Abstract
Transforming growth factor-β/Smad3 signaling plays a critical role in the process of chronic kidney disease (CKD), but targeting Smad3 systematically may cause autoimmune disease by impairing immunity. In this study, we used whole-transcriptome RNA-sequencing to identify the differential gene expression profile, gene ontology, pathways, and alternative splicing related to TGF-β/Smad3 in CKD. To explore common dysregulation of genes associated with Smad3-depednent renal injury, kidney tissues of Smad3 wild-type and knockout mice with immune (anti-glomerular basement membrane glomerulonephritis) and non-immune (obstructive nephropathy)-mediated CKD were used for RNA-sequencing analysis. Totally 1922 differentially expressed genes (DEGs) were commonly found in these CKD models. The up-regulated genes are inflammatory and immune response associated, while decreased genes are material or electron transportation and metabolism related. Only 9 common DEGs were found to be Smad3-dependent in two models, including 6 immunoglobulin genes (Ighg1, Ighg2c, Igkv12-41, Ighv14-3, Ighv5-6 and Ighg2b) and 3 metabolic genes (Ugt2b37, Slc22a19, and Mfsd2a). Our results identify transcriptomes associated with renal injury may represent a common mechanism for the pathogenesis of CKD and reveal novel Smad3 associated transcriptomes in the development of CKD.
Chronic Kidney Disease (CKD) becomes a major problem and healthcare burden in China, which affects 10.8% of the population with high prevalence and low awareness according to a national survey in 20121,2. Renal inflammation and fibrosis are the common manifestations of CKD. Transforming growth factor-β1 (TGF-β1), a profibrogenic cytokine, has an important biological role in the induction of organ inflammation and fibrosis by activating the downstream Smad proteins, especially Smad33,4,5,6. Although Smad3 is a critical transcription factor which conveys signals from TGF-β receptors to the nucleus, targeting Smad3 may cause autoimmune disease by impairing immunity6,7. Thus, alternative approaches to inhibit TGF-β actions should be more precise and can be achieved by identifying the Smad3-dependent transcriptomes in chronic renal injury.
High-throughput RNA sequencing (RNA-Seq) is rapidly emerging as a favorite approach for transcriptome-oriented studies; it permits quantification of genes and products of their processing or partial degradation, and identification of novel coding and non-coding RNA8,9,10. In previous study, we successfully identified long non-coding RNAs in Smad3 wild-type (WT) and knock-out (KO) mouse models of obstructive nephropathy (UUO) and immunologically-induced anti-glomerular basement membrane glomerulonephritis (anti-GBM GN) by using RNA-seq11,12. Compared to WT mice, a number of transcripts including mRNAs and non-coding RNAs in the diseased kidneys from UUO or anti-GBM GN models were significantly altered in Smad3 KO mice. The present study aimed to compare the profiles of Smad3 related DEGs in these two CKD models
Results
RNA-Sequencing of mouse transcriptomes in two CKD models
High-throughput RNA-Seq was used to determine the Smad3-dependent transcriptome profiles of two CKD animal models. Total RNA of kidneys from Smad3 KO and WT mice with UUO or anti-GBM GN were collected for the analysis. As shown in Table 1, after filtering the low quality reads and rRNA sequences from the raw fragments, we obtained over 50 M reads in each sample. Total clean reads were then mapped to mouse genome (mm9) by using Tophat. Approximately, 66.96 ~ 97.62% were successfully mapped to mouse genomic regions by allowing no more than two mismatches in 25 bp segments and only output the alignment results which are no more than at most 10 genomic locations. Among the aligned fragments, 42.65 ~ 87.88% were mapped to unique genomic regions, which were used for further analysis to ensure reliability of analysis results.
Table 1. Mapping overview of RNA-sequencing reads on the mouse genome.
| Sample | All reads | All reads (pairs) | Mapped reads (all) | Mapping rate | Unique mapped | Unique mapped rate |
|---|---|---|---|---|---|---|
| WT-Nor-1 | 51398366 | 25699183 | 25006455 | 97.30% | 21883540 | 85.15% |
| WT-Nor-2 | 52300544 | 26150272 | 25528188 | 97.62% | 22918918 | 87.64% |
| WT-UUO-1 | 51708360 | 25854180 | 18332567 | 70.91% | 13718644 | 53.06% |
| WT-UUO-2 | 51913666 | 25956833 | 24953074 | 96.13% | 22352312 | 86.11% |
| WT-GBM-1 | 52125094 | 26062547 | 24740414 | 94.93% | 21836781 | 83.79% |
| WT-GBM-2 | 54110904 | 27055452 | 26238539 | 96.98% | 22334706 | 82.55% |
| KO-Nor-1 | 53474448 | 26737224 | 25800912 | 96.50% | 22272463 | 83.30% |
| KO-Nor-2 | 53680294 | 26840147 | 25895801 | 96.48% | 21477366 | 80.02% |
| KO-UUO-1 | 54354462 | 27177231 | 22326568 | 82.15% | 15611253 | 57.44% |
| KO-UUO-2 | 51186728 | 25593364 | 24937874 | 97.44% | 22492658 | 87.88% |
| KO-GBM-1 | 54445046 | 27222523 | 18228106 | 66.96% | 11609579 | 42.65% |
| KO-GBM-2 | 51855890 | 25927945 | 24349590 | 93.91% | 20020045 | 77.21% |
Note: WT-Nor:wild-type normal mice; WT-UUO:UUO model in wild-type mice; WT-GBM:anti-GBM GN model in wild-type mice; KO-Nor:Smad3 knockout normal mice; KO-UUO:UUO model in Smad3 knockout mice; KO-GBM: anti-GBM GN model in Smad3 knockout mice.
Analyses of differentially expressed genes
The FPKM value was used to quantifying the mRNA expression. We analyze the profiles of DEGs (differentially expressed genes) between disease and normal control groups. Pearson correlation analysis showed that the FPKM values between these groups highly correlated (r = 0.87–0.92, r < 0.01) (Supplementary Fig. S1).
Comparison of DEGs related to renal injury in two models
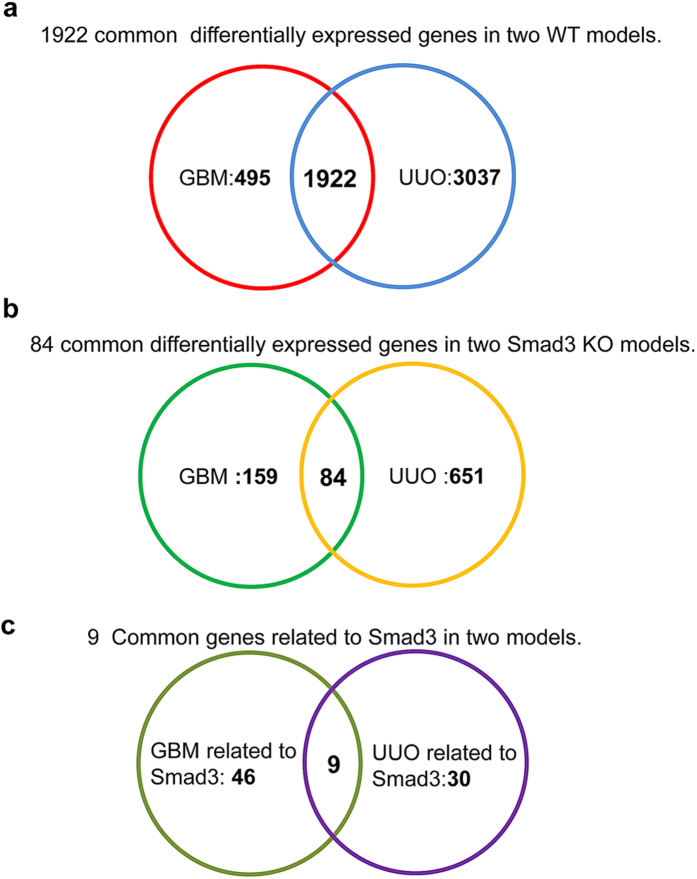
The analysis of DEGs was shown in Fig. 1a and Table 2, compared to WT-Nor (Smad3 wild-type normal control) group, totally 2417 DEGs were found in WT-GBM (Smad3 wild-type anti-GBM GN model) including 1552 up-regulated and 865 down-regulated DEGs which accounting for 64.21% and 35.79% of the total DEGs. In WT-UUO (Smad3 wild-type UUO model),the up- and down-regulated DEGs were 2898 and 2061,accounting for 58.44% and 41.56% of the total 4959 DEGs detected respectively. These findings indicated that the number of DEGs in UUO is significantly larger than the anti-GBM GN models (4959 vs 2417). In addition, 1922 DEGs were commonly found in the wild-type groups of both models including 1203(62.59%) up-regulated and 719(37.41%) down-regulated genes (all the gene ID and fold changes are listed in Supplementary File S1).
