Abstract
Laryngeal cancer (LC) is one of the most prevalent types of head and neck cancer. An increasing interest has been focused on the role of microRNA (miRNAs) in LC development. The study group consisted of 135 larynx cancer patients and 170 cancer-free individuals. Nine polymorphisms of pre-miRNA processing genes, DROSHA (rs6877842), DGCR8 (rs3757, rs417309, and rs1640299), RAN (rs14035), XPO5 (rs11077), DICER1 (rs13078 and rs3742330) and TARBP2 (rs784567), were performed by TaqMan SNP Genotyping Assay. It was found that the frequency of the GT and the TT polymorphic variants of XPO5 gene were higher in LC patients than in controls (p < 0.0001 and p = 0.000183, resp.). In turn, the frequency of the CT genotype of RAN gene was higher in controls than in LC patients (p < 0.0001). The TT and the AG of DICER1 gene (p = 0.034697 for rs13078 and p = 0.0004 for rs3742330) as well as the AG and the GG genotypes of TARBP2 gene (p = 0.008335 and p < 0.0001, resp.) were associated with higher risk of LC occurrence. Our data suggested that polymorphisms of miRNA processing genes might be useful as predictive factors for the LC development.
1. Introduction
Laryngeal cancer (LC) is one of the most prevalent types of head and neck cancer (HNC) in the contemporary world. Despite the improvement of surgery, chemotherapy, and radiotherapy, the 5-year survival rates are less than 60% [1]. In the last years, increasing interest has been focused on the role of microRNA (miRNA) in cancer development and progression. As in other malignancies, miRNA regulates several oncogenes and tumor suppressors, driving the growth, proliferation, metastatic attitude, and drug resistance [2–4].
Among many environmental (like tobacco smoking, alcohol consumption, or Human Papilloma Virus infection) and genetic factors that can cause HNC, it is still unknown whether factors engaged in microRNA (miRNA) processing can be one among the factors that can put an individual at risk of the disease [5, 6]. miRNAs are a class of endogenous, short (approximately 22 nt) noncoding RNAs that have emerged as major regulators of posttranscriptional gene expression. Thus, they can decrease the level of many proteins, despite the high level of transcription. Elements that are crucial for proper working of miRNA are proteins involved in miRNAs processing, because they reorganize the structure of premature pre-miRNAs into fully functioning miRNAs [7].
miRNA genes are first transcribed by RNA II polymerase into primary miRNAs (pri-miRNAs) with several hundred nucleotides. Processing of primary miRNAs (pri-miRNA) by the nuclear RNase III DROSHA within the microprocessor complex also including DGCR8 (DiGeorge syndrome critical region gene 8) produces the 70 to 100 nt pre-miRNAs. The pre-miRNAs are then exported into the cytoplasm by the Exportin-5/Ran-GTP complex and cleaved by DICER as a part of the RNA-induced silencing complex's loading complex including TARBP2 and AGO2. This complex also includes GEMIN3 and GEMIN4 and contributes to both miRNA processing and target gene silencing. miRNAs regulate gene expression in animals and plants through binding to the 3′ untranslated region (UTR) of the mRNAs of their target genes and leading to mRNA cleavage or translation repression [8–10]. Thomson et al. [11] have shown that the repression of mature miRNAs is not consistent with the reductions in the primary miRNA transcripts, suggesting the existence of altered regulations of miRNA processing in human cancers. Aberrant expression of miRNAs contributes to the etiology of many common human diseases, especially cancers [12, 13].
Some changes, like single nucleotide polymorphisms (SNPs), in the structure of genes encoding miR processing proteins may affect their structure or expression level. Although SNPs have been widely implicated in HNC development [14], such evidence is lacking for miRNA biogenesis pathway genes. Thus, the aim of our study is to evaluate the connection of prevalence of LC with SNPs occurring in the following genes: DROSHA (rs6877842), DGCR8 (rs3757, rs417309, and rs1640299), RAN (rs14035), XPO5 (rs11077), DICER1 (rs13078 and rs3742330), and TARBP2 (rs784567). The new data may bring a new knowledge of genetic factors that may affect the risk of LC development. This may be useful as a screening factor that may help to classify the individuals to the group of higher risk and enroll them for the special prophylactic program.
2. Materials and Methods
2.1. Subjects
All subjects included in the study were unrelated white people living in Poland. The study group consisted of 135 patients (118 men and 17 women, mean age: 62 ± 9) with diagnosed head and neck cancer localized in larynx. Control group consisted of 170 cancer-free individuals (149 men and 21 women, mean age: 67 ± 14). Cancer type was confirmed with histopathology examination.
The study cohort included consecutive patients who underwent total or partial laryngectomy operation at Public Central Clinical Hospital in Warsaw, Poland. Macroscopic tumor samples were collected as far as possible from the necrosis area. As controls, noncancerous laryngeal mucosa tissue specimens were obtained from people who had had a total or partial laryngectomy for a benign condition. Tissues' samples obtained during surgery were immediately fixed in 10% buffered formalin for at least 4 h and processed for paraffin embedding.
Prior to examination, the patients and control subjects did not receive medicaments like antibiotics or steroids. Patients enrolled for the examination were analyzed according to cancer staging system of the TNM Classification of Malignant Tumors that describes the extent of cancer in a patient's body: T describes the size of the tumor and whether it has invaded nearby tissue, N describes metastasis to regional lymph nodes, and M indicates the presence of distant metastasis (spread of cancer to another organs).
Using the TNM system the cancer burden can be grouped into following stages: Stage I, T1, N0, M0; Stage II, T2, N0, M0; Stage III, T3, N0, M0 or T1–3, N1, M0; Stage IVA, T4, N0-1, M0 or any T, N2, M0; Stage IVB, any T, N3, M0; and Stage IVC, any T, any N, M1. According to TNM staging, our study included 10 cases of Stage I, 8 cases of Stage II, 55 cases of Stage III, 56 cases of Stage IVA, and 5 cases of Stage IVB. In evaluated patients group there were no cases of distant metastases.
Additionally, the neoplastic grading was also applied: G1, well differentiated (low grade) tumor; G2, moderately differentiated (intermediate grade) tumor; G3, poorly differentiated (high grade) tumor; and G4, undifferentiated (high grade) tumor. Within the patients group subjects were classified as smokers for at least 10 years, 10 to 40 years, and more than 40 years. The smoking attitude of head and neck cancer group was also analyzed for nonsmoking patients, patients smoking up to 20 cigarettes per day, and patients smoking more than 20 cigarettes per day. All patients and controls subjects were recruited from medical units of Head and Neck Neoplasm Surgery Departments, Medical University of Warsaw, Poland. The study was approved by the Ethics Committee of Medical University of Warsaw and written consent was obtained from each patient or healthy subject before enrolling in the study.
2.2. DNA Isolation
DNA was isolated from the formalin-fixed, paraffin-embedded tissues using the BiOstic FFPE Tissue DNA Isolation Kit (MO BIO), according to manufacturer's instructions. 10 mg of paraffin-embedded tissue was used for preparation. Samples were heated at 55°C in an optimized wax melting buffer and Proteinase K to completely digest the tissue. Then, a 90°C heating step was used to remove cross-links in the DNA and ensure successful PCR. The samples were then mixed with a chaotropic salt binding buffer and 100% ethanol for binding to silica filters. Impurities were washed from the column and pure DNA was eluted in a low salt buffer (10 mM Tris, pH 8.0). The purity and amount of DNA were evaluated using Synergy HT microplate reader (BioTek). After extraction, the DNA concentration was measured photometrically and the DNA was diluted to a concentration of 5 ng/μL.
2.3. Genotyping Assay
The aim of this research was to obtain the association of polymorphic variants of genes involved in miRNA processing with the head and neck occurrence risk. In this case-control study, we wanted to evaluate the effects of 9 selected potentially functional single nucleotide polymorphisms (SNPs) in pre-miRNA processing machinery. SNP evaluation was obtained by TaqMan SNP Genotyping Assay with a commercially available primer probe sets (Applied Biosystems, Foster City, CA, USA) and TaqMan Genotyping Master Mix (Applied Biosystems, Foster City, CA, USA) performed on Mx3005P (Agilent Technologies, Santa Clara, CA, USA) according to manufacturer's instructions. Assay IDs were as follows: rs6877842-C_1153852_10, rs3757-C_2539471_1_, rs417309-C_2539468_20, rs1640299-C_7543549_20, rs14035-C_11351340_10, rs11077-C_3109165_1_, rs13078-C_7504801_10, rs3742330-C_27475447_10, and rs784567-C_9576934_20. For TaqMan Genotyping Assay, 5 ng of DNA template was used per reaction well. The mix for every single reaction was prepared as follows: 10 μL TaqMan Genotyping Master Mix (2x), 1 μL TaqMan Genotyping Assay Mix (20x), 9 μL DNase-free, RNase-free water. Then, 20 μL of reaction mix was added to the DNA template and briefly centrifuged to spin down the contents and eliminate air bubbles from the solutions. The reaction thermal profile was as follows: 95°C for 10 min, followed by 50 cycles of 95°C for 15 s and 60°C for 1 min. Representative allelic discrimination plot of genotyping using TaqMan SNP Genotyping Assay is presented in Figure 1. x-axis represents the emission for the A allele-specific probe labeled with 2′-chloro-7′-phenyl-1,4-dichloro-6-carboxyfluorescein (VIC), and y-axis represents the relative fluorescent emission for the B allele-specific probe labeled with 6-carboxyfluorescein (FAM). The resulting cluster plot shows strong fluorescent signals for each allele and clear separation between the three clusters, easily discriminating the two homozygous (diamonds: homozygous AA; squares: homozygous BB) and one heterozygous genotypes (triangle: heterozygous AB). Cross sign represents no template negative controls.
Figure 1.
