Abstract
Dendritic cells (DC) in the thymus have an important role in the establishment of central tolerance by promoting negative selection of autoreactive T cells and regulatory T-cell differentiation. Whereas human DC have recently been studied in various tissues in more detail, thymic DC subsets are still ill-defined. In the present work, we studied the binding of 71 monoclonal antibodies (mAb) submitted to the HLDA10 workshop to human CD123+ plasmacytoid DC and the two subsets of conventional DC (cDC, CD141+ and CD11b+) isolated from thymus tissue of infants undergoing corrective heart surgery. Within the panel, we found mAb binding to thymic pDC and both cDC subsets (for example, anti-Clec12A, TIM-3, Clec4A, CCR5, Axl, FLT3), but most of them additionally reacted with other thymic cell types. MAb directed to CD85h (ILT1) and the C-type lectin Clec7A (now CD369) reacted selectively with both cDC subsets, but not with other cells. Only one mAb directed to CD85g (ILT7) stained thymic pDC in a highly specific manner. Clec9A (DNGR1, now CD370) was the only tested HLDA10 antigen exclusively expressed on thymic CD141+ cDC. The present report summarizes all data obtained.
Dendritic cells (DC) in lymphoid and non-lymphoid tissues are professional antigen-presenting cells that are necessary for pathogen recognition and the initiation of primary T-cell immune responses.1, 2 Although DC in the thymus may also have a role in the protection against certain infections, it is the presentation of self-antigens to the developing T-cell pool that makes DC indispensable for the proper establishment of central tolerance. Thymic DC were not only shown to promote negative selection of autoreactive T cells, but may also participate in the induction of regulatory T cells (reviewed in Klein et al.3 and Hadeiba and Butcher4). As described for various peripheral organs, human thymic DC are most likely heterogeneous regarding their origin and ontogeny, anatomical localization, capacity to take up and process different forms of antigen, and production of various cytokines. It can be anticipated that these differences influence the responses of the maturing T cells interacting with them. Some of these functional differences of thymic DC populations will be reflected by a differential expression of cell surface molecules.
Similar to other tissues, human thymus contains two major types of DC: plasmacytoid DC (pDC, defined as HLA-DR+ cells expressing IL-3Rα (CD123), but lacking expression of CD11c) and conventional DC (cDC, expressing CD11c and high amounts of HLA-DR).5, 6 Within thymic cDC, differential expression of several cell surface molecules, like DC-LAMP, CD14 or CD11b, pointed to the existence of two different subsets also in the human thymus.5, 6, 7 However, an unequivocal classification of thymic cDC was lacking because of the absence of highly specific markers.
To be able to consistently classify human thymic cDC, we compared a number of surface molecules used in the past for cDC subset definition in the peripheral blood or tissues, including CD141 (BDCA-3, thrombomodulin) and CD1c (BDCA-1).8, 9, 10 In these comparisons, we also included molecules that specifically describe functionally different cDC subsets in various mouse tissues, namely XCR1 and SIRPα.11, 12 These comparisons revealed that human thymus also contains CD141+ cDC, which perfectly match the described CD11bneg thymic cDC.7 CD141+ CD11bneg thymic cDC express XCR1, but lack SIRPα (13and data not shown) and thus resemble ‘cross-presenting' DC in peripheral blood.14, 15 Further, we found that all human thymic CD11b+ cDC co-express SIRPα, and lack XCR1 expression, and thus phenotypically resemble human peripheral CD1c+ cDC (13and data not shown). Thus, human thymic cDC can consistently be subdivided into two subsets: CD141+ cDC and SIRPα+ cDC (which are congruent with CD11b+ cDC). For practical reason (monoclonal antibody (mAb) availability, greater flexibility for combination with various HLDA10 mAb formats), CD11b (and not SIRPα) together with CD141 was used for cDC classification in this study. The panel of 71 monoclonal antibodies from the 10th Human Leukocyte Differentiation Antigens workshop (HLDA10) was investigated for binding to freshly isolated human thymic pDC, CD141+ cDC or CD11b+ cDC. In this report, the results obtained are summarized.
