Abstract
One paradigm of oysters as the hyper-accumulators of many toxic metals is the inter-individual variation of metals, but the molecular mechanisms remain very elusive. A comprehensive analysis of the transcriptome of Crassostrea angulata was conducted to reveal the relationship between gene expression and differential Cu body burden in oysters. Gene ontology analysis for the differentially expressed genes showed that the neurotransmitter transporter might affect the oyster behavior, which in turn led to difference in Cu accumulation. The ATP-binding cassette transporters superfamily played an important role in the maintenance of cell Cu homeostasis, vitellogenin and apolipophorin transport, and elimination of excess Cu. Gill and mantle Cu concentrations were significantly reduced after silencing the GABA transporter 2 (GAT2) gene, but increased after the injection of GABA receptor antagonists, suggesting that the function of GABA transporter 2 gene was strongly related to Cu accumulation. These findings demonstrated that GABA transporter can control the action of transmitter GABA in the nervous system, thereby affecting the Cu accumulation in the gills and mantles.
Copper (Cu) is a micronutrient essential for all living organisms, but can also be toxic when present in excess by generating hydroxyl radicals1. Due to human activities, an increasing amount of industrial effluents is released into rivers and coastal waters, and high levels of Cu have been reported in some aquatic ecosystems such as the Restronguet Creek in southwest Cornwall, UK, and the Jiulong River estuary, Fujian, China2,3,4,5. Oysters are one of the most important cultivated mollusk species around the world. Based on the Fishery and Aquaculture Statistics 2012 (FAO Yearbook), oyster production in China amounted to 3,949,000 t, accounting for 83% of the total world production. Due to its sedentary and filter-feeding behavior, high bioaccumulation potential, and easy collection, oysters have been extensively employed in metal contamination monitoring6. Oysters are considered as the hyper-accumulators of various metals, especially for Cu, zinc (Zn), and cadmium (Cd). The classic example is the ‘green-sick’ oyster (an indicative of Cu accumulation) reported in several estuaries around the world7,8. Another example is the blue colored oysters with a Cu concentration as high as 14,000 μg/g5,6.
One paradigm in marine biomonitoring is that metal concentrations in bivalves can vary greatly among different species and different individuals. Biokinetic model has been used to explore the inter- and intra-species differences in metal concentrations in oysters and other aquatic animals9,10,11. Luoma and Rainbow compared the biokinetics of green mussels (a relatively moderate Zn accumulator) and barnacles (a Zn hyper-accumulator) and suggested a negative relationship between the Zn body burden and the efflux rate10. Pan and Wang reported a 5-fold inter-individual variation in Cu body burden in the oyster Saccostrea cucullata with similar age, shell length and dry weight11. Such individual difference was not related to gender, but could be explained by the variation in efflux, with a lower efflux resulting in a very high Cu body burden. The molecular mechanism underlying such inter- and intra-species difference in Cu bioaccumulation is however unknown. Presently, there is an increasing number of transcriptome and proteome analyses in bivalves, which indicated that transcripts or networks of proteins responded to Cu and other metals12,13,14,15,16. These studies focused on identifying the mechanisms responsible for organisms to adapt to or alleviate the damage due to metal (Cu, Zn, and Cd) stress and accumulation in marine organisms. The transcriptome study can provide information of the potential functional genes and gene regulatory mechanisms underlying the inter- and intra-species differences in metal accumulation in marine bivalves.
The aim of this study was therefore to gain insight into the molecular mechanisms of the intra-species differences in Cu body concentrations in oyster Crassostrea angulata and to identify the key genes related to Cu accumulation. We performed a comprehensive analysis of the transcriptome of oysters using gills and mantle tissues. Digital gene expression (DGE) technology was applied to analyze the relationships between gene expression and differential Cu body concentrations. To analyze the differentially expressed genes (DEGs), AgriGO and KEGG functional enrichment analysis was conducted. The function clustering entries related to the behavioral change, Cu iron transport and Cu stored components were identified. According to the functional categories of these DEGs, our analysis focused on the genes involved in lipid transporter activity, Cu ion binding, neurotransmitter activity, and ATPase activity. We focused on GABA transporter 2 (GAT2), which is related to neurotransmitter activity and therefore may play an important role in Cu accumulation in oysters. In addition, we evaluated the changes in Cu body concentration in the oyster resulting from GAT2 gene interference. GABA and GABA receptor agonists or antagonists were injected to validate the neurotransmitter-relevant candidate genes that may play important roles in the intra-species differences in Cu accumulation. To our knowledge, this is the first study that integrates transcriptome and intra-species differences in Cu body concentration in marine bivalves to provide further insights into the mechanism of Cu accumulation.
Results
Cu concentrations in oyster tissues used for DGE analysis
Individuals with the largest differences in Cu body concentration were used for differential gene expression analysis. Sixty individuals collected from an oyster farming site (as G1 and M1) were dissected and measured for Cu concentrations in the gills and mantles. Six oysters with the highest Cu concentrations in their gills and mantles were selected. Their gills were pooled and flagged as G1-H group, and mantles were pooled as M1-H. Similarly, six oysters with the lowest Cu concentrations in their gills and mantles were selected and their gills were pooled and flagged as G1-L group, and mantles were pooled as M1-L. The 60 individuals exposed to Cu (as G2 and M2) were also subjected to the same expression analysis. G2-L and M2-L groups were pooled from 6 oysters’ gills and mantles that showed the lowest Cu body concentrations. G2-H and M2-H groups were pooled from highest Cu body concentration individuals. Figure 1a shows the Cu concentrations in the gills and the mantle used for DGE analysis. Figure 1b,c show their total weights and shell lengths. No visible phenotypic differences were observed, whereas large variations in body Cu concentrations (2.7- to 4.3-fold) were detected. These groups (G1-H vs. G1-L; M1-H vs. M1-L; G2-H vs. G2-L; and M2-H vs. M2-L) showed significantly different Cu concentrations (p <0.01, pair-wise t test). Oysters subjected to a 30-day Cu exposure accumulated higher Cu concentration than the baseline Cu body concentration (e.g., Cu in G2 an M2 groups were higher than that in G1 and M1 groups, Fig. 1a). The results of the DGE analysis therefore showed that individuals within the same population had large variations in Cu concentrations.
Figure 1.
