Abstract
The initiation of plant lateral organs from the shoot apical meristem (SAM) is closely associated with the formation of specialized domains of restricted growth known as the boundaries. These zones are required in separating the meristem from the growing primordia or adjacent organs but play a much broader role in regulating stem cell activity and shoot patterning. Studies have revealed a network of genes and hormone pathways that establish and maintain boundaries between the SAM and leaves. Recruitment of these pathways is shown to underlie a variety of processes during the reproductive phase including axillary meristems production, flower patterning, fruit development, and organ abscission. This review summarizes the role of conserved gene modules in patterning boundaries throughout the life cycle.
Keywords: meristem, lateral organ boundary, organ separation, inflorescence architecture, fruit patterning, flower patterning, abscission, dehiscence
Introduction
The shoot apical meristem (SAM) plays a crucial role in plant development as a continuous source of founder cells for provision of new leaves, shoots, and internodes throughout the life cycle. The SAM is organized into a central zone composed of slowly dividing stem cells, a peripheral zone where lateral organs initiate, and a rib zone that provides cells for internodes (Aichinger et al., 2012). The maintenance of meristems depends on the balance between two antagonistic activities: propagation of stem cells at the center of the meristem and the initiation of organs at the periphery. Boundaries are domains of restricted growth that maintain this balance by separating the meristem from the growing primordia and by forming an interface between organs (Žádníková and Simon, 2014). These interfaces play a critical role by influencing cell fate in adjacent tissues. The best-characterized boundary is the domain that separates leaves from the SAM during the vegetative phase. How principles governing the activity of this boundary apply to other developmental contexts is an important question. For example, boundaries in the leaf control shape and complexity whereas boundaries in the inflorescence have specialized functions such as axillary meristem (AM) production, fruit dehiscence, and organ abscission. Thus, many aspects of plant architecture are dependent on the boundary. In this review, we first describe the genetic control of boundaries during the vegetative phase, and then focus on elaboration of these pathways for specialized functions during the reproductive phase focusing on the model plant species Arabidopsis thaliana (Arabidopsis).
Sam Initiation, Sam Maintenance And Cotyledon Separation
The NAM-ATAF-CUC (NAC)-type CUP-SHAPED COTYLEDON1 (CUC1), CUC2, and CUC3 transcription factors confer boundary identity in higher land plants (Maugarny et al., 2015). These factors initiate the SAM and establish boundaries in conjunction with SHOOT MERISTEMLESS (STM), a three-amino acid loop extension (TALE) class I KNOTTED1-like (KNOX) homeodomain protein [(Figure 1A) and (Hamant and Pautot, 2010; Hay and Tsiantis, 2010)]. CUC-STM forms a conserved module in development that was first identified in embryos (Aida et al., 1999; Takada et al., 2001; Aida et al., 2002). During the globular stage of embryogenesis, CUC1 and CUC2 genes are activated in a narrow band between the presumptive cotyledons (Aida et al., 1999; Takada et al., 2001), where auxin is depleted (Benková et al., 2003) based on positional cues provided by WUSCHEL-RELATED HOMEOBOX (WOX2) and WOX8/STIMPY-LIKE (Lie et al., 2012). CUC1/2 factors activate STM at late globular stage to initiate the meristem and separate the cotyledons (Aida et al., 1999). STM in turn maintains CUC expression (Aida et al., 1999). More recently, STM has also been identified as a direct regulator of CUC1 (Spinelli et al., 2011). This pattern is reinforced by two SWI/SNF chromatin remodeling ATPase complexes BRAMHA (BRM) and SPLAYED (SYD) acting independently of STM: BRM is a positive regulator of all three CUC genes whereas SYD is required for CUC2 expression (Kwon et al., 2006). By late torpedo stage, STM marks the central region of the meristem and is slightly detected in boundaries while expression of CUC genes is restricted to boundaries (Long and Barton, 1998; Aida et al., 1999). Double mutant analyses show that contributions of the different CUC genes are partially redundant. For example, CUC1/2 are essential for meristem initiation while CUC3 plays a more prominent role in organ separation (Vroemen et al., 2003) and AM production (Hibara et al., 2006). CUC1 and CUC2 transcripts are targeted by microRNA164 (miR164) to restrict their expression domain while CUC3 from a different subclade does not contain a miR164 binding site (Laufs et al., 2004; Mallory et al., 2004).
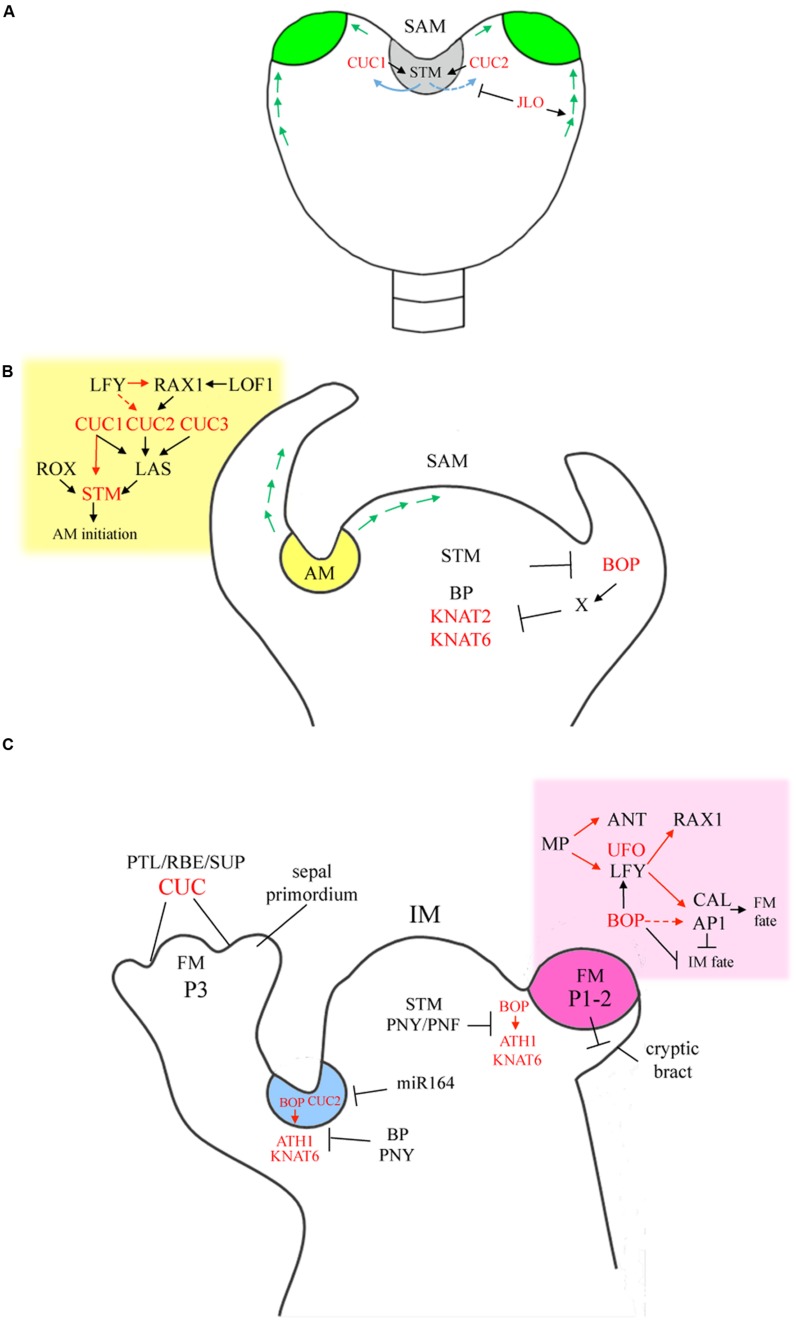
FIGURE 1.
Homologous boundary gene networks controlling biogenesis of shoot apical meristem (SAM), axillary meristems (AMs), and floral meristems (FMs). (A) SAM initiation (gray). Early heart stage is shown. CUC genes are activated in a narrow band of auxin-depleted cells located between the presumptive cotyledons at globular stage. Activation of CUCs in this domain is partly dependent on chromatin remodeling ATPases and various other factors not depicted. Once activated, CUC1 and CUC2 are redundantly required for STM expression to form the presumptive SAM. STM in turn, directly maintains expression of CUC1 and indirectly promotes CUC2 and CUC3 in establishing a feedback loop that ultimately restricts CUC expression to the axils of cotyledons. Reciprocally, STM expression is restricted to the SAM and slightly detected in boundaries. LBD family member JLO promotes PIN1 expression required for formation of auxin maxima and represses STM and KNOX members to allow cotyledon outgrowth. Green arrows, direction of auxin flow. Green, auxin maxima at the cotyledon primordia. (B) SAM maintenance and AM formation (yellow). STM represses BOP1/2 to maintain indeterminacy in the SAM. Conversely, BOP1/2 restrict KNOX expression in the proximal region of leaves to control patterning. Formation of an AM requires depletion of auxin from the leaf axil followed by a burst of CK. CUC1-3 are redundantly required for AM initiation functioning downstream of LFY and RAX1 to promote LAS. LOF1/2 contribute to RAX1 promotion. CUC1, LAS, and ROX activities are required for sustained expression of STM and establishment of the AM. Green arrows, direction of auxin flow. (C) IM activity. PNY and PNF restrict BOP1/2-ATH1-KNAT6 expression to boundary domains flanking the IM essential for meristem maintenance and flowering. FMs (pink). FMs are AMs with determinate fate that form in the axil of leaves whose development are repressed (cryptic bract). Auxin responsive transcription factor MP directly activates ANT and LFY to initiate FM formation. LFY directly promotes the expression of RAX1 and AP1 and CAL whose products confer floral fate. BOPs facilitate establishment of FMs via promotion of LFY expression, activation of AP1, and repression of IM identity genes. UFO is a LFY co-activator also required for formation of boundaries in the flower. Later, CUC factors are required to separate floral organs and maintain boundaries between whorls in association with numerous stage-specific factors including PTL, RBE, and SUP required for localized repression of growth. Inflorescence architecture. BP and PNY are expressed in the stem cortex where they collectively promote internode elongation, stem differentiation, phyllotaxy, and pedicel angle by restricting boundary genes BOP1/2 and downstream effectors ATH1 and KNAT6 to the pedicel axil (blue). Misexpression of these genes in the BP-PNY domain restricts growth, disrupts vascular patterning, and causes ectopic lignification. CUC2 expression is restricted by miR164 to the pedicel axil to maintain internode patterning. FM, floral meristem; IM, inflorescence meristem. P, primordia; stages as indicated. Red lettering, SAM-leaf boundary genes. Red arrows, direct regulation. Dashed line, putative interaction.
Other three-amino acid loop extension (TALE) homeodomain transcription factors contribute redundantly with STM in SAM initiation and maintenance. The TALE superfamily is divided into KNOX and BELL classes, whose members function as heterodimers (Hamant and Pautot, 2010; Hay and Tsiantis, 2010). Formation of KNOX-BELL heterodimers regulates nuclear localization (Cole et al., 2006; Rutjens et al., 2009; Kim et al., 2013) and influences binding site selection (Smith et al., 2002). Within the KNOX subclass, BREVIPEDICELLUS (BP)/KNOTTED1-LIKE FROM A. THALIANA1 (KNAT1) is expressed in the peripheral and rib zones of the SAM (Lincoln et al., 1994) whereas KNAT6 is expressed in boundaries (Belles-Boix et al., 2006). Mutation in BP enhances only the meristem defect of weak stm mutants (Byrne et al., 2002) whereas knat6 mutation also impairs cotyledon separation showing a specific role for KNAT6 in boundaries (Belles-Boix et al., 2006). KNAT2, the fourth KNOX class I member is expressed at the base of the meristem and in lateral organ boundaries but its inactivation does not enhance the meristem defects of weak stm mutants (Byrne et al., 2002; Belles-Boix et al., 2006). KNAT2 role in the SAM remains undetermined.
At least three BELL homeodomain proteins encoded by PENNYWISE (PNY), POUND-FOOLISH (PNF), and ARABIDOPSIS THALIANA HOMEOBOX GENE1 (ATH1) interact with STM to maintain the SAM (Byrne et al., 2003; Kanrar et al., 2006; Rutjens et al., 2009). PNY is expressed in the central zone of the SAM (Smith et al., 2004); PNF is expressed in the central and rib zones of the SAM (Smith et al., 2004), whereas ATH1 is more broadly expressed in the SAM, young leaves, and boundaries and is shown to control patterning in the basal region of shoot organs (Proveniers et al., 2007; Gómez-Mena and Sablowski, 2008). PNY and ATH1 contribute redundantly with STM in SAM initiation and maintenance (Byrne et al., 2003; Kanrar et al., 2006; Rutjens et al., 2009). PNY and PNF maintain the integrity of the central zone since the expression domain of STM is narrower in pny pnf double mutants (Ung et al., 2011). Meristem termination defects in this mutant are attributed to depletion of nuclear localized BELL-STM complexes (Rutjens et al., 2009), but recent data show that PNY and PNF negatively regulate lateral organ boundary genes including ATH1 and KNAT6 expression to maintain SAM function (Khan et al., 2015).
TALE transcription factors repress cellular differentiation in the meristem in part by regulating the abundance of hormones including gibberellins (GA), cytokinins (CK), and brassinosteroids (BR) (Figures 2A,B). A WUSCHEL-CLAVATA (WUS-CLV) feedback loop functions in parallel to keep the stem-cell niche constant in size (Schoof et al., 2000). A high CK: low GA ratio promotes meristem maintenance since high CK sustains cell division and low GA inhibits cell differentiation (Aichinger et al., 2012). Accordingly, meristem activity in stm mutants can be restored by elevating CK biosynthesis and inhibited by elevating GA abundance or signaling or by reducing CK content (Hay et al., 2002; Jasinski et al., 2005; Yanai et al., 2005). KNOX proteins raise CK levels by activating ISOPENTENYL TRANSFERASE7 for CK biosynthesis (Jasinski et al., 2005; Yanai et al., 2005) and lower GA levels by directly inhibiting biosynthetic genes encoding GA20-oxidases (Sakamoto et al., 2001) and activating catabolic genes encoding GA2-oxidases (Bolduc and Hake, 2009). Genes encoding GA2-oxidase are expressed at the boundary between the SAM and leaves confining GA to leaves where growth is taking place (Jasinski et al., 2005; Bolduc and Hake, 2009). WUS contributes to this network by lowering the abundance of ARABIDOPSIS RESPONSE REGULATOR ARR7 and ARR15 (Leibfried et al., 2005) thus increasing sensitivity to CK in the central zone and promoting its own expression (Gordon et al., 2009). BR are a class of growth-promoting hormones recently shown to play a role in meristem maintenance (Tsuda et al., 2014). New work in rice and maize show that KNOX factors in the SAM maintain indeterminacy in part via direct activation of BR catabolism genes thereby downregulating BR signaling in the meristem. Inactivation of three rice orthologs of Arabidopsis catabolic gene BAS1 (PHYTOCHROME B ACTIVATION TAGGED SUPPRESSOR1) results in the premature differentiation of meristematic cells (Tsuda et al., 2014).