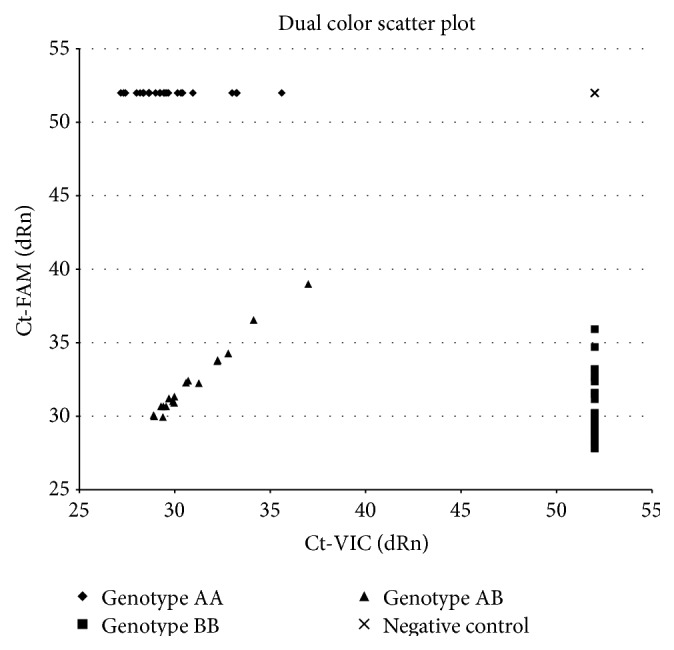
Representative allelic discrimination plot of genotyping using TaqMan SNP Genotyping Assay. Diamonds: homozygous AA; squares: homozygous BB; triangle: heterozygous AB. Cross sign represents no template negative controls.
Evaluation of 20% of randomly chosen samples was performed again with RT-PCR to confirm the previously obtained results of genotyping and the results were 100% concordant.
2.4. Statistical Analysis
Genotype frequencies for each polymorphism were evaluated using the Hardy-Weinberg equilibrium test. Allele frequencies and the prevalence of genotypes were determined for the study and control groups and compared by χ 2 test. If expected frequencies in 2 × 2 contingency table were smaller than 5, Fisher's exact probability test was performed. Significant probability values obtained were analyzed for multiple testing using Bonferroni correction (p value after Bonferroni correction [p corr]). Statistical significance was defined as p < 0.05. All analyses were performed using STATISTICA 6.0 software (StatSoft, Tulsa, OK, USA).
3. Results
3.1. Distributions of Investigated Genotypes in Polish Population
Genotype frequencies for each polymorphism were evaluated using the Hardy-Weinberg equilibrium test. The outcomes of statistical analysis are presented in Table 1. Among LC patients observed genotype frequencies of some of evaluated single nucleotide polymorphisms (rs6877842, rs14035, rs13078, and rs11077) were not in agreement with HWE. It may be due to genetic changes occurring in tumor tissue during carcinogenesis, for example, loss of heterozygosity as it was described previously [15–17] or accumulation of mutations that predispose the individual to larynx cancer development.
Table 1.
The genotype frequencies for each evaluated polymorphism using the Hardy-Weinberg equilibrium test.
| Gene/rs number | Chi-squared test p value for patients group | Chi-squared test p value for control group |
|---|---|---|
| DROSHA/6877842 | 0.537047 | 0.384044 |
| DICER1/3742330 | 0.000000 | 0.000001 |
| DICER1/13078 | 0.987719 | 0.000000 |
| DGCR8/1640299 | 0.021333 | 0.406734 |
| DGCR8/3757 | 0.000000 | 0.000000 |
| DGCR8/417309 | 0.026576 | 0.227311 |
| RAN/14035 | 0.540760 | 0.002413 |
| TARBP2/784567 | 0.006552 | 0.000005 |
| XPO5/11077 | 0.941889 | 0.000001 |
3.2. The Association of miRNA Processing Genes Polymorphisms and Cancer Risk
We performed an analysis comparing the prevalence of polymorphic variants (heterozygotes or polymorphic homozygotes versus wild type homozygotes) of selected miRNA processing genes in healthy subjects and patients with LC (Table 2).
Table 2.
The odds ratio (OR) of the miRNA processing genes single nucleotide polymorphisms in head and neck cancer patients and healthy controls.
| Gene/rs number | Genotype |
Controls number |
Patients number |
Odds ratio | 0.95 confidence intervals | p | |
|---|---|---|---|---|---|---|---|
| (0.95 CI) | |||||||
| Lower limit | Upper limit | ||||||
| DROSHA | |||||||
| 6877842 | CC | 15 | 6 | Ref. | |||
| CG | 79 | 49 | 1.5506 | 0.5638 | 4.2644 | 0.392883 | |
| GG | 76 | 73 | 2.4013 | 0.8836 | 6.526 | 0.078775 | |
|
| |||||||
| DICER1 | |||||||
| 3742330 | AA | 63 | 23 | Ref. | |||
| AG | 103 | 100 | 2.6593 | 1.5326 | 4.6144 | 0.0004 | |
| GG | 4 | 0 | NA | ||||
| 13078 | AA | 10 | 7 | Ref. | |||
| AT | 124 | 46 | 0.53 | 0.1904 | 1.4747 | 0.17013F | |
| TT | 36 | 75 | 2.9762 | 1.0473 | 8.4579 | 0.034697 | |
|
| |||||||
| DGCR8 | |||||||
| 1640299 | GG | 16 | 6 | Ref. | |||
| GT | 93 | 47 | 1.3477 | 0.4949 | 3.6696 | 0.559829 | |
| TT | 61 | 60 | 2.623 | 0.9614 | 7.1562 | 0.053443 | |
| 3757 | AA | 15 | 4 | Ref. | |||
| AG | 119 | 89 | 2.8046 | 0.9 | 8.74 | 0.065196 | |
| GG | 36 | 29 | 3.0208 | 0.9039 | 10.0951 | 0.064411 | |
| 417309 | AA | 8 | 13 | Ref. | |||
| AG | 46 | 32 | 0.4281 | 0.1591 | 1.1516 | 0.088032 | |
| GG | 116 | 67 | 0.3554 | 0.1401 | 0.9015 | 0.024484 | |
|
| |||||||
| RAN | |||||||
| 14035 | CC | 67 | 73 | Ref. | |||
| CT | 93 | 32 | 0.3158 | 0.1876 | 0.5317 | <0.0001 | |
| TT | 10 | 5 | 0.4589 | 0.1492 | 1.4115 | 0.165857 | |
|
| |||||||
| TARBP2 | |||||||
| 784567 | AA | 35 | 7 | Ref. | |||
| AG | 114 | 70 | 3.0702 | 1.2935 | 7.2871 | 0.008335 | |
| GG | 21 | 51 | 12.1429 | 4.661 | 31.6345 | <0.0001 | |
|
| |||||||
| XPO5 | |||||||
| 11077 | GG | 82 | 26 | Ref. | |||
| GT | 44 | 62 | 4.4441 | 2.4723 | 7.9883 | <0.0001 | |
| TT | 34 | 36 | 3.3394 | 1.7544 | 6.3563 | 0.000183 | |
It showed that DROSHA rs6877842 SNP is not associated with higher occurrence of LC. We also evaluated a connection between LC and two SNPs of DGCR8 gene: rs3757, rs417309, and rs1640299. We did not find any statistically significant connection between LC and any of rs3757 as well as rs1640299 polymorphic variants. GG genotype of DGCR8 rs41709 is less common in LC subjects (OR: 0.3554, 0.95 CI: 0.1401–0.9015, and p = 0.024484). After using Bonferroni correction, this association was no longer statistically significant [p corr = 0.220]. Similar trend of lower frequency in patients' group of CT heterozygote was observed in case of RAN rs14035 polymorphism (OR: 0.3158, 0.95 CI: 0.1876–0.5317, and p < 0.0001), and after the Bonferroni correction, the positive association remained [p corr = 0.001]. On the other hand, the occurrence of XPO5 rs11077 of both GT and TT polymorphic variants was higher in LC individuals than in control subjects (OR: 4.4441, 0.95 CI: 2.4723–7.9883, and p < 0.0001 and OR: 3.3394, 0.95 CI: 1.7544–6.3563, and p = 0.000183; resp.), and after the Bonferroni correction, the positive association remained [p corr = 0.001 and p corr = 0.002]. Also both investigated polymorphisms of DICER1 seem to be associated with higher risk of LC occurrence (OR: 2.9762, 0.95 CI: 1.0473–8.4579, and p = 0.034697 [p corr = 0.312] for TT genotype of rs13078 as well as OR: 2.6593, 0.95 CI: 1.5326–4.6144, and p = 0.0004 [p corr = 0.004] for AG genotype of rs3742330). We also found a strong association between the AG and GG genotypes of the TARBP2 rs784567 polymorphism and the risk for LC (OR: 3.0702, 0.95 CI: 1.2935–7.2871, and p = 0.008335 [p corr = 0.075] and OR: 12.1429, 0.95 CI: 4.661–31.6345, and p < 0.0001 [p corr = 0.001], resp.).