Results
Because of the low abundance of DC in thymic tissue (approximately 0.2–0.3%), DC were enriched from digested human thymi by Nycodenz-gradient centrifugation to 1–4% pDC (defined as live CD123+ HLA-DR+ cells) and 6–10% cDC (defined as live CD123neg lineageneg HLA-DR+ CD11cint/high cells). The cDC were further subdivided into CD141+ or CD11b+ cDC (Figure 1). This enrichment allowed an efficient simultaneous analysis of all primary thymic DC subsets. At the same time, gating on lineageneg HLA-DR+ CD11cneg CD141neg CD123neg cells allowed to analyze other HLA-DR+ thymic cells, which were not DC (HLA-DR+ non-DC). Gating on HLA-DRneg, but lineage-positive cells encompassed mainly immature T cells, but also CD56+ NK cells. The results, summarized in Table 1, were highly reproducible, with each of the HLDA10 mAb giving comparable staining profiles on cells of all tested donors. Slight variations between single donors were observed only in the absolute fluorescence intensities or in the percentage of the positive fractions. On the basis of their staining profile, the HLDA10 mAb could be grouped into:
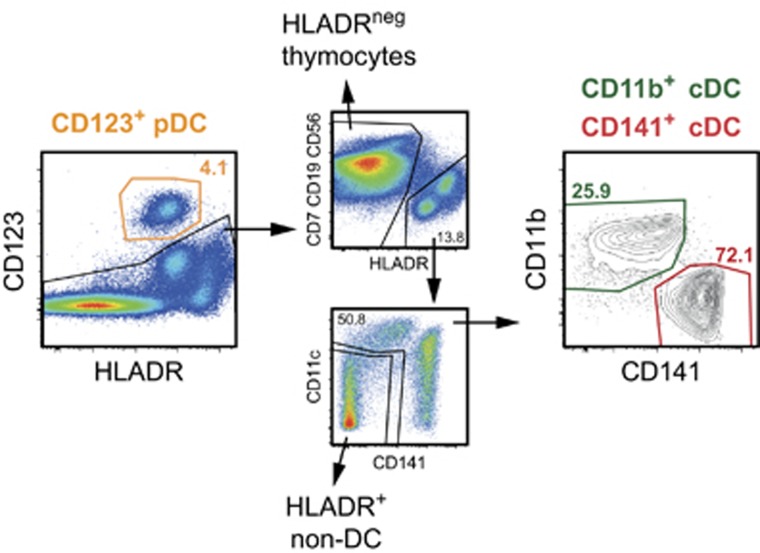
Figure 1.
Definition of primary human thymic DC subsets. Thymic CD123+ HLA-DR+ cells were defined as pDC, and CD123– lin– HLA-DR+ CD11cint/high cells were defined as cDC and further subdivided into CD141+ cDC and CD11b+ cDC. The inset numbers represent cell frequencies of one tissue donor. Prior to DC subset gating, gates were set on live cells defined by scatter characteristics and exclusion of propidium iodide-positive events. Doublets were excluded using forward scatter (FSC) and side scatter (SSC) height versus area characteristics.
Table 1. Reactivity of HLDA10 mAb to human thymic DC populations and other thymic cells.
| Antigen | Clone | Code | Label |
cDC |
pDC | HLADR+non-DC | HLADRneg thymocytes | |
|---|---|---|---|---|---|---|---|---|
| CD141 | CD11b | |||||||
| Axl | FAB154P | 10−50 | PE | ++ | ++ | −/+ | − | − |
| B7-H4 | MIH43 | 10−64 | PE | − | − | − | − | − |
| Calreticulin | FMU−CRT-2 | 10−23 | None | − | + | − | − | − |
| FMU−CRT-8 | 10−29 | None | − | −/+ | − | − | − | |
| FMU−CRT-17 | 10−42 | None (IgM) | − | − | − | − | − | |
| CD1a | 619 | 10−10 | None | +++ | +++ | − | +++ | +++ |
| 010e | 10−03 | None | +++ | +++ | − | +++ | +++ | |
| CD1b | O249 | 10−18 | None | ++ | +++ | − | ++ | +++ |
| CD1c | L161 | 10−26 | None | +++ | +++ | − | +++ | +++ |
| CD85g (ILT7) | 17G10.2 | 10−66 | PE | − | − | ++++ | − | − |
| CD85h (ILT1) | 24 | 10−74 | PE | ++ | +++ | − | − | − |
| CD101 | BB27 | 10−34 | None | ++ | +++ | − | − | +/++ |
| CD135 (Flt-3/Flk-2) | FAB812P | 10−15 | PE | +++ | ++ | +++ | − | − |
| CD195 (CCR5) | HEK/1/85 | 10−76 | PE | ++ | ++++ | ++++ | ++ | * |
| CD245 | DY12 | 10−43 | None | ++ | ++ | + | −/+ | −/+ |
| DY35 | 10−48 | None | −/+ | + | − | − | − | |
| CD273, B7-DC | ANC8D12 | 10−61 | Bio | − | ++ | − | + | − |
| Clec2D/OCIL | FAB3480P | 10−06 | PE | − | ++ | − | + | − |
| Clec4a (DCIR) | 111F8.04 | 10−71 | FITC | − | +++ | ++ | −/+ | − |
| 9E8 | 10−72 | PE | +++ | ++++ | +++ | ++ | + | |