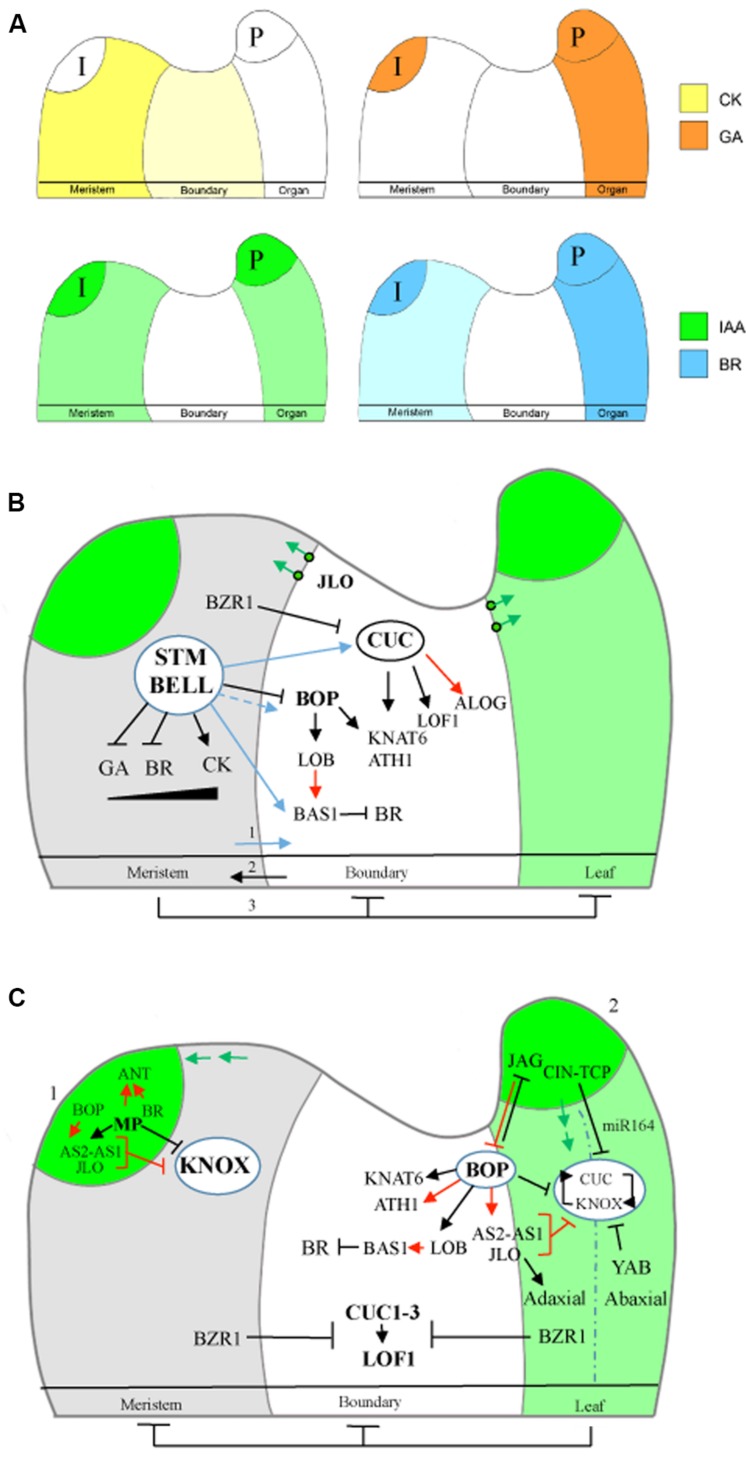
FIGURE 2.
Summary of hormone profiles and genetic interactions that maintain the leaf-SAM boundary. (A) The shoot apex contains: the meristem, the initium, boundaries, and organ primordia. The predicted distribution of key hormones are summarized (see text for details). Meristem, high CK/IAA ratio and low GA/BR ratio promotes indeterminate growth. Primordia, high IAA activates BR and represses CK biosynthesis and GA increases to promote determinate growth. Boundary, depletion for growth-promoting hormones IAA, GA, BR, and CK inhibits cell division allowing separation of meristem-organ compartments. I, Initium; CK, cytokinin; BR, brassinosteroid; GA, gibberellin; and IAA, auxin. (B) Summary of gene networks at the meristem-boundary interface. Meristem, STM maintains indeterminate growth by promoting CK and repressing GA/BR accumulation (black gradient indicates hormone abundance). Auxin, shown in green, marks the site of primordia initiation and distal blade of emerging leaf. (1) KNOX proteins initialize the boundary through promotion of BR catabolic genes (BAS1) and boundary transcription genes including CUC and BOP2 (blue arrows). CUC factors confer boundary identity required for activation of other classes of boundary regulators including BOP, KNAT-BELL, LOF1, ALOG, and LBD members that collectively restrict growth, modulate meristematic activity, and pattern the boundary. PIN1 auxin efflux carriers (green circles) are orientated facing outward such that auxin is drained away from the boundary. Green arrows indicate direction of auxin flow. (2) Boundary genes contribute to meristem maintenance (see text). (3) STM-BELL meristem factors preserve meristem integrity by restricting BOP1/2 and KNAT6 to boundaries. BR-activated transcription factor BZR1 represses CUC/LOF1 in the meristem domain. (C) Summary of interactions at the leaf-boundary interface. (1) Polar auxin transport establishes auxin maxima in the peripheral zone where leaf initiation takes place. Auxin response factor MP initiates primordium formation by repressing KNOX genes, activating ANT members and leaf identity genes including AS1 and stimulating synthesis of BR where BZR1 binds to the ANT promoter as a positive regulator. Boundary genes BOP1/2 and JLO expressed in the organ initial contribute to organ polarity and stable repression of KNOX genes. (2) Primordium outgrowth coincides with synthesis of auxin and repolarization of PIN transports toward the leaf base, which becomes a low IAA/BR domain. BOPs and JLO now restricted to the boundary reinforce this pattern in hormones via regulation of LOB and PIN1, respectively. CIN-TCPs and BZR1 in leaves maintain repression of CUC/LOF1. JAG in the distal blade represses BOPs. BOPs expressed in the proximal petiole domain of leaves maintain organ polarity and repress KNOX and JAG genes required for simple leaf shape indirectly in part via activation of AS2. YAB contributes to the repression of KNOX and CUC in the abaxial domain.
Boundary Formation
The boundary that separates the SAM from the primordia is a domain of restricted growth. This feature relies on depletion of auxin and BR from boundary cells thereby maintaining a low rate of growth relative to surrounding tissues (Figures 2A,C). Spatial regulation of polar auxin transporters establishes a minimum for auxin. This is accomplished in part by PIN-FORMED1 (PIN1) transporters oriented outwardly along the long axis of cells in the plane of the groove such that auxin is drained away from the boundary into the adjacent organ and meristem compartments (Heisler et al., 2010). Striking images of auxin depletion from the adaxial boundary domain of leaf primordia are observed using the auxin concentration sensor DII-Venus or auxin-responsive reporter gene DR5:VENUS (Wang et al., 2014a,b). ABC/PGP (ATP-binding cassette/P-glycoprotein) pumps are a secondary type of auxin transporter. ABC19 in this family depletes auxin from the boundary creating a low-auxin niche necessary for promotion of CUC2 and LOF2 expression (Zhao et al., 2013). An auxin minimum is essential in several boundary-related processes including AM formation (Tian et al., 2014; Wang et al., 2014a,b), differentiation of valve margins in the fruit (Sorefan et al., 2009), and timing of floral organ abscission (Estornell et al., 2013).
The boundary is also a minimum for BR (Bell et al., 2012; Gendron et al., 2012). The LATERAL ORGAN BOUNDARY DOMAIN (LBD) transcription factor LATERAL ORGAN BOUNDARIES (LOB) maintains low levels of BR to inhibit growth at boundaries. Ectopic expression of LOB reduces growth similar to BR defective mutants, while loss of LOB function causes overgrowth of the boundary region and organ fusion (Bell et al., 2012; Gendron et al., 2012). A feedback loop is required in establishing this pattern. Auxin-induced BR in the leaf activates LOB1 (Chung et al., 2011) which in turn directs activation of cytochrome P450 gene BAS1 to inhibit BR accumulation at the boundary (Bell et al., 2012). Two BTB-ankyrin transcriptional co-activators, BLADE-ON-PETIOLE1 (BOP1) and BOP2, reinforce this pattern by promoting LOB expression in the boundary domain (Ha et al., 2007). Fluorescent reporters show that BR-activated transcription factor BRASSINOZOLE-RESISTANT1 (BZR1) fails to accumulate in the nuclei of boundary cells thereby allowing expression of CUC genes which in turn repress growth at the boundary (Gendron et al., 2012).
Emerging data suggest that KNOX activity provides a positional cue in establishing the SAM-leaf boundary (Bolduc et al., 2012; Johnston et al., 2014; Tsuda et al., 2014). Several mechanisms are identified. One study shows that rice KNOX gene Oryza sativa homeobox1 (OSH1) expressed in the meristem and base of emerging leaves facilitates SAM function and boundary formation by lowering BR abundance (Tsuda et al., 2014). Transcriptomic studies in maize focusing on the blade-sheath boundary of leaves further reveal that CUCs, TALEs, and BOPs are downstream targets of KNOTTED1 (KN1) under positive regulation (Bolduc et al., 2012; Johnston et al., 2014). Grasses have a blade-sheath boundary containing hinge-like auricles that control leaf angle and a fringe of epidermal tissue called the ligule whose formation is under the control of boundary genes. Barley UNICULME4 is a BOP homolog required for ligule outgrowth (Tavakol et al., 2015). Maize genes required for ligule development include LIGULELESS2 (LG2) which encodes a TGA bZIP factor (Walsh et al., 1998), LG3/LG4 which are closely related genes to Arabidopsis KNAT2 and KNAT6; and KNOTTED1/ROUGH SHEATH1 homologs of STM or BP (Bolduc et al., 2012; Johnston et al., 2014). Pre-ligule tissue is enriched for homologs of CUC2, BOP1/2, KNAT6, and two BELL genes, whose loci in several cases are bound directly by KN1 as identified through chromatin immunoprecipitation assays (Bolduc et al., 2012; Johnston et al., 2014). These data support a model in which KNOX accumulation at the base of the leaf primordia and auxin accumulation in the distal portion of the primordia provide opposing positional cues in demarcating the blade-sheath boundary (Bolduc et al., 2012; Johnston et al., 2014). Arabidopsis studies showing that STM directly activates CUC1 and indirectly promotes CUC2, CUC3, and BOP2 expression (Spinelli et al., 2011) support this model.
Genetic studies show that CUC genes play a central role in maintaining growth repression in boundaries. Inactivation of any two CUCs leads to ectopic growth at cotyledon boundaries causing fusion along their margins (Aida et al., 1997; Vroemen et al., 2003). GROWTH-REGULATING FACTORS (GRFs) which act as broad regulators of cell proliferation function synergistically with CUCs in this role. Leaf fusion defects in grf1 grf2 grf3 triple mutants are dramatically enhanced by inactivation of GRF4 or CUC genes resulting in cup-shaped cotyledons and embryos that lack a functional SAM (Lee et al., 2015). Two ALOG (Arabidopsis LSH1 and Oryza G1) family members ORGAN BOUNDARY1/LIGHT-DEPENDENT SHORT HYPOCOTYL (OBO1/LSH3) and OBO4/LSH4 are direct targets of CUC1 and thought to repress differentiation of boundary cells (Cho and Zambryski, 2011; Takeda et al., 2011). Another regulator is the MYB transcription factor LATERAL ORGAN FUSION1 (LOF1), which promotes organ separation and meristem maintenance. Inactivation of LOF1 enhances stm-10 meristem termination and organ fusion defects (Lee et al., 2009).
Organ Initiation
One of the earliest steps in initiation of lateral organs is down-regulation of STM at sites of auxin maxima in the peripheral zone of the meristem. Boundary and leaf identity genes are transiently expressed in the same compartment undergoing differentiation. Various studies show that organ initiation and boundary formation are interconnected processes (Heisler et al., 2005; Besnard et al., 2011). Auxin in the distal portion of the primordia controls the localization of boundary genes ultimately restricting their expression to the base of the emerging leaf (Figures 2A,C). In brief, cotyledons and leaves are initiated at auxin maxima generated by polar auxin transport. Polar auxin distribution is dependent on a family of efflux carriers including PIN1 whose membrane localization is controlled by the serine/threonine kinase PINOID. Threshold levels of auxin trigger activation of the auxin-responsive transcription factor MONOPTEROS (MP), which down-regulates STM and activates AINTEGUMENTA (ANT), ANT-like (AIL), and ASYMMETRIC LEAVES1 (AS1) genes to initiate leaf development (Long and Barton, 1998; Besnard et al., 2011; Yamaguchi et al., 2013). As the primordium emerges, PIN1 polarity reverses to generate new auxin peaks coinciding with a narrow band of cells marked by CUC expression (Heisler et al., 2005). Mutations in PIN1, PID, or MP that disrupt auxin transport or signaling lead to expansion of STM and CUC expression to the periphery where they suppress cotyledon outgrowth (Aida et al., 2002; Furutani et al., 2004; Schuetz et al., 2008). Proper distribution of auxin in forming this pattern is dependent on SEUSS and SEUSS-like components of the LEUNIG repressor complex although the mechanism is still unknown (Lee et al., 2014). Auxin in the leaf initial further alters the balance of hormones to favor growth and determinacy. In particular, auxin stimulates BR (Chung et al., 2011) and GA synthesis (Frigerio et al., 2006) and represses CK production (Nordstrom et al., 2004; Besnard et al., 2011). Primordium outgrowth also depends on physical changes in cell wall stiffness (Besnard et al., 2011; Peaucelle et al., 2011). Auxin stimulates the active transport of protons into the extracellular space required in activating enzymes that relax the cell wall and promotes the transcription of remodeling factors including expansions, pectin methylesterase, and hydrolases (Besnard et al., 2011; Peaucelle et al., 2011). These changes are coupled with a shift toward growth isotropy, which facilitates organ outgrowth (Sassi et al., 2014).
Leaf Differentiation
Leaf differentiation requires the maintenance of KNOX repression and the restriction of CUC2/3 expression along the leaf margin. KNOX repression is accomplished by an interacting network of leaf and boundary factors (Figure 2C). A key player in this network is the MYB transcription factor AS1, which acts in a trimeric complex with the LBD transcription factors AS2 and JAGGED LATERAL ORGANS (JLO) (Guo et al., 2008; Rast and Simon, 2012). AS1 and AS2 bind to distinct sites in the BP and KNAT2 promoter where they interact through looping to induce silencing via recruitment of the histone chaperone HIRA and Polycomb-repressive complex2 (Guo et al., 2008; Lodha et al., 2013). STM is also a target of PRC but how this complex is recruited to the promoter is unknown (Lodha et al., 2013). JLO is transiently expressed at sites of organ initiation and resolves to the leaf-meristem boundary during outgrowth. Loss-of-function mutations in JLO impair organ outgrowth and enhance the margin patterning defects of as2 mutants. This phenotype is caused in part by ectopic expression of BP and STM at the base of leaf primordia combined with defects in auxin distribution (Rast and Simon, 2012). JLO promotes PIN expression for auxin build-up at organ initiation sites and later for auxin efflux from the boundary (Bureau et al., 2010; Rast and Simon, 2012; Žádníková and Simon, 2014).
BOP1/2 activity in organ initials partially overlaps with JLO and likewise resolves to the boundary of emerging leaves and petiole domains during outgrowth (Ha et al., 2004; Hepworth et al., 2005; Norberg et al., 2005; Borghi et al., 2007). BOP1/2 have a dual function. They repress genes that confer meristem cell fate and induce genes that promote lateral organ fate and polarity (Ha et al., 2007). BOP1/2 transcripts are first detected in the boundaries of torpedo stage embryos consistent with a function downstream or in parallel with CUCs (Ha et al., 2004). STM represses BOP1/2 to maintain indeterminacy and conversely, BOP1/2 restrict KNOX expression to pattern the leaf (Jun et al., 2010). A prolonged phase of morphogenetic competence in bop1 bop2 petioles coupled with KNOX reactivation results in initiation of ectopic leaflets reminiscent of development in a compound leaf (Ha et al., 2003, 2007; Khan et al., 2014). BOP1 binds directly to the promoter of AS2 likely recruited by a TGA factor (Jun et al., 2010). Synergistic enhancement of meristematic activity in bop1 bop2 as1 and bop1 bop2 as2 petioles shows that BOP1/2 repression of KNOX genes is not entirely via AS1–AS2 and is likely indirect. Leaf patterning defects in bop1 bop2 are also attributed to misexpression of abaxial/adaxial organ polarity determinants and the C2H2 zinc finger transcription factor JAGGED (JAG) which promotes cell proliferation (Norberg et al., 2005; Ha et al., 2007, 2010). JAG is normally restricted to the distal blade where it represses BOP2 to allow extension of the leaf margin (Schiessl et al., 2014).