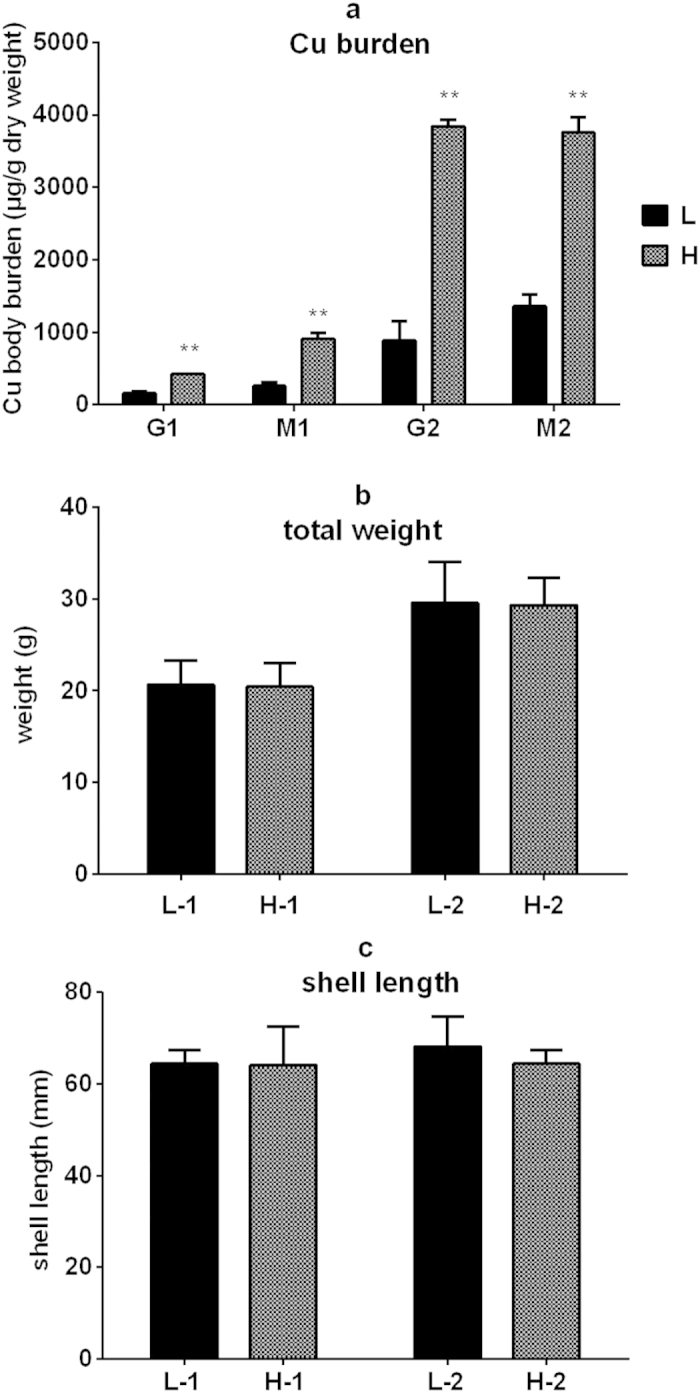
The tissue Cu concentration (a), total body weight (b) and shell length (c) of oysters selected for DGE. G1 and M1: gills and mantle tissues without experimental Cu exposure; G2 and M2: gills and mantle tissues after Cu exposure; L and H represent low and high Cu tissue concentration, respectively. L-1 and H-1: oysters without experimental Cu exposure; L-2 and H-2 oysters after Cu exposure. Data are mean ± s.d. (six replicates); **p <0.01 (pair-wise t test).
Sequencing and identification of differentially expressed genes (DEGs)
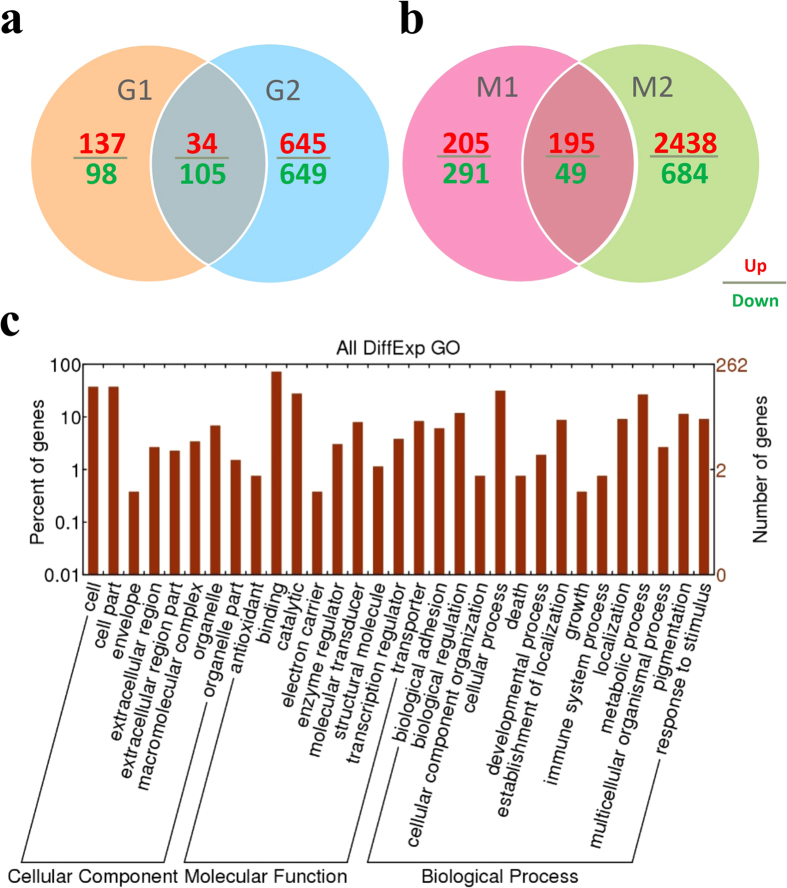
The Illumina Genome Analyzer was used to perform high-throughput Tag-seq analysis of the gills and mantle libraries. The major characteristics of eight libraries are summarized in the Supplementary Table S1. A total of 5.8–6.3 million reads of 50 bp in length was generated for each sample, and the number of mapped reads was within the range of 4.26–4.76 million. The high-quality reads from individual libraries were mapped to the oyster genome; more than 20,960 mapped genes per library were determined simultaneously (Supplementary Table S1 and S2). To identify the transcriptome difference between the oysters that displayed inter-individual variations (up to 4.3-fold) in Cu body burden, the gene expression levels in the G1-H group was compared to those of the G1-L group. Similarly, the M1-H, G2-H, and M2-H groups were compared with the M1-L, G2-L, and M2-L groups, respectively. Furthermore, the DEGs in the gills of groups G1-H and G1-L intersecting with that between G2-H and G2-L (G1-L vs. G1-H ∩ G2-L vs. G2-H) were evaluated. The DEGs of the mantle in the similar treatments were assessed in relation to M1-L vs. M1-H ∩ M2-L vs. M2-H. A total of 139 DEGs between the high and low-Cu concentrations in the gills were identified (Fig. 2a). Among these genes, 34 (24.5%) were up-regulated and 105 (75.5%) were down-regulated in the gills of high-Cu concentration. In addition, 244 DEGs were detected in the mantle tissues (Fig. 2b), among which 195 (79.9%) were upregulated and 49 (20.1%) were down-regulated in the high-Cu mantles. All these genes (Supplementary Table S3) were grouped into 13 functional categories, namely, biological adhesion, biological regulation, development process, cellular process, metabolic process, immune system process, localization, response to stimulus, pigmentation, growth, multicellular organismal process, death, and cellular component organization (Fig. 2c).
Figure 2. Significant differentially expressed genes (DEGs).
(a) DEGs of oyster gills with great variation in the body burden of Cu by Venn diagram: G1-L vs.G1-H ∩ G2-L vs. G2-H. (b) The DEGs in high Cu accumulated mantles compared to low Cu accumulated groups: M1-L vs. M1-H ∩ M2-L vs. M2-H. A FDR of ≤0.001 and an absolute value of the log2 ratio >2 were used as the threshold to determine significant differences in gene expression. (c) Gene ontology analysis of all DEGs in the high-Cu and low-Cu oysters.
Genes related to the individual differences of Cu enrichment
AgriGO and KEGG functional enrichment analysis was used to determine the gene function of these DEGs (Supplementary Tables S4 and S5). The functional categories of these DEGs in the gills were related to lipid transport and neurotransmitter transport (selected functions in Table 1). There were 2 genes that encoded for apolipophorin or similar proteins enriched in lipid transport (Table 1), and were upregulated in the gills of high-Cu concentration. One gene encoding for the excitatory amino acid transporter 1 (EAAT 1) was specifically down-regulated in the gills of high-Cu concentration.