Secondly, we subdivided patients group according to TNM classification. In Tables 3(a) and 3(b), the correlations between the investigated gene polymorphisms and the stage of LC in comparison to healthy subjects were performed. In T1 stage we found a decreased prevalence of AG and GG genotype of DGCR8 rs417309 (OR: 0.087, 0.95 CI: 0.0136–0.5564, and p = 0.011931 [p corr = 0.107] and OR: 0.0517, 0.95 CI: 0.0098–0.2719, and p = 0.001265 [p corr = 0.011], resp., Table 3(a)) as well as RAN rs14035 CT polymorphic variant (OR: 0.2058, 0.95 CI: 0.0415–1.0221, and p = 0.038382 [p corr = 0.345], Table 3(a)). On the other hand, we found an association between T2 in LC patients and high abundance of XPO5 rs11077 GT heterozygote (OR: 4.9697, 0.95 CI: 1.2546–19.686, and p = 0.016786 [p corr = 0.150], Table 3(a)). The most interesting here was a fact of a very big difference between allele distribution in cancer and control subjects during analysis of TARBP2 rs784567 polymorphism. It occurred that GG genotype may put an individual at high risk of T2 LC (OR: 13.3333, 0.95 CI: 1.556–114.2517, and p = 0.005146 [p corr = 0.046], Table 3(a)). Analysis of subjects with T3 stage showed a decrease of CT heterozygote occurrence of RAN rs14035 polymorphism (OR: 0.2401, 0.95 CI: 0.1133–0.509, and p < 0.0001 [p corr = 0.001], Table 3(b)) in LC patients compared to healthy subjects. Further investigation showed also a high abundance of two XPO5 rs11077 genotypes: GT (OR: 4.8455, 0.95 CI: 2.1424–10.9591, and p < 0.0001 [p corr = 0.001], Table 3(b)) and TT (OR: 3.8588, 0.95 CI: 1.5918–9.3545, and p = 0.001873 [p corr = 0.017], Table 3(b)). Similar association was found related to AG DICER1 rs3742330 heterozygote (OR: 2.2242, 0.95 CI: 1.0641–4.649, and p = 0.030873 [p corr = 0.277], Table 3(b)) and GG TARBP2 rs7834567 homozygote (OR: 7.9167, 0.95 CI: 2.3694–26.4517, and p = 0.00027 [p corr = 0.002], Table 3(b)).The heterozygote of RAN rs14035 SNP was also found to be negatively associated with T4 (OR: 0.3902, 0.95 CI: 0.1854–0.8216, and p = 0.011477 [p corr = 0.103], Table 3(b)). On the opposite side, patients with T4 stage of LC were shown to be the carriers of AG DICER1 rs3742330 genotype (OR: 3.0583, 0.95 CI: 1.3452–6.9527, and p = 0.005805 [p corr = 0.052], Table 3(b)) and TARBP2 polymorphic variants (OR: 7.6754, 0.95 CI: 1.0036–58.6983, and p = 0.022201 [p corr = 0.199] for AG and OR: 36.6667, 0.95 CI: 4.6001–292.2647, and p < 0.0001 [p corr = 0.001] for GG, Table 3(b)) more often than control subjects.
Table 3.
An association of the miRNA processing genes single nucleotide polymorphisms with tumor stage of head and neck cancer.
(a).
| Gene/rs number | Genotype |
Controls number |
T1 | T2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients | Odds ratio | 0.95 confidence intervals | p | Patients | Odds ratio | 0.95 confidence intervals | p | |||||
| number | Lower limit | Upper limit | number | Lower limit | Upper limit | |||||||
| DROSHA | ||||||||||||
| 6877842 | CC | 15 | 1 | Ref. | 0 | Ref. | ||||||
| CG | 79 | 5 | 0.9494 | 0.1034 | 8.7141 | 0.6590F | 6 | NA | ||||
| GG | 76 | 4 | 0.7595 | 0.0793 | 7.2769 | 0.5940F | 11 | NA | ||||
|
| ||||||||||||
| DICER1 | ||||||||||||
| 3742330 | AA | 63 | 1 | Ref. | 3 | Ref. | ||||||
| AG | 103 | 8 | 4.8932 | 0.5978 | 40.0546 | 0.0967F | 12 | 2.4466 | 0.6645 | 9.0082 | 0.1669 | |
| GG | 4 | 0 | NA | 0 | NA | |||||||
| 13078 | AA | 10 | 0 | Ref. | 1 | Ref. | ||||||
| AT | 124 | 8 | NA | 8 | 0.6452 | 0.0732 | 5.6871 | 0.5239F | ||||
| TT | 36 | 2 | NA | 8 | 2.2222 | 0.2478 | 19.9312 | 0.4181F | ||||
|
| ||||||||||||
| DGCR8 | ||||||||||||
| 1640299 | GG | 16 | 0 | Ref. | 1 | Ref. | ||||||
| GT | 93 | 2 | NA | 9 | 1.5652 | 0.1854 | 13.2126 | 0.5614F | ||||
| TT | 61 | 4 | NA | 5 | 1.3115 | 0.1429 | 12.0333 | 0.6432F | ||||
| 3757 | AA | 15 | 1 | Ref. | 0 | Ref. | ||||||
| AG | 119 | 7 | 0.8824 | 0.1014 | 7.6744 | 0.6255F | 15 | NA | ||||
| GG | 36 | 1 | 0.4167 | 0.0244 | 7.107 | 0.5166F | 2 | NA | ||||
| 417309 | AA | 8 | 4 | Ref. | 2 | Ref. | ||||||
| AG | 46 | 2 | 0.087 | 0.0136 | 0.5564 | 0.0119F | 8 | 0.6957 | 0.1243 | 3.892 | 0.4909F | |
| GG | 116 | 3 | 0.0517 | 0.0098 | 0.2719 | 0.0012F | 5 | 0.1724 | 0.0288 | 1.0322 | 0.0898F | |
|
| ||||||||||||
| RAN | ||||||||||||
| 14035 | CC | 67 | 7 | Ref. | 9 | Ref. | ||||||
| CT | 93 | 2 | 0.2058 | 0.0415 | 1.0221 | 0.0383F | 6 | 0.4803 | 0.1632 | 1.4139 | 0.1761 | |
| TT | 10 | 1 | 0.9571 | 0.1063 | 8.6223 | 0.7245F | 0 | NA | ||||
|
| ||||||||||||
| TARBP2 | ||||||||||||
| 784567 | AA | 35 | 1 | Ref. | 1 | Ref. | ||||||
| AG | 114 | 6 | 1.8421 | 0.2144 | 15.8246 | 0.4907F | 8 | 2.4561 | 0.2969 | 20.3221 | 0.3494F | |
| GG | 21 | 2 | 3.3333 | 0.2846 | 39.0437 | 0.3346F | 8 | 13.3333 | 1.556 | 114.2517 | 0.0051F | |
|
| ||||||||||||
| XPO5 | ||||||||||||
| 11077 | GG | 82 | 2 | Ref. | 3 | Ref. | ||||||
| GT | 44 | 4 | 3.7273 | 0.6565 | 21.1609 | 0.1274F | 8 | 4.9697 | 1.2546 | 19.686 | 0.0167F | |
| TT | 34 | 3 | 3.6176 | 0.5784 | 22.6277 | 0.1663F | 5 | 4.0196 | 0.9094 | 17.7671 | 0.0634F | |
F p values calculated with Fisher's exact probability test.
(b).
| Gene/rs number | Genotype | T3 | T4 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Controls | Patients | Odds ratio | 0.95 confidence intervals | p | Controls | Patients | Odds ratio | 0.95 confidence intervals | p | ||||
| number | number | Lower limit | Upper limit | number | number | Lower limit | Upper limit | ||||||
| DROSHA | |||||||||||||
| 6877842 | CC | 15 | 4 | Ref. | 15 | 1 | Ref. | ||||||
| CG | 79 | 17 | 0.807 | 0.238 | 2.7364 | 0.4730F | 79 | 21 | 3.9873 | 0.4978 | 31.9386 | 0.1440F | |
| GG | 76 | 32 | 1.5789 | 0.4863 | 5.127 | 0.4424 | 76 | 26 | 5.1316 | 0.6458 | 40.776 | 0.0747F | |
|
| |||||||||||||
| DICER1 | |||||||||||||
| 3742330 | AA | 63 | 11 | Ref. | 63 | 8 | Ref. | ||||||
| AG | 103 | 40 | 2.2242 | 1.0641 | 4.649 | 0.0308 | 103 | 40 | 3.0583 | 1.3452 | 6.9527 | 0.0058 | |
| GG | 4 | 0 | NA | 4 | 0 | NA | |||||||
| 13078 | AA | 10 | 3 | Ref. | 10 | 3 | Ref. | ||||||
| AT | 124 | 15 | 0.4032 | 0.0997 | 1.6304 | 0.1859F | 124 | 15 | 0.4032 | 0.0997 | 1.6304 | 0.1859F | |
| TT | 36 | 35 | 3.2407 | 0.8222 | 12.7733 | 0.0807 | 36 | 30 | 2.7778 | 0.7001 | 11.0216 | 0.1345 | |
|
| |||||||||||||
| DGCR8 | |||||||||||||
| 1640299 | GG | 16 | 2 | Ref. | 16 | 3 | Ref. | ||||||
| GT | 93 | 23 | 1.978 | 0.4245 | 9.2218 | 0.3022F | 93 | 13 | 0.7455 | 0.1908 | 2.9131 | 0.4531 | |
| TT | 61 | 27 | 3.541 | 0.7605 | 16.4881 | 0.0739 | 61 | 24 | 2.0984 | 0.5603 | 7.8588 | 0.2067 | |
| 3757 | AA | 15 | 3 | Ref. | 15 | 0 | Ref. | ||||||
| AG | 119 | 34 | 1.4286 | 0.3905 | 5.2255 | 0.4235F | 119 | 34 | NA | ||||
| GG | 36 | 15 | 2.0833 | 0.5251 | 8.2662 | 0.2317F | 36 | 11 | NA | ||||
| 417309 | AA | 8 | 3 | Ref. | 8 | 4 | Ref. | ||||||
| AG | 46 | 13 | 0.7536 | 0.1745 | 3.2541 | 0.4838F | 46 | 9 | 0.3913 | 0.0968 | 1.5817 | 0.1704F | |
| GG | 116 | 28 | 0.6437 | 0.1604 | 2.5833 | 0.3825F | 116 | 31 | 0.5345 | 0.151 | 1.8918 | 0.2557F | |
|
| |||||||||||||
| RAN | |||||||||||||
| 14035 | CC | 67 | 33 | Ref. | 67 | 24 | Ref. | ||||||
| CT | 93 | 11 | 0.2401 | 0.1133 | 0.509 | <0.0001 | 93 | 13 | 0.3902 | 0.1854 | 0.8216 | 0.0114 | |
| TT | 10 | 3 | 0.6091 | 0.157 | 2.3634 | 0.3528F | 10 | 1 | 0.2792 | 0.0339 | 2.2978 | 0.1914F | |
|
| |||||||||||||
| TARBP2 | |||||||||||||
| 784567 | AA | 35 | 4 | Ref. | 35 | 1 | Ref. | ||||||
| AG | 114 | 31 | 2.3794 | 0.7857 | 7.2059 | 0.1160 | 114 | 25 | 7.6754 | 1.0036 | 58.6983 | 0.0222 | |
| GG | 21 | 19 | 7.9167 | 2.3694 | 26.4517 | 0.00027 | 21 | 22 | 36.6667 | 4.6001 | 292.2647 | <0.0001 | |
|
| |||||||||||||
| XPO5 | |||||||||||||
| 11077 | GG | 82 | 10 | Ref. | 82 | 11 | Ref. | ||||||
| GT | 44 | 26 | 4.8455 | 2.1424 | 10.9591 | <0.0001 | 44 | 24 | 4.0661 | 1.8231 | 9.0689 | 0.000364 | |
| TT | 34 | 16 | 3.8588 | 1.5918 | 9.3545 | 0.001873 | 34 | 12 | 2.631 | 1.0584 | 6.5403 | 0.033306 | |
F p values calculated with Fisher's exact probability test.