Figure 1. Number of total DEGs in two CKD models of WT and Smad3 KO mice.
(a) Venn diagram has shown1922 common differentially expressed genes in two WT models. (b) Venn diagram of 84 common differentially expressed genes in two Smad3 KO models. (c) Venn diagram of 9 common Smad3-related differentially expressed genes in two models. GBM:anti-GBM GN model in wild-type or Smad3 knockout mice; UUO: UUO model in wild-type mice or Smad3 knockout mice.
Table 2. The number of differentially expressed genes in two CKD models.
| Samples | Total | Up (%) | Down (%) |
|---|---|---|---|
| WT-GBM vs WT-Nor | 2417 | 1552 (64.21%) | 865 (35.79%) |
| WT-UUO vs WT-Nor | 4959 | 2898 (58.44%) | 2061 (41.56%) |
| Common in two WT models | 1922 | 1203 (62.59%) | 719 (37.41%) |
| KO-GBM vs KO-Nor | 243 | 117 (48.15%) | 126 (51.85%) |
| KO-UUO vs KO-Nor | 735 | 400 (54.42%) | 335 (45.58) |
| Common in two KO models | 84 | 36 (42.86%) | 48 (57.14%) |
Note: WT-GBM:anti-GBM GN model in wild-type mice; WT-Nor:wild-type normal mice; WT-UUO:UUO model in wild-type mice; KO-GBM:anti-GBM GN model in Smad3 knockout mice; KO-UUO:UUO model in Smad3 knockout mice; KO-Nor:Smad3 knockout normal mice; Up:up-regulated genes; Down:down-regulated genes.
In WT-GBM, Ighg1 (immunoglobulin heavy constant gamma 1, LogFC = 13.29, FDR = 6.93E-30) was largely upregulated when compared to WT-Nor whereas Sftpc (surfactant associated protein C) (LogFC = −11.40, FDR = 5.63E-05) was greatly suppressed. The increase of Ighg1 may be associated with humoral immune response of B cell13. Sftpc is an extremely hydrophobic surfactant protein14,15, deficiency mice of Sftpc leads to inflammation and increased cytokine production16. In WT-UUO, FlnA (filamin, alpha, LogFC = 17.25, FDR = 1.40E-54) was the most up-regulated gene compared to the WT-Nor, which encode a widely expressed actin-binding protein that regulates the reorganization of actin cytoskeleton by interacting with integrins, transmembrane receptor complexes, and second messengers17, whereas, Sftpc (LogFC = −11.37,FDR = 8.30E-05) was significantly downregulated.
Comparison of Smad3-related DEGs in renal injury
Further analysis revealed that, compared with WT mice, many genes were differentially expressed in the kidneys of Smad3 knockout mice during CKD. As shown in Fig. 1b and Table 2, deletion of Smad3 significantly diminished the genes expression diversity in two disease models; the total numbers of DEGs were largely reduced when compared to the WT models. Compared to KO-Nor (normal control of Smad3 knockout mice) group, the total number of DEGs was 243 in KO-GBM (anti-GBM GN model of Smad3 knockout mice) and 735 in KO-UUO (UUO model of Smad3 knockout mice). The up and down-regulated genes were 117 and 126 in KO-GBM, while 400 and 335 were detected in KO-UUO compared to the KO-Nor (Smad3 knockout normal mice). The total common DEGs in two CKD models of Smad3 KO mice were 84, which included 36 up-regulated and 48 down-regulated genes (Table 2). All the comparisons of DEGs in Smad3 KO models were also listed in Supplementary File S1 (including all the gene ID and fold changes).
To identify of Smad3-related genes in two models, DEGs have different expression direction in WT and Smad3 KO mice will be considered as Smad3-related. In these DEGs, 55 genes in anti-GBM GN model and 39 genes in UUO model were Smad3-related and differentially expressed in the WT and Smad3 KO kidneys (Fig. 1c and Table 3). The 9 common Smad3-associated DEGs in two CKD models were including 6 immunoglobulin genes (Ighg1, Ighg2c, Igkv12-41, Ighv14-3, Ighv5-6, and Ighg2b) and 3 metabolic genes (Ugt2b37, Slc22a19, and Mfsd2a).
Table 3. The number of differentially expressed genes in renal injury related to Smad3.
| Comparision | GBM related to Smad3 | UUO related to Smad3 | Common | Gene name |
|---|---|---|---|---|
| Up in WT & Down in KO | 50 | 17 | 6 | Ighg1, Ighg2c, Igkv12-41, Ighv14-3, Ighv5-6, Ighg2b |
| Down in WT & Up in KO | 5 | 22 | 3 | Ugt2b37, Slc22a19, Mfsd2a |
Note: WT:anti-GBM GN or UUO model in wild-type mice; KO:anti-GBM GN or UUO model in Smad3 knockout mice; Up:up-regulated genes; Down:down-regulated genes.
Analysis of Gene Ontology and Pathways
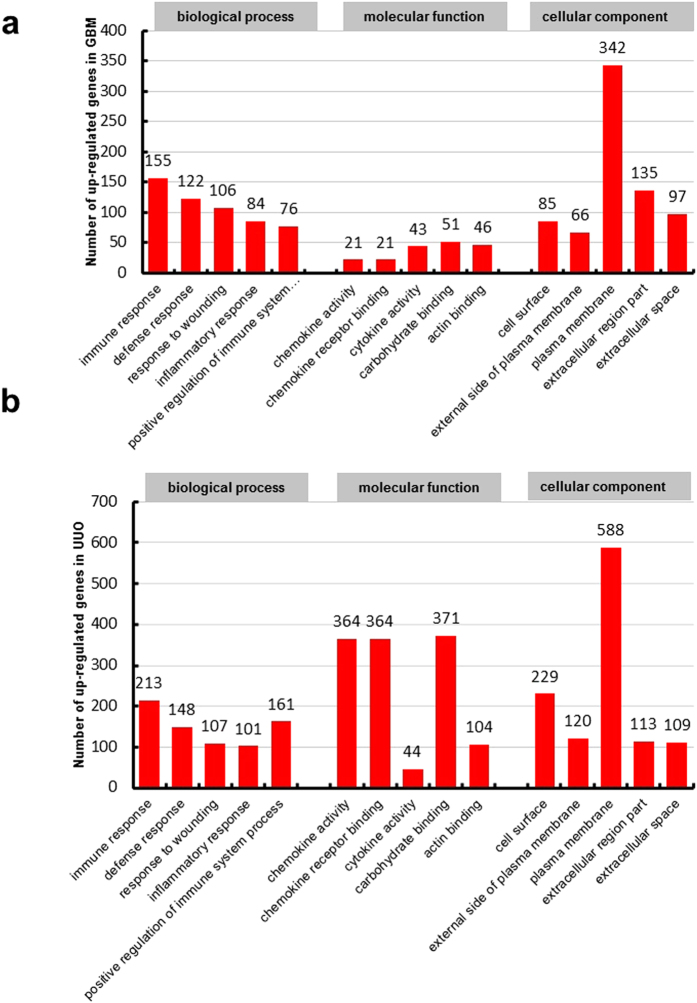
The NIH Database for Annotation, Visualization and Integrated Discovery (DAVID) was used to perform gene functional annotation clustering. Gene Ontology describes attributes of genes in three non-overlapping domains of molecular biology which including biological processes, molecular function and cellular component. We summarized the number of significant GO terms that DEGs enriched in Table 4. Because ‘biological process’ is the most widely used subontology of GO to evaluate the functions of gens, we focus on the Go terms of biological process. According to GO terms for biological process in up-regulated genes, we obtained 171 significant GO terms in WT-GBM and 209 in WT-UUO compared to WT-Nor. In two WT models, common up-regulated genes are enriched in 138 GO terms for biological process. The top 5 GO terms of up-regulated genes with lowest FDR in two WT models were shown in Fig. 2a,b, immune response, defense response, response to wounding, inflammatory response, positive regulation of immune system process were the common GO terms of biological processes in two models.
