Abstract
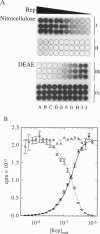
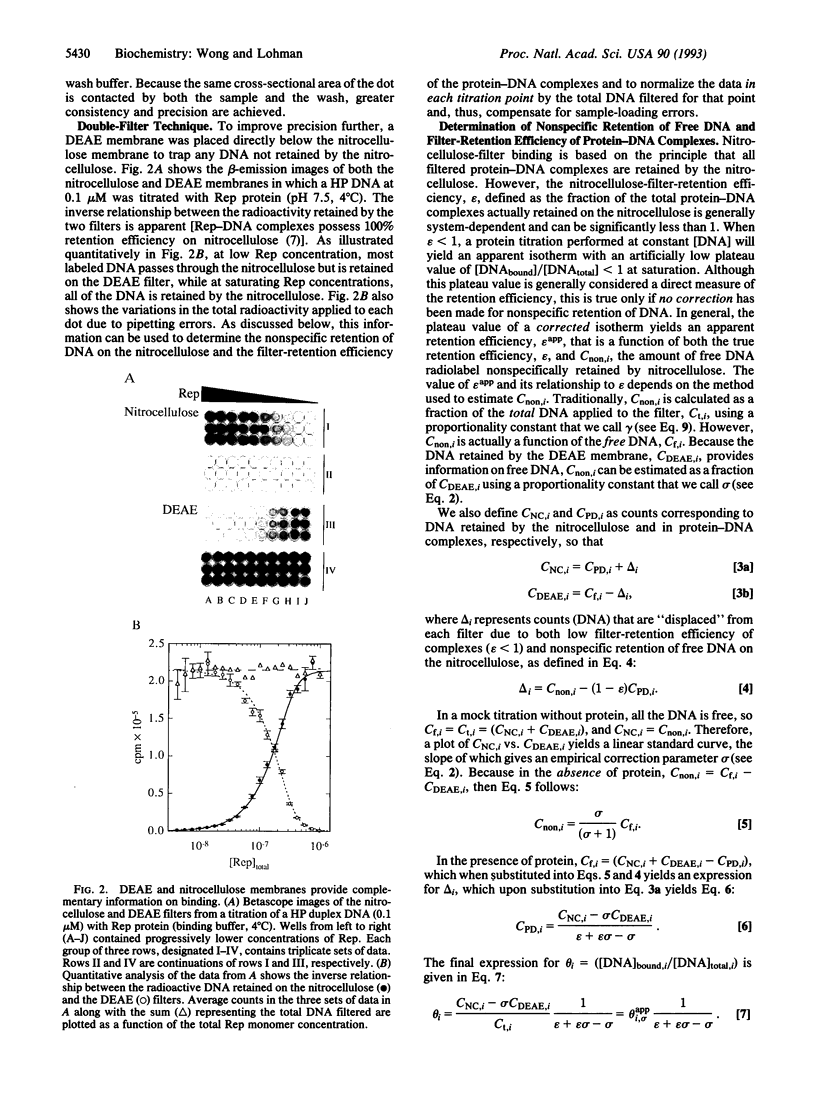
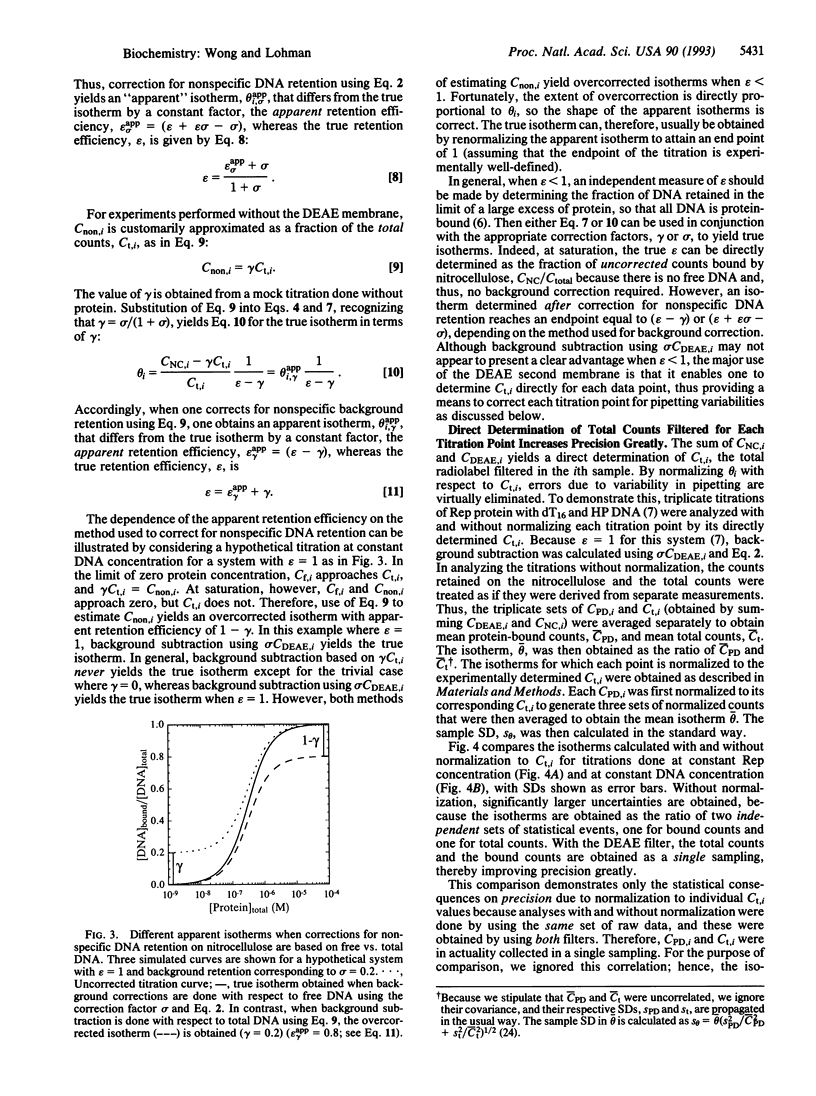
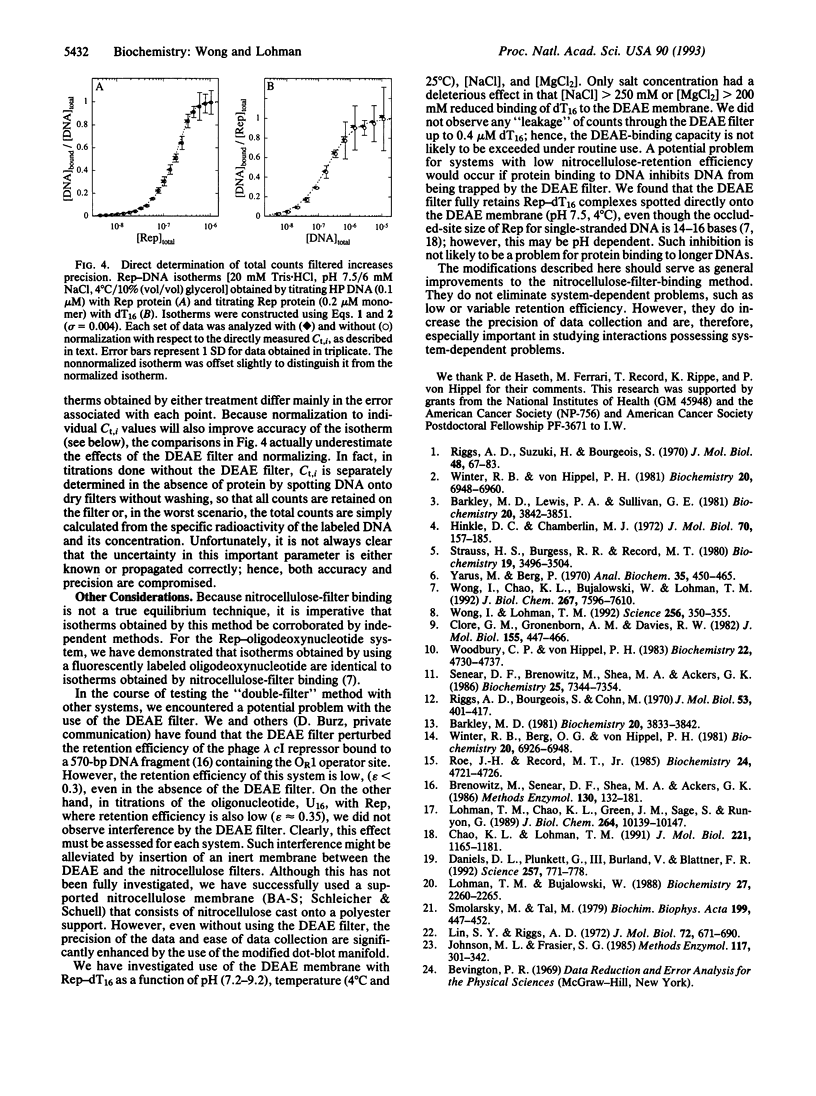
Nitrocellulose-filter binding is a powerful technique commonly used to study protein-nucleic acid interactions; however, its utility in quantitative studies is often compromised by its lack of precision. To improve precision and accuracy, we have introduced two modifications to the traditional technique: the use of a 96-well dot-blot apparatus and the addition of a DEAE membrane beneath the nitrocellulose membrane. Using the dot-blot apparatus, an entire triplicate set of data spanning 20-24 titrant concentrations can be collected on a single 4.5 x 5 inch sheet of nitrocellulose, obviating the need to manipulate separate filters for each titration point. The entire titration can then be quantitated simultaneously with direct two-dimensional beta-emission imaging technology. The DEAE second membrane traps all DNA that does not bind to the nitrocellulose, enabling a direct determination of the total amount of DNA filtered. This measurement improves precision by allowing the amount of DNA retained by the nitrocellulose to be normalized against the total amount of DNA filtered. The DEAE membrane also permits a more accurate quantitation of filter-retention efficiency and nonspecific background retention based on free DNA rather than total DNA filtered. The general approach and methods of analysis to obtain equilibrium binding isotherms are discussed, using as examples our studies of the Escherichia coli Rep protein, a helicase, and its interactions with short oligodeoxynucleotides.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barkley M. D., Lewis P. A., Sullivan G. E. Ion effects on the lac repressor--operator equilibrium. Biochemistry. 1981 Jun 23;20(13):3842–3851. doi: 10.1021/bi00516a027. [DOI] [PubMed] [Google Scholar]
