (55.6) | 0.61 |
| Female | 35 (44.3) | 16 (55.2) | 20 (37.7) | 8(44.4) | |||
| Age | <65 | 32 (40.5) | 14 (48.3) | 0.47 | 22 (41.5) | 4 (22.2) | 0.14 |
| ≥65 | 47 (59.5) | 15 (51.7) | 31 (58.5) | 14 (77.8) | |||
| Tumor location | Colon | 58 (73.4) | 26 (89.7) | 0.072 | 37 (69.8) | 12 (66.7) | 0.80 |
| Rectum | 21 (26.6) | 3(10.3) | 16 (30.2) | 6(33.3) | |||
| Tumor size (mm) | <50 | 48 (60.8) | 14 (48.3) | 0.24 | 28 (52.8) | 8 (4.4.4) | 0.54 |
| ≥50 | 31 (39.2) | 15 (51.7) | 25 (47.2) | 10 (55.6) | |||
| Cell differentiation | WD | 41 (51.9) | 12 (41.4) | 0.33 | 26 (49.1) | 6 (33.3) | 0.25 |
| MD/PD | 38 (48.1) | 17 (58.6) | 27 (50.9) | 12 (66.7) | |||
| Lymphatic invasion | Positive | 24 (30.4) | 12 (41.4) | 0.28 | 19 (35.8) | 3 (16.7) | 0.13 |
| Negative | 55 (69.6) | 17 (58.6) | 34 (64.2) | 15 (83.3) | |||
| Venous invasion | Positive | 59 (74.7) | 5 (17.2) | 0.38 | 37 (69.8) | 12 (66.7) | 0.80 |
| Negative | 20 (25.3) | 24 (82.8) | 16 (30.2) | 6(33.3) | |||
| Preoperative CEA | <5.0 | 41 (51.9) | 16 (55.2) | 0.76 | 29 (54.7) | 7(38.9) | 0.25 |
| ≥5.0 | 38 (48.1) | 13 (44.8) | 24 (45.3) | II (61.1) | |||
| Preoperative CA19-9 | <37 | 69 (87.3) | 24 (82.8) | 0.88 | 43(81.1) | 15 (83.3) | 0.83 |
| ≥37 | 10 (12.3) | 5(17.2) | 10 (18.9) | 3(16.7) | |||
| Tumor stage (UICC) | I | 12 (15.2) | 1(3.4) | 0.17 | 7(13.2) | 3(16.7) | 0.88 |
| II | 34 (43.0) | 17 (58.6) | 19 (35.8) | 7 (38.9) | |||
| III | 33 (41.8) | 11 (37.9) | 27 (50.9) | 8(44.4) | |||
| T stage | T1-2 | 21 (26.6) | 2(6.9) | 0.027 | 9(17.0) | 4 (22.2) | 0.62 |
| T3-4 | 58 (73.4) | 27 (93.1) | 44 (83.0) | 14 (77.8) | |||
| N stage | N0 | 46 (58.2) | 18 (62.1) | 0.72 | 26 (49.1) | 11 (61.1) | 0.38 |
| N1-2 | 33 (41.8) | 11(37.9) | 27 (50.9) | 7(38.9) | |||
| Adjuvant chemotherapy | Yes | 24 30.4) | 9 31.0) | 0.95 | 23 (43.4) | 6 (33.3) | 0.45 |
| No | 55 (69.6) | 20 (69.0) | 30 (56.6) | 12 (66.7) | |||
| Distant Recurrence | Yes | 6 (7.6) | 9 (31.0) | 0.0018 | 7 (13.2) | 7 (38.9) | 0.024 |
| No | 73 (92.4) | 20 (69.0) | 46 (86.8) | 11 (61.1) | |||
WD = well differentiated; MD = moderately differentiated; PD = poorly differentiated.