Table 4. The number of significant GO of differentially expressed genes.
| Regulated direction | GO | WT-GBM vs WT-Nor | WT-UUO vs WT-Nor | Common in WT | KO-GBM vs KO-Nor | KO-UUO vs KO-Nor | Common in KO |
|---|---|---|---|---|---|---|---|
| UP-regulation | BP | 171 | 209 | 138 | 0 | 22 | 0 |
| MF | 9 | 34 | 9 | 0 | 13 | 0 | |
| CC | 12 | 23 | 10 | 1 | 9 | 1 | |
| Down-regulation | BP | 26 | 105 | 26 | 5 | 14 | 1 |
| MF | 16 | 52 | 16 | 5 | 25 | 4 | |
| CC | 16 | 28 | 16 | 4 | 13 | 1 |
Note:WT-GBM:anti-GBM GN model in wild-type mice; WT-Nor:wild-type normal mice; WT-UUO:UUO model in wild-type mice; KO-GBM:anti-GBM GN model in Smad3 knockout mice; KO-UUO:UUO model in Smad3 knockout mice; KO-N-or:Smad3 knockout normal mice; BP:biological processes; MF:molecular function; CC:cellular component.
Figure 2.
Histogram showing the top five significant GO terms (including biological processes, molecular function and cellular component) of up-regulated genes in WT-GBM (a) or WT-UUO models (b) compared to WT-Nor.WT-UUO:UUO model in wild-type mice; WT-GBM:anti-GBM GN model in wild-type mice; WT-Nor:wild-type normal mice.
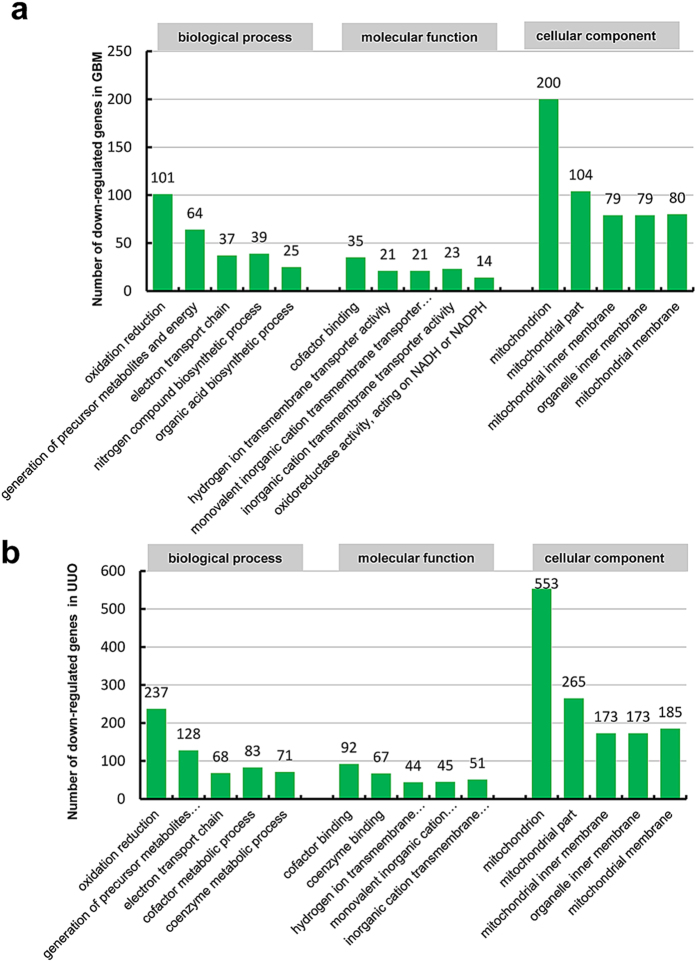
According to the GO analysis results of down-regulated genes in Table 4, 26 significant GO terms for biological process were found in WT-GBM and 105 in WT-UUO compared to WT-Nor respectively and the top 5 GO terms of down-regulated genes in two models were shown in Fig. 3a,b, There were 26 common down-regulated genes enriched GO terms in biological process, oxidation reduction, generation of precursor metabolites and energy, electron transport chain are the most significant GO terms of biological processes in two models.
Figure 3.
Histogram showing the top five significant GO terms (including biological processes, molecular function and cellular component) of down-regulated genes in WT-GBM (a) or WT-UUO models (b) compared to WT-Nor.WT-UUO:UUO model in wild-type mice; WT-GBM:anti-GBM GN model in wild-type mice; WT-Nor:wild-type normal mice.
The number of significant GO terms in Smad3 KO models is much less than WT models. Only a few significant GO terms were found in these two KO models, that may be due to less DEGs were found in KO models. For up-regulated genes, we find only one common significant GO term of cellular component (extracellular region part) in KO-UUO and KO-GBM. For down-regulated genes, one biological process (acute inflammatory response), one cellular component (extracellular space) and 4 molecular function (endopeptidase inhibitor activity, peptidase inhibitor activity, enzyme inhibitor activity, serine-type endopeptidase inhibitor activity) were common significant GO terms in KO-UUO and KO-GBM. Additional details for GO analysis of DEGs in WT and Smad3 knockout models were listed in Supplementary File S2 and S3 (including all the GO terms and gene ID).
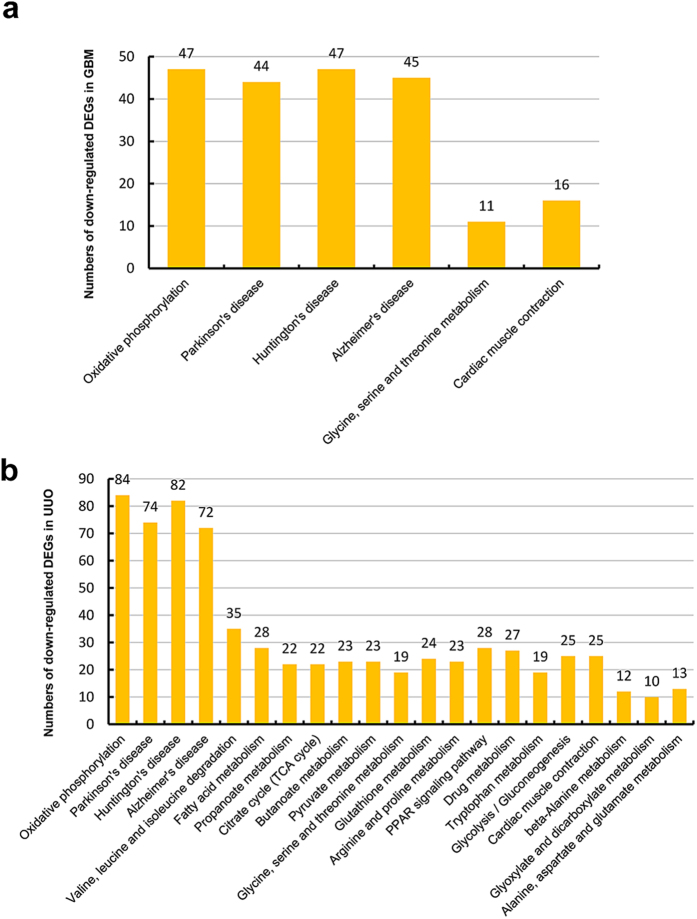
To identify the relationship between DEGs and to uncover the common pathways in renal injury, the DEGs were analyzed by KEGG Pathway. The number of significant pathway was shown in Table 5 and all the KEGG analysis in two models was listed in Supplementary File S4 and S5 (including all the pathway terms and gene ID), In WT-GBM and WT-UUO, the significant KEGG pathways based on DEGs were shown in Figs 4 and 5. For up-regulated genes in two models showed in Fig. 4, the common DEGs in WT-GBM and WT-UUO models were enriched in 13 pathways, such as cell adhesion molecules; cytokine-cytokine receptor interaction; hematopoietic cell lineage; chemokine signaling pathway; complement and coagulation cascades; B cell receptor signaling pathway etc. For down-regulated genes, 6 common pathways in two WT models were: oxidative phosphorylation; Parkinson’s disease; Huntington’s disease; Alzheimer’s disease; glycine, serine and threonine metabolism; cardiac muscle contraction (Fig. 5).