SNP and lymph node status associations are presented in Table 4. RAN rs14035 heterozygote carriers were less likely to have both node positive or negative tumors (OR: 0.3955, 0.95 CI: 0.2264–0.6909, and p = 0.0009 [p corr = 0.008] and OR: 0.131, 0.95 CI: 0.0431–0.3977, and p < 0.0001 [p corr = 0.0009], resp.). Performed analysis indicated also that XPO5 rs11077 polymorphic variants are associated with tumors without lymph node metastases (OR: 4.2177, 0.95 CI: 2.1965–8.099, and p < 0.0001 [p corr = 0.001] for GT and OR: 3.5542, 0.95 CI: 1.7534–7.2043, and p = 0.000311 [p corr = 0.003] for TT). Similar correlation was shown also related to DICER1 rs13078 TT genotype (OR: 3.6111, 0.95 CI: 1.0503–12.416, and p = 0.033112 [p corr = 0.298]), DICER1 rs3742330 AG heterozygote (OR: 2.6041, 0.95 CI: 1.4276–4.7502, and p = 0.001483 [p corr = 0.013]), and TARBP2 rs784567 GG homozygote (OR: 8.5714, 0.95 CI: 3.2371–22.6961, and p < 0.0001 [p corr = 0.001]). XPO5 rs11077 and DICER1 rs3742330 heterozygotes were also more likely to have node positive tumors (OR: 5.0584, 0.95 CI: 1.9743–12.9602, and p = 0.000323 [p corr = 0.003] and OR: 3.5476, 0.95 CI: 1.3057–9.6388, and p = 0.009116 [p corr = 0.082], resp.). DGCR8 rs1640299 polymorphic variants were also correlated with N0 LC stage (OR: 4.4602, 0.95 CI: 1.6527–12.0368, and p = 0.0016 [p corr = 0.014] for GT and OR: 32.787, 0.95 CI: 11.1638–96.2956, and p < 0.0001 [p corr = 0.001] for TT). Moreover, such association was also observed in patients with lymph node metastasis positive tumors (OR: 8.5269, 0.95 CI: 1.0874–66.8658, and p = 0.0159 [p corr = 0.143] for GT and OR: 64.812, 0.95 CI: 8.0116–524.3237, and p < 0.0001 [p corr = 0.0009] for TT). All investigated polymorphisms were not significantly associated with lymph node metastases when we compared groups having lymph node metastases with LC group without them.
Table 4.
An association of the miRNA processing genes single nucleotide polymorphisms with lymph node metastases occurrence in head and neck cancer.
| Gene/rs number | Genotype |
Controls number |
N0 | N1–3 | N1–3 versus N0 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients | Odds ratio | 0.95 confidence intervals | p | Patients | Odds ratio | 0.95 confidence intervals | p | Odds ratio | 0.95 confidence intervals | p | ||||||
| Number | Lower limit | Upper limit | number | Lower limit | Upper limit | Lower limit | Upper limit | |||||||||
| DROSHA | ||||||||||||||||
| 6877842 | CC | 15 | 6 | Ref. | 0 | Ref. | Ref. | |||||||||
| CG | 79 | 36 | 1.1392 | 0.4085 | 3.1769 | 0.8064 | 13 | NA | NA | |||||||
| GG | 76 | 51 | 1.6776 | 0.6104 | 4.611 | 0.3125 | 22 | NA | NA | |||||||
|
| ||||||||||||||||
| DICER1 | ||||||||||||||||
| 3742330 | AA | 63 | 18 | Ref. | 5 | Ref. | Ref. | |||||||||
| AG | 103 | 71 | 2.6041 | 1.4276 | 4.7502 | 0.0014 | 29 | 3.5476 | 1.3057 | 9.6388 | 0.0091 | 1.4704 | 0.4989 | 4.3338 | 0.4839 | |
| GG | 4 | 0 | NA | 0 | NA | NA | ||||||||||
| 13078 | AA | 10 | 4 | Ref. | 3 | Ref. | Ref. | |||||||||
| AT | 124 | 38 | 0.7661 | 0.2273 | 2.5825 | 0.4394F | 8 | 0.2151 | 0.0492 | 0.9401 | 0.0612F | 0.2807 | 0.0523 | 1.506 | 0.1470F | |
| TT | 36 | 52 | 3.6111 | 1.0503 | 12.416 | 0.0331 | 23 | 2.1296 | 0.5292 | 8.5696 | 0.2263F | 0.5897 | 0.122 | 2.85 | 0.3904F | |
|
| ||||||||||||||||
| DGCR8 | ||||||||||||||||
| 1640299 | GG | 16 | 5 | Ref. | 1 | Ref. | Ref. | |||||||||
| GT | 93 | 34 | 4.4602 | 1.6527 | 12.0368 | 0.0016 | 13 | 8.5269 | 1.0874 | 66.8658 | 0.0159 | 1.911 | 0.2035 | 17.9611 | 0.493F | |
| TT | 61 | 43 | 32.787 | 11.1638 | 96.2956 | <0.0001 | 17 | 64.812 | 8.0116 | 524.3237 | <0.0001 | 1.976 | 0.2148 | 18.1876 | 0.474F | |
| 3757 | AA | 15 | 4 | Ref. | 0 | Ref. | Ref. | |||||||||
| AG | 119 | 63 | 1.9853 | 0.6321 | 6.2352 | 0.2334 | 26 | NA | NA | |||||||
| GG | 36 | 23 | 2.3958 | 0.7068 | 8.1213 | 0.1532 | 6 | NA | NA | |||||||
| 417309 | AA | 8 | 11 | Ref. | 2 | Ref. | Ref. | |||||||||
| AG | 46 | 25 | 0.4427 | 0.1589 | 1.2335 | 0.1138 | 7 | 0.6087 | 0.1067 | 3.4736 | 0.4382F | 1.54 | 0.2746 | 8.6355 | 0.4822F | |
| GG | 116 | 49 | 0.3072 | 0.1165 | 0.8104 | 0.0131 | 18 | 0.6207 | 0.122 | 3.1588 | 0.4176F | 2.0204 | 0.4077 | 10.013 | 0.3118F | |
|
| ||||||||||||||||
| RAN | ||||||||||||||||
| 14035 | CC | 67 | 51 | Ref. | 22 | Ref. | Ref. | |||||||||
| CT | 93 | 28 | 0.3955 | 0.2264 | 0.6909 | 0.0009 | 4 | 0.131 | 0.0431 | 0.3977 | <0.0001 | 0.3312 | 0.1037 | 1.0573 | 0.0541 | |
| TT | 10 | 4 | 0.5255 | 0.1559 | 1.7718 | 0.2942 | 1 | 0.3045 | 0.0369 | 2.5153 | 0.2253 | 0.5795 | 0.0612 | 5.4857 | 0.5363 | |
|
| ||||||||||||||||
| TARBP2 | ||||||||||||||||
| 784567 | AA | 35 | 7 | Ref. | 0 | Ref. | Ref. | |||||||||
| AG | 114 | 50 | 2.193 | 0.9124 | 5.2711 | 0.0741 | 20 | NA | NA | |||||||
| GG | 21 | 36 | 8.5714 | 3.2371 | 22.6961 | <0.0001 | 15 | NA | NA | |||||||
|
| ||||||||||||||||
| XPO5 | ||||||||||||||||
| 11077 | GG | 82 | 19 | Ref. | 7 | Ref. | Ref. | |||||||||
| GT | 44 | 43 | 4.2177 | 2.1965 | 8.099 | <0.0001 | 19 | 5.0584 | 1.9743 | 12.9602 | 0.0003 | 1.1993 | 0.432 | 3.3294 | 0.7290 | |
| TT | 34 | 28 | 3.5542 | 1.7534 | 7.2043 | 0.0003 | 8 | 2.7563 | 0.9264 | 8.2008 | 0.0599F | 0.7755 | 0.2407 | 2.4981 | 0.6713 | |
F p values calculated with Fisher's exact probability test.
We also analyzed the relation between miRNA processing genes polymorphic variants and the stage of cancer (Tables 5(a) and 5(b)). Some of them were associated with higher prevalence of LC in particular stage of the disease. It was found that DICER1 rs3742330 AG heterozygote is connected with LC Stages III and IV (OR: 2.3243, 0.95 CI: 1.0828–4.9893, and p = 0.0276 [p corr = 0.248] and OR: 2.9971, 0.95 CI: 1.4173–6.3378, and p = 0.0031 [p corr = 0.028], resp., Table 5(b)) and DICER1 rs13078 TT genotype is associated with LC Stage III (OR: 9.1667, 0.95 CI: 1.1122–75.5513, and p = 0.01450 [p corr = 0.131], Table 5(b)). Moreover, TARBP2 rs784567 AG heterozygote is connected to higher occurrence of LC Stage IV (OR: 9.8246, 0.95 CI: 1.2953–74.5144, and p = 0.0075 [p corr = 0.068], Table 5(b)) and GG polymorphic variant is related to both LC Stage III and Stage IV (OR: 6, 0.95 CI: 1.9403–18.5537, and p = 0.0009 [p corr = 0.008] and OR: 3.3333, 0.95 CI: 5.4718–343.1724, and p < 0.0001 [p corr = 0.001], resp., Table 5(b)). XPO5 rs11077 GT and TT genotypes are also associated with LC Stage III (OR: 4.9697, 0.95 CI: 2.1259–11.6175, and p < 0.0001 [p corr = 0.001] and OR: 4.2876, 0.95 CI: 1.7271–10.644, and p = 0.001 [p corr = 0.009], resp., Table 5(b)) and LC Stage IV (OR: 4.1573, 0.95 CI: 1.9643–8.799, and p = 0.0001 [p corr = 0.0009] and OR: 2.5973, 0.95 CI: 1.1054–6.1026, and p = 0.0254 [p corr = 0.221], resp., Table 5(b)).