Table 1. Gene ontology/pathway terms for selected genes regulated ≥two-fold with differentially expressed Cu body concentrations in the gills/mantles (low concentration vs. high concentration).
| Gill | Expression pattern | TypeA | G1-L Vs G1-H |
G2-L Vs G2-H |
Description | ||
|---|---|---|---|---|---|---|---|
| FCB | FDRC | FCB | FDRC | ||||
| Lipid transport | P | ||||||
| OYG_10014469_10040256 | Upregulated | 1.602509 | 1.13E-12 | 2.328729 | 5.9E-143 | Apolipoprotein | |
| OYG_10018340_10030734 | Upregulated | 1.236194 | 1.52E-10 | 1.943109 | 6.53E-78 | hypothetical protein CGI_10018340 [Crassostreagigas] | |
| Neurotransmitter transport | P | ||||||
| OYG_10017841_10008053 | Downregulated | −2.12145 | 1.17E-23 | −5.3088 | 0 | Excitatory amino acid transporter 1 | |
| Mantle | Expression pattern | TypeA | M1-L Vs M1-H | M1-L Vs M1-H | Description | ||
| FCB | FDRC | FCB | FDRC | ||||
| Lipid transport | P | ||||||
| OYG_10015914_10025506 | Upregulated | 1.067678 | 0.000617 | 2.090331 | 6.7E-14 | Vitellogenin/VTG | |
| OYG_10014469_10040256 | Upregulated | 0.895977 | 1.1E-07 | 4.766276 | 2.1E-221 | apolipoprotein | |
| Neurotransmitter transport | P | ||||||
| OYG_10000052_10033075 | Upregulated | 2.372763 | 1.16E-09 | 1.69651 | 1.48E-11 | Proline transporter | |
| OYG_10026098_10002797 | Downregulated | −2.31727 | 2.01E-15 | −1.46292 | 8.04E-05 | Glycine transporter 2 | |
| OYG_10028335_10036542 | Upregulated | 1.373422 | 5.73E-31 | 1.053064 | 3.2E-21 | GABA transporter 2/GAT2 | |
| Vesicle | C | ||||||
| OYG_10027950_10024008 | Upregulated | 1.075756 | 7.24E-13 | 2.202442 | 4.56E-49 | ATP-binding cassette sub-family A member 3/ABCA3 | |
| Copper ion binding | F | ||||||
| OYG_10020885_10039411 | Upregulated | 1.499383 | 3.5E-32 | 1.701766 | 1.9E-09 | L-ascorbate oxidase | |
| OYG_10017371_10010587 | Upregulated | 1.107054 | 0.002036 | 1.43978 | 1.89E-11 | Laccase-2 | |
| ATPase activity | F | ||||||
| OYG_10027950_10024008 | Upregulated | 1.075756 | 7.24E-13 | 2.202442 | 4.56E-49 | ATP-binding cassette sub-family A member 3/ABCA3 | |
| OYG_10000886_10006267 | Upregulated | 2.263479 | 1E-06 | 2.052857 | 1.65E-06 | Multidrug resistance protein 1/ABCB1 | |
| OYG_10002457_10026801 | Upregulated | 1.792962 | 4.94E-10 | 1.89126 | 1.07E-09 | Multidrug resistance protein 3/ABCB3 | |
| Response to chemical stimulus | P | ||||||
| OYG_10000886_10006267 | Upregulated | 2.263479 | 1E-06 | 2.052857 | 1.65E-06 | Multidrug resistance protein 1/ABCB1 | |
| OYG_10002457_10026801 | Upregulated | 1.792962 | 4.94E-10 | 1.89126 | 1.07E-09 | Multidrug resistance protein 3/ABCB3 | |
| OYG_10022618_10024381 | Upregulated | 1.365183 | 1.95E-22 | 2.727761 | 1.19E-08 | Porphobilinogen deaminase | |
| OYG_10004131_10027936 | Upregulated | 1.406247 | 5.84E-09 | 1.229159 | 4.59E-06 | UDP-glucose 6-dehydrogenase | |
AType: P, biological process F, molecular function C, Cellular Component.
BFC: log2 fold change.
CFDR: false discovery rate.
The functional categories of the high-Cu enrichment mantles genes were related to lipid transport, neurotransmitter transport, vesicle, response to chemical stimulus, ATPase activity, and copper ion binding (Table 1). In the lipid transport group, 2 genes encoding vitellogenin (VTG) or apolipophorin (apoLp) proteins were upregulated. In the neurotransmitter transport group, genes encoding the proline transporter and GABA transporter 2 were also identified. The gene encoding the protein, namely the ATP-binding cassette sub-family A member 3 (ABCA3), had been implicated in vesicle transport. Two genes encoding copper ion-binding proteins were specifically upregulated in the high-Cu enrichment mantles. There were four genes that were enriched in response to the chemical stimulus group, namely those encoding for the multidrug resistance protein 1 (ABCB1), multidrug resistance protein 3 (ABCB3), porphobilinogen deaminase, and UDP-glucose 6-dehydrogenase. Genes encoded in ATPase activity function included ABCB1, ABCB3 and ABCA3.
Validation of DGE data by quantitative RT-PCR
To validate the quantitative data of the DGE libraries, we quantified the expression levels of 12 selected genes from the gills or mantle DGE libraries by qRT-PCR. The expressions of these genes measured by qRT-PCR agreed well with the Tag-seq analysis pattern (Supplementary Fig. S1).
Inhibition of Cu enrichment by interference of GAT genes
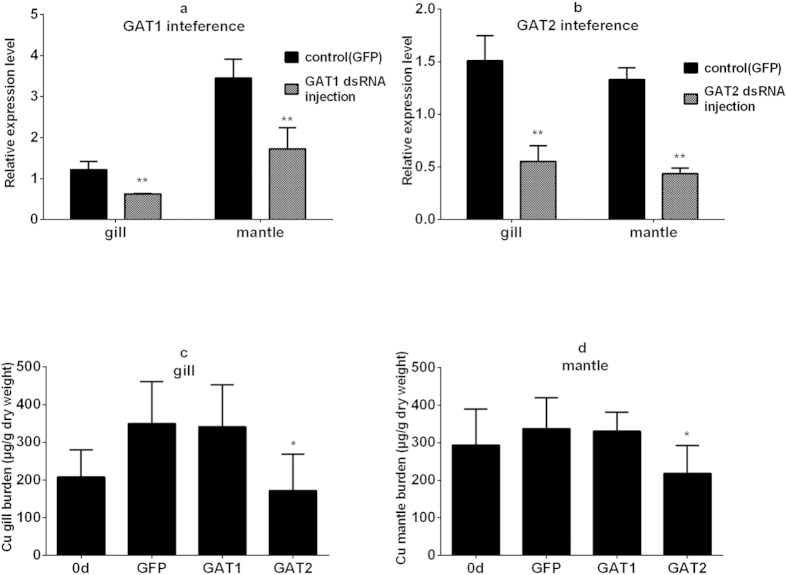
The function clustering entries related to the behavior, Cu iron transport and Cu stored components were the potential candidates in explaining the Cu individual variation in oysters. The neurotransmitter transport may regulate the neurotransmitters and eventually the oysters’ behavior. In the mantle, expression of GAT2 was significantly increased in high-Cu accumulating individuals compared to the low-Cu accumulating oysters. The GABA transporters (GAT1, GAT2, GAT3, and BGT1) all potentially controlled the action of transmitter GABA in the nervous system17. To gain more insight into the roles of GAT2 in mediating Cu accumulation, we further conducted RNA interference experiments. Using qRT-PCR, we monitored the GAT1 and GAT2 transcript abundance 48 h after the first dsRNA injection. In this protocol, the receptor transcript abundance in the dsGAT1 and dsGAT2 injected oyster gills and mantles decreased significantly by 33–51% as compared to the control oysters (Fig. 3a,b). The gills and mantles were also collected and their Cu concentrations were analyzed after a series of four injections to further determine the roles of the GAT1/GAT2 gene in Cu accumulation. Compared to the tissue Cu concentration before injection (0 d column), the control batch showed an increase in the gills and mantles (Fig. 3c,d), indicating that the oysters were in the process of Cu accumulation. Surprisingly, GAT2-silenced oysters showed a significant reduction in Cu concentration in both the gills and mantles (Fig. 3c,d), thus the silencing of the GABA transport-related GAT2 gene may reduce the Cu accumulation.