Table 5. The number of significant KEGG pathway of differentially expressed genes.
| Regulated direction | WT-GBM vs WT-Nor | WT-UUO vs WT-Nor | Common in WT | KO-GBM vs KO-Nor | KO-UUO vs KO-Nor | Common in KO |
|---|---|---|---|---|---|---|
| UP-regulation | 17 | 19 | 13 | 1 | 4 | 0 |
| Down-regulation | 6 | 21 | 6 | 0 | 8 | 0 |
Note: WT-GBM:anti-GBM GN model in wild-type mice; WT-Nor:wild-type normal mice; WT-UUO:UUO model in wild-type mice; KO-GBM:anti-GBM GN model in Smad3 knockout mice; KO-UUO:UUO model in Smad3 knockout mice; KO-Nor:Smad3 knockout normal mice.
Figure 4.
Histogram showing all the significant KEGG pathways of up-regulated genes in WT-GBM (a) or WT-UUO models (b) compared to WT-Nor.WT-UUO:UUO model in wild-type mice;WT-GBM:anti-GBM GN model in wild-type mice; WT-Nor:wild-type normal mice.
Figure 5.
Histogram showing all the significant KEGG pathways of down-regulated genes in WT-GBM (a) or WT-UUO models (b) compared to WT-Nor. WT-UUO:UUO model in wild-type mice; WT-GBM:anti-GBM GN model in wild-type mice; WT-Nor:wild-type normal mice.
As same as the results of GO analysis, only a few significant KEGG pathways based on DEGs were found in KO-GBM and KO-UUO models compared to KO-Nor. As listed in Supplementary File S4 and S5, only one pathway (Complement and coagulation cascades) in KO-GBM, 4 pathways (ECM-receptor interaction, Focal adhesion, Cytokine-cytokine receptor interaction, Chemokine signaling pathway) in KO-UUO were found significantly in up-regulated genes related to renal injury. For down-regulated genes, 8 pathways (Oxidative phosphorylation, Parkinson’s disease, Alzheimer’s disease, Huntington’s disease, etc) were found in KO-UUO models, but no significant pathway was found in KO-GBM models. None of the KEGG pathway was found regulated by Smad3, which may be largely due to less DEGs were found in KO models.
Dysregulation of splicing in CKD mouse transcriptome
Alternative splicing (AS) is a mechanism, by which more than one mRNA transcripts are generated from the same mRNA precursor due to variations in the incorporation of coding regions, giving rise to functionally different proteins. We used our RNA-Seq data to detect AS events and genes with alterations in AS between samples genome-widely. Five type of AS models were detected including skipping exon (SE), retention intron (RI), alternative 5′ splice site (A5SS), alternative 3′ splice site (A3SS), mutually exclusive exons (MEX) in our results. All the significant AS genes and events in WT and Smad3 knockout groups were listed in Supplementary File S6. The distribution of AS in disease models compared to normal groups was shown in Fig. 6, the number of significant AS events in WT and Smad3-KO model were listed respectively. SE, and RI are the most prevalent type of AS events in all groups, accounting for about 70% of all detected AS events. It is interesting that the distribution of AS events were changed in the Smad3-KO groups, characterized by the increasing gene number of MXE, compared to WT. In order to identify the functional role and relationship between AS genes, these genes were further analyzed by DAVID GO (Supplementary File S7). and KEGG pathway (Supplementary File S8). Two GO terms of CC (mitochondrial part and mitochondrion) were significant in WT-UUO models, but none in WT-GBM was found. One GO terms of BP (transmembrane transport) and 2 GO terms of MF (coenzyme binding and cofactor binding) were significant in KO-GBM models, while 6 MF (nucleotide binding, adenyl nucleotide binding, etc) in KO-UUO models. No significant KEGG pathway was found in all the groups.
Figure 6. Pie charts showing the number ofthe significant AS genes and events in WT and Smad3 knockout groups.
(a) WT-GBM compared toWT-Nor. (b) WT-UUOcompared to WT-Nor. (c) KO-GBM compared toKO-Nor. (d) KO-UUO compared to KO-Nor. WT-GBM:anti-GBM GN model in wild-type mice; WT-Nor:wild-type normal mice; WT-UUO:UUO model in wild-type mice; KO-GBM:anti-GBM GN model in Smad3 knockout mice; KO-UUO:UUO model in Smad3 knockout mice; KO-Nor:Smad3 knockout normal mice; SE:SkippingExon; MEX: Mutually Exclusive Exons; A5SS: Alternative 5′ splice site; A3SS: Alternative 3′ splice site; RI: Retained Intron
Real-time Quantitative PCR and Western blot Validation
Validation of DEGs in different models was performed by using real-time qPCR. As shown in Table 6, two groups of DEGs were included: DEGs in wild type mice of UUO and anti-GBM GN models; DEGs in Smad3 knockout mice of UUO and GBM models. Two of most up- and down-regulated genes in each group were selected for validation. Although mRNA variation in fold changes was different between qPCR and RNA-seq, the expression patterns were coincident between the results of these two techniques. Some of the DEGs were also selected for validation at protein level by Western blot. As shown in Fig. 7, the protein level of Flna and Trem1 were also significantly up-regulated in WT-UUO compared to WT-Nor, while Pvalb was down-regulated. We also detect some classical markers in UUO models for justification, the extracelluar matrix protein Collagen type III (Col.III) and Collagen type I (Col.I) were upregulated as our previous reports.