| FAB1748P | 10−13 | PE | +++ | ++++ | +++ | ++ | + | |
| Clec4D (Dectin-3) | 9B9 | 10−78 | PE | − | ++ | − | − | − |
| FAB2806P | 10−21 | PE | − | ++ | − | − | − | |
| Clec5A/MDL-1 | FAB238P | 10−28 | PE | − | +++ | − | − | − |
| Clec5C/NKp80 | FAB1900P | 10−31 | PE | − | − | − | − | * |
| Clec7A (Dectin-1) | 15 E 2 | 10−79 | PE | ++++ | ++++ | − | − | − |
| FAB1859P | 10−35 | PE | ++++ | ++++ | − | − | − | |
| GE2 | 10−01 | Bio | ++ | +++ | − | − | − | |
| Clec8A/LOX-1 | FAB1798P | 10−40 | PE | − | + | + | + | − |
| Clec9A (DNGR1) | 8F9 | 10−02 | None | +++ | − | − | − | − |
| 8F9 | 10−65 | PE | ++++ | − | − | − | − | |
| 9A11 | 10−09 | Bio | ++++ | − | − | − | − | |
| FAB6049P | 10−45 | PE | ++++ | ++ | − | − | − | |
| Clec12A | 50C1 | 10−73 | PE | ++++ | ++++ | +++ | − | + |
| HB3 | 10−17 | None | ++++ | +++ | ++ | − | −/+ | |
| FAB2946P | 10−51 | PE | ++++ | ++++ | +++ | − | − | |
| Clec13A/CD302 | FAB637P | 10−54 | PE | − | +++ | − | − | − |
| Clec14A | FAB7436P | 10−57 | PE | + | − | − | − | − |
| DC-SIGN like | 118A8.05 | 10−83 | FITC | − | ++ | − | − | − |
| DORA | 104A10.01 | 10−77 | FITC | − | − | − | − | − |
| FAT1 cadherin | FMU−FAT1−7 | 10−16 | None | − | + | − | − | − |
| FMU−FAT−6 | 10−08 | None | − | −/+ | − | − | − | |
| FDF03 | 36H2 | 10−84 | FITC | − | +++ | − | − | − |
| FPR1 | FAB3744P | 10−47 | PE | − | +++ | − | − | − |
| FPRL1/FPRL/2 | FAB3479P | 10−36 | PE | − | ++ | − | − | − |
| GARP | ANC10G10 | 10−63 | Bio | − | − | − | − | − |
| ANC8C9 | 10−62 | Bio | − | − | − | − | + | |
| IL-1RAcP | AY19 | 10−53 | None | ++ | +++ | + | ++ | +++ |
| IL-13 Ra2 | FMU−IL−13RA2−14 | 10−41 | None | − | ++ | − | − | − |
| FMU−IL−13RA2−7 | 10−30 | None | − | + | − | − | − | |
| FMU−IL−13RA2−8 | 10−37 | None | − | + | − | −/+ | − | |
| LPAP | CL3 | 10−04 | None | − | −/+ | − | − | − |
| CL4 | 10−11 | None | − | −/+ | − | − | − | |
| CL7 | 10−19 | None | − | ++ | − | + | − | |
| MAIR II | TX45 | 10−80 | PE | + | ++++ | ++ | + | − |
| P2X7 | L4 | 10−70 | None | + | +++ | ++ | ++ | ++ |
| Tetanus toxoid | CMRF−81 | 10−85 | Bio | − | − | − | − | − |
| Tie-2 | FAB3131P | 10−56 | PE | − | ++ | − | − | − |
| TIM-1 | 1D12 | 10−67 | PE | − | −/+ | − | − | − |
| FAB1750P | 10−14 | PE | − | ++ | − | − | − | |
| Tim-3 | F38−2E2 | 10−75 | PE | ++++ | ++++ | +++ | ++ | + |
| FAB2365P | 10−24 | PE | ++++ | ++++ | ++++ | ++ | ++ | |
| Tim-4 | 9F4 | 10−81 | PE | − | − | − | − | − |
| Trem-2 | FAB17291P | 10−07 | PE | − | +++ | ++ | − | + |
| TSLP-R | 1B4 | 10−68 | PE | − | − | + | − | + |
| ULBP-3 | FAB1517P | 10−52 | PE | + | + | + | + | ++ |
| Unknown | BGA69 | 10−38 | None | + | + | − | − | − |
| Unknown | CMRF−44 | 10−82 | None (IgM) | + | ++ | − | +++ | − |
| Unknown | CMRF−56 | 10−69 | None | ++ | +++ | − | ++++ | + |
| Unknown | MDR64 | 10−59 | None | − | −/+ | − | − | − |
| Vimentin | SC5 | 10−55 | None (IgM) | + | ++ | − | +++ | − |
Abbreviations: cDC, conventional dendritic cells; DC, dendritic cells; FITC, fluorescein isothiocyanate; HLDA10, 10th Human Leukocyte Differentiation Antigens workshop; mAb, monoclonal antibody; pDC, plasmacytoid dendritic cells; PE, phycoerythrin.