Figure 3. GAT1/GAT2 RNA interference.
The efficiency of GAT1 (a) and GAT2 (b) RNA interference in gill and mantle following RNAi treatment. Each column represents mean ± s.d. (n=3); **p <0.01 (pair-wise t test).Cu concentration in the gills (c) and mantles (d) following GAT1/GAT2 RNA interference. Each column represents mean ± s.d. (n=8); *p <0.05 (pair-wise t test).
Functional analysis of GABA in Cu accumulation by injection of GABA and GABA receptor agonists and antagonists
GAT2 is a promising candidate for peripheral GABA uptake and is capable of rapidly absorbing the synaptic cleft and extracellular fluid of GABA, resulting in the termination of the GABA synaptic transmission process18. To further confirm the effect of GAT2 on gill and mantle Cu accumulation through the regulation of GABA, we conducted another Cu accumulation study by GABA as well as GABA receptor agonist and antagonist injection. Compared to the tissue Cu concentrations before injection (0 d column), the control group displayed an increase in Cu accumulation in the gills and mantles (Fig. 4a,b). GABA, GABA receptor agonists (muscimol, baclofen), and a single GABAA /GABAB receptor antagonist (bicuculline or phaclofen) injection had no significant effect on Cu accumulation in the gills. However, gill Cu concentrations increased significantly after GABAA and GABAB receptor antagonists (bicuculline + phaclofen) treatments (Fig. 4a). These drugs imparted a similar effect on mantles, except for GABA. The Cu concentrations of the mantles were significantly reduced after GABA injection (Fig. 4b).
Figure 4. GABA, GABA receptor agonists and antagonists effect on Cu accumulation in the oysters.
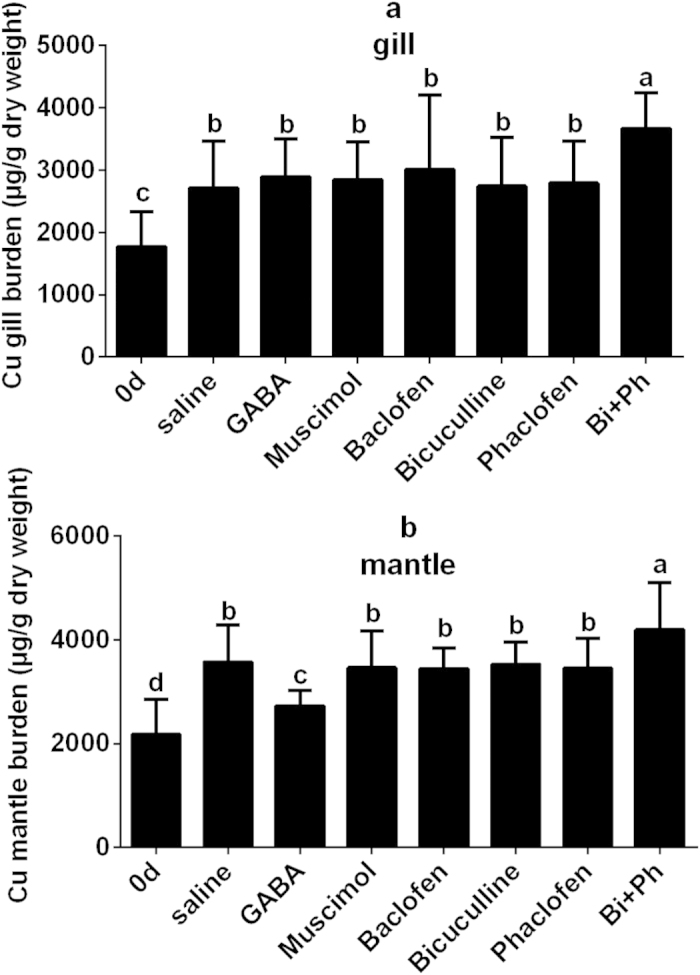
(a) Gill Cu burden after GABA, GABA receptor agonists and antagonists injection. (b) Mantle Cu burden after GABA, GABA receptor agonists and antagonists injection. Each column represents mean ± s.d. (n=12); p <0.05 (one-way ANOVA followed by LSD and Duncan post-hoc comparisons). Different letters represent significant difference between two treatments.
Discussion
Phenotypic differences resulting from gene expression variations have been observed in various species19. In the present study, two individual variation groups were constructed, and the intersection of the two sets of DGE results generated from these groups were used to validate the results. Earlier studies suggested that biodynamics played important roles in Cu accumulation, inter-species and intra-species differences in Cu body burdens in oysters5,10,11,20. The present study showed that genes enriched in neurotransmitter transport functional category were differentially expressed in the mantles and gills (Table 1). Specifically, the EAAT1 gene was down-regulated in the high-Cu gills. EAAT1 is responsible in terminating the excitatory signal by removing (or absorbing) glutamate, which is the principal excitatory neurotransmitter secreted by the neuronal synaptic cleft into neuroglia and neurons21. In contrast, the GAT2 gene was up-regulated in the high-Cu mantle. GAT2 is capable of rapidly absorbing the synaptic cleft and extracellular fluid of GABA, resulting in the termination of the GABA synaptic transmission process 18,22. GABA is the major inhibitory neurotransmitter of both vertebrates and invertebrates, and may play a signaling role in peripheral organs. However, GABA has been shown to elicit both inhibitory and excitatory actions in the central neurons in mollusca23. To gain further insight into the roles of GAT2 in mediating Cu accumulation, we interfered both the GAT1 gene, which was not differentially expressed, and the GAT2 by dsRNA injection to detect the changes in oyster Cu accumulation. The oysters harboring an interfered GAT2 gene showed a significant reduction in mantle and gill Cu concentrations as compared to the control group (Fig. 3). This finding suggested that the differences in GAT2 transcriptional expression could lead to changes in Cu concentrations. It is noteworthy that GAT2-silenced oysters even showed a reduction in Cu concentration compared to the initial tissue concentration before injection (0 d column). Thus, Cu efflux was presumably higher than the Cu uptake after GAT2 gene interference. We then further extended the Cu accumulation study by GABA, as well as GABA receptor agonist and antagonist injection (Fig. 4). Injection of a GABAA receptor antagonist combined with a GABAB receptor antagonist significantly facilitated the mantle and gill Cu bioaccumulation, whereas a single receptor antagonist did not produce any significant effect. These data suggested that the oysters improved their Cu bioaccumulation when GABA was suppressed. In addition, the GABAA and GABAB receptor were both involved in this regulatory process. GABA significantly reduced the mantle Cu concentration, whereas GABA showed no significant effect on gill and receptor agonists showed no significant effect on gill and mantle Cu accumulation. Previous studies indicated that the longest duration of behavioral change caused by GABA and receptor agonists injection in mollusca was 200 min24. The Cu accumulation ability in the gills was weaker than that in the mantles (Fig. 4, 0 d and saline groups). Unlike mantles, the action time of GABA may not be enough as compared to the whole Cu exposure time in gills. Therefore, there was no significant Cu burden change after GABA injection in gills. Similarly, the dose and injection frequency of the receptor agonist were not sufficient to generate a significant or persistent change in oyster behavior. It thus appeared that the differences in GAT2 transcriptional expression regulated the GABA, resulting in changes in Cu accumulation in oysters. Such effect was probably caused by the behavioral changes observed with GABA administration.