Table 6. Validation of RNA-sequencing results by real-time PCR.
| Comparison | Gene | Regulated direction | FC by qRT-PCR | p-value by qRT-PCR | LogFC by RNA-seq | FDR by RNA-seq |
|---|---|---|---|---|---|---|
| WT-GBM vs WT-Nor | Ighg1 | Up | 161.1160 | 0.0640 | 13.2864 | 6.93E–30 |
| Igkc | Up | 2.2981 | 0.0020 | 9.9349 | 4.04E–32 | |
| Cox6a2 | Down | 0.3434 | 0.0456 | −5.6157 | 5.25E–10 | |
| Angptl7 | Down | 0.2022 | 0.0043 | −5.6949 | 4.17E–46 | |
| Sftpc | Down | 0.0235 | 0.0246 | −11.4023 | 5.63E–05 | |
| WT-UUO vs WT-Nor | Il1rn | Up | 105.1993 | 0.0006 | 10.5668 | 7.98E–26 |
| Trem1 | Up | 4.0638 | 0.0185 | 8.7348 | 1.19E–18 | |
| Flna | up | 6.2702 | 0.0070 | 17.2533 | 1.40E–54 | |
| Pvalb | Down | 0.0051 | 0.0000 | −6.9307 | 2.20E–13 | |
| Ugt1a9 | Down | 0.4222 | 0.0001 | −6.2263 | 1.81E–11 | |
| Sftpc | Down | 0.0591 | 0.0270 | −11.3729 | 8.30E–05 | |
| KO-GBM vs KO-Nor | Rpl29 | Up | 1.8738 | 0.0331 | 7.0270 | 9.69E–16 |
| Ceacam2 | Up | 6.3943 | 0.0179 | 5.4004 | 1.91E–13 | |
| Igj | Down | 0.1877 | 0.0023 | −3.5747 | 2.70E–05 | |
| Hrg | Down | 0.3559 | 0.0379 | −8.21951 | 4.07E–04 | |
| KO-UUO vs KO-Nor | Col11a1 | Up | 5.5698 | 0.0071 | 6.9993 | 5.48E–07 |
| Ccl7 | Up | 23.4986 | 0.0055 | 7.228708 | 4.57E–02 | |
| Cyp3a11 | Down | 0.4963 | 0.0165 | −7.8656 | 5.78E–08 | |
| Serpina1a | Down | 0.7025 | 0.0470 | −8.8200 | 2.54E–20 |
Note: WT-GBM:anti-GBM GN model in wild-type mice; WT-Nor:wild-type normal mice; WT-UUO:UUO model in wild-type mice; KO-GBM:anti-GBM GN model in Smad3 knockout mice; KO-UUO:UUO model in Smad3 knockout mice; KO-Nor:Smad3 knockout normal mice.
Figure 7. (a) Western blot analysis show protein expression levels of filamin alpha (Flna), collagen III (Col. III), collagen I (Col. I), Trem1, Pvalb.
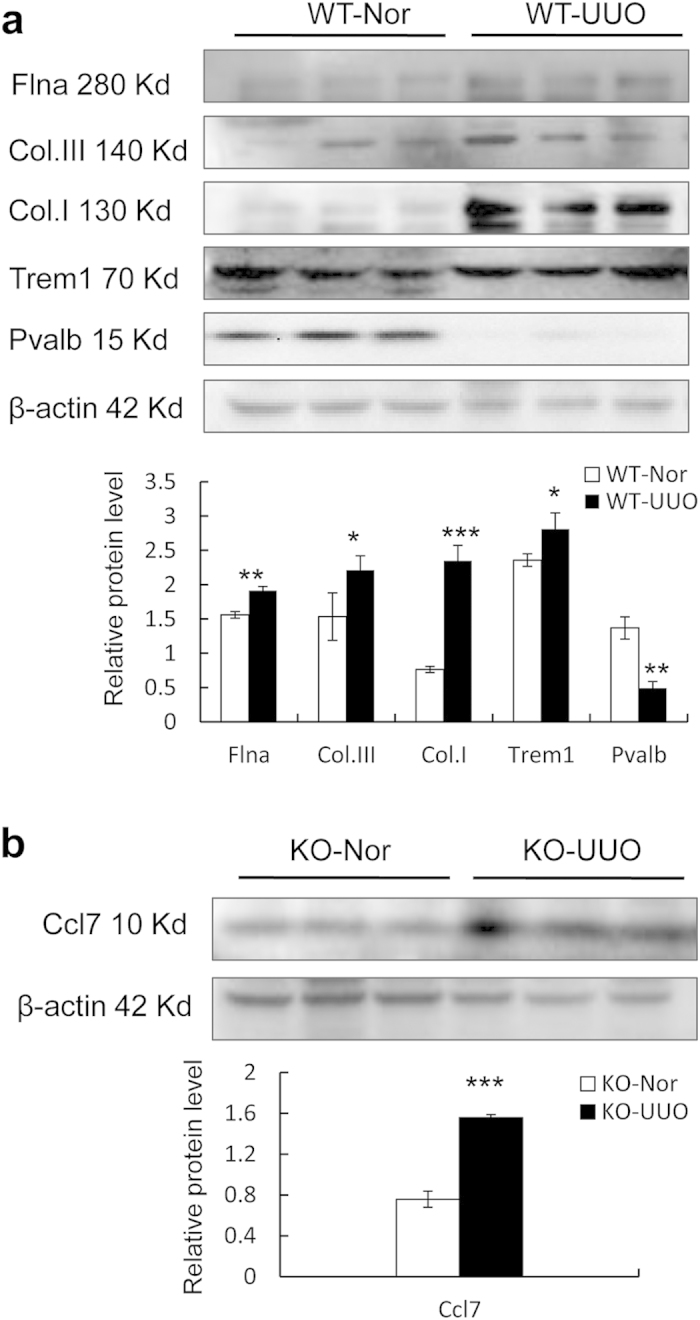
(b) Ccl7 expression by western blot analysis. Each bar represents the mean ± SD for at least 6 animals. *p < 0.05, **p < 0.01, ***p < 0.001 vs WT-Nor or KO-Nor.WT-Nor:wild-type normal mice; WT-UUO:UUO model in wild-type mice; KO-Nor:Smad3 knockout normal mice; KO-UUO:UUO model in Smad3 knockout mice.
Discussion
Most CKD patients regardless the initial causes undergo a final common pathway to interstitial fibrosis and glomerulosclerosis. Immunologically-mediated anti-GBM GN and non-immunologically-associated UUO models are two classic models closely related to patients with CKD and were commonly used to investigate the mechanism of CKD. In anti-GBM GN model, a severe and rapidly progressive crescentic GN was developed, including the presence of immunoglobulin complexes and immune cells in the kidney which may cause irreversible chronic renal injury18. UUO model generates progressive obstructive nephropathy marked by renal interstitial fibrosis and also hemodynamic and metabolic changes19. Although these two typical CKD models are quite different of phenotypic symptoms, they also have some common mechanisms, such as the infiltration of immune cells and excess deposition of the extracellular matrix in the diseased kidney.
We have previously found that TGF-β/Smad3 is essential in the process of CKD, especially in renal fibrosis5. Therefore, addressing the pathogenesis of the renal injury in terms of Smad3-targeted molecules will help to develop a specific therapy for CKD. In our previous study, we found that transcripts including microRNAs and long non-coding RNAs are participated in the gene regulation during the CKD development11,12,20,21,22,23. Recently, a research group using deep sequencing to obtain the gene expression profiles of 14 microdissected renal tubule segments from the proximal tubule through the inner medullary collecting duct of normal rat kidneys24. Different from their study, the present study, functional annotation-based analysis of DEGs (differentially expressed genes) in CKD revealed two dominant themes: 1) common dysregulation of genes associated with renal injury in two CKD models; and 2) genes associated with Smad3 in renal injury. RNA-Seq was used for screening the common DEGs between two CKD models, and bioinformatics was used to analyze the expression variations, gene function and alternative splicing of DEGs, in order to further identify Smad3-related genes and biological pathways that regulate the pathogenesis of chronic renal injury.
In our study, to identify the signaling molecules and their putative functions in the process of CKD, the mouse kidney tissues of anti-GBM GN at day 10 and UUO at day 5 were selected for RNA-sequencing, which are relative earlier time points in the period of chronic kidney injury, so as to understand the biological changes happened during the pathogenesis and progression of CKD. In the kidney tissue of anti-GBM GN at day 10, a large number of genes related to immune response were significantly increased, while transcripts of surfactant associated protein, channel protein and metabolic enzymes were decreased. Among them, 155 up-regulated genes could be successfully clustered into GO terms of immune response, 101 down-regulated genes ranked into oxidation reduction. In comparison of WT-GBM and WT-Nor, immunoglobulin genes are mostly up-regulated, such as Ighg1, Ighg2c, Igkv, which are closely related to the antibody production of B cell, and have the potential to promote cell growth and proliferation25,26.