High RAD54B expression is an independent risk factor for distant recurrence
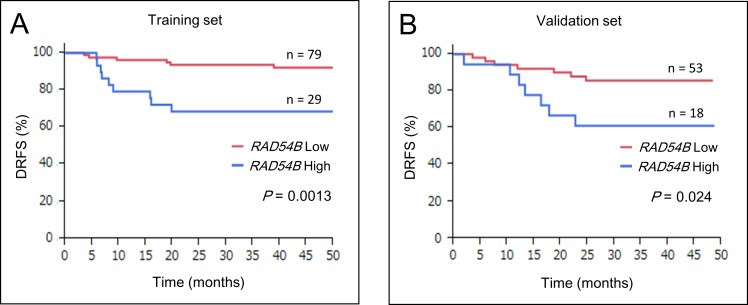
Kaplan–Meier survival analyses of 108 stage I–III CRC patients in the training set revealed that the RAD54B high group had inferior DRFS compared with the low group (estimated 3-year DRFS was 93.5% in the low group and 68.5% in the high group; P = 0.0013; Figure 2A). Multivariate analysis using the Cox proportional-hazards model showed that high RAD54B expression was the only independent prognostic factor associated with DRFS in the training set (hazard ratio, 4.31; 95% CI, 1.53–13.1; P = 0.0060; Table 4).
Figure 2. DRFS of RAD54B high and low groups.
A. Kaplan–Meier survival analyses of 108 stage I–III CRC patients in the training set revealed that the RAD54B high group had inferior DRFS compared with the low group (P = 0.0013). B. Kaplan–Meier survival analyses of 71 stage I–III CRC patients in the validation set revealed that the RAD54B high group had inferior DRFS compared with the low group (P = 0.024).
Table 4. Univariate and multivariate analysis of the factors associated with DRFS.
| Training set | Univariate analysis | HR | Multivariate analysis | P value |
|---|---|---|---|---|
| Factors | P value | 95% CI | ||
| RAD54B (High vs Low group) | 0.0013 | 4.31 | 1.53 to 13.1 | 0.0060 |
| T stage (T1-2 vs T3-4) | 0.14 | 2.96 | 0.57 to 54.4 | 0.23 |
| N stage (N0 vs N1-2) | 0.14 | 2.44 | 0.88 to 7.23 | 0.088 |
| Cell differentiation (WD vs MD/PD) | 0.073 | - | - | - |
| Lymphatic invasion (positive vs negative) | 0.090 | - | - | - |
| Venous invasion (positive vs negative) | 0.33 | - | - | - |
| Preoperative CEA (<5.0 vs ≥5.0) | 0.51 | - | - | - |
| Preoperative CA19-9 (<37 vs ≥37) | 0.89 | - | - | - |
| Age (<65 vs ≥65) | 0.37 | - | - | - |
| Location (Colon vs Rectum) | 0.94 | - | - | - |
| Sex (Male vs Female) | 0.34 | - | - | - |
| Tumor size (mm) (<50 vs ≥50) | 0.49 | - | - | - |
| Validation set | Univariate analysis | HR | Multivariate analysis | P value |
|---|---|---|---|---|
| Factors | P value | 95% CI | ||
| RAD54B (High vs Low group) | 0.024 | 3.627 | 1.23 to 10.7 | 0.021 |
| T stage (T1-2 vs T3-4) | 0.18 | 3.432 | 0.66 to 63.1 | 0.17 |
| N stage (N0 vs N1-2) | 0.13 | 2.182 | 0.74 to 7.21 | 0.16 |
| Cell differentiation (WD vs MD/PD) | 0.34 | - | - | - |
| Lymphatic invasion (positive vs negative) | 0.54 | - | - | - |
| Venous invasion (positive vs negative) | 0.094 | - | - | - |
| Preoperative CEA (<5.0 vs ≥5.0) | 0.073 | - | - | - |
| Preoperative CA19-9 (<37 vs ≥37) | 0.19 | - | - | - |
| Age (<65 vs ≥65) | 0.41 | - | - | - |
| Location (Colon vs Rectum) | 0.74 | - | - | - |
| Sex (Male vs Female) | 0.71 | - | - | - |
| Tumor size (mm) (<50 vs ≥50) | 0.84 | - | - | - |
WD = well differentiated; MD = moderately differentiated; PD = poorly differentiated; HR = hazard ratio; CI = confidence interval.