CINCINNATA (CIN)-like TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP) factors are another class that contribute to negative regulation of CUC and KNOX activity in leaves to promote organ outgrowth and simple leaf shape (Koyama et al., 2007, 2010). Consistent with this view, CIN-TCPs are predominantly expressed in leaves and depleted from the boundary (Tian et al., 2014). Several mechanisms are identified. First, TCP3 directly promotes miR164, which targets CUC1 and CUC2 transcripts for cleavage. Second, TCP3 directly promotes AS1 whose product represses CUC3 and KNOX expression (Koyama et al., 2010). In addition, TCP3 targets auxin inducible genes that repress SAM function and cause cotyledon fusion when overexpressed (Koyama et al., 2010). Lastly, TCP4 binds to CUC2 and inhibits its activity by blocking the formation of homo-dimers and hetero-dimers with CUC3. TCP4 also impairs CUC3 transactivation ability (Rubio-Somoza et al., 2014).
Leaf Shape
Variations in the KNOX-PIN-CUC module play a central role in controlling leaf shape and complexity. The leaf margin, due to its meristematic feature, is particularly sensitive to alterations in this module. In simple leaf species such as Arabidopsis, CUC2, and CUC3 expression is restricted to the sinus of serrations along the leaf margin while CUC1 expression is not detected (Nikovics et al., 2006; Hasson et al., 2011). The balance between CUC2 and miRNA164 transcripts controls the degree of leaf serration (Nikovics et al., 2006). CUC3 contributes to leaf shape at a later stage (Hasson et al., 2011). Similar to the primordia initiation, the formation of serrations depends on auxin. To explain how serrations form on a leaf margin, Bilsborough et al. (2011) proposed a model in which two feedback loops work in concert. In the first loop, PIN1 convergence in the leaf margin generates an auxin maximum, reinforced by auxin feedback on PIN1. In the second loop, CUC2 acts non-cell autonomously to promote growth through the generation of PIN1-dependent auxin maxima and contributes to tooth outgrowth (Kawamura et al., 2010). Auxin in turn downregulates CUC2 restricting expression to regions between serrations where growth is repressed.
The rachis of a compound leaf is likewise sensitive to alterations in KNOX-PIN-CUC expression. While simple leaves have a single undivided blade in which KNOX repression is continuous, compound leaves have a divided blade consisting of pairs of leaflets attached to a central rachis. This morphology is associated with an extended primary morphogenesis phase during which reactivation of KNOX genes begins the cycle by promoting auxin accumulation thereby directing leaflet initiation on the rachis (Di Giacomo et al., 2013). Down-regulation of tomato BOPa (one of three homologs) further enhances leaf complexity by extending the window for rachis responsiveness to auxin (Ichihashi et al., 2014). BOPa fulfills this function in part by forming a complex with LIGHT-DEPENDENT SHORT HYPOCOTYL3b (an ALOG family member) that represses tomato KNATM encoded by KD1/PETROSELINUM to modulate KNOX activity (Ichihashi et al., 2014). KNATM, a mini-KNOX lacking the homeodomain, modulates KNOX-BELL activity by competing for BELL binding partners (Kimura et al., 2008; Magnani and Hake, 2008). Analysis of the maize KN1 cistrome confirms that a majority of directly regulated genes are involved in auxin signaling, biosynthesis, and transport including PIN1 (Bolduc et al., 2012). Several legume species such as Medicago truncatula and pea use orthologs of Arabidopsis LEAFY (LFY) as an alternate source of meristem activity but remain reliant on CUC2 function for creation of auxin peaks required in leaflet initiation (Nikovics et al., 2006; Blein et al., 2008; Efroni et al., 2010). These data illustrate that variations in the KNOX-PIN-CUC module cause diversity in leaf patterning. Recruitment of this same module at later stages governs AM formation, gynoecium, and ovule development (Ishida et al., 2000; Hibara et al., 2006; Scofield et al., 2007; Raman et al., 2008).
New Frontiers
In the next part of the review, we examine the role of boundary genes during the reproductive phase. Boundaries in the inflorescence determine plant architecture through the separation of organs and the distribution of flowers on the stem but also constitute a source of AM for production of branches and flowers. Boundaries are also sites where abscission and dehiscence take place. A number of genes are recurrently expressed in these boundaries including KNOX-BELL, BOP, and CUC/miR164 regulators. Similar to the role of TALE factors in the SAM, PNY and PNF preserve meristem integrity essential for flowering by excluding BOP1/2/KNAT6-ATH1 from the meristem. Inflorescence architecture is likewise controlled by restricting CUC2 and BOP1/2-KNAT6/ATH1 to boundaries at the base of floral shoots. CUC-STM factors play a conserved role in formation of new meristems, including AMs that give rise to lateral branches and flowers and meristematic tissues internal to the fruit. CUC factors also play a critical role in separation of floral organs and ovules in developing flowers. TALE factors including BP and PNY preserve formation of meristematic replum tissue in fruits whereas BOP1/2-KNAT6/2, which are expressed in adjacent valve margin boundary tissues potentially contribute to dehiscence. This same network of TALE and BOP factors regulates abscission. How these conserved modules are integrated during reproductive development is now discussed.
Axillary Meristems
The boundary located between the stem and the leaf base constitutes a source of AMs, which can remain dormant or produce secondary inflorescences and flowers (Figure 1B). The specification and the development of AMs involves numerous transcription factors and is modulated by hormones such as auxin, CK, BR, and strigolactones (Janssen et al., 2014). Other hormones may also be involved based on specific patterns of enrichment for abscisic acid and ethylene or depletion of jasmonic acid (JA) responsive genes in the boundary domain (Tian et al., 2014). Similar to the SAM, formation of AMs requires CUC-STM factors whose activity at the boundary is dependent on auxin and CK.
Recent studies in Arabidopsis and tomato show that the establishment of a stem cell niche in leaf axils requires auxin depletion followed by pulse of CK (Wang et al., 2014a,b). Manipulation of the auxin gradient using chemical inhibitors of auxin transport or mutations in auxin transport machinery including PIN1 or PID showed that disruption of auxin minima strongly inhibits AM initiation (Wang et al., 2014a,b). STM, which is a marker of AMs in the mature leaf axil (Grbic and Bleecker, 2000; Long and Barton, 2000), fails to accumulate in a strong pid-9 mutant indicating that its expression is dependent on an auxin minimum (Wang et al., 2014a). CK perception and signaling is enhanced in leaf axils prior to AM initiation and the TCS::GFP (two-component output sensor) synthetic reporter used to visualize CK response indicates that a pulse of CK follows the auxin minimum and is required to stimulate AM production (Wang et al., 2014b). STM may contribute to this pulse based on its CK promoting activity in the SAM (Jasinski et al., 2005; Yanai et al., 2005). Arabidopsis mutants affected in CK perception (histidine kinase receptor mutants) or CK signaling (ARR-B type transcription factor mutants) show reduced AM production whereas overproduction of CK restores AM initiation in a rax1 mutant (Wang et al., 2014b).
REGULATOR OF AXILLARY MERISTEM1 (RAX1) is a MYB transcription factor that specifies AMs in redundancy with RAX2 and RAX3 (Keller et al., 2006; Muller et al., 2006). One regulator of RAX1 is the MYB transcription factor LOF1 which also functions to promote AM and organ separation (Lee et al., 2009). RAX1 acts through CUC2 and is required in conjunction with CUC3 and the GRAS-domain protein LATERAL SUPPRESSOR (LAS) to maintain STM expression in AMs. RAX1 maintains the boundary zone through the repression of GA similarly to STM in the SAM (Keller et al., 2006; Muller et al., 2006). LAS and RAX1 promote AMs via the bHLH transcription factor REGULATOR OF AM FORMATION (ROX; Yang et al., 2012). LFY, which is involved in flower specification has recently been shown to promote AM proliferation through its direct target RAX1 and potentially others (Chahtane et al., 2013). Redundant pathways mask this role as mutations in LFY combined with mutations affecting various pathways including meristem, auxin signaling, floral transition and patterning, or boundary genes such BOP1/2 show defects in AM formation (Chahtane et al., 2013). LFY further contributes to meristem emergence via CK signaling potentially through its interaction with WUS. The negative regulator of CK signaling, ARABIDOPSIS RESPONSE REGULATOR7 (ARR7), was found to be up-regulated in lfy-12 inflorescences, and LFY interacts directly with the ARR7 promoter (Moyroud et al., 2011; Winter et al., 2011; Chahtane et al., 2013). LFY may also act through CUC2 and the auxin signaling pathway as genes from this pathway are bound by LFY (Moyroud et al., 2011; Winter et al., 2011; Yamaguchi et al., 2013). Thus, meristem emergence results from the convergence of LFY and LAS pathways.
Genetic studies show that CUC genes contribute redundantly and differently to AM initiation and boundary maintenance with CUC3 playing a prominent role (Hibara et al., 2006). Inactivation of CUC1 and CUC2 has no effect on AM initiation and an effect in cuc2 mutants is seen only in the absence of CUC3 (Hibara et al., 2006). Further studies show a contribution of CUC1/2 to AMs. Overexpression of miR164 dramatically reduces the initiation of AMs in the cuc3-2 mutant, and reciprocally, miR164-resistant versions of CUC1 and CUC2 form extra accessory side shoots (Raman et al., 2008). Key downstream targets of the CUC pathway include STM and LAS required in AM establishment (Raman et al., 2008), with LAS being a direct target of CUC2 (Tian et al., 2014). STM expression is missing in the adaxial boundary domain of cuc3-2 mutants and LAS expression is reduced in miR164 overexpressing lines, which diminishes STM expression (Greb et al., 2003; Raman et al., 2008).
Translatome analysis of LAS-expressing boundary and AS1-expressing leaf primordia cells coupled with genome-scale mapping of transcription factor binding sites reveals that CUC2 and LAS are regulatory hubs for AM initiation (Tian et al., 2014). This work identifies the auxin-induced APETALA2 domain transcription factor DORNRÖSCHEN as a direct activator of CUC2. This work also identifies SQUAMOSA PROMOTER BINDING PROTEIN-LIKE9 and 15 as repressors of LAS and CUC2 that regulate AM initiation likely in response to environmental signals (Tian et al., 2014).
Separation of axillary shoots from subtending leaves and formation of accessory side shoots requires LOF1 and LOF2 acting downstream of CUC genes (Lee et al., 2009; Gendron et al., 2012). Lof1 defects in organ separation are enhanced by mutation of the closely related LOF2 whose expression is more widespread but dependent on LOF1 at the boundary (Lee et al., 2009). STM expression in AMs is reduced in lof1 lof2 double mutants (Lee et al., 2009). Overexpression of a cysteine-rich signaling peptide TAXIMIN1 mimics the phenotype of lof1 lof2 double mutants (Colling et al., 2015). Interestingly, this phenotype is not due to a reduction of LOF1/2 or other boundary transcripts suggesting an independent mechanism (Colling et al., 2015). A peptide signaling cascade has not been previously linked to formation of boundaries in plants.
Studies in other species show this hierarchy to be highly conserved (Janssen et al., 2014). Tomato GOBLET (GOB) encodes a NAC-domain transcription factor similar to CUC2 (Berger et al., 2009); LAS is an ortholog of LAS (Schumacher et al., 1999; Greb et al., 2003); and BLIND is an ortholog of RAX1 (Schmitz et al., 2002). Remarkably these same genes are regulators of leaf complexity. Homologous genes have also been identified in cereals as regulators of tillering and panicle architecture. LAX PANICLE1 in rice and BARREN STALK in maize encode bHLH proteins orthologous to ROX in Arabidopsis. Mutants in these genes show a reduction in panicle branches and spikelets and fail to form AMs during the vegetative phase resulting in a reduction in tillers (Komatsu et al., 2003; Gallavotti et al., 2004; Oikawa and Kyozuka, 2009). These proteins sustain early proliferation of the AM by forming a boundary between the meristem and axillary bud (Oikawa and Kyozuka, 2009; Yang et al., 2012). Barley CUL4 is a BOP homolog required for tiller formation (Tavakol et al., 2015). BOP1/2 are required for production of various determinate axillary shoots including stipules, nectaries, and flowers in dicots (Khan et al., 2014). BOP expression is down-regulated at an early stage of indeterminate IM formation and moves to the boundary between the meristem and AM demonstrating a transient role similar to ROX (Xu et al., 2010; Yang et al., 2012). The contribution of CUL4 in AMs production suggests a partial conservation of BOP function in monocots and dicots.
Inflorescence Architecture
The maintenance of boundaries during stem growth is critical in preserving plant architecture. Ectopic expression of boundary genes prevents the elongation and proper differentiation of stem internodes resulting in aberrant phyllotaxy. This is illustrated by clustering of flowers on the stems of plants expressing a miR164-resistant version of CUC2 (Peaucelle et al., 2007). The restriction of CUC2 expression to the floral stem axil by miR164 in the IM is required to maintain the boundary between the pedicel and the stem (Figure 1C). The TALE transcription factors BP and PNY constitute another set of architecture determinants (Figure 1C). These factors are required to maintain internode patterning during stem growth and radial patterning in both primary and secondary phases of stem development (Smith and Hake, 2003). BP and PNY are expressed in the stem cortex and adjacent vascular tissues and form a boundary between the IM and lateral organs (Smith and Hake, 2003). Mutations in BP lead to short compact internodes, downward pointing siliques, and precocious outgrowth of paraclades (Douglas et al., 2002; Venglat et al., 2002). Vascular bundles in bp mutants are often irregular in size and/or spacing. Bundles tend to be underdeveloped with xylem elements reduced or lacking in lignin (Smith and Hake, 2003). Mutations in PNY cause shortened internodes and clusters of flowers on stems and partial loss of apical dominance (Byrne et al., 2003; Smith and Hake, 2003). These phenotypes are enhanced in the double mutant showing that BP-PNY have partially overlapping functions in specification of boundaries during internode growth (Smith and Hake, 2003). Genetic and transcriptome studies indicate that PNY modulates the activity of plant cell wall modifying enzymes required in loosening cell walls to allow organ initiation and internode elongation (Peaucelle et al., 2011; Etchells et al., 2012). BP regulates an overlapping set of genes and prevents premature deposition of lignin in elongating stems by direct repression of genes in the lignin biosynthetic pathway (Mele et al., 2003; Wang et al., 2006).
Genetic and expression studies show that bp and pny inflorescence defects are caused by the localized misexpression of lateral organ boundary genes KNAT6, ATH1, BOP1/2 and to a lesser extent KNAT2 in stems (Ragni et al., 2008; Khan et al., 2012a,b). Inactivation of BOP1/2 or KNAT6 or ATH1 fully rescues pny defects to restore wild type inflorescence architecture. Similarly, inactivation of BOP1/2 or KNAT6 in combination with KNAT2 or ATH1 rescues bp defects in internode elongation and pedicel orientation. The regular pattern of vascular bundles and the pattern of lignin deposition in stems during secondary growth are reestablished in these mutants (Khan et al., 2012a,b). BOP1/2 require the functions of these downstream genes to exert changes in inflorescence architecture suggesting a linear pathway (Khan et al., 2012a,b). Further analysis of this module shows that BOP1 directly activates ATH1 whereas activation of KNAT6 is indirect (Khan et al., 2015). BP/STM are recently shown to promote xylem differentiation in the cambium through the repression of BOP1 and BOP2 (Liebsch et al., 2014). Thus, restriction of the BOP1/2-ATH1-KNAT6 boundary module by BP-PNY is critical for plant architecture. Recent data reveal that BP directly represses KNAT2 and KNAT6 expression by recruiting the chromatin remodeling ATPase BRAHMA to the promoter (Zhao et al., 2015).