It was previously reported that a number of genes related to immune response and fibrosis were increased on day 7 in anti-GBM GN model, and transcripts of growth factors, channel protein and metabolic enzymes were decreased on around day 11 to 1627. It is consistent with our results, supporting the reliability of RNA-sequencing. For further understanding of the biological functions and interactions of these genes, pathway-based analysis was conducted based on the KEGG Pathway database. Results of KEGG pathway were similar with the GO analysis, the significant enriched pathways for up-regulated genes in WT-GBM were cell adhesion molecules, cytokine receptor interaction, ECM-receptor interaction, while down-regulated DEGs were enriched in the pathways of oxidative phosphorylation, Parkinson’s disease, Huntington’s disease, Alzheimer’s disease.
In the UUO model, the mostly-induced genes in WT-UUO were coding for cytoskeleton proteins, such as FlnA (filamin, alpha) and moesin, which are important to the remodeling of cytoskeleton and modulation of cell shape and migration28. In addition, a number of inflammatory and immune response genes are also up-regulated in UUO. This speculation consistent with the GO analysis, up-regulated genes at the UUO day 5 could be successfully clustered into GO terms of immune response, response to wounding, inflammatory response, positive regulation of immune system process, defense response, and cell adhesion. For down-regulated genes, they were enriched in GO terms of oxidation reduction, generation of precursor metabolites and energy, electron transport chain, cofactor metabolic process, coenzyme metabolic process, fatty acid metabolic process, transmembrane transport. According to the pathway analysis of WT-UUO, by comparing to WT-Nor, genes associated with cytokine-cytokine receptor interaction, chemokine signaling pathway, focal adhesion, ECM-receptor interaction were significantly upreguated, whereas, those related to the pathway of oxidative phosphorylation Parkinson’s disease, Huntington’s disease, Alzheimer’s disease were downregulated.
As expected, alterations of DEGs found in these two models exhibited common mechanisms in the development of renal injury. In both anti-GBM GN and UUO models, the functions of commonly up-regulated genes were associated with cell adhesion, inflammation and immune response at the early stage of CKD. KEGG pathway network analysis showed that genes belong to cell adhesion molecules, cytokine-cytokine receptor interaction, hematopoietic cell lineage, chemokine signaling pathway, complement and coagulation cascades, B cell receptor signaling pathway were commonly up-regulated. In contrast, the commonly down-regulated genes are related to material or electron transportation and metabolism, their KEGG pathway were oxidative phosphorylation, Parkinson’s disease, Huntington’s disease, Alzheimer’s disease, glycine, serine and threonine metabolism, cardiac muscle contraction. Although, Parkinson’s disease (PD), Huntington’s disease (HD), Alzheimer’s disease (AD) are neurodegenerative disorders cause serious movement or cognitive disability, the proteins in these pathways also involved in multiple cell functions and DEGs enriched in these pathways are potentially participated in renal disease. In PD pathway, mutations or altered expression of proteins in this pathway contributes to the damage and subsequent loss of dopaminergic neurons through common mechanisms including proteasome dysfunction, mitochondrial impairment, and oxidative stress29,30. The mutation of proteins in HD pathway also may lead to abnormal endocytosis, augmented p53 mediates mitochondrial dysfunction, as well as Ca2+ signaling disorder31,32,33. AD associated proteins have various pathological effects on cell and organelle function through activation of cell surface death receptors, disrupting mitochondria function, and triggering calcium dysfunction34,35.
These results imply that progressive renal fibrosis may begin with inflammatory response after injury, whereas, remodeling of the cytoskeleton in renal cells was the initial stage of fibrosis. At this stage, the inflammatory and immune system is activated, accompanied with the cytoskeleton reconstruction and the extracellular matrix deposition. At the same time, the normal cell functions, such as metabolism, oxidative phosphorylation, material transportation and some signal transduction were dramatically suppressed.
To further investigate the pathogenic role of Smad3 at the early stage of chronic renal injury, two CKD models were performed on Smad3 knockout mice. Our results revealed that, compared to WT mice, deletion of Smad3 largely altered the gene expression profiles of these two diseases. It is consistent with our previous observations that fibrogenesis was prevented in Smad3 KO UUO mice and a loss of Treg and enhanced Th-2 and Th-17-mediated renal injury in anti-GBM GN mice lacking Smad336. In anti-GBM GN and UUO models, the number of DEGs related to Smad3 was 55 and 39 respectively. Most of the Smad3-related DEGs in anti-GBM model have putative functions in immune response, since 46 immunoglobulin genes were found in the total of 55 DEGs. In addition, Smad3-related DEGs in UUO model were belonged to immune response, but also exhibited distinct biological functions that were enriched in organic anion transporter and metabolism. Only 9 common DEGs were related to Smad3 in these two models, including 6 immunoglobulin genes (Ighg1, Ighg2c, Igkv12-41, Ighv14-3, Ighv5-6 and Ighg2b) and 3 metabolic genes (Ugt2b37, Slc22a19, and Mfsd2a). Ugt2b37 (UDP-glucuronosyltransferases) is a metabolic enzymes involved in the metabolism of key endogenous compounds including bilirubin, bile acids, fatty acids, steroid hormones, thyroid hormones and fat soluble vitamins37. Slc22a19 belongs to organic anion transporter family, also named organic anion transporter 5 (Oat5), is a renal organic anion/dicarboxylates exchanger under physiological conditions38,39. Mfsd2a (major facilitator super family domain containing 2a), usually considers as the major transporter for docosahexaenoic acid (DHA) and a key regulator of ‘blood-brain barrier'40,41. but also found functions in modulating cell cycle and matrix attachment by inducing a G1 block, and then suppress tumor cells by impairing adhesion and migration42. These common DEGs related to Smad3 are all have potential roles in renal injury.
Alternative splicing (AS) is a regulated mechanism during gene expression, during this process, particular exons of a gene may be included within or excluded from the final processed mRNA produced from that gene. Since Smad3 can alter the gene expression profile, we also use our RNA-Seq data to detect AS events and genes with alterations in AS between CKD and normal groups. The results shown that the distribution of AS events were changed in the Smad3-KO groups, characterized by the increasing gene number of MXE, indicated that deficiency of Smad3 may have impact on gene splicing.
Regrettably, no significance common pathway and very rare significant GO term were found in the CKD models lacking of Smad3. This may largely be due to limited DEGs that were found in the Smad3 KO models. By the RNA-seq based transcriptome analysis, we found the central role for Smad3 in the immune response of B cell, which can regulate the transcription of immunoglobulin. The results could explain the phenomenon that blocks renal fibrosis by targeting Smad3 may cause autoimmune diseases5,43.
To summarize, our transcriptome analysis of two mice CKD models suggested that increased expression of inflammation and immune response related genes, and cytoskeletons may be as biological markers at the early stage of chronic renal injury. Furthermore, we demonstrated that Smad3 is a critical transcription factor in the immune response, which can regulate the transcription of immunoglobulins. It implies the important role of Smad3 in the immune-mediated glomerulonephritis. Besides, there are some limitations to this study since the UUO and anti-GBM GN models may not be able to completely recapitulate the human CKD phenotype. The sample size used to perform RNA-sequencing is too small, and the usage of whole kidney tissue for RNA-sequencing reduces sensitivity for detecting cell type specific changes of transcripts, particularly in those with low abundance. Nevertheless, our findings from this study identify the downstream signaling molecules of Smad3 may have potential functions in renal injury; it may provide precise and potential therapeutic targets for CKD.