RAD54B expression in the validation set
Next, the RAD54B expression cut-off value of 3.63 determined in the training set was applied to the independent cohort of 89 CRC patients for validation. Seventy-one stage I–III CRC patients (excluding stage IV patients) in the validation set were stratified into the RAD54B low (n = 53) and high (n = 18) expression groups. Table 3 shows that clinicopathological factors including UICC stage, T stage, and N stage were not significantly different in the two groups. During the median follow-up period of 37.0 (28.5–46.0) months, seven patients (13.2%) in the RAD54B low group (n = 53) and seven patients (38.9%) in the high group (n = 18) developed distant recurrence (Table 3, P = 0.024). Kaplan–Meier survival analyses revealed that the RAD54B high group had inferior DRFS compared with the low group with (estimated 3-year DRFS was 85.7% in the low group and 61.1% in the high group; P = 0.024; Figure 2B). Multivariate analysis using the Cox proportional-hazards model showed that high RAD54B expression was an independent prognostic factor associated with DRFS even in the validation set (hazard ratio, 3.63; 95% CI, 1.23–10.7; P = 0.021; Table 4).
RAD54B expression is inversely correlated with CDKN1A expression
We next studied the relationship between RAD54B and p53 functions in clinical CRC samples. Because p53 activates CDKN1A gene transcription and its expression reflects the functional status of p53 protein [25, 26], the CDKN1A expression was measured and compared with that of RAD54B. This analysis was limited to samples without any p53 hotspot mutations observed in CRC because most of such p53 mutants lack the normal transcriptional activities [27, 28]. Using sequencing analysis of the p53 gene, 92 samples without p53 hotspot mutations were extracted from the combined training and validation set (n = 212). Our data showed that CDKN1A expression tends to decrease remarkably in CRC samples which expression of RAD54B are within an approximately 3-fold increase compared with normal mucosa. Particularly, in samples with RAD54B expression values ranging between 1.00–2.95, Spearman's rank-order correlation demonstrated a significant weakly negative correlation between CDKN1A and RAD54B expression (ρ = −0.29, P = 0.027; Figure 3A).
Figure 3. Scatter plot analysis between RAD54B and CDKN1A expression.
A. A weakly significant negative correlation was found between RAD54B and CDKN1A expression values in wild-type p53 samples with RAD54B expression values ranging between 1.00–2.95 (ρ = −0.29, P = 0.027). B. A moderately significant negative correlation was found between RAD54B and CDKN1A expression values in wild-type p53 samples with Arg72 variant, provided RAD54B expression values were under 6.00 (ρ = −0.44, P = 0.0099).
The degradation of p53 caused by RAD54B requires the interaction between p53 and MDM2 [13]. A study reported that p53 has a sequence polymorphism resulting in either Pro or Arg at amino acid position 72, and that although both variants can equivalently transactivate CDKN1A, the Arg72 variant undergoes ubiquitination markedly better than the Pro72 variant via greater interaction with MDM2 [29]. Therefore we analyzed the sequence of amino acid position 72 and the correlation between RAD54B and CDKN1A expression. The analysis limited to 34 samples with Arg72 in 92 samples without p53 hotspot mutations revealed that there was a more remarkably significant negative correlation between RAD54B and CDKN1A expression provided RAD54B expression values were under 6.00 (ρ = −0.44, P = 0.0099; Figure 3B).
DISCUSSION
The present study is the first report demonstrating the utility of RAD54B as a prognostic biomarker in CRC patients. Human RAD54B was identified in 1999 as a homolog of RAD54. Biochemical studies have shown that RAD54B plays an important role in the DNA repairing system by HR [6-9]. However, few studies have focused on the clinical importance of RAD54B thus far. In this study, we confirmed that RAD54B expression was elevated in the majority of the CRC tissues compared with corresponding normal mucosa (116/123 in the training set, 94.3%). Based on this finding, we examined the impact of RAD54B expression on the clinical outcome of CRC patients. Our results revealed that CRC patients with high RAD54B expression had significantly inferior DRFS compared with those with low RAD54B expression. Multivariate analyses using Cox proportional-hazards regression model demonstrated that high RAD54B expression was an independent risk factor for distant recurrence in stage I–III CRC patients in both the training and validation sets. These results clearly demonstrate the prognostic value of RAD54B expression in CRC patients.