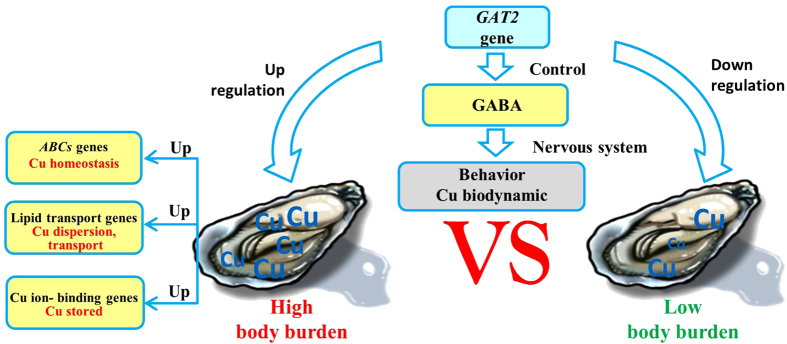
GABA administration can cause a variety of behavioral changes that may be manifested by the combined effects on the neural circuitry for feeding activity, locomotion, respiration, male mating, and defensive behavior in some species of mollusks. For example, GABA induced the movements of buccal and radular muscles in Lymnaea stagnalis, resulting in an excitatory effect on the feeding pattern and motor activity24,25. In oysters Crassostrea virginica, GABA worked at both the cerebral ganglion and visceral ganglia as an inhibitory ganglionic neurotransmitter to inhibit HT neurons that innervated the gills and speed up the beating of the lateral gill cilia26. The feeding activity, adductor muscle, and mantle movement of oysters may also be regulated by GABA. The filtration activity is closely related to the dissolved uptake of Cu11, and the adductor/mantle muscles can control the extent of contact between soft tissues and seawater using shell. Therefore, when the GAT2 gene was upregulated in some individuals, inhibitory and excitatory actions in the central neurons were elicited by GABA suppressed. We therefore inferred that neurotransmitters, especially GABA, which was regulated by GAT2, can change the oyster behavior, which may have an impact on the biodynamics of Cu (Fig. 5).
Figure 5. The sketch map of important functional genes related to the inter-individual variations of Cu in oysters.
Excess Cu can induce redox reactions by generating reactive oxygen species (ROS)27. Most animals utilize the protective ROS-scavenging enzymes such as superoxide dismutase (SOD) and catalase (CAT) to deal with the elevated levels of ROS. The expression of proteins or genes involved in antioxidative stress (SOD, CAT, extracellular superoxide dismutase: ECSOD, cytochrome c oxidase, glutathione transferase: GST) can be stimulated by acute or chronic Cu exposure6,28. Similarly, genes encoding for heat shock proteins (HSPs), which play a vital role in the transport, folding, and assembly of proteins, are induced by various casual agents such as metals12,29,30. According to our data, all these genes (SOD, CAT, ECSOD, GST, and HSPs) were detected in the eight DGE libraries, but were either not or differentially expressed in very small quantity (Supplementary Table S6). Therefore, difference of oxidative stress among oysters with 4-fold difference of Cu body burdens was not obvious. It was possible that they might use transporter proteins and detoxification enzymes to remove the excessive Cu and maintain Cu homeostasis within cells31.
Previous studies suggested that the ATP-binding cassette transporter (ABC) superfamily was important in the maintenance of cellular metal homeostasis. ABC was composed of eight subfamilies and interacted with a wide range of chemicals including metals by pumping them across the cell membrane. These cellular activities were also called multidrug resistance32,33,34. ABCB1 is a drug efflux transporter of the ABCB subfamily functioning in cellular efflux and intracellular resistance to metals. Recently, ABCB1 was reported as a metal (Cu, Zn, and Cd) efflux transporter in the copepod Tigriopus japonicus, Asiatic clam Corbicula fluminea, and earthworm Eisenia fetida35,36,37. We also found that high-Cu mantles harbored these upregulated genes involved in vesicular transport, response to chemical stimulus, and ATPase activity (Table 1). The genes within these 3 groups encode for ABCB1, ABCB3, and ABCA3 all belonged to the ABC superfamily, suggesting that it played an important role in oysters by maintaining the cellular Cu homeostasis.
Metallothioneins (MTs) with thiol groups (–SH) of cysteine residues were important in metal detoxification by binding with metals in aquatic invertebrates, including bivalves38,39,40. However, some studies have shown that Cu imparted no effect on MTs in oysters, and MTLP played a minor role in binding Cu in oysters. Cellular debris was the main subcellular fraction for Cu binding11,41. In the present study, the genes encoding MTs were detected, but were not differentially expressed in the gills or mantles (Supplementary Table S6). Among the high-Cu mantle upregulated genes, two genes in the Cu ion-binding gene functional category were enriched (Table 1) and encoded as the protein L-ascorbate oxidase and laccase-2. Interestingly, laccases contained four Cu-binding sites and catalyzed the oxidation of a wide variety of phenolic substrates found in various plants, fungi, and microorganisms, but they were generally not found in aquatic invertebrates42,43,44. The upregulated expression indicated that laccase-2 may potentially bind with Cu for either detoxification or export. Consequently, MTs may not be the major detoxification in the oysters, and excessive Cu may be stored in other components such as laccase.
Our study also detected upregulated genes enriched in lipid transport functional category (Table 1). These encoded VTG and apoLp proteins have been used as biomarkers for metal exposure45,46. Cu exposure of Paracyclopina nana resulted in the time-dependent upregulation of VTG transcription47. VTG is a large precursor protein of egg yolk vitellin present in oviparous vertebrates and invertebrates48. In several insect species, VTGs are primarily synthesized in the fat bodies in sex-, tissue-, and stage-specific manners. After synthesis, they are secreted into the hemolymph and then sequestered by competent oocytes via receptor-mediated endocytosis49,50. VTG functions as a carrier protein for inorganic phosphates, lipids, carbohydrates, and metals, in addition to its role as a yolk precursor molecule51. Ghosh and Thomas reported that VTGs in estradiol-treated male red drum (Sciaenopsocellatus) contained calcium, magnesium, zinc, iron, and Cu, and most of the Cd had been incorporated in the ovaries, suggesting that this was an important route for metal accumulation in ovarian tissues52. In our study, VTGs and apoLp genes were upregulated in the gills and mantle of high-Cu oysters, and might play an important role in the transport or dispersion of excess Cu.
Our results showed that the neurotransmitter transport genes especially GAT2 gene can regulate the inhibitory and excitatory actions elicited by GABA. The resulting behavioral changes may have an effect on Cu absorption and biodynamics, and led to individual variations in Cu accumulation. In the high-Cu oysters, ABCB1, ABCB3, and ABCA3 maintained the cellular Cu homeostasis to protect from Cu toxicity; VTGs might combine Cu with other molecules and transport or eliminate the excess Cu, while excessive Cu may be stored in components such as laccase-2 (Fig. 5). These potential genes should be further examined to confirm their functions not only in transcriptional level but also in translational level.