Flower Initiation And Patterning
Floral inductive signals acting on the SAM cause restructuring to form the IM. Completion of this process requires the PNY and PNF BELL members. In pny pnf mutants, apices support the production of leaves, but internode elongation and flower initiation are blocked (Smith et al., 2004; Kanrar et al., 2008; Lal et al., 2011). Recent data show that this block is due to misexpression of BOP1/2 and its downstream effectors KNAT6 and ATH1 which prevent accumulation of floral meristem identity genes including LFY, CAULIFLOWER (CAL), and APETALA1 (AP1) required for flower production (Khan et al., 2015). PNY in this network directly represses BOP1/2 to maintain its expression at boundaries. One study shows that ectopic BOP1/2 expression reduces responsiveness to FT by lowering the abundance of its binding partner FD (Andrés et al., 2015). Transcript profiling of BOP1 overexpressing plants further identifies promotion of JA as a potential mechanism for inhibiting accumulation of SQUAMOSA PROMOTER BINDING-LIKE PROTEINS and counteracting responsiveness to GAs (Khan et al., 2015). Thus, the setting of lateral boundaries by PNY and PNF via the restriction of BOP1/2-ATH1-KNAT6 expression is critical for meristem integrity and specification of flowers.
The floral meristem constitutes an AM whose rapid proliferation represses outgrowth of the subtending leaf (Long and Barton, 2000). Initiation of flowers is an auxin-dependent process similar to that in leaves (Figure 1C). Mutations in MP or PIN1 result in naked IM “pins” lacking flowers due to misexpression of meristem/organ/boundary markers including STM, LFY, CUC, and ANT throughout the peripheral zone (Vernoux et al., 2000; Hay et al., 2006; Schuetz et al., 2008). MP integrates auxin and floral signals by directly activating ANT/AIL6 which triggers proliferation in combination with LFY which activates flower development (Yamaguchi et al., 2013). Interestingly, MP does not bind to the LFY promoter during the vegetative stage indicating that binding is stage-specific (Yamaguchi et al., 2013). LFY reinforces this loop via direct activation of genes in the auxin pathway, direct activation of RAX1, and direct activation of AP1 and CAL whose products confer floral fate (Wagner et al., 1999; William et al., 2004; Winter et al., 2011; Yamaguchi et al., 2013).
BOP1/2 and UNUSUAL FLORAL ORGANS (UFO) are boundary regulators that facilitate LFY function. Genetic studies reveal that BOPs play a supporting role in the promotion of LFY expression (Karim et al., 2009), proliferation of the floral meristem, and determinacy in part through direct activation of AP1 (Xu et al., 2010). Several of these functions are shared with UFO (Norberg et al., 2005; Xu et al., 2010; Risseeuw et al., 2013). Outgrowth of the floral meristem is delayed in bop1 bop2, ufo-1, and lfy mutants or absent in bop1 bop2 lfy-1 triple mutants resulting in barren axils (Levin and Meyerowitz, 1995; Wilkinson and Haughn, 1995; Norberg et al., 2005; Xu et al., 2010). Inactivation of BOP1/2 or UFO CAL greatly enhances the floral branching defect in ap1 mutants caused by derepression of CK biosynthesis in sepal axils leading to ectopic FM initiation and loss of shoot determinacy (Levin and Meyerowitz, 1995; Xu et al., 2010; Han et al., 2014). UFO is the F-box subunit of an SCF-based E3 ubiquitin ligase complex which binds to LFY and functions as a transcriptional co-activator (Lee et al., 1997; Chae et al., 2008). Paradoxically, UFO stimulates LFY activity by directing ubiquitination of its transcriptional activation domain thus marking the protein for turnover which is required for maximal induction of target genes (Chae et al., 2008). Similar functions are shown for BOP and UFO orthologs in a variety of species (Khan et al., 2014; Vlad et al., 2014).
Arabidopsis flowers are composed of sepals, petals, stamens, and carpels arranged in four concentric whorls. LFY is responsible for this patterning by activating three sets of homeotic genes that function combinatorially according to the ABC model (Lohmann and Weigel, 2002). In ufo mutants, petals and stamens are reduced or absent and organs are fused or chimeric indicating disrupted boundaries in the flower (Levin and Meyerowitz, 1995; Wilkinson and Haughn, 1995). In bop1 bop2 mutants, sepal-to-petal conversions and sepal-whorl organ fusions are localized to the abaxial side of flowers where BOP1/2 are transcribed during late stage 2 (Hepworth et al., 2005; Xu et al., 2010). Inactivation of BOP1/2 or UFO in a weak lfy background creates a strong lfy phenotype indicating closely related functions for these genes (Levin and Meyerowitz, 1995; Wilkinson and Haughn, 1995; Xu et al., 2010). UFO is activated in the dome of stage 2 flowers and resolves to a cup-shaped domain around STM-expressing cells in the central zone of stage 3 flowers (Lee et al., 1997; Samach et al., 1999) and is possibly involved in creating a boundary. Embryo expression of UFO is dependent on STM (Long and Barton, 1998) but the mutant has no obvious defects during this stage due to genetic redundancy.
Boundaries in the flower are maintained by various other boundary genes and stage-specific factors including the zinc-finger repressors RABBIT EARS (RBE; Takeda et al., 2004), and SUPERMAN (SUP; Sakai et al., 1995; Nibau et al., 2011) and HANABA TARANU, a GATA3-type transcriptional repressor (Zhao et al., 2004; Nawy et al., 2010). CUC genes are expressed between organ primordia and at the edges of whorls where they repress growth required to separate the floral organs and maintain boundaries between whorls (Ishida et al., 2000; Takada et al., 2001). ATH1 controls basal floral organ boundaries and functions downstream of CUC genes (Gómez-Mena and Sablowski, 2008). LOF1 (Gomez et al., 2011), OBO1/LSH3, and OBO4/LSH4 (Cho and Zambryski, 2011) functioning downstream of CUC1 are likely to contribute based on their expression patterns or overexpression phenotypes in the flower. Floral boundary defects are also observed in bop1 bop2 and jlo flowers (Hepworth et al., 2005; Rast and Simon, 2012). Analogous to leaves, AS1–AS2, and JAG repress boundary genes including CUC1 and CUC2 to promote sepal and petal development (Xu et al., 2008).
PETAL LOSS (PTL) encodes a stage-specific trihelix transcription factor that represses growth in inter-sepal boundaries. In ptl mutants, petals are often absent, or show changes in shape, polarity, and fusion with sepals (Griffith et al., 1999). Sepal fusion is increased in ptl cuc mutants consistent with analysis showing that CUCs suppress upward growth of inter-sepal tissue at the boundary whereas PTL limits overgrowth of the inter-sepal zone required for petal initiation (Lampugnani et al., 2012, 2013). Petal initiation is highly sensitive to perturbations in growth and auxin distribution because primordia arise from 2 to 3 founder cells located in close proximity to the inter-sepal boundary (Lampugnani et al., 2012, 2013).
Fruit Patterning And Dehiscence
The Arabidopsis fruit is derived from the gynoecium, which consists of two fused carpels representing modified leaves (Ferrándiz et al., 2010). The carpels (termed valves after fertilization) are joined to a central replum whose internal surface or carpel margin meristem provides ovules and a septum with transmitting tract. Valve margins are a specialized lateral organ boundary that forms at the valve/replum interface. They ensure the release of the seeds. Thus, three patterning elements define the transverse axis of the mature fruit: valves, valve margins, and replum. Many interactions defining the SAM-leaf boundary are conserved in ovule and fruit development (Ferrándiz et al., 2010; Reyes-Olalde et al., 2013; Arnaud and Pautot, 2014) and (Figures 3 and 4).
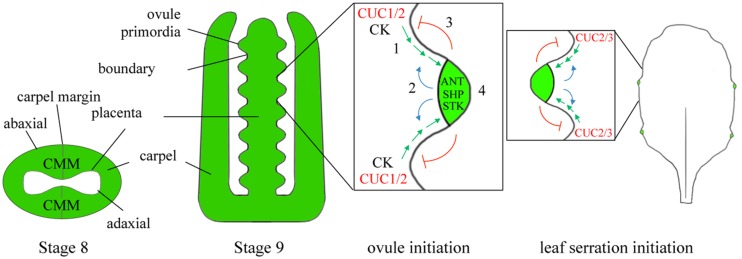
FIGURE 3.
Homologous networks for ovule initiation by the carpel marginal meristem (CMM) and patterning of leaf margin. Two fused carpels in the center of the flower form a tubular structure that constitutes the gynoecium (stages 8 and 9). The inner fused surfaces of the carpels (equivalent to an adaxial leaf surface) form a ridge of meristematic tissue called the CMM that gives rise to ovules and septum. Ovules are initiated in a process that resembles creation of serrations on the leaf margin where CUC2 is required for leaf serration and CUC3 promotes serration growth (see text). (1) CUC1/2 in the placenta together with CK promote the formation of an auxin maximum. (2) Auxin positive feedback reinforces flow of auxin to the primordia tip. (3) Auxin negative feedback restricts CUC1/2 expression to the base of the ovule. (4) Once auxin reaches threshold levels, it switches on ANT which promotes outgrowth of the shoot and MADS box genes SHP1/2 and STK which confer ovule identity. CUC2/3 expression overlaps between ovule primordia and is required for ovule separation (not shown). Adapted from (Cucinotta et al., 2014). Red lettering, SAM-leaf boundary genes.
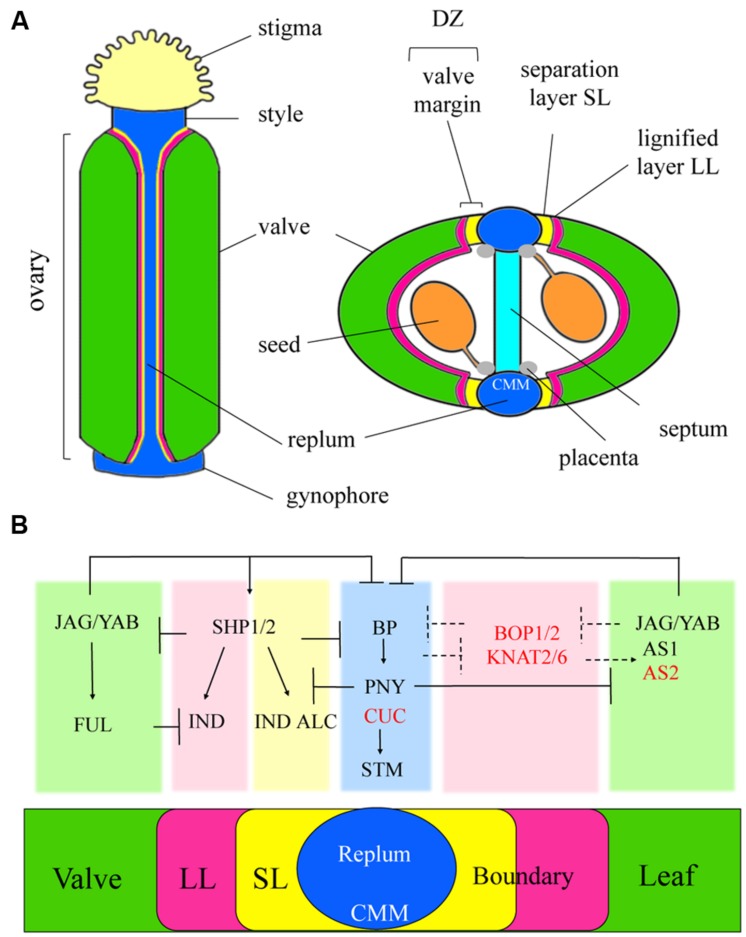
FIGURE 4.
Schematic of an Arabidopsis fruit and summary of networks for fruit patterning compared to the SAM-leaf boundary. (A) The Arabidopsis fruit derived from two fused carpel valves that represent modified leaves. Valve margins are a lateral organ boundary specialized for dehiscence that joins the carpels to a meristematic tissue called the replum. The internal surface of the replum or CMM provides septum and placenta that gives rise to ovules that develop into seeds at fertilization. Differentiation of the valve margins requires GA, CK, and depletion of auxin (see text). When the fruit is mature, the valve margin differentiates to form the dehiscence zone (DZ) comprising two layers: a separation layer (SL) where the fruit will open and a lignified layer (LL) continuous with the lining of the fruit that provides tension required in spring-like opening of the fruit for seed dispersal. (B) Similar to their role in leaves, JAG and YAB factors together with AS1–AS2 are required in restricting expression of meristematic genes BP and PNY to the central replum domain and in restricting valve margin identity genes to the boundary junction. FUL is a stage-specific factor that confers valve identity and functions similarly to AS1–AS2 and JAB/YAB to correctly position the replum and valve margin identity domains. Red lettering, SAM-leaf boundary genes: CUC1/2 activate STM required in formation of the CMM and BOP1/2 and KNAT2/6 expressed in the valve margin of fruit are predicted to antagonize BP-PNY activity in the replum. Dashed arrows, hypothetical interactions.
The carpel marginal meristem (CMM) forms internally at the junction between fused carpels, which is homologous to the marginal meristem of leaves (Pautot et al., 2001). The CMM produces two outgrowths fused centrally to form the septum and is flanked on both sides by the placenta that gives rise to ovules. CUC genes promote fusion of the carpel margins and CMM initiation via the activation of STM similar to their role in SAM (Kamiuchi et al., 2014). Defects in either of these genes impair placental function leading to a reduction in ovules (Endrizzi et al., 1996; Ishida et al., 2000; Scofield et al., 2007). CK also plays an important role in promoting meristematic activity. Visualization of CK using the synthetic TCS::GFP reporter shows CK in the CMM of young gynoecia. Increased levels of CK enhance replum size while decreased levels reduce replum size (Marsch-Martinez et al., 2012). Following establishment of the CMM, CUC1/2 transcripts are detected in the placenta where together with CK they control the localization of PIN1 transporters in creating auxin maxima required for ovule initiation (Bencivenga et al., 2012; Galbiati et al., 2013; Cucinotta et al., 2014). In mutants that overexpress CK, the number of ovules increases (Bartrina et al., 2011). Conversely, where there is a reduction in CK response or defects in auxin synthesis, transport or signaling, the number of ovules decreases (Galbiati et al., 2013). This process is comparable to formation of serrations on the leaf margin (Bilsborough et al., 2011). Once an ovule is initiated, auxin and BR converge to activate ANT for proliferation of the ovule primordia (Galbiati et al., 2013; Huang et al., 2013). Auxin accumulation in the distal tip of the ovule ultimately restricts CUC1/2 expression to boundaries in the ovule (Ishida et al., 2000). CUC3 and CUC2 are later expressed between ovule primordia overlapping in a few cells where they are redundantly required for ovule separation (Goncalves et al., 2015). Cuc3 single mutants show rare fused ovules with defects more severe in the cuc3 cuc2 double mutant. LOF1 may also play a role based on its expression pattern in the inner medial ridges, the placenta, and at the base of the ovules marking these domains as lateral organ boundaries (Gomez et al., 2011). MADS-box transcription factors AGAMOUS, SHATTERPROOF1/2 (SHP1/2), and SEEDSTICK (STK) confer ovule identity (Ishida et al., 2000; Pinyopich et al., 2003; Galbiati et al., 2013).
After fertilization, the ovules develop into seeds and the fruit enlarges. At maturity, the valve margins undergo secondary differentiation to form the dehiscence zone where the fruit opens. The dehiscence zone has two cell layers: a separation layer adjacent to the replum and a lignified layer. The separation layer produces enzymes that break down the middle lamella, a layer of pectin that cements cell walls together. The lignified layer is continuous with the inner layer of the fruit and is required for spring-like opening of the fruit (Ferrándiz et al., 2010).