Table 5.
An association of the miRNA processing genes single nucleotide polymorphisms with stage of larynx cancer.
(a).
| Gene/rs number | Genotype |
Controls number |
Stage I | Stage II | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients | Odds ratio | 0.95 confidence intervals | p ∗ | Patients | Odds ratio | 0.95 confidence intervals | p ∗ | |||||
| number | Lower limit | Upper limit | number | Lower limit | Upper limit | |||||||
| DROSHA | ||||||||||||
| 6877842 | CC | 15 | 1 | Ref. | 0 | Ref. | ||||||
| CG | 79 | 5 | 0.9494 | 0.1034 | 8.7141 | 0.6590 | 3 | NA | ||||
| GG | 76 | 4 | 0.7895 | 0.0824 | 7.5674 | 0.6067 | 5 | NA | ||||
|
| ||||||||||||
| DICER1 | ||||||||||||
| 3742330 | AA | 63 | 1 | Ref. | 2 | Ref. | ||||||
| AG | 103 | 8 | 4.8932 | 0.5978 | 40.0546 | 0.0967 | 4 | 1.2233 | 0.2177 | 6.8735 | 0.5907 | |
| GG | 4 | 0 | NA | 0 | NA | |||||||
| 13078 | AA | 10 | 0 | Ref. | 1 | Ref. | ||||||
| AT | 124 | 8 | NA | 6 | 0.4839 | 0.0529 | 4.4237 | 0.4409 | ||||
| TT | 36 | 2 | NA | 1 | 0.2778 | 0.0159 | 4.8457 | 0.4095 | ||||
|
| ||||||||||||
| DGCR8 | ||||||||||||
| 1640299 | GG | 16 | 0 | Ref. | 0 | Ref. | ||||||
| GT | 93 | 2 | NA | 6 | NA | |||||||
| TT | 61 | 4 | NA | 2 | NA | |||||||
| 3757 | AA | 15 | 1 | Ref. | 0 | Ref. | ||||||
| AG | 119 | 7 | 0.8824 | 0.1014 | 7.6744 | 0.6255 | 6 | NA | ||||
| GG | 36 | 1 | 0.4167 | 0.0244 | 7.107 | 0.5166 | 2 | NA | ||||
| 417309 | AA | 8 | 4 | Ref. | 1 | Ref. | ||||||
| AG | 46 | 2 | 0.087 | 0.0136 | 0.5564 | 0.0119 | 4 | 0.6957 | 0.0686 | 7.0533 | 0.5767 | |
| GG | 116 | 3 | 0.0517 | 0.0098 | 0.2719 | 0.0012 | 2 | 0.1379 | 0.0113 | 1.689 | 0.1993 | |
|
| ||||||||||||
| RAN | ||||||||||||
| 14035 | CC | 67 | 7 | Ref. | 2 | Ref. | ||||||
| CT | 93 | 2 | 0.2058 | 0.0415 | 1.0221 | 0.0383 | 5 | 1.8011 | 0.3392 | 9.5644 | 0.3884 | |
| TT | 10 | 1 | 0.9571 | 0.1063 | 8.6223 | 0.7245 | 0 | NA | ||||
|
| ||||||||||||
| TARBP2 | ||||||||||||
| 7845672 | AA | 35 | 1 | Ref. | 0 | Ref. | ||||||
| AG | 114 | 6 | 1.8421 | 0.2144 | 15.8246 | 0.4907 | 3 | NA | ||||
| GG | 21 | 2 | 3.3333 | 0.2846 | 39.0437 | 0.3346 | 5 | NA | ||||
|
| ||||||||||||
| XPO5 | ||||||||||||
| 11077 | GG | 82 | 2 | Ref. | 1 | Ref. | ||||||
| GT | 44 | 4 | 3.7273 | 0.6565 | 21.1609 | 0.1274 | 4 | 7.4545 | 0.8082 | 68.7565 | 0.06 | |
| TT | 34 | 3 | 3.6176 | 0.5784 | 22.6277 | 0.1663 | 3 | 7.2353 | 0.7267 | 72.0402 | 0.0865 | |
∗All p values are for Fisher's exact probability test due to low sample sizes.
(b).
| Gene/rs number | Genotype |
Controls number |
Stage III | Stage IV | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients | Odds ratio | 0.95 confidence intervals | p | Patients | Odds ratio | 0.95 confidence intervals | p | |||||
| number | Lower limit | Upper limit | number | Lower limit | Upper limit | |||||||
| DROSHA | ||||||||||||
| 6877842 | CC | 15 | 4 | Ref. | 1 | Ref. | ||||||
| CG | 79 | 16 | 0.7595 | 0.2227 | 2.5903 | 0.4371F | 24 | 4.557 | 0.572 | 36.3009 | 0.1034F | |
| GG | 76 | 31 | 1.5296 | 0.4703 | 4.9749 | 0.4795 | 33 | 6.5132 | 0.8259 | 51.3637 | 0.0341F | |
|
| ||||||||||||
| DICER1 | ||||||||||||
| 3742330 | AA | 63 | 10 | Ref. | 10 | Ref. | ||||||
| AG | 103 | 38 | 2.3243 | 1.0828 | 4.9893 | 0.0276 | 49 | 2.9971 | 1.4173 | 6.3378 | 0.0031 | |
| GG | 4 | 0 | NA | 0 | NA | |||||||
| 13078 | AA | 10 | 1 | Ref. | 4 | Ref. | ||||||
| AT | 124 | 14 | 1.129 | 0.1344 | 9.4878 | 0.6945F | 17 | 0.3427 | 0.0967 | 1.215 | 0.1004F | |
| TT | 36 | 33 | 9.1667 | 1.1122 | 75.5513 | 0.01450F | 38 | 2.6389 | 0.7591 | 9.1733 | 0.1006F | |
|
| ||||||||||||
| DGCR8 | ||||||||||||
| 1640299 | GG | 16 | 2 | Ref. | 4 | Ref. | ||||||
| GT | 93 | 21 | 1.8065 | 0.3856 | 8.4634 | 0.3533F | 18 | 0.0968 | 0.0165 | 0.5686 | 0.0112F | |
| TT | 61 | 26 | 3.4098 | 0.731 | 15.9056 | 0.0836F | 26 | 0.5 | 0.0841 | 2.9719 | 0.3691F | |
| 3757 | AA | 15 | 2 | Ref. | 0 | Ref. | ||||||
| AG | 119 | 33 | 2.0798 | 0.4526 | 9.5571 | 0.2709F | 42 | NA | ||||
| GG | 36 | 10 | 2.0833 | 0.4068 | 10.6686 | 0.3073F | 11 | NA | ||||
| 417309 | AA | 8 | 4 | Ref. | 3 | Ref. | ||||||
| AG | 46 | 13 | 0.5652 | 0.1467 | 2.178 | 0.3094F | 13 | 0.7536 | 0.1745 | 3.2541 | 0.4838F | |
| GG | 116 | 25 | 0.431 | 0.1204 | 1.5436 | 0.1704F | 35 | 0.8046 | 0.2025 | 3.1973 | 0.4996F | |
|
| ||||||||||||
| RAN | ||||||||||||
| 14035 | CC | 67 | 32 | Ref. | 29 | Ref. | ||||||
| CT | 93 | 11 | 0.2476 | 0.1166 | 0.5262 | 0.0001 | 14 | 0.3478 | 0.1708 | 0.7081 | 0.0028 | |
| TT | 10 | 3 | 0.6281 | 0.1617 | 2.4405 | 0.3715F | 1 | 0.231 | 0.0283 | 1.8892 | 0.1277F | |
|
| ||||||||||||
| TARBP2 | ||||||||||||
| 784567 | AA | 35 | 5 | Ref. | 1 | Ref. | ||||||
| AG | 114 | 28 | 1.7193 | 0.6174 | 4.788 | 0.2942 | 32 | 9.8246 | 1.2953 | 74.5144 | 0.0075 | |
| GG | 21 | 18 | 6 | 1.9403 | 18.5537 | 0.0009 | 26 | 43.3333 | 5.4718 | 343.1724 | <0.0001 | |
|
| ||||||||||||
| XPO5 | ||||||||||||
| 11077 | GG | 82 | 9 | Ref. | 13 | Ref. | ||||||
| GT | 44 | 24 | 4.9697 | 2.1259 | 11.6175 | <0.0001 | 29 | 4.1573 | 1.9643 | 8.799 | 0.0001 | |
| TT | 34 | 16 | 4.2876 | 1.7271 | 10.644 | 0.001 | 14 | 2.5973 | 1.1054 | 6.1026 | 0.0254 | |
F p values calculated with Fisher's exact probability test.
On the other hand, performed analysis shows also that some of the investigated polymorphic variants are less common in patients with LC than in healthy people. Such phenomena were observed in patients with Stage III LC and Stage IV, where, respectively, DGCR8 rs417309 AG and GG genotypes (OR: 0.087, 0.95 CI: 0.0136–0.5564, and p = 0.0119 [p corr = 0.107] and OR: 0.0517, 0.95 CI: 0.0098–0.2719, and p = 0.0012 [p corr = 0.011], Table 5(b)) as well as rs1640299 GT heterozygote (OR: 0.0968, 0.95 CI: 0.0165–0.5686, and p = 0.0112 [p corr = 0.101], Table 5(b)) were less likely to be found in cancer subjects than in control group. RAN rs14035 CT polymorphic variant was also associated with decreased prevalence in LC patients with Stage I (OR: 0.2058, 0.95 CI: 0.0415–1.0221, and p = 0.0383 [p corr = 0.345], Table 5(a)), Stage III (OR: 0.2476, 0.95 CI: 0.1166–0.5262, and p = 0.0001 [p corr = 0.0009], Table 5(b)), and Stage IV (OR: 0.3478, 0.95 CI: 0.1708–0.7081, and p = 0.0028 [p corr = 0.025], Table 5(b)).