We also analyzed RAD51 expression in the same training set. Although RAD51 is another central component in the HR pathway [15-18], the association of RAD51 expression and cancer prognosis remains controversial. Several studies reported that RAD51 overexpression was associated with poor prognosis in various cancers such as non-small-cell lung carcinoma, head cancers, esophageal squamous cell carcinoma, breast cancer, melanoma, and CRC [19-24]. In contrast, two studies targeting human glioblastoma and breast cancer reported the opposite results that high RAD51 expression was associated with better prognostic outcome [30, 31]. In our study, RAD51 expression was elevated in 60.2% (74/123 in the training set) of CRC samples compared with normal mucosa. This percentage was similar to previous studies [19-24]. However, RAD51 expression was not predictive of disease prognosis such as DRFS in our cohort. Our result implies that the prognostic ability of RAD54B might be better than that of RAD51 in CRC patients. On the other hand, the discrepancy of our results compared with previous reports might arise from the difference in the analysis method, e.g., RAD51 expression was examined using immunohistochemistry in most previous studies, whereas we assessed RAD51 expression using a real-time PCR.
Meanwhile, the difference of clinical influence between RAD54B and RAD51 expression implies another pathway other than the HR, where RAD54B functions as a negative prognostic factor. Based on our in vitro study that RAD54B enhances p53 protein degradation via the MDM2–MDMX ubiquitin–ligase complex [13], we analyzed CDKN1A expression, a target gene of p53, to elucidate the biological role of RAD54B in clinical CRC tissues. The analysis limited to CRC samples with Arg72 showed a significant inverse correlation between RAD54B and CDKN1A expression. This result partially supports the mechanism that RAD54B functions via the degradation of p53 protein function in clinical CRC tissues. However, we could not detect a significant correlation between RAD54B and CDKN1A expression in samples with remarkably high RAD54B expression, implying the presence of another unknown mechanism causing poor prognosis.
Several studies reported that RAD51 overexpression induced tumor resistance to ionizing radiation and anticancer drugs in vitro [32-34]. As for RAD54B, we previously found that xenografts derived from RAD54B knock out HCT116 cells grew more slowly than that from wild-type HCT116 cells after either oxaliplatin or 5-FU treatment in nude mice [13]. These results suggests the possibility that the effect of chemotherapy attenuates by RAD54B overexpression, and suggests the potential therapeutic target such as blocking the RAD54B overexpression to reinforce the effect of the chemotherapy. Unfortunately, in the present study, we were not able to prove statistically the association between RAD54B expression and the effect of adjuvant chemotherapy consisting mainly of 5-FU based regimens probably because of our limited number of patients. We will further examine the effect of RAD54B expression on chemotherapy by accumulating more CRC patients.
Limitations to the present study should be noted. First, the samples were obtained from a single institution and the sample size was not large enough for subgroup analysis. Second, the follow-up period may have been relatively short. Third, this was a retrospective study. A large, prospective cohort study would be desirable to determine the clinical usefulness of RAD54B more accurately.
In conclusion, this study is the first report clinically demonstrating the importance of RAD54B as a prognostic biomarker in CRC patients. An increased expression level of RAD54B may serve as an independent predictor of poor outcome in CRC patients treated with surgical resection, particularly for distant metastasis. Postoperative recurrence risk stratification by RAD54B expression will be beneficial for more individual cancer strategies.
MATERIALS AND METHODS
Collection of tissue samples and clinical data
A total of 212 consecutive CRC samples were analyzed. All patients undergoing surgical resection were pathologically diagnosed with CRC at University of Tokyo Hospital, Tokyo, Japan. Samples obtained between 2008 and 2010 were included in the training set (n = 123), whereas those obtained between 2011 and 2012 were included in the validation set (n = 89). To prevent the influence of preoperative therapy on the targeted gene expression in resected specimens, patients receiving any preoperative chemotherapy or radiotherapy were excluded from the study.
A detailed database containing clinical and pathological information was provided for statistical analysis, and survival data were acquired from clinical charts. The cancer histological grade and clinical stage were identified in accordance with the 7th edition of the TNM classification of Union for International Cancer Control (UICC). The study protocol was approved by the ethics committee of The University of Tokyo Hospital, and written informed consent was obtained from all participating patients.
Total RNA extraction and cDNA synthesis
Tumor tissue and corresponding normal mucosa were first immersed in RNAlater Tissue Protect Tubes (QIAGEN) overnight at 4°C and stored at −20°C until analysis.