The valve margin expresses a pair of stage-specific MADS-box transcription factors encoded by SHP1/2 and boundary genes KNAT2/6 and BOP1/2 that are activated later during carpel development (Liljegren et al., 2000; Ragni et al., 2008; Khan et al., 2012b). SHP1/2 confer valve margin identity via activation of downstream bHLH transcription factors INDEHISCENT (IND) and ALCATRAZ (ALC) (Liljegren et al., 2004). IND is required for differentiation of lignified and separation layers of the valve margin whereas ALC/SPATULA (SPT) are required for differentiation of the separation layer (Rajani and Sundaresan, 2001; Liljegren et al., 2004; Girin et al., 2011; Groszmann et al., 2011). IND leads to the depletion of auxin in valve margins by relocating PIN1 transporters (Sorefan et al., 2009). IND also promotes GA production, which releases ALC and SPT proteins from DELLA repression allowing formation of a productive complex to specify the separation layer (Arnaud et al., 2010). The auxin and GA pathways seem to be independent, since the auxin minimum is maintained in GA deficient mutants. Visualization of CK in mature gynoecium shows CK in valve margins, and this localization depends on IND and SHP1/2 activity (Marsch-Martinez et al., 2012). Interestingly, CK restores valve margins in shp1 shp2 and ind mutants indicating that CK functions downstream of these regulators. In contrast, a complementary pattern is observed for auxin with a synthetic DR5 reporter detected only in replum and valves. Thus, CK promotes valve margins. CK may contribute to the depletion of auxin in valve margins via the localization of PIN transporters (Marsch-Martinez et al., 2012) but it is unknown if the depletion of auxin is required for the accumulation of CK in valve boundaries or if the valve margin regulators IND and SHP1/2 activate this pathway.
The formation and relative size of external domains in the fruit: valves, valve margin, and replum are governed by antagonistic interactions analogous to those at the leaf-boundary-SAM interface (González-Reig et al., 2012). JAG and YABBY (YAB) members FILAMENTOUS (FIL)/YAB3 also found in leaves activate the MADS-box gene FRUITFULL (FUL), which is required for elongation and differentiation of the valves, and SHP1/2, which confer valve-margin identity (Dinneny et al., 2005). FUL in turn represses SHP/IND/ALC to set the valve margin boundary. Fruits in a ful mutant are constricted and ectopically lignified due to the misexpression of valve margin genes (Ferrándiz et al., 2000; Liljegren et al., 2004). AS1–AS2 and JAG/FIL/YAB3 reprise their roles in the leaf by restricting BP expression to the replum. In as1 or as2 mutants or in jag fil yab3 triple mutants, valve width is reduced and the replum is expanded due to an increase in BP expression (Alonso-Cantabrana et al., 2007; González-Reig et al., 2012). BP, which interacts with PNY, also known as REPLUMLESS (Roeder et al., 2003), activates its expression, and contributes redundantly with PNY in maintaining the replum in part by repressing valve and valve-margin identity genes (Alonso-Cantabrana et al., 2007). Thus, inactivation of JAG/FIL/YAB3 or SHP/IND genes partially rescues replum formation in a pny mutant (Roeder et al., 2003; Dinneny et al., 2005; Alonso-Cantabrana et al., 2007). Other factors identified are APETALA2 which prevents overgrowth of the replum and valve margin by repressing BP/PNY and valve margin identity genes (Ripoll et al., 2011) and the zinc finger transcription factor NO TRANSMITTING TRACT which promotes replum development by activating BP (Marsch-Martinez et al., 2014).
The role of boundary genes in fruit patterning and dehiscence is worth exploring. Mutations in SHP1/2 or the different stage-specific bHLH genes block dehiscence (Ferrándiz et al., 2010) but this is not case for KNAT2/6 or BOP1/2. Nevertheless, inactivation of these genes restores replum formation in pny mutants showing that the antagonistic interaction between BOP1/2- KNAT6/2 and PNY also control fruit patterning (Ragni et al., 2008; Khan et al., 2012b). Based on the role of this module in other boundary contexts, BOP1/2 and KNAT2/6 likely contribute to repression of BP/PNY to control replum size, specialization of cells in the separation layer, and formation of lignified cell layers (McKim et al., 2008; Khan et al., 2012a,b). Similar interactions among meristem and boundary genes regulate abscission.
Abscission
Abscission zones (AZs) are typically located at lateral organ boundaries in the plant at the base of leaves, floral organs, or seeds [(Estornell et al., 2013) and Figure 5]. In Arabidopsis, AZs comprised of small, densely cytoplasmic cells form simultaneously with the boundary for detachment of floral organs and seeds (McKim et al., 2008). At anthesis, AZ cells acquire competence to respond to abscission signals and secrete cell-wall modifying and hydrolyzing enzymes that degrade the middle lamella between two adjacent cell files. Ethylene, JA, and abscisic acid are promotive signals for abscission whereas auxin, GA, and BRs are inhibitory (for reviews: Estornell et al., 2013; Niederhuth et al., 2013; Kim, 2014). Depletion of auxin from the AZ is shown to improve sensitivity to ethylene in controlling the timing of abscission (Estornell et al., 2013) reminiscent of other boundaries.
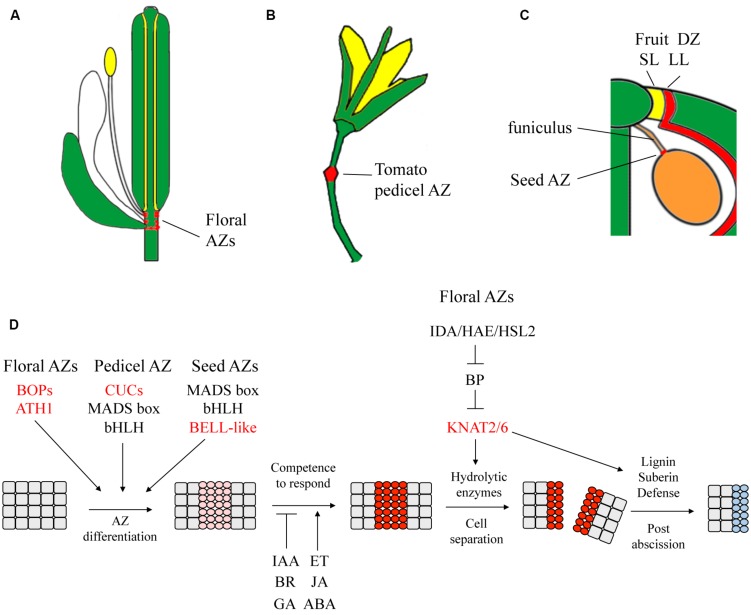
FIGURE 5.
Summary of pathways that control the development of abscission zones (AZs). Placement of AZs (in red) are shown for (A) Arabidopsis flower, (B) tomato pedicel, and (C) Arabidopsis seed and dehiscence zone. (D) The current accepted model for abscission defines four key stages: differentiation of AZ cells (pink), AZ cells acquire competence to respond to hormone inductive signals (red), activation of abscission and detachment (red), and differentiation of a protective layer over the scar (blue). Red lettering, SAM-leaf boundary genes. Dashed lines, hypothetical pathway. IAA, auxin; BR, brassinosteroid; GA, gibberellin; ET, ethylene; CK, cytokinin; JA, jasmonic acid. Adapted from (Leibfried et al., 2005; Niederhuth et al., 2013).
Boundary genes are required for both differentiation and separation phases of abscission. A variety of plant species lacking BOP activity fail to form an AZ (McKim et al., 2008; Wu et al., 2012; Couzigou et al., 2015). BOP1/2 quite possibly perform this function via ATH1 and KNAT2/KNAT6. Inactivation of ATH1 has a mild abscission defect in which formation of the stamen AZs is delayed. A functional AZ eventually develops and organs detach (Gómez-Mena and Sablowski, 2008). Reprising its role in leaves, AS1 positions the medial sepal and petal AZs in Arabidopsis via restriction of BP activity, which in turn restricts expression of the HAESA receptor-like kinase (Gubert et al., 2014). This was discovered through isolation of a new allele, as1-22, which shows a delayed abscission defect (Gubert et al., 2014).
The activation of floral organ abscission involves the peptide INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) and the receptor like kinase HAESA and HAESA-LIKE2 (HAE-HSL2) signaling pathway. Low levels of IDA and HAESA transcripts are also expressed in the mature dehiscence zones of the fruit (Stenvik et al., 2008). Activation of cell separation by IDA-HAE/HSL2 signaling antagonizes BP activity leading to up-regulation of KNAT2 and KNAT6 and accumulation of cell-wall modification and hydrolytic enzymes that mediate separation (Shi et al., 2011). Waxes, suberin, lignin, and pathogenesis-related genes are also induced in protecting exposed cells from dehydration and pathogen attack (Estornell et al., 2013; Kim, 2014). Plants overexpressing BOP1 show significant enrichment for genes involved in lignin biosynthesis, stress and pathogen resistance (Khan et al., 2012a, 2015) suggesting a potential role in post-abscission events at the boundary.
Not all AZs form at a lateral organ boundary (Estornell et al., 2013). The pedicel AZ in tomato is a well-studied example in which a small groove in the floral pedicel leads to differentiation of a “joint” where abscission takes place. Two MADS box proteins JOINTLESS and MACROCALYX form a complex that regulates formation of the pedicel AZ together with LAS required in AM production (Schumacher et al., 1999; Nakano et al., 2012). CUC2 homolog GOB has been proposed to be involved in the regulation of the onset of abscission based on its expression in tomato pedicel AZs (Nakano et al., 2013). Tomato KNAT6 (TKN3), KNAT2 (TKN4), and the KNATM homolog KD1 are all highly expressed in pedicel AZs similar to Arabidopsis. Silencing of KD1 delays abscission by increasing auxin content and overexpression of KD1 has the opposite effect (Ma et al., 2015). Hormonal control and transcript profiling of activated AZs in Arabidopsis and tomato are very similar despite these apparent differences (Estornell et al., 2013; Ito and Nakano, 2015).
An AZ at the base of the seed allows detachment from the funiculus. In Arabidopsis, specification of this AZ requires the MADS-box protein SEEDSTICK and the bHLH factor HECATE3 (Pinyopich et al., 2003; Ogawa et al., 2009). BOP1/2 do not seem to be expressed in the ovule nor is there an obvious requirement for the IDA-HAE/HLS2 signaling pathway (Estornell et al., 2013). This suggests a greater alignment with processes that control dehiscence. The role of PNY has yet to be investigated in seed dehiscence. Interestingly, domesticated japonica rice is selected for a promoter mutation in the PNY/RPL homolog qSHATTERING1 that depletes expression from the abscission layer to inhibit seed shatter (Konishi et al., 2006). qSH1 and a related gene SH5 are required for development of the pedicel AZ and inhibition of these genes reduces shatter by promoting lignin biosynthesis (Yoon et al., 2014). Overexpression of wheat TaqSH1 in Arabidopsis delays abscission and down regulates abscission-promoting genes suggesting that TaqSH1 can function as an upstream regulator of the IDA-HAESA-KNAT pathway (Zhang et al., 2013). Remarkably, the same mutation in qSH1 was found in Brassica rapa which exhibit a narrow replum compared to Arabidopsis (Arnaud et al., 2011). Collectively, these data show remarkable overlap between separation processes involved in pod shatter, floral organ abscission, and seed dehiscence in which lateral organ boundary genes play a key role.
Concluding Remarks
In this review, we illustrate the importance of boundaries throughout development. Studies have revealed a number of genes including CUCs, TALEs, and BOP that play a recurring role thoughout the life cycle. During the reproductive phase, their activities are embedded within specialized networks required for inflorescence, flower, and fruit development. How these pathways are integrated is only partly understood. Many questions also remain concerning the role of these genes in the SAM. Molecular links between CUC genes that confer boundary identity and BOP-TALE factors are not well-established. Recent studies in monocots have shed light on the role of KNOX transcription factors in initiating boundaries but application of this model to dicots is not yet confirmed. While significant progress has been made in understanding how KNOX factors regulate hormone abundance, such links are still largely missing for CUC and BOP factors at the boundary. These factors also repress growth and cell division but few targets have been identified to date and their hierarchy is unclear%. Identification of transcriptional targets is key to understanding how these factors pattern the boundary. Understanding how these networks translate to boundaries during reproductive development is still in its infancy. In particular, the influence of BR and the role of JLO is not yet explored. The function of BOP and TALE factors in fruit and in abscission is also unclear. Finally, the contribution of these networks to the activity of lateral meristems responsible for secondary growth in stems and roots is only partly understood. Transcriptome analysis and further exploration of these factors in boundaries in other species will establish the extent to which the mode of action of these factors is conserved in development.
Wolfgang von Goethe in 1790 first proposed that plants are formed from repeating units called phytomers (Bossinger, 2009). As developmental contexts are further explored this notion can be extended to the molecular level. Work in the last decade in a variety of species makes it obvious that simple patterns established with formation of the embryonic plant underlie pattern development at all stages of plant development. As such, there is much to be learned from drawing parallels and insights from different developmental contexts. Lateral organ boundaries are a particularly intriguing part of the puzzle as they regulate the balance between meristem activity and growth and represent a fundamental gatekeeper of meristem function in plants.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Special thanks to Bronwyn Rowland for illustrations. We acknowledge Aude Maugarny, Owen Rowland, Adina Popescu, and Nicolas Arnaud for critical comments on the manuscript.
Footnotes
Funding. The IJPB benefits from the support of the Labex Saclay Plant Sciences-SPS (ANR-10-LABX-0040-SPS). This work was supported by a Natural Sciences and Engineering Research Council Discovery Grant (no. 327195 to SH). This work was supported by the ANR CHARMFUL program (N° ANR-11BSV2 005 02 to VP).