- Barkley M. D. Salt dependence of the kinetics of the lac repressor-operator interaction: role of nonoperator deoxyribonucleic acid in the association reaction. Biochemistry. 1981 Jun 23;20(13):3833–3842. doi: 10.1021/bi00516a026. [DOI] [PubMed] [Google Scholar]
- Berg O. G., Winter R. B., von Hippel P. H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981 Nov 24;20(24):6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- Brenowitz M., Senear D. F., Shea M. A., Ackers G. K. Quantitative DNase footprint titration: a method for studying protein-DNA interactions. Methods Enzymol. 1986;130:132–181. doi: 10.1016/0076-6879(86)30011-9. [DOI] [PubMed] [Google Scholar]
- Chao K. L., Lohman T. M. DNA-induced dimerization of the Escherichia coli Rep helicase. J Mol Biol. 1991 Oct 20;221(4):1165–1181. doi: 10.1016/0022-2836(91)90926-w. [DOI] [PubMed] [Google Scholar]
- Clore G. M., Gronenborn A. M., Davies R. W. Theoretical aspects of specific and non-specific equilibrium binding of proteins to DNA as studied by the nitrocellulose filter binding assay. Co-operative and non-co-operative binding to a one-dimensional lattice. J Mol Biol. 1982 Mar 15;155(4):447–466. doi: 10.1016/0022-2836(82)90481-8. [DOI] [PubMed] [Google Scholar]
- Daniels D. L., Plunkett G., 3rd, Burland V., Blattner F. R. Analysis of the Escherichia coli genome: DNA sequence of the region from 84.5 to 86.5 minutes. Science. 1992 Aug 7;257(5071):771–778. doi: 10.1126/science.1379743. [DOI] [PubMed] [Google Scholar]
- Hinkle D. C., Chamberlin M. J. Studies of the binding of Escherichia coli RNA polymerase to DNA. I. The role of sigma subunit in site selection. J Mol Biol. 1972 Sep 28;70(2):157–185. doi: 10.1016/0022-2836(72)90531-1. [DOI] [PubMed] [Google Scholar]