Total RNA was extracted from tissue samples using TRIzol Reagent (Life Technologies) and subsequently treated with DNase I (TAKARA BIO INC). Complementary DNA (cDNA) was synthesized from total RNA using PrimeScript RT Master Mix (TAKARA BIO INC) and used as a template for real-time PCR and cDNA sequencing. Each procedure was performed according to the manufacturer's instructions.
Real-time PCR
Gene expression of RAD54B, RAD51, CDKN1A, and TATA-binding protein (TBP) were analyzed by real-time PCR using a 7500 Fast Real-Time PCR system (Life Technologies), with KAPA SYBR FAST qPCR Master Mix (KAPA Biosystems), according to the manufacturer's instructions. The following sets of primers were used: RAD54B, 5′-TCCAGGTCTGAATGAAGAGATTAC-3′ and 5′-TCTAGTACTTTCTTCACTAGGCAG-3′ RAD51, 5′-TATCCAGGACATCACTGCCA-3′ and 5′-GGTGAAGGAAAGGCCATGTA-3′ CDKN1A, 5′-GACTCTCAGGGTCGAAAACGG-3′ and 5′-GCGGATTAGGGCTTCCTCTTG-3′ and TBP, 5′-TGCTGCGGTAATCATGAGGATA-3′ and 5′-TGAAGTCCAAGAACTTAGCTGGAA-3′.
All measurements were performed in triplicates and mean cycle threshold (Ct) values of both tumor tissue and corresponding normal mucosa were calculated for the expression analysis. Gene expression analysis was performed using the ΔΔCt method (RAD54B and RAD51) or the standard curve method (CDKN1A) [35]. TBP was used as an internal control to normalize gene expression values for each gene expression analysis.
The ΔCt value represents the difference between the Ct value of the target gene (RAD54B and RAD51) and that of TBP in the same sample, whereas the ΔΔCt value indicates the difference between the average ΔCt value of the tumor tissue and that of the corresponding normal mucosa. The targeted gene expression value was obtained as 2−ΔΔCt and used in the following analyses.
cDNA sequencing of the p53 gene
For sequencing of p53 gene transcripts, cDNA of the tumor tissue was amplified with PrimeSTAR HS DNA Polymerase (TAKARA BIO INC). Primer sets covering the regions from the first ATG to Gln167 and from Arg156 to Pro301 were used to detect the amino acid polymorphism at position 72 and CRC hot-spot mutations, respectively [29, 36]. Direct cDNA sequencing of each PCR product was performed using BigDye terminator v3.1 (Life Technologies) on a 3100 Genetic Analyzer (Life Technologies). The following sets of primers were used: ATG–Gln167, 5′-GTGACACGCTTCCCTGGAT-3′ and 5′-CTCACAACCTCCGTCATGTG-3′ and Arg156–Pro301, 5′-GTGCAGCTGTGGGTTGATT-3′ and 5′-CAGTGCTCGCTTAGTGCTCC-3′.
Statistical analysis
All statistical analyses were conducted using JMP pro version 10 software packages (SAS Institute). Continuous variables were presented as medians (inter-quartile ranges), and they were analyzed using Kruskal–Wallis test (multiple groups) or Mann–Whitney U test (two groups). Categorical variables were presented as numbers (%) and analyzed using Pearson's chi-squared test or Fisher's exact test, as appropriate. For measuring the strength of association between expression values of targeted genes, Spearman's rank-order correlation was used. Kaplan–Meier survival analysis was adopted for DRFS rate analysis, and survival differences between patients groups were evaluated using the log-rank test. DRFS was defined as the time from surgery to distant metastasis. Multivariable analyses for DRFS were performed using the Cox proportional-hazards regression model, and results were presented as hazard ratios with 95% confidence intervals. Probability values (P) < 0.05 were considered statistically significant.
Footnotes
CONFLICTS OF INTEREST
There are no conflicts of interest to declare.