References
- Aichinger E., Kornet N., Friedrich T., Laux T. (2012). Plant stem cell niches. Annu. Rev. Plant Biol. 63 615–636. 10.1146/annurev-arplant-042811-105555 [DOI] [PubMed] [Google Scholar]
- Aida M., Ishida T., Fukaki H., Fujisawa H., Tasaka M. (1997). Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9 841–857. 10.1105/tpc.9.6.841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M., Ishida T., Tasaka M. (1999). Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126 1563–1570. [DOI] [PubMed] [Google Scholar]
- Aida M., Vernoux T., Furutani M., Traas J., Tasaka M. (2002). Roles of PIN-FORMED1 and MONOPTEROS in pattern formation of the apical region of the Arabidopsis embryo. Development 129 3965–3974. [DOI] [PubMed] [Google Scholar]
- Alonso-Cantabrana H., Ripoll J. J., Ochando I., Vera A., Ferrándiz C., Martínez-Laborda A. (2007). Common regulatory networks in leaf and fruit patterning revealed by mutations in the Arabidopsis ASYMMETRIC LEAVES1 gene. Development 134 2663–2671. 10.1242/dev.02864 [DOI] [PubMed] [Google Scholar]
- Andrés F., Romera-Branchat M., Martìnez-Gallegos R., Patel V., Schneeberger K., Jang S., et al. (2015). Floral induction in Arabidopsis thaliana by FLOWERING LOCUS T requires direct repression of BLADE-ON-PETIOLE genes by homeodomain protein PENNYWISE. Plant Physiol. 169 2187–2199. 10.1104/pp.15.00960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud N., Girin T., Sorefan K., Fuentes S., Wood T. A., Lawrenson T., et al. (2010). Gibberellins control fruit patterning in Arabidopsis thaliana. Genes Dev. 24 2127–2132. 10.1101/gad.593410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud N., Lawrenson T., Østergaard L., Sablowski R. (2011). The same regulatory point mutation changed seed-dispersal structures in evolution and domestication. Curr. Biol. 21 1215–1219. 10.1016/j.cub.2011.06.008 [DOI] [PubMed] [Google Scholar]
- Arnaud N., Pautot V. (2014). Ring the BELL and tie the KNOX: roles for TALEs in gynoecium development. Front. Plant Sci. 5:93 10.3389/fpls.2014.00093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartrina I., Otto E., Strnad M., Werner T., Schmulling T. (2011). Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 23 69–80. 10.1105/tpc.110.079079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell E. M., Lin W. C., Husbands A. Y., Yu L., Jaganatha V., Jablonska B., et al. (2012). Arabidopsis LATERAL ORGAN BOUNDARIES negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc. Natl. Acad. Sci. U.S.A. 109 21146–21151. 10.1073/pnas.1210789109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belles-Boix E., Hamant O., Witiak S. M., Morin H., Traas J., Pautot V. (2006). KNAT6: an Arabidopsis homeobox gene involved in meristem activity and organ separation. Plant Cell 18 1900–1907. 10.1105/tpc.106.041988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencivenga S., Simonini S., Benkova E., Colombo L. (2012). The transcription factors BEL1 and SPL are required for cytokinin and auxin signaling during ovule development in Arabidopsis. Plant Cell 24 2886–2897. 10.1105/tpc.112.100164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E., Michneiwicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., et al. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115 591–602. 10.1016/S0092-8674(03)00924-3 [DOI] [PubMed] [Google Scholar]
- Berger Y., Harpaz-Saad S., Brand A., Melnik H., Sirding N., Alvarez J. P., et al. (2009). The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development 136 823–832. 10.1242/dev.031625 [DOI] [PubMed] [Google Scholar]
- Besnard F., Vernoux T., Hamant O. (2011). Organogenesis from stem cells in planta: multiple feedback loops integrating molecular and mechanical signals. Cell. Mol. Life Sci. 68 2885–2906. 10.1007/s00018-011-0732-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilsborough G. D., Runions A., Barkoulas M., Jenkins H. W., Hasson A., Galinha C., et al. (2011). Model for the regulation of Arabidopsis thaliana leaf margin development. Proc. Natl. Acad. Sci. U.S.A. 108 3424–3429. 10.1073/pnas.1015162108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blein T., Pulido A., Vialette-Guiraud A., Nikovics K., Morin H., Hay A., et al. (2008). A conserved molecular framework for compound leaf development. Science 322 1835–1839. 10.1126/science.1166168 [DOI] [PubMed] [Google Scholar]
- Bolduc N., Hake S. (2009). The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell 21 1647–1658. 10.1105/tpc.109.068221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N., Yilmaz A., Mejia-Guerra M. K., Morohashi K., O’Connor D., Grotewold E., et al. (2012). Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev. 26 1685–1690. 10.1101/gad.193433.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghi L., Bureau M., Simon R. (2007). Arabidopsis JAGGED LATERAL ORGANS is expressed in boundaries and coordinates KNOX and PIN activity. Plant Cell 19 1795–1808. 10.1105/tpc.106.047159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossinger G. (2009). “Segments (Phytomers),” in Encyclopedia of Life Sciences, ed. Hetherington A. M. (Chichester: John Wiley and Sons; ). 10.1002/9780470015902.a0002093.pub2 [DOI] [Google Scholar]
- Bureau M., Rast M. I., Illmer J., Simon R. (2010). JAGGED LATERAL ORGAN (JLO) controls auxin dependent patterning during development of the Arabidopsis embryo and root. Plant Mol. Biol. 74 479–491. 10.1007/s11103-010-9688-2 [DOI] [PubMed] [Google Scholar]
- Byrne M. E., Groover A. T., Fontana J. R., Martienssen R. A. (2003). Phyllotactic pattern and stem cell fate are determined by the Arabidopsis homeobox gene BELLRINGER. Development 130 3941–3950. 10.1242/dev.00620 [DOI] [PubMed] [Google Scholar]
- Byrne M. E., Simorowski J., Martienssen R. A. (2002). ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129 1957–1965. [DOI] [PubMed] [Google Scholar]
- Chae E., Tan Q. K.-G., Hill T. A., Irish V. F. (2008). An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development 135 1235–1245. 10.1242/dev.015842 [DOI] [PubMed] [Google Scholar]
- Chahtane H., Vachon G., Le Masson M., Thevenon E., Perigon S., Mihajlovic N., et al. (2013). A variant of LEAFY reveals its capacity to stimulate meristem development by inducing RAX1. Plant J. 74 678–689. 10.1111/tpj.12156 [DOI] [PubMed] [Google Scholar]
- Cho E., Zambryski P. C. (2011). ORGAN BOUNDARY1 defines a gene expressed at the junction between the shoot apical meristem and lateral organs. Proc. Natl. Acad. Sci. U.S.A. 108 2154–2159. 10.1073/pnas.1018542108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y., Maharjan P. M., Lee O., Fujioka S., Jang S., Kim B., et al. (2011). Auxin stimulates DWARF4 expression and brassinosteroid biosynthesis in Arabidopsis. Plant J. 66 564–578. 10.1111/j.1365-313X.2011.04513.x [DOI] [PubMed] [Google Scholar]
- Cole M., Nolte C., Werr W. (2006). Nuclear import of the transcription factor SHOOT MERISTEMLESS depends on heterodimerization with BLH proteins expressed in discrete subdomains of the shoot apical meristem of Arabidopsis thaliana. Nucleic Acids Res. 35 1281–1292. 10.1093/nar/gkl016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colling J., Tohge T., De Clercq R., Brunoud G., Vernoux T., Fernie A. R., et al. (2015). Overexpression of the Arabidopsis thaliana signalling peptide TAXIMIN1 affects lateral organ development. J. Exp. Bot. 66 5337–5349. 10.1093/jxb/erv291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couzigou J. M., Magne K., Mondy S., Cosson V., Clements J., Ratet P. (2015). The legume NOOT-BOP-COCH-LIKE genes are conserved regulators of abscission, a major agronomical trait in cultivated crops. New Phytol. 10.1111/nph.13634 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Cucinotta M., Colombo L., Roig-Villanova I. (2014). Ovule development, a new model for lateral organ formation. Front. Plant Sci. 5:117 10.3389/fpls.2014.00117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giacomo E., Lanneli M., Frugis G. (2013). TALE and shape: how to make a leaf different. Plants 2 317–342. 10.3390/plants2020317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny J. R., Weigel D., Yanofsky M. F. (2005). A genetic framework for fruit patterning in Arabidopsis thaliana. Development 132 4687–4696. 10.1242/dev.02062 [DOI] [PubMed] [Google Scholar]
- Douglas S. J., Chuck G., Dengler R. E., Pelecanda L., Riggs C. D. (2002). KNAT1 and ERECTA regulate inflorescence architecture in Arabidopsis. Plant Cell 14 547–558. 10.1105/tpc.010391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I., Eshed Y., Lifschitz E. (2010). Morphogenesis of simple and compound leaves: a critical review. Plant Cell 22 1019–1032. 10.1105/tpc.109.073601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endrizzi K., Moussian B., Haecker A., Levin J. Z., Laux T. (1996). The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 10 967–979. 10.1046/j.1365-313X.1996.10060967.x [DOI] [PubMed] [Google Scholar]
- Estornell L. H., Agusti J., Merelo P., Talon M., Tadeo F. R. (2013). Elucidating mechanisms underlying organ abscission. Plant Sci. 19 48–60. 10.1016/j.plantsci.2012.10.008 [DOI] [PubMed] [Google Scholar]
- Etchells J. P., Moore L., Jiang W. Z., Prescott H., Capper R., Saunders N. J., et al. (2012). A role for BELLRINGER in cell wall development is supported by loss-of-function phenotypes. BMC Plant Biol. 12:212 10.1186/1471-2229-12-212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrándiz C., Fourquin C., Prunet N., Scutt C. P., Trehin C., Vialette-Giraud A. (2010). “Carpel Development,” in Advances in Botanical Research, eds Kader J.-C., Delseny M. (Burlington, MA: Academic Press; ), 1–73. [Google Scholar]
- Ferrándiz C., Liljegren S. J., Yanofsky M. F. (2000). Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 289 436–438. 10.1126/science.289.5478.436 [DOI] [PubMed] [Google Scholar]
- Frigerio M., Alabadi D., Perez-Gomez J., Garcia-Carcel L., Phillips A. L., Hedden P., et al. (2006). Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol. 142 553–563. 10.1104/pp.106.084871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani M., Vernoux T., Traas J., Kato T., Tasaka M., Aida M. (2004). PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development 131 5021–5030. 10.1242/dev.01388 [DOI] [PubMed] [Google Scholar]
- Galbiati F., Sinha Roy D., Simonini S., Cucinotta M., Ceccato L., Cuesta C., et al. (2013). An integrative model of the control of ovule primordia formation. Plant J. 76 446–455. 10.1111/tpj.12309 [DOI] [PubMed] [Google Scholar]
- Gallavotti A., Zhao Q., Kyozuka J., Meeley R. B., Ritter M. K., Doebley J. F., et al. (2004). The role of barren stalk1 in the architecture of maize. Nature 432 630–635. 10.1038/nature03148 [DOI] [PubMed] [Google Scholar]
- Gendron J. M., Liu J. S., Fan M., Bai M. Y., Wenkel S., Springer P. S., et al. (2012). Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109 21152–21157. 10.1073/pnas.1210799110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girin T., Paicu T., Stephenson P., Fuentes S., Korner E., O’Brien M., et al. (2011). INDEHISCENT and SPATULA interact to specify carpel and valve margin tissue and thus promote seed dispersal in Arabidopsis. Plant Cell 23 3641–3653. 10.1105/tpc.111.090944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez M. D., Urbez C., Perez-Amador M. A., Carbonell J. (2011). Characterization of constricted fruit (ctf) mutant uncovers a role for AtMYB117/LOF1 in ovule and fruit development in Arabidopsis thaliana. PLoS ONE 6:e18760 10.1371/journal.pone.0018760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Mena C., Sablowski R. (2008). Arabidopsis thaliana HOMEOBOX GENE1 establishes the basal boundaries of shoot organs and controls stem growth. Plant Cell 20 2059–2072. 10.1105/tpc.108.059188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves B., Hasson A., Belcram K., Cortizo M., Morin H., Nikovics K., et al. (2015). A conserved role for CUP-SHAPED COTYLEDON genes during ovule development. Plant J. 83 732–742. 10.1111/tpj.12923 [DOI] [PubMed] [Google Scholar]
- González-Reig S., Ripoll J. J., Vera A., Yanofsky M. F., Martinez-Laborda A. (2012). Antagonistic gene activities determine the formation of pattern elements along the mediolateral axis of the Arabidopsis fruit. PLoS Genet. 8:e1003020 10.1371/journal.pgen.1003020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S. P., Chickarmane V. S., Ohno C., Meyerowitz E. M. (2009). Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. U.S.A. 106 16529–16534. 10.1073/pnas.0908122106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbic V., Bleecker A. B. (2000). Axillary meristem development in Arabidopsis thaliana. Plant J. 21 215–223. 10.1046/j.1365-313x.2000.00670.x [DOI] [PubMed] [Google Scholar]
- Greb T., Clarenz O., Schafer E., Muller D., Herrero R., Schmitz G., et al. (2003). Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 17 1175–1187. 10.1101/gad.260703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith M. E., da Silva Conceição A., Smyth D. R. (1999). PETAL LOSS gene regulates initiation and orientation of second whorl organs in the Arabidopsis flower. Development 126 5635–5644. [DOI] [PubMed] [Google Scholar]
- Groszmann M., Paicu T., Alvarez J. P., Swain S. M., Smyth D. R. (2011). SPATULA and ALCATRAZ, are partially redundant, functionally diverging bHLH genes required for Arabidopsis gynoecium and fruit development. Plant J. 68 816–829. 10.1111/j.1365-313X.2011.04732.x [DOI] [PubMed] [Google Scholar]
- Gubert C. M., Christy M. E., Ward D. L., Groner W. D., Liljegren S. J. (2014). ASYMMETRIC LEAVES1 regulates abscission zone placement in Arabidopsis flowers. BMC Plant Biol. 14:195 10.1186/s12870-014-0195-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Thomas J., Collins G., Timmermans M. C. P. (2008). Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 20 48–58. 10.1105/tpc.107.056127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha C. M., Jun J. H., Fletcher J. C. (2010). Control of Arabidopsis leaf morphogenesis through regulation of the YABBY and KNOX families of transcription factors. Genetics 186 197–206. 10.1534/genetics.110.118703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha C. M., Jun J. H., Nam H. G., Fletcher J. (2004). BLADE-ON-PETIOLE1 encodes a BTB/POZ domain protein required for leaf morphogenesis in Arabidopsis thaliana. Plant Cell Physiol. 45 1361–1370. 10.1093/pcp/pch201 [DOI] [PubMed] [Google Scholar]
- Ha C. M., Jun J. H., Nam H. G., Fletcher J. C. (2007). BLADE-ON-PETIOLE 1 and 2 control Arabidopsis lateral organ fate through regulation of LOB domain and adaxial-abaxial polarity genes. Plant Cell 19 1809–1825. 10.1105/tpc.107.051938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha C. M., Kim G.-T., Kim B. C., Jun J. H., Soh M. S., Ueno Y., et al. (2003). The BLADE-ON-PETIOLE1 gene controls leaf pattern formation through the modulation of meristematic activity in Arabidopsis. Development 130 161–172. 10.1242/dev.00196 [DOI] [PubMed] [Google Scholar]
- Hamant O., Pautot V. (2010). Plant development: a TALE story. C. R. Biol. 333 371–381. 10.1016/j.crvi.2010.01.015 [DOI] [PubMed] [Google Scholar]
- Han Y., Zhang C., Yang H., Jiao Y. (2014). Cytokinin pathway mediates APETALA1 function in the establishment of determinate floral meristems in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111 6840–6845. 10.1073/pnas.1318532111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson A., Plessis A., Blein T., Adroher B., Grigg S., Tsiantis M., et al. (2011). Evolution and diverse roles of the CUP-SHAPED COTYLEDON genes in Arabidopsis leaf development. Plant Cell 23 54–68. 10.1105/tpc.110.081448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A., Barkoulas M., Tsiantis M. (2006). ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf develoment in Arabidopsis. Development 133 3955–3961. 10.1242/dev.02545 [DOI] [PubMed] [Google Scholar]