Methods
Mouse Model of UUO and Anti-GBM GN
The CKD models of UUO and anti-GBMGN were induced in both sexes of C57BL/6J wild-type (WT) and Smad3 knockout (KO) mice at 8 to 10 weeks of age, as previously described3,44. Briefly, for the UUO model, the left ureter from Smad3 WT or KO mice (n = 8 per group, both sexes, 8weeks of age, 22 to 25 g body weight) was ligated and the UUO kidney was harvested at day 5 after surgery for further analysis. For the anti-GBMGN model, Smad3 WT or KO mice were preimmunized with sheep IgG in Freund’s complete adjuvant(Sigma Chemical Co, St. Louis, MO), followed 5 days later by i.v. administration of sheep anti-mouse GBM immunoglobulin at a dose of 60 mg/g of body weight (termed day 0). Diseased mice were euthanized at day 10 for further analysis. Groups of age- and weight-matched normal Smad3 WT or KO mice were used as normal controls. The experimental procedures for both models were performed following the approved protocol by the Animal Experimentation Ethics Committee at The Chinese University of Hong Kong and the experimental methods were carried out in accordance with the approved guidelines.
Sample collection and RNA isolation
Kidney tissues of Smad3 KO/WT were collected from normal, UUO model at day 5, and anti-GBM GN model at day 10 for RNA sequencing (n = 2 in each group). Total RNA from kidneys was isolated by using Trizol reagent (Life Technologies, Carlsbad, CA, USA) in accordance with the manufacturer’s instructions, and then followed by an additional DNase I digestion to remove genomic DNA contamination. RNA quality and purity was checked by using the ND-1000 Nanodrop. And then total RNA samples were used to remove highly abundant ribosomal RNAs before sequencing.
RNA sequencing and reads quantifying, mapping, assembling
The sequencing was done by Illumina Genome Analyzer (serial number Hiseq2000) as previously reported11,12. Both library building and sequencing were performed by Beijing Genomics Institute (BGI), Shenzhen, China. Low-quality reads, over 50% of the sequences reads (including adapters) has quality score ≤10 and N > 5%, were discarded. Ribosome RNA sequences were filtered from the raw fragments. Those left clean reads were mapped to genome (UCSC genome browser, Version: July.2007, mm9) by using Tophat[1.3.0.Linux_x86_64 (bowtie-0.12.7)] and then assembled with Cufflinks (1.3.0.Linux_x86_64)45. The sequencing data obtained were available at the Gene Expression Omnibus Web site (http://www.ncbi.nlm.nih.gov/geo/) under accession GSE55808. All the analysis results of gene expression are also provided as a webpage (http://songyanglab.sysu.edu.cn/Transcriptome_analysis_in_renal_injury).
Analysis of differentially expressed genes
The FPKM (expected number of fragments per kilobase of transcript sequence per millions base pairs sequenced) were used to Quantifying the mRNA expression, which measure of read density reflects the molar concentration of a transcript in the starting sample by normalizing for RNA length and for the total read number in the measurement46. The genes with false discovery rate (FDR = adjusted p-values) <0.05 and fold change (FC)>2 were considered to indicate significant up-regulation; FDR<0.05 and FC<0.5 were considered as significant down-regulation; FC between 0.5 and 2 were considered as no significance. The DEGs were analyzed by software edgeR (Release 3.1)47. All statistical analyses were performed in R. For the identification of Smad3-related genes, DEG has different expression direction in WT and Smad3 KO mice will be considered as Smad3-related, such as genes up-regulated in WT but suppressed in Smad3 KO, or down-regulated in WT but up-regulated in Smad3 KO. Pearson’s correlation coefficient based on transcript abundance(FPKM)were used to measure the gene expression similarity between samples.
Gene Ontology and Pathway Analysis
Significantly up and down-regulated genes were analyzed for enrichment of gene ontology (GO) terms (biological processes, molecular function and cellular component) and KEGG Pathway by using DAVID bioinformatics resources48. For GO term and Pathway analysis, Probabilities were evaluated by Bonferroni correction and FDR values less than 0.05 were considered as significant.
Detection and quantification of alternative splicing variants
The Multivariate Analysis of Transcript Splicing (MATS,Release 2.0.0) was used to detect differently alternative splicing events across samples49. The Ensembl annotation was supplied to STAR to ensure the sensitivity of aligning spliced fragments to known junctions50. Generally, the alternative splicing events were classified into seven different types according to the structures of exons. These seven types include skipping exon (SE), retention intron (RI), alternative 5′ splice site (A5SS), alternative 3′ splice site (A3SS), mutually exclusive exons (MEX), alternative first exons (AFE), and alternative last exons (ALE) as described by Wang et al.51,52. Because of the unperfected algorithms for AFE and ALE, only the remaining five alternative splicing models listed above were analyzed and presented in this study. We detected the differently alternative splicing events by using the cuff-off criteria as FDR <0.1 to filter the result of MATS. The genes has differential expression of splicing variants were used for gene Ontology and pathway analysis as described above.
Quantitative real-time PCR (qPCR)
Gene expression was quantified by real-time PCR with SYBR green Permix Kit(Life Technologies, Carlsbad, CA). Quantitative real-time PCR was carried out on ABI7900 system using the following program: 50 °C for 2 min 95 °C for 10 min; 40 cycles of 95 °C for 15 sec, 58 °C for 15 sec, and 72 °C for 30 sec, and then dissociation steps. Primers for all the tested DEGs and β-actin, an endogenous control for normalization, are listed in Supplementary Table 1. The relative levels of target genes were calculated by using 2−ΔΔCt method53. Statistical analyses were performed using one-way ANOVA followed by Newman-Keuls multiple comparison test from GraphPad Prism 5.0 (Graph Pad Software, San Diego, CA).
Western blot
Protein from kidney tissues was extracted by using the protein lysis buffer(Cell Signaling,Danvers, MA, USA) and then quantified by the Bradford assay (Bio-Rad, USA) . An equal amount of protein was separated on SDS-polyacrylamide gels and transferred onto nitrocellulose membranes (Amersham Biosciences, USA). After blocking non-specific binding with 5% skim milk in TBST for 1 h at room temperature, membranes were then incubated overnight at 4 °C with the primary antibody against Flna (Cell Signaling, Danvers, MA, USA), Col. III and Col. I (Southernbiotech, USA), Trem1 and Pvalb (boster,Wuhan, China), β-actin(Santa Cruz, USA), followed by horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature. The ratio for the protein examined was normalized against β-actin and was expressed as the mean ± SD.
Additional Information
How to cite this article: Zhou, Q. et al. Identification of Genes Associated with Smad3-dependent Renal Injury by RNA-seq-based Transcriptome Analysis. Sci. Rep. 5, 17901; doi: 10.1038/srep17901 (2015).
Supplementary Material
Acknowledgments
This work was supported by grants from TheNational Basic Research Program of China (2012CB517705-2012CB517706), National Natural Science Foundation (81130012 and 81470942), and Foundation for Young teachers of Sun Yat-sen University, China (15ykpy13),The Research Grant Council of Hong Kong (GRF 468711, CUHK3/CRF/12R,TBS T12-402/13N),and the Focused Investment Scheme A program from the Chinese University of Hong Kong.
Footnotes
Author Contributions Q.Z. and X.R.H. performed the experimentand wrote the manuscript; Y.X analyzed the data; P.M.KT. revised manuscript; H.Y.L. and X.Y. designed, supervised and revised the article.