- Lin S. Y., Riggs A. D. Lac repressor binding to non-operator DNA: detailed studies and a comparison of eequilibrium and rate competition methods. J Mol Biol. 1972 Dec 30;72(3):671–690. doi: 10.1016/0022-2836(72)90184-2. [DOI] [PubMed] [Google Scholar]
- Lohman T. M., Bujalowski W. Negative cooperativity within individual tetramers of Escherichia coli single strand binding protein is responsible for the transition between the (SSB)35 and (SSB)56 DNA binding modes. Biochemistry. 1988 Apr 5;27(7):2260–2265. doi: 10.1021/bi00407a002. [DOI] [PubMed] [Google Scholar]
- Lohman T. M., Chao K., Green J. M., Sage S., Runyon G. T. Large-scale purification and characterization of the Escherichia coli rep gene product. J Biol Chem. 1989 Jun 15;264(17):10139–10147. [PubMed] [Google Scholar]
- Riggs A. D., Bourgeois S., Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. J Mol Biol. 1970 Nov 14;53(3):401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Suzuki H., Bourgeois S. Lac repressor-operator interaction. I. Equilibrium studies. J Mol Biol. 1970 Feb 28;48(1):67–83. doi: 10.1016/0022-2836(70)90219-6. [DOI] [PubMed] [Google Scholar]
- Roe J. H., Record M. T., Jr Regulation of the kinetics of the interaction of Escherichia coli RNA polymerase with the lambda PR promoter by salt concentration. Biochemistry. 1985 Aug 27;24(18):4721–4726. doi: 10.1021/bi00339a002. [DOI] [PubMed] [Google Scholar]
- Senear D. F., Brenowitz M., Shea M. A., Ackers G. K. Energetics of cooperative protein-DNA interactions: comparison between quantitative deoxyribonuclease footprint titration and filter binding. Biochemistry. 1986 Nov 18;25(23):7344–7354. doi: 10.1021/bi00371a016. [DOI] [PubMed] [Google Scholar]