- Hay A., Kaur H., Phillips A., Hedden P., Hake S., Tsiantis M. (2002). The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr. Biol. 12 1557–1565. 10.1016/S0960-9822(02)01125-9 [DOI] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2010). KNOX genes: versatile regulators of plant development and diversity. Development 137 3153–3165. 10.1242/dev.030049 [DOI] [PubMed] [Google Scholar]
- Heisler M. G., Hamant O., Krupinski P., Uyttewaal M., Ohno C., Jönsson H., et al. (2010). Alignment between PIN1 polarity and microtubule orientation in the shoot apical meristem reveals a tight coupling between morphogenesis and auxin transport. PLoS Biol. 8:e1000516 10.1371/journal.pbio.1000516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler M. G., Ohno C., Das P., Sieber P., Reddy G. V., Long J. A., et al. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15 1899–1911. 10.1016/j.cub.2005.09.052 [DOI] [PubMed] [Google Scholar]
- Hepworth S. R., Zhang Y., Mckim S., Li X., Haughn G. W. (2005). BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell 17 1434–1448. 10.1105/tpc.104.030536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibara K., Karim M. R., Takada S., Taoka K., Furutani M., Aida M., et al. (2006). Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18 2946–2957. 10.1105/tpc.106.045716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H. Y., Jiang W. B., Hu Y. W., Wu P., Zhu J. Y., Liang W. Q., et al. (2013). BR signal influences Arabidopsis ovule and seed number through regulating related genes expression by BZR1. Mol. Plant 6 456–469. 10.1093/mp/sss070 [DOI] [PubMed] [Google Scholar]
- Ichihashi Y., Aguilar-Martinez J. A., Farhi M., Chitwood D. H., Kumar R., Millon L. V., et al. (2014). Evolutionary developmental transcriptomics reveals a gene network module regulating interspecific diversity in plant leaf shape. Proc. Natl. Acad. Sci. U.S.A. 111 2616–2621. 10.1073/pnas.1402835111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T., Aida M., Takada S., Tasaka M. (2000). Involvement of CUP-SHAPED COTYLEDON genes in gynoecium and ovule development in Arabidopsis thaliana. Plant Cell Physiol. 41 60–67. 10.1093/pcp/41.1.60 [DOI] [PubMed] [Google Scholar]
- Ito Y., Nakano T. (2015). Development and regulation of pedicel abscission in tomato. Front. Plant Sci. 6:442 10.3389/fpls.2015.00442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen B. J., Drummond R. S., Snowden K. C. (2014). Regulation of axillary shoot development. Curr. Biol. 17 28–35. [DOI] [PubMed] [Google Scholar]
- Jasinski S., Piazza P., Craft J., Hay A., Woolley L., Rieu I., et al. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15 1560–1565. 10.1016/j.cub.2005.07.023 [DOI] [PubMed] [Google Scholar]
- Johnston R., Wang M., Sun Q., Sylvester A. W., Hake S., Scanlon M. J. (2014). Transcriptomic analyses indicate that maize ligule development recapitulates gene expression patterns that occur during lateral organ initiation. Plant Cell 26 4718–4732. 10.1105/tpc.114.132688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun J. H., Ha C. M., Fletcher J. C. (2010). BLADE-ON-PETIOLE1 coordinates organ determinacy and axial polarity in Arabidopsis by directly activating ASYMMETRIC LEAVES2. Plant Cell 22 62–76. 10.1105/tpc.109.070763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiuchi Y., Yamamoto K., Furutani M., Tasaka M., Aida M. (2014). The CUC1 and CUC2 genes promote carpel margin meristem formation during Arabidopsis gynoecium development. Front. Plant Sci. 5:165 10.3389/fpls.2014.00165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanrar S., Bhattacharya M., Arthur B., Courtier J., Smith H. M. (2008). Regulatory networks that function to specify flower meristems require the function of homeobox genes PENNYWISE and POUND-FOOLISH in Arabidopsis. Plant J. 54 924–937. 10.1111/j.1365-313X.2008.03458.x [DOI] [PubMed] [Google Scholar]
- Kanrar S., Onguka O., Smith H. M. (2006). Arabidopsis inflorescence architecture requires the activities of KNOX-BELL homeodomain heterodimers. Planta 224 1163–1173. 10.1007/s00425-006-0298-9 [DOI] [PubMed] [Google Scholar]
- Karim M. R., Hirota A., Kwiatkowska D., Tasaka M., Aida M. (2009). A role for Arabidopsis PUCHI in floral meristem identity and bract suppression. Plant Cell 21 1360–1372. 10.1105/tpc.109.067025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura E., Horiguchi G., Tsukaya H. (2010). Mechanisms of leaf tooth formation in Arabidopsis. Plant J. 62 429–441. 10.1111/j.1365-313X.2010.04156.x [DOI] [PubMed] [Google Scholar]
- Keller T., Abbott J., Moritz T., Doerner P. (2006). Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 18 598–611. 10.1105/tpc.105.038588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M., Ragni L., Tabb P., Salasini B. C., Chatfield S., Datla R., et al. (2015). Repression of lateral organ boundary genes by PENNYWISE and POUND-FOOLISH is essential for meristem maintenance and flowering in Arabidopsis thaliana. Plant Physiol. 169 2166–2186. 10.1104/pp.15.00915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M., Tabb P., Hepworth S. R. (2012a). BLADE-ON-PETIOLE1 and 2 regulate Arabidopsis inflorescence architecture in conjunction with homeobox genes KNAT6 and ATH1. Plant Signal. Behav. 7 788–792. 10.4161/psb.20599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M., Xu M., Murmu J., Tabb P., Liu Y., Storey K., et al. (2012b). Antagonistic interaction of BLADE-ON-PETIOLE1 and 2 with BREVIPEDICELLUS and PENNYWISE regulates Arabidopsis inflorescence architecture. Plant Physiol. 158 946–960. 10.1104/pp.111.188573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M., Xu H., Hepworth S. R. (2014). BLADE-ON-PETIOLE genes: setting boundaries in development and defense. Plant Sci. 21 157–171. 10.1016/j.plantsci.2013.10.019 [DOI] [PubMed] [Google Scholar]
- Kim D., Cho Y. H., Ryu H., Kim Y., Kim T. H., Hwang I. (2013). BLH1 and KNAT3 modulate ABA responses during germination and early seedling development in Arabidopsis. Plant J. 75 755–766. 10.1111/tpj.12236 [DOI] [PubMed] [Google Scholar]
- Kim J. (2014). Four shades of detachment: regulation of floral organ abscission. Plant Signal. Behav. 9 e976154 10.4161/15592324.2014.976154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S., Koenig D., Kang J., Yoong F. Y., Sinha N. (2008). Natural variation in leaf morphology results from mutation of a novel KNOX gene. Curr. Biol. 18 672–677. 10.1016/j.cub.2008.04.008 [DOI] [PubMed] [Google Scholar]
- Komatsu K., Maekawa M., Ujiie S., Satake Y., Furutani I., Okamoto H., et al. (2003). LAX and SPA: major regulators of shoot branching in rice. Proc. Natl. Acad. Sci. U.S.A. 100 11765–11770. 10.1073/pnas.1932414100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi S., Izawa T., Lin S. Y., Ebana K., Fukuta Y., Sasaki T., et al. (2006). An SNP caused loss of seed shattering during rice domestication. Science 312 1392–1396. 10.1126/science.1126410 [DOI] [PubMed] [Google Scholar]
- Koyama T., Furutani M., Tasaka M., Ohme-Takagi M. (2007). TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19 473–484. 10.1105/tpc.106.044792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Mitsuda N., Seki M., Shinozaki K., Ohme-Takagi M. (2010). TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22 3574–3588. 10.1105/tpc.110.075598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C. S., Hibara K., Pfluger J., Bazhani S., Metha S., Metha H., et al. (2006). A role for chromatin remodeling in regulation of CUC gene expression in the Arabidopsis cotyledon boundary. Development 133 3223–3230. 10.1242/dev.02508 [DOI] [PubMed] [Google Scholar]
- Lal S., Pacis L. B., Smith H. M. (2011). Regulation of the SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE genes/microRNA156 module by the homeodomain proteins PENNYWISE and POUND-FOOLISH in Arabidopsis. Mol. Plant 4 1123–1132. 10.1093/mp/ssr041 [DOI] [PubMed] [Google Scholar]
- Lampugnani E. R., Kilinc A., Smyth D. R. (2012). PETAL LOSS is a boundary gene that inhibits growth between developing sepals in Arabidopsis thaliana. Plant J. 71 724–735. 10.1111/j.1365-313X.2012.05023.x [DOI] [PubMed] [Google Scholar]
- Lampugnani E. R., Kilinc A., Smyth D. R. (2013). Auxin controls petal initiation in Arabidopsis. Development 140 185–194. 10.1242/dev.084582 [DOI] [PubMed] [Google Scholar]
- Laufs P., Peaucelle A., Morin H., Traas J. (2004). MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131 4311–4322. 10.1242/dev.01320 [DOI] [PubMed] [Google Scholar]
- Lee B. H., Jeon J. O., Lee M. M., Kim J. H. (2015). Genetic interaction between GROWTH-REGULATING FACTOR and CUP-SHAPED COTYLEDON in organ separation. Plant Signal. Behav. 10 e988071 10.4161/15592324.2014.988071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. K., Geisler M., Springer P. S. (2009). LATERAL ORGAN FUSION1 and LATERAL ORGAN FUSION2 function in lateral organ separation and axillary meristem formation in Arabidopsis. Development 136 2423–2432. 10.1242/dev.031971 [DOI] [PubMed] [Google Scholar]
- Lee I. H., Wolfe D. S., Nilsson O., Weigel D. (1997). A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS. Curr. Biol. 7 95–104. 10.1016/S0960-9822(06)00053-4 [DOI] [PubMed] [Google Scholar]
- Lee J. E., Lampugnani E. R., Bacic A., Golz J. F. (2014). SEUSS and SEUSS-LIKE 2 coordinate auxin distribution and KNOXI activity during embryogenesis. Plant J. 80 122–135. 10.1111/tpj.12625 [DOI] [PubMed] [Google Scholar]
- Leibfried A., To J. P., Busch W., Stehling S., Kehle A., Demar M., et al. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438 1172–1175. 10.1038/nature04270 [DOI] [PubMed] [Google Scholar]
- Levin J. Z., Meyerowitz E. (1995). UFO: an Arabidopsis gene involved in both floral meristem and floral organ development. Plant Cell 7 529–548. 10.1105/tpc.7.5.529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie C., Kelsom C., Wu X. (2012). WOX2 and STIMPY-LIKE/WOX8 promote cotyledon boundary formation in Arabidopsis. Plant J. 72 674–682. [DOI] [PubMed] [Google Scholar]
- Liebsch D., Sunaryo W., Holmlund M., Norberg M., Zhang J., Hall H. C., et al. (2014). Class I KNOX transcription factors promote differentiation of cambial derivatives into xylem fibers in the Arabidopsis hypocotyl. Development 141 4311–4319. 10.1242/dev.111369 [DOI] [PubMed] [Google Scholar]
- Liljegren S. J., Ditta G. S., Eshed Y., Savidge B., Bowman J. L., Yanofsky M. F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404 766–770. 10.1038/35008089 [DOI] [PubMed] [Google Scholar]
- Liljegren S. J., Roeder A. H., Kempin S. A., Gremski K., Ostergaard L., Guimil S., et al. (2004). Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell 116 843–853. 10.1016/S0092-8674(04)00217-X [DOI] [PubMed] [Google Scholar]
- Lincoln C., Long J., Yamaguchi J., Serikawa K., Hake S. (1994). A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6 1859–1876. 10.1105/tpc.6.12.1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodha M., Marco C. F., Timmermans M. C. P. (2013). The ASYMMETRIC LEAVES complex maintains repression of KNOX homeobox genes via direct recruitment of Polycomb-repressive complex2. Genes Dev. 27 596–601. 10.1101/gad.211425.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann J. U., Weigel D. (2002). Building beauty: the genetic control of floral patterning. Dev. Cell 2 135–142. 10.1016/S1534-5807(02)00122-3 [DOI] [PubMed] [Google Scholar]
- Long J. A., Barton M. K. (1998). The development of apical embryonic pattern in Arabidopsis. Development 125 3027–3035. [DOI] [PubMed] [Google Scholar]
- Long J., Barton M. K. (2000). Initiation of axillary and floral meristems in Arabidopsis. Dev. Biol. 218 341–353. 10.1006/dbio.1999.9572 [DOI] [PubMed] [Google Scholar]
- Ma C., Meir S., Xiao L., Tong J., Liu Q., Reid M. S., et al. (2015). A KNOTTED1-LIKE HOMEOBOX protein regulates abscission in tomato by modulating the auxin pathway. Plant Physiol. 167 844–853. 10.1104/pp.114.253815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani E., Hake S. (2008). KNOX lost the OX: the Arabidopsis KNATM gene defines a novel class of KNOX transcriptional regulators missing the homeodomain. Plant Cell 20 875–887. 10.1105/tpc.108.058495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A. C., Dugas D. V., Bartel D. P., Bartel B. (2004). MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr. Biol. 14 1035–1046. 10.1016/j.cub.2004.06.022 [DOI] [PubMed] [Google Scholar]
- Marsch-Martinez N., Ramos-Cruz D., Irepan Reyes-Olalde J., Lozano-Sotomayor P., Zuniga-Mayo V. M., De Folter S. (2012). The role of cytokinin during Arabidopsis gynoecia and fruit morphogenesis and patterning. Plant J. 72 222–234. 10.1111/j.1365-313X.2012.05062.x [DOI] [PubMed] [Google Scholar]
- Marsch-Martinez N., Zuniga-Mayo V. M., Herrera-Ubaldo H., Ouwerkerk P. B., Pablo-Villa J., Lozano-Sotomayor P., et al. (2014). The NTT transcription factor promotes replum development in Arabidopsis fruits. Plant J. 80 69–81. 10.1111/tpj.12617 [DOI] [PubMed] [Google Scholar]
- Maugarny A., Gonçalves B., Arnaud N., Laufs P. (2015). “CUC transcription factors: to the meristem and beyond,” in Plant Transcription Factors: Evolutionary, Structural and Functional Aspects, ed. Gonzalez D. H. (Amsterdam: Elsevier; ). [Google Scholar]
- McKim S. M., Stenvik G. E., Butenko M. A., Kristiansen W., Cho S. K., Hepworth S. R., et al. (2008). The BLADE-ON-PETIOLE genes are essential for abscission zone formation in Arabidopsis. Development 135 1537–1546. 10.1242/dev.012807 [DOI] [PubMed] [Google Scholar]
- Mele G., Ori M., Sato Y., Hake S. (2003). The knotted1-like homeobox gene BREVIPEDICELLUS regulates cell differentiation by modulating metabolic pathways. Genes Dev. 17 2088–2093. 10.1101/gad.1120003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyroud E., Minguet E. G., Ott F., Yant L., Pose D., Monniaux M., et al. (2011). Prediction of regulatory interactions from genome sequences using a biophysical model for the Arabidopsis LEAFY transcription factor. Plant Cell 23 1293–1306. 10.1105/tpc.111.083329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller D., Schmitz G., Theres K. (2006). BLIND homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 18 586–597. 10.1105/tpc.105.038745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T., Fujisawa M., Shima Y., Ito Y. (2013). Expression profiling of tomato pre-abscission pedicels provides insights into abscission zone properties including competence to respond to abscission signals. BMC Plant Biol. 13:40 10.1186/1471-2229-13-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T., Kimbara J., Fujisawa M., Kitagawa M., Ihashi N., Maeda H., et al. (2012). MACROCALYX and JOINTLESS interact in the transcriptional regulation of tomato fruit abscission zone development. Plant Physiol. 158 439–450. 10.1104/pp.111.183731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawy T., Bayer M., Mravec J., Friml J., Birnbaum K. D., Lukowitz W. (2010). The GATA factor HANABA TARANU is required to position the proembryo boundary in the early Arabidopsis embryo. Dev. Cell 19 103–113. 10.1016/j.devcel.2010.06.004 [DOI] [PubMed] [Google Scholar]
- Nibau C., Di Stilio V. S., Wu H. M., Cheung A. Y. (2011). Arabidopsis and Tobacco SUPERMAN regulate hormone signalling and mediate cell proliferation and differentiation. J. Exp. Bot. 62 949–961. 10.1093/jxb/erq325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederhuth C. E., Cho S. K., Seitz K., Walker J. C. (2013). Letting go is never easy: abscission and receptor-like protein kinases. J. Integr. Plant Biol. 55 1251–1263. 10.1111/jipb.12116 [DOI] [PubMed] [Google Scholar]
- Nikovics K., Blein T., Peaucelle A., Ishida T., Morin H., Aida M., et al. (2006). The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 18 2929–2945. 10.1105/tpc.106.045617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norberg M., Holmlund M., Nilsson O. (2005). The BLADE ON PETIOLE genes act redundantly to control the growth and development of lateral organs. Development 132 2203–2213. 10.1242/dev.01815 [DOI] [PubMed] [Google Scholar]