3.3. miRNA Processing Genes Polymorphisms in relation to Smoking Status
We performed stratified analysis to estimate the interaction between miRNA processing genes single nucleotide polymorphisms and cigarette smoking (Tables 6–8, Supplementary Materials available online at http://dx.doi.org/10.1155/2015/298378). We divided patients into subgroups based on their smoking status: duration of smoking habit (nonsmokers, subject who have been smoking for less than 10 years, subjects who have been smoking in range between 20 and 40 years, and those who have been smoking for more than 40 years) as well as number of cigarettes smoked per day (nonsmokers, subjects who smoke less than 20 cigarettes daily, and those who smoke more than 20 cigarettes a day). We did not observe any elevated frequency of studied SNPs polymorphic variants correlating with cigarette smoking among LC patients in any of investigated cases. The relationship between smoking habits and susceptibility of LC occurrence in some cases was impossible to calculate due to low frequency of the genotypes.
4. Discussion
In this case-control study of 135LC patients and 170 cancer-free controls in a Polish population, we investigated the associations between SNPs of miRNA biosynthesis genes and risk of LC. This is the first report considering the association of the risk of larynx cancer and SNPs of the following polymorphisms: DROSHA (rs6877842), DGCR8 (rs3757, rs417309, and rs1640299), RAN (rs14035), XPO5 (rs11077), DICER1 (rs13078 and rs3742330), and TARBP2 (rs784567). The rs3742330 AG and rs13078 TT genotypes of DICER1 are correlated with increased risk of larynx cancer. In turn, the rs14035 RAN CT heterozygote and DGCR8 rs417309 GG genotype were significantly inversely associated with the presence of larynx cancer. In addition, the rs3742330 of DICER1, rs784567 of TARBP2, rs417309 of DGCR8, and rs14035 of RAN as well as rs11077 of XPO5 are associated with the LC progression depending on the tumor size. Furthermore, the DGCR8 rs1640299, DICER1 rs3742330 and rs13078, RAN rs14035, and XPO5 rs11077 as well as rs784567 of TARBP2 genes single nucleotide polymorphisms have demonstrated an association with tumor progression depending on the lymph node metastases. The observed genotype frequencies of rs6877842 and rs417309 alleles were not in agreement with HWE. The above information about genotype and allele frequencies of rs6877842 and rs417309 is consistent with NCBI data that shows that rs6877842 C allele frequency is about 0.018 and rs417309 A allele frequency is lower than 0.08 in population of European descent (NCBI SNP database, access date: January 13th 2015) [18, 19].
Our findings suggest, for the first time, that potentially functional polymorphisms of genes encoding proteins of miRNA processing may play a role in the tumors arising at larynx. The rs6877842 and the rs784567 polymorphisms are located in the promoter of DROSHA and TRBP2 genes, respectively, and hence can affect the level of protein expression. The remaining genes polymorphisms are located in the 3′-UTR, which is the binding site of miRNAs; thereby they may affect the efficiency of miRNA processing [20–23].
Our study shows that carriers of evaluated genotypes of miRNAs processing genes may be put at higher risk of larynx cancer development with high probability of lymph nodes occurrence. On the other hand, it appeared that RAN rs14035 CT polymorphic variant may possess a kind of protective effect on individuals, because a relatively small number of LC patients were carriers of this heterozygote, even in comparison with healthy subjects. To the best of our knowledge, polymorphisms evaluated in our study were not analyzed in the context of larynx cancer before. There is also a very little information about the connection of investigated genes with head and neck cancer. On the other hand, SNPs of miRNAs processing genes are widely analyzed in other types of cancer; however, these pieces of data are inconsistent.
In the present association study, we found that DROSHA polymorphism was not associated with the risk of larynx cancer. DROSHA is a member of RNase III superfamily and is an important nuclease that executes the initial step in miRNA processing by transforming pri-miRNA to pre-miRNA. RNA interference of DROSHA resulted in accumulation of pri-miRNA and reduction of pre-miRNA and mature RNA [24]. There were some studies describing the role of DROSHA in cancer [25]. We have found only few papers investigating role of DROSHA rs6877842 SNP in T-cell lymphoma [26, 27], esophageal cancer [28], and idiopathic ovarian insufficiency [29]. Tian et al.'s data showed that among patients with T-cell lymphoma carriers of GC genotype in Chinese population have higher complete remission rate compared with those carrying CC genotype (OR: 0.07, 0.95 CI: 0.01–0.072, and p = 0.026) [26]. It was also confirmed by Li et al. who proved that variant allele of this polymorphism also increased the overall survival of T-cell lymphoma patients compared to the wild type genotype (HR: 0.27, 0.95 CI: 0.11–0.67, and p = 0.005) [19]. Additionally, a haplotype analysis of DROSHA rs6877842 in Korean patients suggests that ∗∗∗ACTA is associated with higher POI [13]. In case of esophageal cancer there was no connection between prevalence of investigated polymorphism and tumor occurrence. Although there is no information about connection between head and neck cancers and rs6877842, there are some proofs of dysregulation of miRNA processing genes at the expression level. Guo et al. showed the mean level of DROSHA and DICER mRNA was significantly downregulated in nasopharyngeal cancer (NPC) tissue specimens and cell lines when compared with controls. Low expression of DICER and DROSHA protein was significantly correlated with shorter progression-free survival and overall survival of NPC patients [30].
DICER1 is an enzyme responsible for the cleavage of miRNA precursors and has previously been implicated in the oncogenic process of several cancers [31–33]. DICER1 and transactivation-responsive RNA-binding protein mediate pre-miRNA processing. A recent study indicated that DICER1 functions as a haploinsufficient tumor suppressor in cancer [34]. Indeed, lower levels of DICER1 mRNA have been associated with decreased cancer survival [35]. Evidence indicates that DICER1 may play crucial roles in the tumorigenesis of different cancers. Some studies showed that lower levels of DICER1 mRNA expression were associated with the development of lung cancer [36], colon cancer [33], and ovarian cancer [35]. However, studies also demonstrated that elevated expression levels of DICER1 were correlated with increased cell proliferation of oral cancer cells [37]. Moreover, Gao and colleagues showed that the expression of DICER was significantly higher in the laryngeal squamous cell carcinoma (LSCC) than in the polyp tissue specimens. DICER expression level was significantly associated with the TNM stage. Survival analyses also revealed a strong association between tumor DICER expression and the survival of the patients with LSCC [38]. On the other hand, as it was mentioned before, Guo et al.'s study showed decreased mRNA expression in nasopharyngeal cancer [30]. These analyses show that upregulation or downregulation of DICER mRNA expression may depend on the site of tumor appearance. We have found an association between risk of laryngeal cancer and two DICER1 SNPs: rs13078 and rs3742330. It was also found that rs13078 (HR = 1.66; 0.95 CI: 1.09–2.52; p = 0.02) was associated with the risk of death of patients with colorectal adenocarcinoma [39]. Lin et al. performed an analysis of correlation between survival and recurrence in patients with renal cell carcinoma and DICER SNPs [40]. In haplotype analysis, haplotypes of DICER showed significant association with RCC survival. Specifically, compared with the AT haplotype (in order of rs3742330 and rs13078), the haplotype AA had HR of 1.51 (95% CI = 0.99–2.31) and the haplotype GA was associated with increased HR of 2.04 (95% CI = 1.00–4.15). Similarly, in diplotype analysis, using the AT-AT as the reference group, the diplotype AT-GT was associated with 2.86-fold increased risk (95% CI = 1.11–7.34; p = 0.03) and the diplotype AA-AA was at 3.48-fold increased risk (95% CI = 1.21–9.97; p = 0.02). Another study showed that patients with at least one variant allele of SNP rs3742330 in DICER had a significantly increased risk of oral premalignant lesions (OR, 2.09; 95% CI, 1.03–4.24) [41]. On the other hand, there was no statistically significant association between rs3742330 and rs13078 SNPs and esophageal cancer [28].
DGCR8 is a double-stranded RNA-binding protein that functions as the noncatalytic subunit of the microprocessor complex and facilitates RNA cleavage by the RNase III protein DROSHA. In vitro knockdown of DROSHA, DGCR8, and DICER1 impaired miRNA processing and thereby promoted oncogenic transformation in mouse lung cancer cells and tumor development in vivo [42]. Han et al. [43] reported that RNASEN and DGCR8 regulate each other posttranscriptionally and that DGCR8 stabilizes RNASEN via protein-protein interactions. Because of the direct effect of DGCR8 and RNASEN on miRNA biogenesis and the associations between miRNA expression and cancer development the variations in either gene might affect head and neck cancer occurrence. In our study we evaluated the role of three DGCR8 SNPs in larynx cancer: rs3757, rs417309, and rs1640299. Other researchers also evaluated the role of these polymorphisms in context of renal cell carcinoma and esophageal cancer. Horikawa et al. and Lin et al. also evaluated these polymorphisms referred to renal cell carcinoma, but SNPs as well as haplotype analysis did not reveal any statistically significant correlations [40, 44]. Study performed by Ye et al. also did not show any association between these polymorphisms and esophageal cancer [28]. No linkage was also presented between the occurrence of cervical cancer and rs3757 polymorphic variants [45]. However, Jiang et al. have shown that the rs417309 polymorphism of DGCR8 gene was associated with an increased breast cancer risk (OR = 1.50; 95% confidence interval (CI): 1.16–1.93). Besides, using luciferase assay, they have found that the variant A allele of rs417309 compared to allele G elevates DGCR8 protein expression [46]. Because the rs417309 polymorphism is located in the 3′-UTR, which is the binding site of miR-106b and miR-579, it might affect the miRNAs maturation. Gong et al. have indicated that the expression of miR-106b was dramatically increased in breast cancer tissues compared to in healthy tissue [47]. Additionally, it was shown that the expression of DGCR8 was upregulated in several types of cancer [48, 49]. Therefore, rs417309 may impair the binding of miRNAs such as miR-106b with DGCR8 and disrupt the process of miRNAs maturation and consequently play an important role in the tumorigenesis.