− binding of the mAb was similar to negative control.
*a distinct subpopulation of 5–10% was stained by the mAb.
−/+ 10–25% of the respective cell subset was recognized/shifted by the mAb in one or two individuals only.
+ 10–25% of the respective cell subset was recognized/shifted by the mAb.
++ 25–60% of the cell subset was recognized/shifted by the mAb.
+++ 60–95% of the cell subset was recognized/shifted by the mAb.
++++ 95–100% of the cell subset was recognized the mAb.
Values represent the average of three thymus donors.
mAb binding to all primary thymic DC populations (pDC, CD141+ cDC and CD11b+ cDC);
mAb binding only to cDC but not to pDC;
mAb preferentially binding to a subpopulation of CD11b+ thymic cDC;
mAb exclusively binding to CD141+ cDC;
mAb exclusively binding to pDC;
mAb that gave only marginal staining on at least one DC population;
mAb that did not bind to any of the primary human thymic DC subsets.
HLDA10 mAb recognizing antigen expressed on all human thymic DC populations
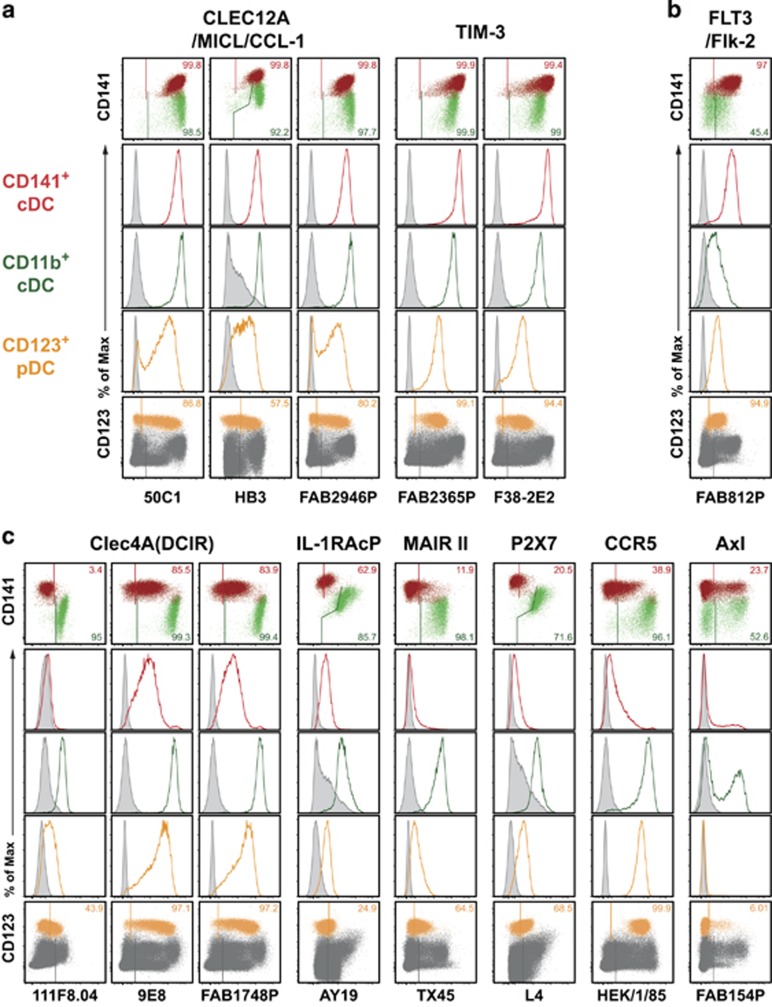
From all 71 tested HLDA10 mAb, 13 mAb bound their antigen on pDC and on both cDC subsets. Of these, Clec12A (now CD371, 3 test mAb) and TIM-3 (now CD366, 2 test mAb) showed a similar expression level on both cDC subsets and a lower expression on pDC (Figure 2a). The receptor tyrosine kinase FLT3 (CD135), reported to be required for DC development, was found to be one of the few HLDA10-defined molecules clearly expressed at higher levels on thymic CD141+ cDC, as compared with CD11b+ cDC (Figure 2b). In contrast, mAb recognizing either Clec4A (DCIR, now CD367), the interleukin-1 receptor accessory protein IL-1RacP (IL-1 R3), the myeloid-associated immunoglobulin-like receptor II (MAIR II), the purinergic receptor P2X7 or the chemokine receptor CCR5 (CD195) gave a clearly higher signal with CD11b+ thymic cDC. The mAb recognizing Axl receptor tyrosine kinase stained subpopulations of both cDC subsets and a small fraction of around 10% of pDC (Figure 2c). Of all HLDA10 mAb considered in this chapter, only the anti-FLT3 and anti-Axl mAb showed a selective binding to thymic DC, while all other mAb bound also other thymic cell populations (compare Table 1).