REFERENCES
- 1.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 2.Hoskins JM, Goldberg RM, Qu P, Ibrahim JG, McLeod HL. UGT1A1*28 genotype and irinotecan-induced neutropenia: dose matters. Journal of the J Natl Cancer Inst. 2007;99:1290–1295. doi: 10.1093/jnci/djm115. [DOI] [PubMed] [Google Scholar]
- 3.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 4.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, Rivera F, Kocakova I, Ruff P, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Eng J Med. 2013;369:1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 5.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 6.Hiramoto T, Nakanishi T, Sumiyoshi T, Fukuda T, Matsuura S, Tauchi H, Komatsu K, Shibasaki Y, Inui H, Watatani M, Yasutomi M, Sumii K, Kajiyama G, et al. Mutations of a novel human RAD54 homologue, RAD54B, in primary cancer. Oncogene. 1999;18:3422–3426. doi: 10.1038/sj.onc.1202691. [DOI] [PubMed] [Google Scholar]
- 7.Miyagawa K, Tsuruga T, Kinomura A, Usui K, Katsura M, Tashiro S, Mishima H, Tanaka K. A role for RAD54B in homologous recombination in human cells. EMBO J. 2002;21:175–180. doi: 10.1093/emboj/21.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarai N, Kagawa W, Fujikawa N, Saito K, Hikiba J, Tanaka K, Miyagawa K, Kurumizaka H, Yokoyama S. Biochemical analysis of the N-terminal domain of human RAD54B. Nucleic Acids Res. 2008;36:5441–5450. doi: 10.1093/nar/gkn516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanaka K, Kagawa W, Kinebuchi T, Kurumizaka H, Miyagawa K. Human Rad54B is a double-stranded DNA-dependent ATPase and has biochemical properties different from its structural homolog in yeast, Tid1/Rdh54. Nucleic Acids Res. 2002;30:1346–1353. doi: 10.1093/nar/30.6.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helleday T. Homologous recombination in cancer development, treatment and development of drug resistance. Carcinogenesis. 2010;31:955–960. doi: 10.1093/carcin/bgq064. [DOI] [PubMed] [Google Scholar]
- 11.Krejci L, Altmannova V, Spirek M, Zhao X. Homologous recombination and its regulation. Nucleic Acids Res. 2012;40:5795–5818. doi: 10.1093/nar/gks270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson LH, Schild D. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat Res. 2001;477:131–153. doi: 10.1016/s0027-5107(01)00115-4. [DOI] [PubMed] [Google Scholar]
- 13.Yasuhara T, Suzuki T, Katsura M, Miyagawa K. Rad54B serves as a scaffold in the DNA damage response that limits checkpoint strength. Nat Commun. 2014;5:5426. doi: 10.1038/ncomms6426. [DOI] [PubMed] [Google Scholar]
- 14.Grunda JM, Fiveash J, Palmer CA, Cantor A, Fathallah-Shaykh HM, Nabors LB, Johnson MR. Rationally designed pharmacogenomic treatment using concurrent capecitabine and radiotherapy for glioblastoma; gene expression profiles associated with outcome. Clinl Cancer Res. 2010;16:2890–2898. doi: 10.1158/1078-0432.CCR-09-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol Mol Biol Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heyer WD, Li X, Rolfsmeier M, Zhang XP. Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res. 2006;34:4115–4125. doi: 10.1093/nar/gkl481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ceballos SJ, Heyer WD. Functions of the Snf2/Swi2 family Rad54 motor protein in homologous recombination. Biochim Biophys Acta. 2011;1809:509–523. doi: 10.1016/j.bbagrm.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benson FE, Baumann P, West SC. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 19.Qiao GB, Wu YL, Yang XN, Zhong WZ, Xie D, Guan XY, Fischer D, Kolberg HC, Kruger S, Stuerzbecher HW. High-level expression of Rad51 is an independent prognostic marker of survival in non-small-cell lung cancer patients. Br J Cancer. 2005;93:137–143. doi: 10.1038/sj.bjc.6602665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Connell PP, Jayathilaka K, Haraf DJ, Weichselbaum RR, Vokes EE, Lingen MW. Pilot study examining tumor expression of RAD51 and clinical outcomes in human head cancers. Int J Oncol. 2006;28:1113–1119. [PubMed] [Google Scholar]