RAN is a unique member of the Ras superfamily of GTPases, which is essential to the transportation of pre-miRNAs from nucleus to cytoplasm through the nuclear pore complex in a GTP-dependent manner [50]. PolymiRTS database suggests that the ancestral allele lies in a binding site for miR-575, which is disrupted by the derived allele that in addition creates a binding site for miR-182∗. Although these are in silico results, they raise the possibility that in addition to affecting cancer risk through the disruption of miRNA nuclear export a more intricate pathway may be involved that includes miRNA regulation. Some studies have investigated the associations between rs14035 polymorphism of this gene and risk of several cancers. Evaluation of role of rs14035 in hepatocellular carcinoma showed no statistically significant differences (p < 0.05) between patients and healthy controls [51]. There was also no association with the risk of renal cell carcinoma [44] or oral premalignant lesions [52]. On the other hand, data showed an association between occurrence of recessive variant of investigated RAN polymorphism and esophageal cancer (p = 0.024) [28]. rs14035 was also evaluated as a predictor of clinical outcomes in colorectal adenocarcinoma patients [34]. Among 117 patients with Stage II disease who received 5-FU based chemotherapy, the most significant association with recurrence was conferred by the variant allele of RAN:rs14035 in a dose-dependent manner (per allele HR = 2.32; 95% CI, 1.28 to 4.21; p = 0.005; q = 0.06). For Stage II diseases, RAN:rs14035 was associated with overall survival with high significance in patients receiving surgery and adjuvant fluoropyrimidine treatment.
Xpo5 mediates nuclear export of pre-miRNA in a RAN GTP-dependent manner by binding to pre-miRNA and RAN GTPase in the nucleus [53]. XPO5 is found in the nuclear membrane and mediates the transport of pre-miRNA between the nucleic and cytoplasmic compartments so as to adjust the whole miRNA expression level. Knock-down of XPO5 expression leads to reduced miRNA levels [53]. A mutated and inactive XPO5 resulted in reduced miRNA processing and decreased miRNA target inhibition; the restored XPO5 seemed as a tumor suppressor to reverse the impaired export of pre-miRNA in colon cancer [54]. The miR-SNP of rs11077 of XPO5 has been associated with the risk of esophageal cancer (OR = 1.84, 95% CI: 1.16–2.93, and p = 0.010) as well as the overall survival in myeloma and lymphoma [28, 55, 56]. The AC genotype of rs11077, which carries C or A allele, was significantly associated with a better chemotherapy response in patients with non-small cell lung (p = 0.001). In addition, rs11077 was independently associated with overall survival in advanced NSCLC patients through multivariate analysis (relative risk 0.457; 95% confidence interval: 0.251–0.831; p = 0.010) [57]. The altered XPO5 expression may affect the miRNAs, leading to overall downregulation of miRNA expression profiles and thereby mediates the hepatocellular carcinoma survival. rs11077 CC genotype shows association with reduced Renilla expression in a Renilla luciferase 3′UTR reporter system. It implies that this SNP could modify XPO5 expression so as to result in overall expression of miRNA in multiple myeloma cells [35].
TARBP2 (trans-activation-responsive RNA binding protein 2) is a component of the miRNA loading complex (composed of DICER1, AGO2, and TRBP2) required for the formation of RISC. Melo and colleagues [58] identified two frameshift mutations in TARBP2 that introduce premature stop codons, resulting in reduced TRBP expression. One function of TRBP is regulating DICER1 stability; thus these mutations resulted in reduced DICER1 expression and lower miRNA production and were associated with higher cellular proliferation levels [58]. It has been shown that the variant allele of rs784567, which is located in the promoter of the TRBP gene, was associated with neither a risk of bladder cancer (p = 0.07) [21] nor renal cell carcinoma [40] or oral premalignant lesions [41]. Patients with Hodgkin Lymphoma that were carriers of both XPO5 AA/CC and TARBP2 TT/TC genotypes had the shortest disease free survival (p = 0.008) and overall survival (p = 0.008). On the other hand, rs784567 (HR = 1.59; 95% CI, 1.03 to 2.43; p = 0.04) was also associated with the risk of death in colorectal cancer but lost significance after adjusting for multiple comparison [33]. Given the differential cell of origin for cancers and differential cell type specificity of miRNA transcriptomes, it is reasonable to assume that the effects of miR-SNP will be modulated in a cell type-specific manner.
Several potential limitations of the present study warrant considerations. First of all, a relatively small sample size may limit the statistical power of our study, especially in stratification analysis by tumor sites. Secondly, it is a hospital-based case-control study and inherent selection bias cannot be completely excluded. Thirdly, since the intensity and duration of drinking were absent in this study, it was difficult to do future analysis for such exposure variables. Thus, larger, well-designed epidemiological studies with ethnically diverse populations are warranted to confirm and expand our findings.
However, our findings provide new information about the relationship of genes involved in the microRNAs maturation and the development of larynx cancer. The results indicate that polymorphic variants of these genes may affect not only the development of cancer but also disease progression. Further studies confirming our results with larger study group can help to discover new diagnostic markers of LC, which greatly facilitate the initiation of treatment and consequently a better prognosis for the patient. Furthermore, our results may also have a large impact on the clinical studies, because they provide information about cancer progression.
5. Conclusion
In light of recent reports, the role of microRNAs in cancer development is considerable. The miRNAs are involved in all aspects of cancer biology, such as proliferation, apoptosis, migration, and angiogenesis. Our results suggest that rs3742330 of DICER1, rs13078 of DICER1, and rs784567 of TARBP2 as well as rs11077 of XPO5 might be associated with a risk of laryngeal cancer occurrence in the Polish population. Additionally, rs417309 of DGCR8, rs3742330 and rs13078 of DICER1, and rs784567 of TARBP2 as well as rs11077 of XPO5 are associated with the progression of LC depending on the tumor size and lymph node metastases. However, further epidemiologic studies with larger subject numbers should be performed to confirm and expand our results. In addition, the results of our study warrant further functional studies to elucidate the mechanisms by which polymorphisms of miRNA machinery genes affect LC development.
Supplementary Material
The Tables 6–8 provide an assessment of association between miRNA processing genes single nucleotide polymorphisms and smoking cigarettes. Table 6 shows the relationship of these SNPs with smoking status, Table 7 with duration of smoking, while Table 8 number of cigarettes smoked per day. Performed analysis did not show any statistically significant differences in the frequency of studied SNPs polymorphic variants correlating with cigarette smoking among LC patients.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Ferlay J., Steliarova-Foucher E., Lortet-Tieulent J., et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. European Journal of Cancer. 2013;49(6):1374–1403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 2.Chang S. S., Wei W. J., Smith I., et al. MicroRNA alterations in head and neck squamous cell carcinoma. International Journal of Cancer. 2008;123(12):2791–2797. doi: 10.1002/ijc.23831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hui A. B. Y., Lenarduzzi M., Krushel T., et al. Comprehensive MicroRNA profiling for head and neck squamous cell carcinomas. Clinical Cancer Research. 2010;16(4):1129–1139. doi: 10.1158/1078-0432.CCR-09-2166. [DOI] [PubMed] [Google Scholar]
- 4.Ramdas L., Giri U., Ashorn C. L., et al. miRNA expression profiles in head and neck squamous cell carcinoma and adjacent normal tissue. Head & Neck. 2009;31(5):642–654. doi: 10.1002/hed.21017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shah A. A., Jeffus S. K., Stelow E. B. Squamous cell carcinoma variants of the upper aerodigestive tract: a comprehensive review with a focus on genetic alterations. Archives of Pathology and Laboratory Medicine. 2014;138(6):731–744. doi: 10.5858/arpa.2013-0070-ra. [DOI] [PubMed] [Google Scholar]
- 6.Vigneswaran N., Williams M. D. Epidemiologic trends in head and neck cancer and aids in diagnosis. Oral and Maxillofacial Surgery Clinics of North America. 2014;26(2):123–141. doi: 10.1016/j.coms.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ha M., Kim V. N. Regulation of microRNA biogenesis. Nature Reviews Molecular Cell Biology. 2014;15(8):509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 8.Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 10.Davis-Dusenbery B. N., Hata A. Mechanisms of control of microRNA biogenesis. Journal of Biochemistry. 2010;148(4):381–392. doi: 10.1093/jb/mvq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomson J. M., Newman M., Parker J. S., Morin-Kensicki E. M., Wright T., Hammond S. M. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes & Development. 2006;20(16):2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romero-Cordoba S. L., Salido-Guadarrama I., Rodriguez-Dorantes M., Hidalgo-Miranda A. miRNA biogenesis: biological impact in the development of cancer. Cancer Biology and Therapy. 2014;15(11):1444–1455. doi: 10.4161/15384047.2014.955442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rupaimoole R., Wu S. Y., Pradeep S., et al. Hypoxia-mediated downregulation of miRNA biogenesis promotes tumour progression. Nature Communications. 2014;5, article 5202 doi: 10.1038/ncomms6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cadoni G., Boccia S., Petrelli L., et al. A review of genetic epidemiology of head and neck cancer related to polymorphisms in metabolic genes, cell cycle control and alcohol metabolism. Acta Otorhinolaryngologica Italica. 2012;32(1):1–11. [PMC free article] [PubMed] [Google Scholar]
- 15.Adamson R., Jones A. S., Field J. K. Loss of heterozygosity studies on chromosome 17 in head and neck cancer using microsatellite markers. Oncogene. 1994;9(7):2077–2082. [PubMed] [Google Scholar]
- 16.Chen Y., Chen C. DNA copy number variation and loss of heterozygosity in relation to recurrence of and survival from head and neck squamous cell carcinoma: a review. Head and Neck. 2008;30(10):1361–1383. doi: 10.1002/hed.20861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abou-Elhamd K.-E. A., Habib T. N., Moussa A.-E., Badawy B. S. The role of genetic susceptibility in head and neck squamous cell carcinoma. European Archives of Oto-Rhino-Laryngology. 2008;265(2):217–222. doi: 10.1007/s00405-007-0436-4. [DOI] [PubMed] [Google Scholar]
- 18. rs6877842 NCBI SNP database, 2015, http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=6877842.