- Smolarsky M., Tal M. Novel method for measuring polyuridylic acid binding to ribosomes. Biochim Biophys Acta. 1970 Feb 18;199(2):447–452. doi: 10.1016/0005-2787(70)90087-0. [DOI] [PubMed] [Google Scholar]
- Strauss H. S., Burgess R. R., Record M. T., Jr Binding of Escherichia coli ribonucleic acid polymerase holoenzyme to a bacteriophage T7 promoter-containing fragment: selectivity exists over a wide range of solution conditions. Biochemistry. 1980 Jul 22;19(15):3496–3504. doi: 10.1021/bi00556a014. [DOI] [PubMed] [Google Scholar]
- Winter R. B., von Hippel P. H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 2. The Escherichia coli repressor--operator interaction: equilibrium measurements. Biochemistry. 1981 Nov 24;20(24):6948–6960. doi: 10.1021/bi00527a029. [DOI] [PubMed] [Google Scholar]
- Wong I., Chao K. L., Bujalowski W., Lohman T. M. DNA-induced dimerization of the Escherichia coli rep helicase. Allosteric effects of single-stranded and duplex DNA. J Biol Chem. 1992 Apr 15;267(11):7596–7610. [PubMed] [Google Scholar]
- Wong I., Lohman T. M. Allosteric effects of nucleotide cofactors on Escherichia coli Rep helicase-DNA binding. Science. 1992 Apr 17;256(5055):350–355. doi: 10.1126/science.256.5055.350. [DOI] [PubMed] [Google Scholar]
- Woodbury C. P., Jr, von Hippel P. H. On the determination of deoxyribonucleic acid-protein interaction parameters using the nitrocellulose filter-binding assay. Biochemistry. 1983 Sep 27;22(20):4730–4737. doi: 10.1021/bi00289a018. [DOI] [PubMed] [Google Scholar]
- Yarus M., Berg P. On the properties and utility of a membrane filter assay in the study of isoleucyl-tRNA synthetase. Anal Biochem. 1970 Jun;35(2):450–465. doi: 10.1016/0003-2697(70)90207-1. [DOI] [PubMed] [Google Scholar]