Figure 2.
HLDA10 mAb binding to both thymic cDC and pDC. A larger number of HLDA10 mAb recognized both cDC subsets and pDC, and to a variable extent also other thymic cell populations. Dot plots show stainings as overlays of CD141+ (red) versus CD11b+ (green) cDC (top row), or pDC (orange) within total thymocytes (dark gray) (bottom row). Inset numbers give the proportion of the respective DC populations stained. Gates were set according to background controls (FMO for fluorophore-labeled mAb, secondary reagent controls for IgM or biotinylated mAb or isotype controls for unlabeled IgG mAb). Histogram overlays (middle rows) show signals of the panel mAb obtained with the color-coded thymic DC populations (open histograms) in comparison with the background staining (filled-in gray). The molecules recognized by these mAb could be grouped into being (a) similarly expressed on both cDC subsets, (b) expressed to a higher degree on CD141+ cDC and (c) expressed to a higher degree on CD11b+ cDC. The reactivity with other thymic cell populations can be seen in Table 1. Shown are representative data from one out of three thymic tissues.
HLDA10 mAb recognizing antigen on both thymic cDC populations, but not on pDC
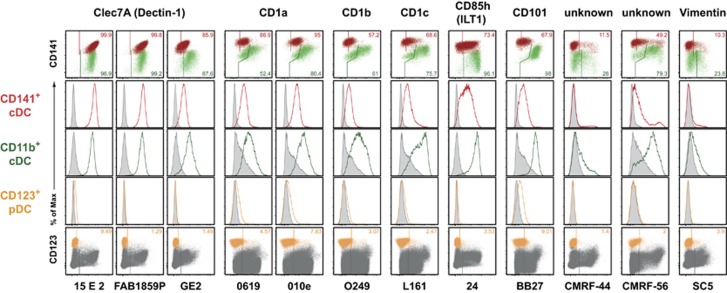
Twelve of the HLDA10 mAb were found to bind both thymic CD141+ and CD11b+ cDC populations, but not pDC (Figure 3). Expression of Clec7A (Dectin-1) could be detected exclusively on thymic cDC, and not on other thymic cell populations with all three HLDA10 mAb clones. Of these, clone GE2 (biotin-conjugated) gave a slightly stronger signal on CD11b+ thymic cDC. CD1a, CD1b and CD1c were broadly expressed on various thymic cell populations, with the exception of pDC (Figure 3 and Table 1). CD1a was similarly detected with two mAb clones on the majority of both CD141+ and CD11b+ cDC subsets. MAb recognizing CD1b, CD1c, CD85h (ILT1) or CD101 generally gave a higher signal with CD11b+ cDC compared with CD141+ DC. Interestingly, in one of three thymus samples, no staining of CD101 on CD141+ cDC was detectable, but still a strong expression on CD11b+ DC was detected. The mAb clones CMRF-44 and CMRF-56 (recognizing both unknown antigens), and also the anti-Vimentin mAb SC5, stained subpopulations on both cDC subsets, but did not stain pDC. The marginal signal obtained with mAb directed to CD1a and CD101 on pDC was interpreted as background; further studies would be warranted to clarify this point. In summary, of the mAb discussed in this chapter, only anti-Clec7A and CD85h mAb exclusively stained thymic cDC; all other mAb also stained other thymic cell populations (Table 1).
Figure 3.