- Nordstrom A., Tarkowski P., Tarkowska D., Norbaek R., Astot C., Dolezal K., et al. (2004). Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: a factor of potential importance for auxin-cytokinin-regulated development. Proc. Natl. Acad. Sci. U.S.A. 101 8039–8044. 10.1073/pnas.0402504101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M., Kay P., Wilson S., Swain S. M. (2009). ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are Polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 21 216–233. 10.1105/tpc.108.063768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa T., Kyozuka J. (2009). Two-step regulation of LAX PANICLE1 protein accumulation in axillary meristem formation in rice. Plant Cell 21 1095–1108. 10.1105/tpc.108.065425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pautot V., Dockx J., Hamant O., Kronenberger J., Grandjean O., Jublot D., et al. (2001). KNAT2: evidence for a link between KNOTTED-like genes and carpel development. Plant Cell 13 1719–1734. 10.1105/tpc.13.8.1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peaucelle A., Braybrook S. A., Le Guillou L., Bron E., Kuhlemeier C., Höfte H. (2011). Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr. Biol. 21 1720–1726. 10.1016/j.cub.2011.08.057 [DOI] [PubMed] [Google Scholar]
- Peaucelle A., Morin H., Traas J., Laufs P. (2007). Plants expressing a miR164-resistant CUC2 gene reveal the importance of post-meristematic maintenance of phyllotaxy in Arabidopsis. Development 134 1045–1050. 10.1242/dev.02774 [DOI] [PubMed] [Google Scholar]
- Pinyopich A., Ditta G. S., Savidge B., Liljegren S. J., Baumann E., Wisman E., et al. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424 85–88. 10.1038/nature01741 [DOI] [PubMed] [Google Scholar]
- Proveniers M., Rutjens B., Brand M., Smeekens S. (2007). The Arabidopsis TALE homeobox gene ATH1 controls floral competency through positive regulation of FLC. Plant J. 52 899–913. 10.1111/j.1365-313X.2007.03285.x [DOI] [PubMed] [Google Scholar]
- Ragni L., Belles-Boix E., Gunl M., Pautot V. (2008). Interaction of KNAT6 and KNAT2 with BREVIPEDICELLUS and PENNYWISE in Arabidopsis inflorescences. Plant Cell 20 888–900. 10.1105/tpc.108.058230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajani S., Sundaresan V. (2001). The Arabidopsis myc/bHLH gene ALCATRAZ enables cell separation in fruit dehiscence. Curr. Biol. 11 1914–1922. 10.1016/S0960-9822(01)00593-0 [DOI] [PubMed] [Google Scholar]
- Raman S., Greb T., Peaucelle A., Blein T., Laufs P., Theres K. (2008). Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J. 55 65–76. 10.1111/j.1365-313X.2008.03483.x [DOI] [PubMed] [Google Scholar]
- Rast M. I., Simon R. (2012). Arabidopsis JAGGED LATERAL ORGANS acts with ASYMMETRIC LEAVES2 to coordinate KNOX and PIN expression in shoot and root meristems. Plant Cell 24 2917–2933. 10.1105/tpc.112.099978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Olalde J. I., Zuniga-Mayo V. M., Chavez Montes R. A., Marsch-Martinez N., De Folter S. (2013). Inside the gynoecium: at the carpel margin. Trends Plant Sci. 18 644–655. 10.1016/j.tplants.2013.08.002 [DOI] [PubMed] [Google Scholar]
- Ripoll J. J., Roeder A. H., Ditta G. S., Yanofsky M. F. (2011). A novel role for the floral homeotic gene APETALA2 during Arabidopsis fruit development. Development 138 5167–5176. 10.1242/dev.073031 [DOI] [PubMed] [Google Scholar]
- Risseeuw E., Venglat P., Xiang D., Komendant K., Daskalchuk T., Babic V., et al. (2013). An activated form of UFO alters leaf development and produces ectopic floral and inflorescence meristems. PLoS ONE 8:e83807 10.1371/journal.pone.0083807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder A. H., Ferrándiz C., Yanofsky M. F. (2003). The role of the REPLUMLESS homeodomain protein in patterning the Arabidopsis fruit. Curr. Biol. 13 1630–1635. 10.1016/j.cub.2003.08.027 [DOI] [PubMed] [Google Scholar]
- Rubio-Somoza I., Zhou C.-M., Confraria A., Martinho C., Von Born P., Baena-Gonzalez E., et al. (2014). Temporal control of leaf complexity by miRNA-regulated licensing of protein complexes. Curr. Biol. 24 2714–2719. 10.1016/j.cub.2014.09.058 [DOI] [PubMed] [Google Scholar]
- Rutjens B., Bao D., Van Eck-Stouten E., Brand M., Smeekens S., Proveniers M. (2009). Shoot apical meristem function in Arabidopsis requires the combined activities of three BEL1-like homeodomain proteins. Plant J. 58 641–654. 10.1111/j.1365-313X.2009.03809.x [DOI] [PubMed] [Google Scholar]
- Sakai H., Medrano L., Meyerowitz E. (1995). Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature 378 199–203. 10.1038/378199a0 [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Kamiya N., Ueguchi-Tanaka M., Iwahori S., Matsuoka M. (2001). KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev. 15 581–590. 10.1101/gad.867901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A., Klenz J. E., Kohalmi S. E., Risseeuw E., Haughn G. W., Crosby W. L. (1999). The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J. 20 433–445. 10.1046/j.1365-313x.1999.00617.x [DOI] [PubMed] [Google Scholar]
- Sassi M., Ali O., Boudon F., Cloarec G., Abad U., Cellier C., et al. (2014). An auxin-mediated shift toward growth isotropy promotes organ formation at the shoot meristem in Arabidopsis. Curr. Biol. 24 2335–2342. 10.1016/j.cub.2014.08.036 [DOI] [PubMed] [Google Scholar]
- Schiessl K., Muino J. M., Sablowski R. (2014). Arabidopsis JAGGED links floral organ patterning to tissue growth by repressing Kip-related cell cycle inhibitors. Proc. Natl. Acad. Sci. U.S.A. 111 2830–2835. 10.1073/pnas.1320457111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz G., Tillmann E., Carriero F., Fiore C., Cellini F., Theres K. (2002). The tomato BLIND gene encodes a MYB transcription factor that controls the formation of lateral meristems. Proc. Natl. Acad. Sci. U.S.A. 99 1064–1069. 10.1073/pnas.022516199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H., Lenhard M., Haecker A., Mayer K. F., Jurgens G., Laux T. (2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100 635–644. 10.1016/S0092-8674(00)80700-X [DOI] [PubMed] [Google Scholar]
- Schuetz M., Berleth T., Mattsson J. (2008). Multiple MONOPTEROS-dependent pathways are involved in leaf initiation. Plant Physiol. 148 870–880. 10.1104/pp.108.119396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher K., Schmitt T., Rossberg M., Schmitz G., Theres K. (1999). The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc. Natl. Acad. Sci. U.S.A. 96 290–295. 10.1073/pnas.96.1.290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield S., Dewitte W., Murray J. A. H. (2007). The KNOX gene SHOOT MERISTEMLESS is required for the development of reproductive meristematic tissues in Arabidopsis. Plant J. 50 767–781. [DOI] [PubMed] [Google Scholar]
- Shi C. L., Stenvik G. E., Vie A. K., Bones A. M., Pautot V., Proveniers M., et al. (2011). Arabidopsis class I KNOTTED-like homeobox proteins act downstream in the IDA-HAE/HSL2 floral abscission signaling pathway. Plant Cell 23 2553–2567. 10.1105/tpc.111.084608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. M., Hake S. (2003). The interaction of two homeobox genes, BREVIPEDICELLUS and PENNYWISE, regulates internode patterning in the Arabidopsis inflorescence. Plant Cell 15 1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. M. S., Boschke I., Hake S. (2002). Selective interaction of plant homeodomain proteins mediates high DNA-binding affinity. Proc. Natl. Acad. Sci. U.S.A. 99 9579–9584. 10.1073/pnas.092271599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. M. S., Campbell B. C., Hake S. (2004). Competence to respond to floral inductive signals requires the homeobox genes PENNYWISE and POUND-FOOLISH. Curr. Biol. 14 812–817. 10.1016/j.cub.2004.04.032 [DOI] [PubMed] [Google Scholar]
- Sorefan K., Girin T., Liljegren S. J., Ljung K., Robles P., Galván-Ampudia C. S., et al. (2009). A regulated auxin minimum is required for seed dispersal in Arabidopsis. Nature 459 583–586. 10.1038/nature07875 [DOI] [PubMed] [Google Scholar]
- Spinelli S. V., Martin A. P., Viola I. L., Gonzalez D. H., Palatnik J. F. (2011). A mechanistic link between STM and CUC1 during Arabidopsis development. Plant Physiol. 156 1894–1904. 10.1104/pp.111.177709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik G. E., Tandstad N. M., Guo Y., Shi C. L., Kristiansen W., Holmgren A., et al. (2008). The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20 1805–1817. 10.1105/tpc.108.059139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S., Hibara K., Ishida T., Tasaka M. (2001). The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 128 1127–1135. [DOI] [PubMed] [Google Scholar]
- Takeda S., Hanano K., Kariya A., Shimizu S., Zhao L., Matsui M., et al. (2011). CUP-SHAPED COTYLEDON1 transcription factor activates the expression of LSH4 and LSH3, two members of the ALOG gene family, in shoot organ boundary cells. Plant J. 66 1066–1077. 10.1111/j.1365-313X.2011.04571.x [DOI] [PubMed] [Google Scholar]
- Takeda S., Matsumoto N., Okada K. (2004). RABBIT EARS, encoding a SUPERMAN-like zinc finger protein, regulates petal development in Arabidopsis thaliana. Development 131 425–434. 10.1242/dev.00938 [DOI] [PubMed] [Google Scholar]
- Tavakol E., Okagaki R., Verderio G., Shariati J. V., Hussien A., Bilgic H., et al. (2015). The barley Uniculme4 gene encodes a BLADE-ON-PETIOLE-like protein that controls tillering and leaf patterning. Plant Physiol. 168 164–174. 10.1104/pp.114.252882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C., Zhang X., He J., Yu H., Wang Y., Shi B., et al. (2014). An organ boundary-enriched gene regulatory network uncovers regulatory hierarchies underlying axillary meristem initiation. Mol. Syst. Biol. 10 755 10.15252/msb.20145470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K., Kurata N., Ohyanagi H., Hake S. (2014). Genome-wide study of KNOX regulatory network reveals brassinosteroid catabolic genes important for shoot meristem function in rice. Plant Cell 26 3488–3500. 10.1105/tpc.114.129122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ung N., Lal S., Smith H. M. S. (2011). The role of PENNYWISE and POUND-FOOLISH in the maintenance of the shoot apical meristem in Arabidopsis. Plant Physiol. 156 605–614. 10.1104/pp.110.171462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venglat S. P., Dumonceaux T., Rozwadowski K., Parnell L., Babic V., Keller W., et al. (2002). The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 99 4730–4735. 10.1073/pnas.072626099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T., Kronenberger J., Grandjean O., Laufs P., Traas J. (2000). PIN-FORMED 1 regulates cell fate at the periphery of the shoot apical meristem. Development 127 5157–5165. [DOI] [PubMed] [Google Scholar]
- Vlad D., Kierzkowski D., Rast M. I., Vuolo F., Dello Ioio R., Galinha C., et al. (2014). Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science 343 780–783. 10.1126/science.1248384 [DOI] [PubMed] [Google Scholar]
- Vroemen C. W., Mordhorst A. P., Albrecht C., Kwaaitaal M. A. C. J., De Vries S. C. (2003). The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15 1563–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D., Sablowski R. W., Meyerowitz E. M. (1999). Transcriptional activation of APETALA1 by LEAFY. Science 285 582–584. 10.1126/science.285.5427.582 [DOI] [PubMed] [Google Scholar]
- Walsh J., Waters C. A., Freeling M. (1998). The maize gene liguleless2 encodes a basic leucine zipper protein involved in the establishment of the leaf blade-sheath boundary. Genes Dev. 12 208–218. 10.1101/gad.12.2.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Kohlen W., Rossmann S., Vernoux T., Theres K. (2014a). Auxin depletion from the leaf axil conditions competence for axillary meristem formation in Arabidopsis and tomato. Plant Cell 26 2068–2079. 10.1105/tpc.114.123059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wang J., Shi B., Yu T., Qi J., Meyerowitz E. M., et al. (2014b). The stem cell niche in leaf axils is established by auxin and cytokinin in Arabidopsis. Plant Cell 26 2055–2067. 10.1105/tpc.114.123083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Xu W., Ma L., Fu Z., Deng X., Li J., et al. (2006). Requirement of KNAT1/BP for the development of abscission zones in Arabidopsis thaliana. J. Integr. Plant Biol. 48 15–26. 10.1111/j.1744-7909.2005.00085.x-i1 [DOI] [Google Scholar]
- Wilkinson M. D., Haughn G. W. (1995). UNUSUAL FLORAL ORGANS controls meristem identity and organ primordia fate in Arabidopsis. Plant Cell 7 1485–1499. 10.2307/3870137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- William D. A., Su Y., Smith M. R., Lu M., Baldwin D. A., Wagner D. (2004). Genomic identification of direct target genes of LEAFY. Proc. Natl. Acad. Sci. U.S.A. 101 1775–1780. 10.1073/pnas.0307842100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter C. M., Austin R. S., Blanvillain-Baufume S., Reback M. A., Monniaux M., Wu M. F., et al. (2011). LEAFY target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Dev. Cell 20 430–443. 10.1016/j.devcel.2011.03.019 [DOI] [PubMed] [Google Scholar]
- Wu X. M., Yu Y., Han L. B., Li C. L., Wang H. Y., Zhong N. Q., et al. (2012). The tobacco BLADE-ON-PETIOLE2 gene mediates differentiation of the corolla abscission zone by controlling longitudinal cell expansion. Plant Physiol. 159 835–850. 10.1104/pp.112.193482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Li Z., Zhu Y., Wang H., Ma H., Dong A., et al. (2008). Arabidopsis genes AS1, AS2, and JAG negatively regulate boundary-specifying genes to promote sepal and petal development. Plant Physiol. 146 566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Hu T., Mckim S. M., Murmu J., Haughn G. W., Hepworth S. R. (2010). Arabidopsis BLADE-ON-PETIOLE1 and 2 promote floral meristem fate and determinacy in a previously undefined pathway targeting APETALA1 and AGAMOUS-LIKE24. Plant J. 63 974–989. 10.1111/j.1365-313X.2010.04299.x [DOI] [PubMed] [Google Scholar]
- Yamaguchi J., Wu M.-F., Winter C. M., Berns M. C., Nole-Wilson S., Yamaguchi A., et al. (2013). A molecular framework for auxin-mediated initiation of flower primordia. Dev. Cell 24 271–282. 10.1016/j.devcel.2012.12.017 [DOI] [PubMed] [Google Scholar]
- Yanai O., Shani E., Dolezal K., Tarkowski P., Sablowski R., Sandberg G., et al. (2005). Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 15 1566–1571. 10.1016/j.cub.2005.07.060 [DOI] [PubMed] [Google Scholar]
- Yang F., Wang Q., Schmitz G., Muller D., Theres K. (2012). The bHLH protein ROX acts in concert with RAX1 and LAS to modulate axillary meristem formation in Arabidopsis. Plant J. 71 61–70. 10.1111/j.1365-313X.2012.04970.x [DOI] [PubMed] [Google Scholar]
- Yoon J., Cho L. H., Kim S. L., Choi H., Koh H. J., An G. (2014). The BEL1-type homeobox gene SH5 induces seed shattering by enhancing abscission-zone development and inhibiting lignin biosynthesis. Plant J. 79 717–728. 10.1111/tpj.12581 [DOI] [PubMed] [Google Scholar]
- Žádníková P., Simon R. (2014). How boundaries control plant development. Curr. Opin. Plant Biol. 17 116–125. 10.1016/j.pbi.2013.11.013 [DOI] [PubMed] [Google Scholar]
- Zhang L., Liu D., Wang D., Hang R., Geng S., Wu L., et al. (2013). Over expression of the wheat BEL1-like gene TaqSH1 affects floral organ abscission in Arabidopsis thaliana. J. Plant Biol. 56 98–105. 10.1007/s12374-012-0438-7 [DOI] [Google Scholar]
- Zhao H., Liu L., Mo H., Qian L., Cao Y., Cui S., et al. (2013). The ATP-binding cassette transporter ABCB19 regulates postembryonic organ separation in Arabidopsis. PLoS ONE 8:e60809 10.1371/journal.pone.0060809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Yang S., Chen C. Y., Li C., Shan W., Lu W., et al. (2015). Arabidopsis BREVIPEDICELLUS interacts with the SWI2/SNF2 chromatin remodeling ATPase BRAHMA to regulate KNAT2 and KNAT6 expression in control of inflorescence architecture. PLOS Genet. 11:e1005125 10.1371/journal.pgen.1005125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Medrano L., Ohashi K., Fletcher J. C., Yu H., Sakai H., et al. (2004). HANABA TARANU is a GATA transcription factor that regulates shoot apical meristem and flower development in Arabidopsis. Plant Cell 16 2586–2600. 10.1105/tpc.104.024869 [DOI] [PMC free article] [PubMed] [Google Scholar]