Materials and Methods
Oysters and Cu measurement
Fujian oysters, C. angulata, of similar body weight (15.7–24.5 g) and shell length (5.5–7.5 cm) were collected from an oyster farming site in Gangkou village (23° 42’ N, 117° 19’ E), Zhaoan Bay, Fujian Province, China. All the experimental individuals came from the same group of nine months old. The culture site was considered to be relatively free from anthropogenic Cu influences. The measured Cu concentrations in the seawater from the Zhaoan Bay were 0.39–1.37 μg/L, and in the sediments were 11.9–34.2 μg/g53. Sixty individuals were directly dissected and measured for Cu concentrations in the gills and mantles by inductively coupled plasma-mass spectrometry (ICP-MS), using methods described in previous study5,6. Specifically, six oysters with the highest Cu concentrations in their gills or mantles were pooled and flagged as G1-H group and M1-H, respectively. Six oysters with the lowest Cu concentrations in their gills or mantles were pooled and flagged as G1-L group and M1-L, respectively (Fig. 1).
To avoid the effects of environmental factors on the DGE results, oysters were also exposed to Cu (CuCl2). Before the exposure, oysters were kept in tanks containing natural seawater and acclimated for 7 days. The seawater was filtered through 0.45-μm filter (salinity: 28–30%; 26–28 °C). During the exposure, the seawater was renewed daily, and the oysters were fed commercial clean algal powders of Chlorella at a rate of approximately 2% of their soft tissue dry weight per day. A stock solution of 10 mg/L Cu (CuCl2) was prepared with Milli-Q-filtered water. Three groups of 60 individuals per tank (80 L) were exposed to 30 μg Cu/L for 30 days. At the end of exposure, 60 individuals (20 individuals per tank) were randomly selected. Similarly, the G2-L and M2-L groups were selected from 6 oysters that had the lowest Cu body concentrations, whereas G2-H and M2-H groups had the highest Cu body concentrations (Fig. 1).
DGE library construction and sequencing
Approximately 100 mg of tissue from each of the six samples per group was collected for individual RNA extraction. Total RNA was extracted separately using Trizol reagent (Gibco BRL, USA), quantified using NanoDrop ND-2000 (Thermo Scientific, Les Ulis, France), and assessed by agarose gel electrophoresis.
Eight tag libraries from the non-exposed oysters (G1-H, G1-L, M1-H, and M1-L) and exposed oysters (G2-H, G2-L, M2-H, and M2-L) were prepared for DGE, which was performed in parallel using the Digital Gene Expression Tag Profile Kit (Illumina). The samples were prepared following the protocol described in the previous study16. The mRNA was enriched by using the oligo (dT) magnetic beads, further fragmented into short fragments, and mixed with the fragmentation buffer. Then, the first strand of cDNA was synthesized by using random hexamer primers based on these fragments. The second strand was synthesized using buffer, dNTPs, RNase H, and DNA polymerase I. After the double-stranded cDNA was purified with magnetic beads, end reparation and 3’-end single nucleotide A (adenine) addition was then performed. Finally, sequencing adaptors were ligated to the strands. Also, the fragments were enriched by PCR amplification. During the QC step, Agilent 2100 Bioanalyzer and ABI StepOnePlus Real-Time PCR System were used to quantify the sample library. Finally, the suitable library products were sequenced using IlluminaHiSeqTM 2000. Raw sequence data are available in the NCBI’s Gene Expression Omnibus (GEO) database under the accession number GSE65653.
Aligning DGE tags to the reference transcriptome
To map the DGE tags, we filtered all the sequenced raw data with Perl scripts to remove low-quality tags (i.e., tags with unknown nucleotide “N”), empty tags (i.e., no tag sequence between the adaptors), and tags with only one copy number (which might have resulted from sequencing errors). C. angulata was considered a subspecies of Crassostrea gigas54. We also collected 80889 mRNA sequences of C. angulata from the NCBI and used C. gigas genome as database to assess the sequence identity between the two species. There were 68376 mRNA (84.5%) with more than 70% identity mapping to the database; 66522 mRNA had more than 90% identity. Such comparison suggested that the two species have a high sequence identity. For tag annotation, the clean tags containing CATG and 21-bp tag sequences were mapped to the C. gigas genome sequences (GEO: GSE31012) with tophat2, allowing not more than one nucleotide mismatch55. The clean tags were designated as unambiguous clean tags. For gene expression analysis, the number of unambiguous clean tags for each gene was calculated and normalized to the number of transcripts per million clean tags.
DGE library characteristics and functional annotation of differentially expressed genes
Statistical analysis of the frequency of each tag in different cDNA libraries with Cufflinks was performed to compare gene expression patterns in different samples56. False discovery rate (FDR) was used to determine the threshold of the P value in multiple tests and analyses. A FDR of ≤0.001 and an absolute value of the log2 ratio >2 were used as the threshold to determine significant differences in gene expression. We obtained the DEGs in high Cu accumulated groups (G1-H, M1-H, G2-H, and M2-H) compared to low Cu accumulated groups (G1-L,M1-L, G2-L, and M2-L). The DEGs were used for Gene Ontology (GO) and KEGG Orthology (KO) enrichment analyses using GOstats. These GO terms were assigned to query protein sequences, which generated a broad overview of groups of genes catalogued in the three ontology vocabularies (cellular component, biological process, and molecular function). The data presented herein represented a GO analysis at level 2, illustrating general functional categories. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were assigned to the assembled sequences using GOstats to link the online KEGG Annotation Server (http://www.genome.jp/kegg/kaas/), which provides KO assignments and pathway mapping.
Quantitative real-time PCR (qRT-PCR)
To validate the quantitative data of DGE libraries, we quantified the expression levels of 20 selected genes from the gills or mantle DGE libraries by qRT-PCR. The total RNA samples used for the qRT-PCR assay were similar to that used in the DGE experiments. Gene-specific primers were designed according to the reference Unigene sequences using the Primer Premier 5.0 (Supplementary Table S7). The first-strand cDNA was synthesized using a PrimeScripttm RT reagent Kit (TaKaRa, Dalian, China). qRT-PCR was performed according to the manufacturer’s specifications (ThermoDyNAmo Flash SYBR Green qPCR Kit, Thermo Scientific, US) on the ABI 7500 Fast Real-Time PCR System (Applied Biosystems®, US). The elongation factor 1α gene (EF1α, GenBank ID: BQ426516) was used as a normalizer (Supplementary Table S7), and the relative expression levels of genes were presented in terms of 2-ΔΔCT.
dsRNA preparation and injection into oysters
According to the Pacific oyster sequence (GenBank Accession No.: EKC23164.1; EKC37107.1), GAT1 and GAT2 fragments of C. angulata cDNA were PCR amplified using total RNA extracted from the mantle as template. PCR fragments were sub-cloned into pMD19T-GAT1 and pMD19T-GAT2 vectors (Takara) and sequenced. pMD19T-EGFP was constructed using a 289-bp EGFP gene (EU716633.1) fragment from a pEGFP-N1 plasmid. All dsRNA synthesis assays were performed as previously described57.
Oysters were first acclimated in tanks containing natural seawater (salinity = 27–30%; 22–25 °C) for 7 days, afterwards 6 oysters were randomly chosen and dissected, and their gills and mantles were measured for Cu concentrations. Then the oysters were divided into 3 experiment groups (green fluorescent protein: GFP control group, GAT1, and GAT2), and were exposed to Cu at 10 μg/L for 12 days (15 individuals in 50-L tank). The shells of these oysters were drilled near the adductor muscle using a microbit (1–2 mm) to facilitate dsRNA injection. Before the experiment, the injection site was verified by injecting a similar volume of 1% methylene blue solution to determine the distribution of injected dye in the tissue58. After 30 min, the injected dye was distributed across the soft tissues. All the drugs were applied by means of single injection into the adductor muscle every three days, for a total of four times. The injection volume was 50 μL of saline solution with 100 μg of dsRNA. Approximately 48 h after the first injection, 3 individuals that were randomly selected from each group were dissected and used for qRT-PCR analysis to determine the effect of the injected dsRNA. The qRT-PCR protocol was the same as previously described and the EF1α, β-actin were used as normalizers. At the end of the experiment, 6 individuals were dissected and the Cu concentrations in the gills and mantles were measured. The primers used in the experiment are presented in Supplementary Table S8.