HLDA10 mAb binding to both thymic cDC subsets but not to pDC. HLDA10 mAb reacting with cDC but not pDC. Dot plots show stainings as overlays of CD141+ (red) versus CD11b+ (green) cDC (top row), or pDC (orange) within total thymocytes (dark gray) (bottom row). Inset numbers give the proportion of the respective DC populations stained. Gates were set according to background controls (FMO for fluorophore-labeled mAb, secondary reagent controls for IgM or biotinylated mAb or isotype controls for unlabeled IgG mAb). Histogram overlays (middle rows) show signals of the panel mAb obtained with the color-coded thymic DC populations (open histograms) in comparison with the background staining (filled-in gray). The minimal signals with mAb to CD1a, CD1b, CD1c and CD101 on pDC were regarded as background. The reactivity with other thymic cell populations can be seen in Table 1. Shown are representative data from one out of three thymic tissues.
HLDA10 mAb preferentially recognizing antigen on a subpopulation of thymic CD11b+ cDC
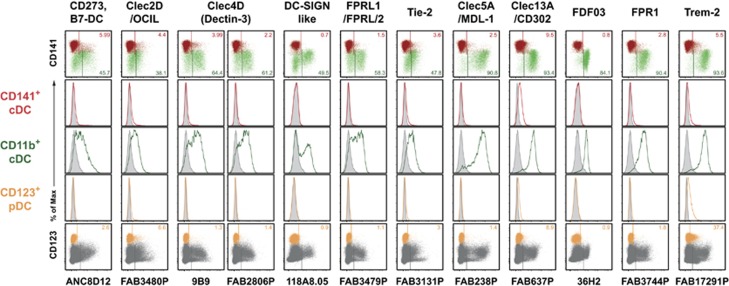
MAb of this category predominantly stained the CD11b+ thymic cDC subset. MAb directed to CD273 (B7-DC), Clec2D, Clec4D (Dectin-3, now CD368), DC-SIGN like, the formyl peptide receptor 2 FPRL1 and the tyrosine kinase Tie-2 only bound to a subpopulation of the CD11b+ subset (Figure 4). Expression of Clec5A (MDL-1), Clec13A (CD302), FDF03 (PILRα) and the formyl peptide receptor 1 (FPR1) could be detected on the large majority of CD11b+ cDC, but still not on the entire population. All of the mAb listed above selectively bound CD11b+ DC, with the exception of of mAb directed to CD273 and Clec2D, which in addition stained thymic HLA-DR+ non-DC (Table 1). Trem-2 (mAb FAB17291P) was found on almost all CD11b+ cDC, barely on CD141+ cDC, but also on a substantial subpopulation of pDC (Figure 4).
Figure 4.
HLDA10 mAb with preferential binding to CD11b+ cDC. A number of panel mAb recognized mainly or selectively molecules on CD11b+ cDC. Dot plots show stainings as overlays of CD141+ (red) versus CD11b+ (green) cDC (top row), or pDC (orange) within total thymocytes (dark gray) (bottom row). Inset numbers give the proportion of the respective DC populations stained. Gates were set according to background controls (FMO for fluorophore-labeled mAb, secondary reagent controls for IgM or biotinylated mAb or isotype controls for unlabeled IgG mAb). Histogram overlays (middle rows) show signals of the panel mAb obtained with the color-coded thymic DC populations (open histograms) in comparison with the background staining (filled-in gray). The reactivity with other thymic cell populations can be seen in Table 1. Shown are representative data from one out of three thymic tissues.
HLDA10 mAb recognizing antigen expressed exclusively on thymic CD141+ cDC
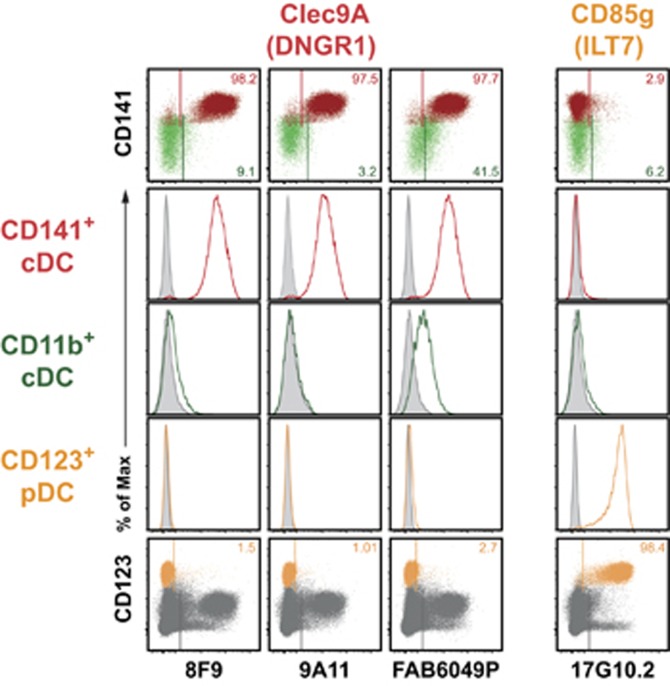
MAb clones 8F9 or 9A11, directed to the C-type lectin Clec9A (DNGR1, now CD370), were the only mAb of the HLDA10 panel selectively recognizing CD141+ thymic cDC (and no other thymic cells, Figure 5 and Table 1). The third anti-Clec9A clone FAB6049P in addition gave a signal with CD11b+ cDC (Figure 5). This discrepancy needs further evaluation.