- 21.Li Y, Yu H, Luo RZ, Zhang Y, Zhang MF, Wang X, Jia WH. Elevated expression of Rad51 is correlated with decreased survival in resectable esophageal squamous cell carcinoma. J Surg Oncol. 2011;104:617–622. doi: 10.1002/jso.22018. [DOI] [PubMed] [Google Scholar]
- 22.Tennstedt P, Fresow R, Simon R, Marx A, Terracciano L, Petersen C, Sauter G, Dikomey E, Borgmann K. RAD51 overexpression is a negative prognostic marker for colorectal adenocarcinoma. Int J Cancer. 2013;132:2118–2126. doi: 10.1002/ijc.27907. [DOI] [PubMed] [Google Scholar]
- 23.Barbano R, Copetti M, Perrone G, Pazienza V, Muscarella LA, Balsamo T, Storlazzi CT, Ripoli M, Rinaldi M, Valori VM, Latiano TP, Maiello E, Stanziale P, et al. High RAD51 mRNA expression characterize estrogen receptor-positive/progesteron receptor-negative breast cancer and is associated with patient's outcome. Int J Cancer. 2011;129:536–545. doi: 10.1002/ijc.25736. [DOI] [PubMed] [Google Scholar]
- 24.Jewell R, Conway C, Mitra A, Randerson-Moor J, Lobo S, Nsengimana J, Harland M, Marples M, Edward S, Cook M, Powell B, Boon A, de Kort F, et al. Patterns of expression of DNA repair genes and relapse from melanoma. Clinl Cancer Res. 2010;16:5211–5221. doi: 10.1158/1078-0432.CCR-10-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cox LS. Multiple pathways control cell growth and transformation: overlapping and independent activities of p53 and p21Cip1/WAF1/Sdi1. J Pathol. 1997;183:134–140. doi: 10.1002/(SICI)1096-9896(199710)183:2<134::AID-PATH960>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 26.Boulaire J, Fotedar A, Fotedar R. The functions of the cdk-cyclin kinase inhibitor p21WAF1. Pathol Biol. 2000;48:190–202. [PubMed] [Google Scholar]
- 27.Pavletich NP, Chambers KA, Pabo CO. The DNA-binding domain of p53 contains the four conserved regions and the major mutation hot spots. Genes Dev. 1993;7:2556–2564. doi: 10.1101/gad.7.12b.2556. [DOI] [PubMed] [Google Scholar]
- 28.Kato S, Han SY, Liu W, Otsuka K, Shibata H, Kanamaru R, Ishioka C. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci U S A. 2003;100:8424–8429. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dumont P, Leu JI, Della Pietra AC, 3rd, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33:357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 30.Soderlund K, Skoog L, Fornander T, Askmalm MS. The BRCA1/BRCA2/Rad51 complex is a prognostic and predictive factor in early breast cancer. Radiother Oncol. 2007;84:242–251. doi: 10.1016/j.radonc.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 31.Welsh JW, Ellsworth RK, Kumar R, Fjerstad K, Martinez J, Nagel RB, Eschbacher J, Stea B. Rad51 protein expression and survival in patients with glioblastoma multiforme. Int J Radiat Oncol Biol Phys. 2009;74:1251–1255. doi: 10.1016/j.ijrobp.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 32.Vispe S, Cazaux C, Lesca C, Defais M. Overexpression of Rad51 protein stimulates homologous recombination and increases resistance of mammalian cells to ionizing radiation. Nucleic Acids Res. 1998;26:2859–2864. doi: 10.1093/nar/26.12.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slupianek A, Schmutte C, Tombline G, Nieborowska-Skorska M, Hoser G, Nowicki MO, Pierce AJ, Fishel R, Skorski T. BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol Cell. 2001;8:795–806. doi: 10.1016/s1097-2765(01)00357-4. [DOI] [PubMed] [Google Scholar]
- 34.Hansen LT, Lundin C, Spang-Thomsen M, Petersen LN, Helleday T. The role of RAD51 in etoposide (VP16) resistance in small cell lung cancer. Int J Cancer. 2003;105:472–479. doi: 10.1002/ijc.11106. [DOI] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Soussi T, Asselain B, Hamroun D, Kato S, Ishioka C, Claustres M, Beroud C. Meta-analysis of the p53 mutation database for mutant p53 biological activity reveals a methodologic bias in mutation detection. Clin Cancer Res. 2006;12:62–69. doi: 10.1158/1078-0432.CCR-05-0413. [DOI] [PubMed] [Google Scholar]