GABA, GABA receptor agonist, and antagonist injection
Gamma-amino butyric acid (GABA), GABAA, and GABAB receptor agonists (muscimol, baclofen), and their antagonists (bicuculline, phaclofen) were purchased from Sigma Chemical Co. (St. Louis, MO, USA). They were prepared just before use and dissolved in saline solution.
Oyster acclimation, injection protocol, and daily administration were the same as previously described. Seven experimental groups (each with 20 individuals in 80-L waters) were exposed to Cu for 12 days. All the drugs were injected into the adductor muscle of the oysters every three days, for a total of four times. The drugs and doses used in the injection for each experimental group were as follows: GABA (15 μg/g body weight), muscimol (10 μg/g body weight), baclofen (15 μg/g body weight), bicuculline (8 μg/g body weight), phaclofen (2 μg/g body weight), bicuculline (8 μg/g body weight) + phaclofen (2 μg/g body weight), and saline. These drugs and doses were used based on the preliminary experiments and earlier study24. At the end of the experiment, 12 individuals were dissected and the Cu concentrations in the gills and mantles were also analyzed.
Additional Information
How to cite this article: Shi, B. et al. Transcriptome analysis of the key role of GAT2 gene in the hyper-accumulation of copper in the oyster Crassostrea angulata. Sci. Rep. 5, 17751; doi: 10.1038/srep17751 (2015).
Supplementary Material
Acknowledgments
This study was funded by the National Basic Research Program of China (2010 CB 126403), the Special Fund for Ocean Scientific Research in the Public Interest (201305016), a Key Project from the National Natural Science Foundation (21237004) and the Earmarked Fund for Modern Agro-industry Technology Research System (CARS-48).
Footnotes
References
- Yuan M., Li X. H., Xiao J. H. & Wang S. P. Molecular and functional analyses of COPT/Ctr-type copper transporter-like gene family in rice. Bmc Plant Biol. 11, 69 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. D. et al. Heavy metal distribution in sediment profiles of the Pearl River estuary, South China. Appl. Geochem. 15, 567–581 (2000). [Google Scholar]
- Pan K. & Wang W.-X. Trace metal contamination in estuarine and coastal environments in China. Sci. Total. Environ. 421, 3–16 (2012). [DOI] [PubMed] [Google Scholar]
- Bryan G. W. & Langston W. J. Bioavailability, accumulation and effects of heavy metals in sediments with special reference to United Kingdom estuaries: a review. Environ. Pollut. 76, 89–131 (1992). [DOI] [PubMed] [Google Scholar]
- Wang W.-X. et al. Copper and zinc contamination in oysters: Subcellular distribution and detoxification. Environ. Toxicol. Chem. 30, 1767–1774 (2011). [DOI] [PubMed] [Google Scholar]
- Luo L., Ke C., Guo X., Shi B. & Huang M. Metal accumulation and differentially expressed proteins in gill of oyster (Crassostrea hongkongensis) exposed to long-term heavy metal-contaminated estuary. Fish & shellfish Immunol. 38, 318–329 (2014). [DOI] [PubMed] [Google Scholar]
- Han B. C. & Hung T. C. Green Oysters Caused by Copper Pollution on the Taiwan Coast. Environ. Pollut. 65, 347–362 (1990). [DOI] [PubMed] [Google Scholar]
- Sarasquete C. et al. Histochemical distribution and accumulation of trace metals in the heart of green and normal Crassostrea angulata specimens from different southwest Spanish coasts. Eur. J. Histochem. 41, 139–148 (1997). [PubMed] [Google Scholar]
- Cain D. J. & Luoma S. N. Influence of Seasonal Growth, Age, and Environmental Exposure on Cu and Ag in a Bivalve Indicator, Macoma-Balthica, in San-Francisco Bay. Mar Ecol Prog Ser 60, 45–55 (1990). [Google Scholar]
- Luoma S. N. & Rainbow P. S. Why is metal bioaccumulation so variable? Biodynamics as a unifying concept. Environ. Sci. Technol. 39 (7), 1921–1931 (2005). [DOI] [PubMed] [Google Scholar]
- Pan K. & Wang W.-X. Biodynamics to explain the difference of copper body concentrations in five marine bivalve species. Environ. Sci. Technol. 43, 2137–2143 (2009). [DOI] [PubMed] [Google Scholar]
- Zhang G. et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54 (2012). [DOI] [PubMed] [Google Scholar]
- Bigot A., Minguez L., Giamberini L. & Rodius F. Early defense responses in the freshwater bivalve Corbicula fluminea exposed to copper and cadmium: transcriptional and histochemical studies. Environ. Toxicol. 26, 623–632 (2011). [DOI] [PubMed] [Google Scholar]
- Ivanina A. V., Sokolov E. P. & Sokolova I. M. Effects of cadmium on anaerobic energy metabolism and mRNA expression during air exposure and recovery of an intertidal mollusk Crassostrea virginica. Aquat. Toxicol. 99, 330–342 (2010). [DOI] [PubMed] [Google Scholar]
- Neave M. J. et al. The transcriptome and proteome are altered in marine polychaetes (Annelida) exposed to elevated metal levels. J. Proteomics 75, 2721–2735 (2012). [DOI] [PubMed] [Google Scholar]
- Meng X. L. et al. De novo characterization of Japanese scallop Mizuhopecten yessoensis transcriptome and analysis of its gene expression following cadmium exposure. Plos One 8, e64485, 10.1371/journal.pone.0064485 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y. et al. Deletion of the gamma-aminobutyric acid transporter 2 (GAT2 and SLC6A13) gene in mice leads to changes in liver and brain taurine contents. J. Biol. Chem. 287, 35733–35746 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti F., Minelli A. & Melone M. GABA transporters in the mammalian cerebral cortex: localization, development and pathological implications. Brain Res. Rev. 45, 196–212 (2004). [DOI] [PubMed] [Google Scholar]
- Huang L. et al. Comparative transcriptome sequencing of tolerant rice introgression line and its parents in response to drought stress. BMC genomics 15, 1026 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.-X. Comparison of metal uptake rate and absorption efficiency in marine bivalves. Environ. Toxicol. Chem. 20, 1367–1373 (2001). [PubMed] [Google Scholar]
- Zerangue N. & Kavanaugh M. P. Flux coupling in a neuronal glutamate transporter. Nature 383, 634–637 (1996). [DOI] [PubMed] [Google Scholar]
- Gadea A. & Lopez-Colome A. M. Glial transporters for glutamate, glycine, and GABA: II. GABA transporters. J. Neurosci. Res. 63, 461–468 (2001). [DOI] [PubMed] [Google Scholar]
- Walker R. J. Transmitters and modulators in Mollusca (ed. Willows A. O. D. ) 279–485 (Academic, 1986). [Google Scholar]
- Romanova E. V., Rubakhin S. S. & Rozsa K. S. Behavioral changes induced by GABA-receptor agonists in Lymnaea stagnalis L. Gen. Pharmacol-Vasc. S. 27, 1067–1071 (1996). [DOI] [PubMed] [Google Scholar]
- Ferguson G. P. & Benjamin P. R. The whole-body withdrawal response of Lymnaea stagnalis. II. Activation of central motoneurones and muscles by sensory input. J. Exp. Biol. 158, 97–116 (1991). [DOI] [PubMed] [Google Scholar]