Figure 5.
HLDA10 mAb with selective recognition of thymic CD141+ DC or pDC. Mab of the HLDA10 panel reacting with only one DC subset. Dot plots show stainings as overlays of CD141+ (red) versus CD11b+ (green) cDC (top row), or pDC (orange) within total thymocytes (dark gray) (bottom row). Inset numbers give the proportion of the respective DC populations stained. Gates were set according to background controls (FMO for fluorophore-labeled mAb, secondary reagent controls for IgM or biotinylated mAb or isotype controls for unlabeled IgG mAb). Histogram overlays (middle rows) show stainings of the HLDA10 mAb on the color-coded thymic DC populations (open histograms) in comparison with the background staining (filled-in gray). MAb 8F9 and 9A11 directed to Clec9A/DNGR1 selectively recognized CD141+ DC, while the third clone FAB6049P exhibited additional reactivity with CD11b+ cDC. One mAb (clone 17G10.2) directed to CD85g selectively stained pDC. Shown are representative data from one out of three thymic tissues.
HLDA10 mAb recognizing antigen expressed exclusively on pDC
Mab clone 17G10.2, specific for CD85g (ILT7), exclusively recognized thymic pDC. Other thymic cell populations, including cDC, did not give any signal (Figure 5 and Table 1).
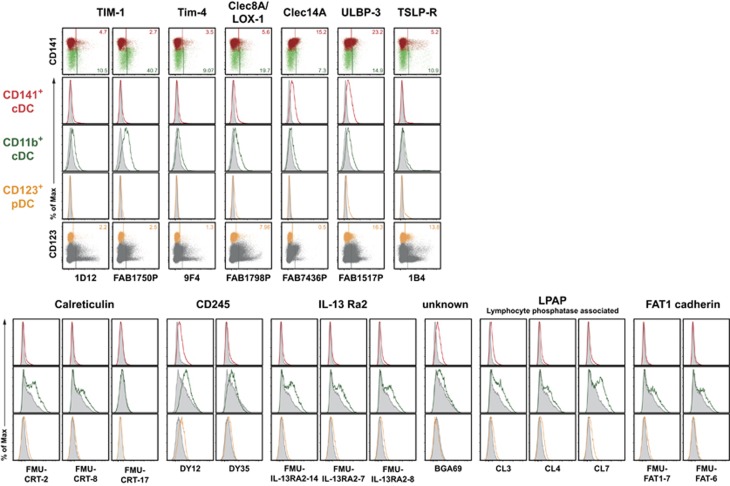
HLDA10 mAb giving only marginal staining with at least one DC population
Although the majority of the tested HLDA10 mAb was able to clearly detect cell surface molecules on human thymic DC, some HLDA10 mAb showed only week staining on at least one human thymic DC subset; mAb recognizing TIM-1 (HAVCR1, now CD365), TIM-4, Clec8A (LOX-1), Clec14A and ULBP-3 belonged to this group (Figure 6, upper row). Expression of the TSLP-R was detected only on minor subpopulations of both thymic cDC subsets (5–10% of each subset) and on approximately 20% of pDC (Figure 6, upper panels). For a number of HLDA10 mAb provided as unlabeled IgG antibodies (CD245, IL-13 Ra2, LPAP (lymphocyte phosphatase-associated) or FAT1 cadherin), specific binding could not easily be distinguished from background signal, especially on CD11b+ cDC (Figure 6, lower panels).
Figure 6.