References
- Chen W. et al. Prevalence and risk factors associated with chronic kidney disease in an adult population from southern China. Nephrol Dial Transplant 24, 1205–1212 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. et al. Prevalence of chronic kidney disease in China: a cross-sectional survey. Lancet 379, 815–822 (2012). [DOI] [PubMed] [Google Scholar]
- Huang X. R., Chung A. C., Wang X. J., Lai K. N. & Lan H. Y. Mice overexpressing latent TGF-beta1 are protected against renal fibrosis in obstructive kidney disease. Am J Physiol Renal Physiol 295, F118–127 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. R. et al. Smad3 mediates cardiac inflammation and fibrosis in angiotensin II-induced hypertensive cardiac remodeling. Hypertension 55, 1165–1171 (2010). [DOI] [PubMed] [Google Scholar]
- Lan H. Y. & Chung A. C. TGF-beta/Smad signaling in kidney disease. Semin Nephrol 32, 236–243 (2012). [DOI] [PubMed] [Google Scholar]
- Meng X. M. et al. Smad2 protects against TGF-beta/Smad3-mediated renal fibrosis. J Am Soc Nephrol 21, 1477–1487 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan H. Y. Diverse Roles of TGF-beta/Smads in Renal Fibrosis and Inflammation. Int J Biol Sci 7, 1056–1067 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L. & Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5, 621–628 (2008). [DOI] [PubMed] [Google Scholar]
- Ozsolak F. & Milos P. M. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet 12, 87–98 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Gerstein M. & Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10, 57–63 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q. et al. Identification of Novel Long Noncoding RNAs Associated with TGF-beta/Smad3-Mediated Renal Inflammation and Fibrosis by RNA Sequencing. Am J Pathol 184, 409–417 (2013). [DOI] [PubMed] [Google Scholar]
- Zhou Q., Huang X. R., Yu J., Yu X. & Lan H. Y. Long Noncoding RNA Arid2-IR Is a Novel Therapeutic Target for Renal Inflammation. Mol Ther 23, 1034–1043 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerri R. L. et al. Effects of lactation and pregnancy on gene expression of endometrium of Holstein cows at day 17 of the estrous cycle or pregnancy. J Dairy Sci 95, 5657–5675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warr R. G. et al. Low molecular weight human pulmonary surfactant protein (SP5): isolation, characterization, and cDNA and amino acid sequences. Proc Natl Acad Sci USA 84, 7915–7919 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tredano M. et al. Mutation of SFTPC in infantile pulmonary alveolar proteinosis with or without fibrosing lung disease. Am J Med Genet A 126A, 18–26 (2004). [DOI] [PubMed] [Google Scholar]
- Glasser S. W. et al. Persistence of LPS-induced lung inflammation in surfactant protein-C-deficient mice. Am J Respir Cell MolBiol 49, 845–854 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J. W. et al. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron 21, 1315–1325 (1998). [DOI] [PubMed] [Google Scholar]
- Kurts C., Panzer U., Anders H. J. & Rees A. J. The immune system and kidney disease: basic concepts and clinical implications. Nat Rev Immunol 13, 738–753 (2013). [DOI] [PubMed] [Google Scholar]
- Chevalier R. L., Forbes M. S. & Thornhill B. A. Ureteral obstruction as a model of renal interstitial fibrosis and obstructive nephropathy. Kidney Int 75, 1145–1152 (2009). [DOI] [PubMed] [Google Scholar]
- Chung A. C., Huang X. R., Meng X. & Lan H. Y. miR-192 mediates TGF-beta/Smad3-driven renal fibrosis. J Am Soc Nephrol 21, 1317–1325 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W. et al. TGF-beta/Smad3 signaling promotes renal fibrosis by inhibiting miR-29. J Am Soc Nephrol 22, 1462–1474 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Chung A. C., Chen H. Y., Meng X. M. & Lan H. Y. Smad3-mediated upregulation of miR-21 promotes renal fibrosis. J Am Soc Nephrol 22, 1668–1681 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q. et al. TGF-{beta}-induced MiR-491-5p expression promotes Par-3 degradation in rat proximal tubular epithelial cells. J BiolChem 285, 40019–40027 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. W., Chou C. L. & Knepper M. A. Deep Sequencing in Microdissected Renal Tubules Identifies Nephron Segment-Specific Transcriptomes. J Am SocNephrol 26, doi: 10.1681/ASN.2014111067 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan B., Zheng S., Liu C. & Xu Y. Suppression of IGHG1 gene expression by siRNA leads to growth inhibition and apoptosis induction in human prostate cancer cell. Mol Biol Rep 40, 27–33(2013). [DOI] [PubMed] [Google Scholar]
- Qiu Y. et al. Immunoglobulin G expression and its colocalization with complement proteins in papillary thyroid cancer. Mod Pathol 25, 36–45 (2012). [DOI] [PubMed] [Google Scholar]
- Kim J. H. et al. Gene expression profiling of anti-GBM glomerulonephritis model: the role of NF-kappaB in immune complex kidney disease. Kidney Int 66, 1826–1837 (2004). [DOI] [PubMed] [Google Scholar]
- Grimbert P. et al. The Filamin-A is a partner of Tc-mip, a new adapter protein involved in c-maf-dependent Th2 signaling pathway. Mol Immunol 40, 1257–1261 (2004). [DOI] [PubMed] [Google Scholar]
- Gandhi S. & Wood N. W. Molecular pathogenesis of Parkinson’s disease. Hum Mol Genet 14 Spec No. 2, 2749–2755 (2005). [PubMed] [Google Scholar]
- Vila M. & Przedborski S. Targeting programmed cell death in neurodegenerative diseases. Nat Rev Neurosci 4, 365–375 (2003). [DOI] [PubMed] [Google Scholar]
- Bae B. I. et al. p53 mediates cellular dysfunction and behavioral abnormalities in Huntington’s disease. Neuron 47, 29–41 (2005). [DOI] [PubMed] [Google Scholar]
- Bossy-Wetzel E., Petrilli A. & Knott A. B. Mutant huntingtin and mitochondrial dysfunction. Trends Neurosci 31, 609–616 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T. S. et al. Disturbed Ca2+ signaling and apoptosis of medium spiny neurons in Huntington’s disease. Proc Natl Acad Sci USA 102, 2602–2607 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezprozvanny I. & Mattson M. P. Neuronal calcium mishandling and the pathogenesis of Alzheimer’s disease. Trends Neurosci 31, 454–463 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson M. P. Pathways towards and away from Alzheimer’s disease. Nature 430, 631–639 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inazaki K. et al. Smad3 deficiency attenuates renal fibrosis, inflammation, and apoptosis after unilateral ureteral obstruction. Kidney Int 66, 597–604 (2004). [DOI] [PubMed] [Google Scholar]
- Rowland A., Miners J. O. & Mackenzie P. I. The UDP-glucuronosyltransferases: their role in drug metabolism and detoxification. Int J Biochem Cell Biol 45, 1121–1132 (2013). [DOI] [PubMed] [Google Scholar]
- Anzai N. et al. Functional characterization of rat organic anion transporter 5 (Slc22a19) at the apical membrane of renal proximal tubules. J Pharmacol Exp Ther 315, 534–544 (2005). [DOI] [PubMed] [Google Scholar]
- Youngblood G. L. & Sweet D. H. Identification and functional assessment of the novel murine organic anion transporter Oat5 (Slc22a19) expressed in kidney. Am J Physiol Renal Physiol 287, F236–244 (2004). [DOI] [PubMed] [Google Scholar]
- Ben-Zvi A. et al. Mfsd2a is critical for the formation and function of the blood-brain barrier. Nature 509, 507–511 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L. N. et al. Mfsd2a is a transporter for the essential omega-3 fatty acid docosahexaenoic acid. Nature 509, 503–506 (2014). [DOI] [PubMed] [Google Scholar]
- Spinola M. et al. MFSD2A is a novel lung tumor suppressor gene modulating cell cycle and matrix attachment. Mol Cancer 9, 62 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung A. C. et al. Advanced glycationend-products induce tubular CTGF via TGF-beta-independent Smad3 signaling. J Am Soc Nephrol 21, 249–260 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. R., Chung A. C., Zhou L., Wang X. J. & Lan H. Y. Latent TGF-beta1 protects against crescentic glomerulonephritis. J Am Soc Nephrol 19, 233–242 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L. & Salzberg S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J. & Smyth G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G. Jr. et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4, P3 (2003). [PubMed] [Google Scholar]
- Shen S. et al. MATS: a Bayesian framework for flexible detection of differential alternative splicing from RNA-Seq data. Nucleic Acids Res 40, e61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin A. J., Clark F. & Smith C. W. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol 6, 386–398 (2005). [DOI] [PubMed] [Google Scholar]
- Wang Z. & Burge C. B. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14, 802–813 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.