- Mathieu S., Sylvain D., Walden F., Catapane E. & Carroll M. GABA is an inhibitory neurotransmitter in ganglia of the bivalve mollusc, Crassostrea virginica. FASEB J. 28 (1 Supplement), 1059.4 (2014). [Google Scholar]
- Manzl C., Enrich J., Ebner H., Dallinger R. & Krumschnabel G. Copper-induced formation of reactive oxygen species causes cell death and disruption of calcium homeostasis in trout hepatocytes. Toxicology 196, 57–64 (2004). [DOI] [PubMed] [Google Scholar]
- Craig P. M., Wood C. M. & McClelland G. B. Oxidative stress response and gene expression with acute copper exposure in zebrafish (Danio rerio). Am. J. Physiol-Reg I. 293, R1882–R1892 (2007). [DOI] [PubMed] [Google Scholar]
- Basu N. et al. Heat shock protein genes and their functional significance in fish. Gene 295, 173–183 (2002). [DOI] [PubMed] [Google Scholar]
- Han D. M. R. et al. Proteomic Analysis of the Copper Ion-Induced Stress Response in a Human Embryonic Carcinoma Cell Line. Int. J. Toxicol. 31, 397–406 (2012). [DOI] [PubMed] [Google Scholar]
- Epel D. et al. Efflux transporters: Newly appreciated roles in protection against pollutants. Environ. Sci. Technol. 42, 3914–3920 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobna Z. et al. Metabolism of arsenic in human liver: the role of membrane transporters. Arch. Toxicol. 84, 3–16 (2010). [DOI] [PubMed] [Google Scholar]
- Long Y., Li Q., Wang Y. H. & Cui Z. B. MRP proteins as potential mediators of heavy metal resistance in zebrafish cells. Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol. 153, 310–317 (2011). [DOI] [PubMed] [Google Scholar]
- Bosnjak I. et al. Multidrug efflux transporters limit accumulation of inorganic, but not organic, mercury in sea urchin embryos. Environ. Sci. Technol. 43, 8374–8380 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard M., Baudrimont M., Boudou A. & Bourdineaud J. P. Induction of a multixenobiotic resistance protein (MXR) in the Asiatic clam Corbicula fluminea after heavy metals exposure. Aquat. Toxicol. 67, 347–357 (2004). [DOI] [PubMed] [Google Scholar]
- Bosnjak I. et al. First evidence of the P-glycoprotein gene expression and multixenobiotic resistance modulation in earthworm. Arh. Hig. Rada. Toksiko. 65, 67–75 (2014). [DOI] [PubMed] [Google Scholar]
- Jeong C. B. et al. Functional characterization of P-glycoprotein in the intertidal copepod Tigriopus japonicus and its potential role in remediating metal pollution. Aquat. Toxicol. 156, 135–147 (2014). [DOI] [PubMed] [Google Scholar]
- Amiard J., Amiardtriquet C., Barka S., Pellerin J. & Rainbow P. Metallothioneins in aquatic invertebrates: Their role in metal detoxification and their use as biomarkers. Aquat. Toxicol. 76, 160–202 (2006). [DOI] [PubMed] [Google Scholar]
- Choi C. Y., An K. W., Nelson E. R. & Habibi H. R. Cadmium affects the expression of metallothionein (MT) and glutathione peroxidase (GPX) mRNA in goldfish, Carassius auratus. Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol. 145, 595–600 (2007). [DOI] [PubMed] [Google Scholar]
- Ng T. Y. T., Rainbow P. S., Amiard-Triquet C., Amiard J. C. & Wang ,. W.-X. Metallothionein turnover, cytosolic distribution and the uptake of Cd by the green mussel Perna viridis. Aquat. Toxicol. 84, 153–161 (2007). [DOI] [PubMed] [Google Scholar]
- Lanston W. J., Bebianno M. J. & Burt G. R. Metal handling strategies in molluscs In Metal metabolism in aquatic environments (ed. Langston W. J. & Bebianno M. J. ) 219–283 (Springer, 1998). [Google Scholar]
- Call H. P. & Mucke I. History, overview and applications of mediated lignolytic systems, especially laccase-mediator-systems (Lignozym(R)-process). J. Biotechnol. 53, 163–202 (1997). [Google Scholar]
- Cantarella G., Galli C. & Gentili P. Free radical versus electron-transfer routes of oxidation of hydrocarbons by laccase/mediator systems catalytic or stoichiometric procedures. J. Mol. Catal. B: Enzym. 22, 135–144 (2003). [Google Scholar]
- Majeau J. A., Brar S. K. & Tyagi R. D. Laccases for removal of recalcitrant and emerging pollutants. Bioresour. Technol. 101, 2331–2350 (2010). [DOI] [PubMed] [Google Scholar]
- Falfushynska H. I., Gnatyshyna L. L. & Stoliar O. B. Effect of in situ exposure history on the molecular responses of freshwater bivalve Anodonta anatina (Unionidae) to trace metals. Ecotoxicol. Environ. Saf. 89, 73–83 (2013). [DOI] [PubMed] [Google Scholar]
- Zorita I., Ortiz-Zarragoitia M., Soto M. & Cajaraville M. P. Biomarkers in mussels from a copper site gradient (Visnes, Norway): An integrated biochemical, histochemical and histological study. Aquat. Toxicol. 78, S109–S116 (2006). [DOI] [PubMed] [Google Scholar]
- Hwang D.-S. et al. Molecular characterization and expression of vitellogenin (Vg) genes from the cyclopoid copepod, Paracyclopina nana exposed to heavy metals. Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol. 151, 360–368 (2010). [DOI] [PubMed] [Google Scholar]
- Shu Y. H. et al. Molecular characterization and expression pattern of Spodoptera litura (Lepidoptera: Noctuidae) vitellogenin, and its response to lead stress. J. Insect. Physiol. 55, 608–616 (2009). [DOI] [PubMed] [Google Scholar]
- Sappington T. W. & Raikhel A. S. Molecular characteristics of insect vitellogenins and vitellogenin receptors. Insect. Biochem. Molec. 28, 277–300 (1998). [DOI] [PubMed] [Google Scholar]
- Park K. & Kwak I.-S. Assessment of potential biomarkers, metallothionein and vitellogenin mRNA expressions in various chemically exposed benthic Chironomus riparius larvae. Ocean Sci. J. 47, 435–444 (2012). [Google Scholar]
- Taborsky G. Iron-binding by phosvitin and its conformational consequences. J. Biol. Chem. 255, 2976–2984 (1980). [PubMed] [Google Scholar]
- Ghosh P. & Thomas P. Binding of metals to red drum vitellogenin and incorporation into oocytes. Mar. Environ. Res. 39, 165–168 (1995). [Google Scholar]
- Zhong S. et al. Study on assessment and categorization for the eco-environmental quality of shellfish culture regions in Zhaoan Bay, Fujian province. J. Fujian. Fish. 34, 268–277 (2012). [Google Scholar]
- Wang H., Qian L., Liu X., Zhang G. & Guo X. Classification of a common cupped oyster from southern China. J. Shellfish Res. 29, 857–866 (2010). [Google Scholar]
- Kim D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priya T. A. J., Li F. H., Zhang J. Q., Yang C. J. & Xiang J. H. Molecular characterization of an ecdysone inducible gene E75 of Chinese shrimp Fenneropenaeus chinensis and elucidation of its role in molting by RNA interference. Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol. 156, 149–157 (2010). [DOI] [PubMed] [Google Scholar]
- Sim Y.-B. et al. Effect of GABA receptor agonists or antagonists injected spinally on the blood glucose level in mice. Neurochem. Res. 38, 1055–1062 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.