- 19. rs417309 NCBI SNP database, 2015, http://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=417309.
- 20.Cao W.-M., Gao Y., Yang H.-J., et al. Germline mutations of DICER1 in Chinese women with BRCA1/BRCA2-negative familial breast cancer. Genetics and Molecular Research. 2014;13(4):10754–10760. doi: 10.4238/2014.december.18.16. [DOI] [PubMed] [Google Scholar]
- 21.Yang H., Dinney C. P., Ye Y., Zhu Y., Grossman H. B., Wu X. Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer. Cancer Research. 2008;68(7):2530–2537. doi: 10.1158/0008-5472.can-07-5991. [DOI] [PubMed] [Google Scholar]
- 22.Lin R.-J., Lin Y.-C., Chen J., et al. microRNA signature and expression of Dicer and Drosha can predict prognosis and delineate risk groups in neuroblastoma. Cancer Research. 2010;70(20):7841–7850. doi: 10.1158/0008-5472.can-10-0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho S. H., Ko J. J., Kim J. O., et al. 3′-uTR polymorphisms in the MiRNA machinery genes DROSHA, DICER1, RAN, and XPO5 are associated with colorectal cancer risk in a korean population. PLoS ONE. 2015;10(7) doi: 10.1371/journal.pone.0131125.e0131125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee Y., Ahn C., Han J., et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 25.Foulkes W. D., Priest J. R., Duchaine T. F. DICER1: mutations, microRNAs and mechanisms. Nature Reviews. Cancer. 2014;14(10):662–672. doi: 10.1038/nrc3802. [DOI] [PubMed] [Google Scholar]
- 26.Tian X., Zhang B., Li X., et al. Association of microRNA-related genes (DROSHA, DICER1 and GEMIN4) polymorphisms with T-cell lymphoma prognosis. Zhonghua Xue Ye Xue Za Zhi. 2014;35(5):408–413. doi: 10.3760/cma.j.issn.0253-2727.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Li X., Tian X., Zhang B., Chen J. Polymorphisms in microRNA-related genes are associated with survival of patients with T-cell lymphoma. Oncologist. 2014;19(3):243–249. doi: 10.1634/theoncologist.2013-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye Y., Wang K. K., Gu J., et al. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prevention Research. 2008;1(6):460–469. doi: 10.1158/1940-6207.capr-08-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rah H., Jeon Y. J., Lee B. E., et al. Association of polymorphisms in microRNA machinery genes (DROSHA, DICER1, RAN, and XPO5) with risk of idiopathic primary ovarian insufficiency in Korean women. Menopause. 2013;20(10):1067–1073. doi: 10.1097/GME.0b013e3182883907. [DOI] [PubMed] [Google Scholar]
- 30.Guo X., Liao Q., Chen P., et al. The microRNA-processing enzymes: drosha and Dicer can predict prognosis of nasopharyngeal carcinoma. Journal of Cancer Research and Clinical Oncology. 2012;138(1):49–56. doi: 10.1007/s00432-011-1058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selever J., Gu G., Lewis M. T., et al. Dicer-mediated upregulation of BCRP confers tamoxifen resistance in human breast cancer cells. Clinical Cancer Research. 2011;17(20):6510–6521. doi: 10.1158/1078-0432.ccr-11-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han L., Zhang A., Zhou X., et al. Downregulation of Dicer enhances tumor cell proliferation and invasion. International Journal of Oncology. 2010;37(2):299–305. doi: 10.3892/ijo-0000678. [DOI] [PubMed] [Google Scholar]
- 33.Dong Y., Yu J., Ng S. S. M. MicroRNA dysregulation as a prognostic biomarker in colorectal cancer. Cancer Management and Research. 2014;6:405–422. doi: 10.2147/cmar.s35164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar M. S., Pester R. E., Chen C. Y., et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes and Development. 2009;23(23):2700–2704. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merritt W. M., Lin Y. G., Han L. Y., et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. The New England Journal of Medicine. 2008;359(25):2641–2650. doi: 10.1056/nejmoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karube Y., Tanaka H., Osada H., et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Science. 2005;96(2):111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jakymiw A., Patel R. S., Deming N., et al. Overexpression of dicer as a result of reduced let-7 microRNA levels contributes to increased cell proliferation of oral cancer cells. Genes Chromosomes and Cancer. 2010;49(6):549–559. doi: 10.1002/gcc.20765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao C., Li X., Tong B., et al. Up-regulated expression of Dicer reveals poor prognosis in laryngeal squamous cell carcinoma. Acta Oto-Laryngologica. 2014;134(9):959–963. doi: 10.3109/00016489.2014.920962. [DOI] [PubMed] [Google Scholar]
- 39.Lin M., Gu J., Eng C., et al. Genetic polymorphisms in microRNA-related genes as predictors of clinical outcomes in colorectal adenocarcinoma patients. Clinical Cancer Research. 2012;18(14):3982–3991. doi: 10.1158/1078-0432.CCR-11-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin J., Horikawa Y., Tamboli P., Clague J., Wood C. G., Wu X. Genetic variations in microRNA-related genes are associated with survival and recurrence in patients with renal cell carcinoma. Carcinogenesis. 2010;31(10):1805–1812. doi: 10.1093/carcin/bgq168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clague J., Lippman S. M., Yang H., et al. Genetic variation in microRNA genes and risk of oral premalignant lesions. Molecular Carcinogenesis. 2010;49(2):183–189. doi: 10.1002/mc.20588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar M. S., Lu J., Mercer K. L., Golub T. R., Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nature Genetics. 2007;39(5):673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 43.Han J., Pedersen J. S., Kwon S. C., et al. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136(1):75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horikawa Y., Wood C. G., Yang H., et al. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clinical Cancer Research. 2008;14(23):7956–7962. doi: 10.1158/1078-0432.ccr-08-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mi Y., Wang L., Zong L., Pei M., Lu Q., Huang P. Genetic variants in microRNA target sites of 37 selected cancer-related genes and the risk of cervical cancer. PLoS ONE. 2014;9(1) doi: 10.1371/journal.pone.0086061.e86061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang Y., Chen J., Wu J., et al. Evaluation of genetic variants in microRNA biosynthesis genes and risk of breast cancer in Chinese women. International Journal of Cancer. 2013;133(9):2216–2224. doi: 10.1002/ijc.28237. [DOI] [PubMed] [Google Scholar]
- 47.Gong C., Qu S., Lv X.-B., et al. BRMS1L suppresses breast cancer metastasis by inducing epigenetic silence of FZD10. Nature Communications. 2014;5, article 5406 doi: 10.1038/ncomms6406. [DOI] [PubMed] [Google Scholar]
- 48.Zhang X., Cairns M., Rose B., et al. Alterations in miRNA processing and expression in pleomorphic adenomas of the salivary gland. International Journal of Cancer. 2009;124(12):2855–2863. doi: 10.1002/ijc.24298. [DOI] [PubMed] [Google Scholar]
- 49.Sand M., Skrygan M., Georgas D., et al. Expression levels of the microRNA maturing microprocessor complex component DGCR8 and the RNA-induced silencing complex (RISC) components argonaute-1, argonaute-2, PACT, TARBP1, and TARBP2 in epithelial skin cancer. Molecular Carcinogenesis. 2012;51(11):916–922. doi: 10.1002/mc.20861. [DOI] [PubMed] [Google Scholar]
- 50.Bohnsack M. T., Czaplinski K., Görlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10(2):185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu S., An J., Lin J., et al. Single nucleotide polymorphisms of microRNA processing machinery genes and outcome of hepatocellular carcinoma. PLoS ONE. 2014;9(3) doi: 10.1371/journal.pone.0092791.e92791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roy R., De Sarkar N., Ghose S., et al. Association between risk of oral precancer and genetic variations in microRNA and related processing genes. Journal of Biomedical Science. 2014;21, article 48 doi: 10.1186/1423-0127-21-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lund E., Güttinger S., Calado A., Dahlberg J. E., Kutay U. Nuclear export of microRNA precursors. Science. 2004;303(5654):95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 54.Melo S. A., Moutinho C., Ropero S., et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18(4):303–315. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 55.de Larrea C. F., Navarro A., Tejero R., et al. Impact of MiRSNPs on survival and progression in patients with multiple myeloma undergoing autologous stem cell transplantation. Clinical Cancer Research. 2012;18(13):3697–3704. doi: 10.1158/1078-0432.ccr-12-0191. [DOI] [PubMed] [Google Scholar]
- 56.Navarro A., Muñoz C., Gaya A., et al. MiR-SNPs as markers of toxicity and clinical outcome in hodgkin lymphoma patients. PLoS ONE. 2013;8(5) doi: 10.1371/journal.pone.0064716.e64716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ding C., Li C., Wang H., Li B., Guo Z. A miR-SNP of the XPO5 gene is associated with advanced non-small-cell lung cancer. OncoTargets and Therapy. 2013;6:877–881. doi: 10.2147/ott.s48284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Melo S. A., Ropero S., Moutinho C., et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nature Genetics. 2009;41(3):365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Tables 6–8 provide an assessment of association between miRNA processing genes single nucleotide polymorphisms and smoking cigarettes. Table 6 shows the relationship of these SNPs with smoking status, Table 7 with duration of smoking, while Table 8 number of cigarettes smoked per day. Performed analysis did not show any statistically significant differences in the frequency of studied SNPs polymorphic variants correlating with cigarette smoking among LC patients.