HLDA10 mAb giving only marginal staining on at least one thymic DC population. MAb of the HLDA10 panel with low or unclear reactivity. Dot plots show stainings as overlays of CD141+ (red) versus CD11b+ (green) cDC (top row), or pDC (orange) within total thymocytes (dark gray) (bottom row). Inset numbers give the proportion of the respective DC populations stained. Gates were set according to background controls (FMO for fluorophore-labeled mAb, secondary reagent controls for IgM or biotinylated mAb or isotype controls for unlabeled IgG mAb). Histogram overlays (middle rows) show stainings of the HLDA10 mAb on the color-coded thymic DC populations (open histograms) in comparison with the background staining (filled-in gray). Shown are representative data from one out of three thymic tissues.
HLDA10 mAb not recognizing human thymic cells
The provided HLDA10 mAb directed to B7-H4 (clone MIH43), DORA (104A10.01), GARP (ANC10G10, ANC8C9), Tetanus toxoid (CMRF-81) and the mAb MDR64 (flow cytometry data not shown) did not stain any of the tested Nycodenz-enriched thymic cell populations. The anti-Clec5C mAb FAB1900P did stain HLA-DRneg thymocytes, but failed to bind to any of the primary thymic DC subsets (see Table 1).
Discrepancies in the staining pattern of HLDA10 mAb
In most cases, different HLDA10 mAb clones directed to the same antigen gave similar staining results. Nevertheless, a few exceptions were seen. For example, binding of the anti-Clec4A mAb clones 9E8 and FAB1748P (coupled to PE) resulted in a very bright staining of CD11b+ cDC and pDC, and also gave a clearly detectable signal with CD141+ cDC, whereas clone 111F8.04 (conjugated to fluorescein isothiocyanate) did not stain CD141+ cDC; in this case, the observed difference most likely results from a lower affinity of clone 111F8.04 and/or a lower brightness of fluorescein isothiocyanate (Figure 2c). The differences in the staining characteristics of the various Clec9A mAb clones (Figure 5) were discussed in the respective section. The anti-Clec12A clone FAB2946P selectively stained DC and no other tested cell population, while clones 50C1 and HB3 also recognized a small population of HLA-DRneg thymocytes. The unlabeled IgG mAb FMU-CRT-2 and FMU-CRT-8, which should recognize calreticulin, slightly stained CD11b+ DC, but the anti-calreticulin IgM clone FMU-CRT-17 did not stain any thymic cell population. Whether the listed discrepancies were due to differences in the avidity of given mAb clones for their antigen, resulted from the use of different secondary reagents, or simply reflected higher unspecific binding, require further evaluation.
Discussion
All results provided here were obtained with DC isolated by mechanical and enzymatical disruption of thymic tissue obtained from three children undergoing surgery for congenital heart defects at the age of 2–8 months; these infants were otherwise healthy. Nycodenz enrichment, performed because of the low frequency of DC in the thymus, allowed to analyze all DC subsets (pDC, CD141+ cDC and CD11b+ cDC) with the HLDA10 mAb in parallel. At the same time, this enrichment reduced the proportion of thymic lymphocytes (developing T cells, B and NK cells, all defined via lineage markers) in the preparation. The Nycodenz-enriched cell preparation also contained HLA-DR+ cells which were negative for CD11c, CD141 and CD123, and were thus not regarded as DC. As a general caveat, we cannot exclude that some small, higher-density thymic population was lost during the DC isolation and enrichment procedure used.
On the basis of the characterization of thymic DC subsets by Vandenabeele et al.,7 expression of CD123 was used to define pDC. As described in the introduction, CD141 and CD11b were used to subdivide thymic cDC, which were defined as CD123neg lineageneg HLA-DR+ CD11cint/high cells. It has been very recently shown that this phenotypic subdivision also reflects functional differences between these two human thymic cDC subsets.13
All flow cytometry stainings described in this report were carried out using a concentration of the tested mAb recommended by the HLDA10 workshop. This concentration may not give optimal stainings in terms of signal-to-noise ratio and specificity in all instances. Because of this technical limitation, a more detailed analysis of antigens that are apparently expressed at low levels is warranted.
Furthermore, we cannot be certain whether the expression pattern obtained with the HLDA10 panel on thymic DC from infants will completely match the pattern on adult thymocytes. In this context, however, it is interesting to note that the observed expression characteristics of several HLDA10 antigens resembled the pattern described for peripheral DC in adults (see references below).
A substantial proportion of the tested HLDA10 mAb not only bound thymic DC, but also other cells. This can be concluded for mAb recognizing CD1a, CD1b and CD1c, CD101, CD195 (CCR5), IL-1RacP, P2X7, TIM-3, Vimentin, and the unknown antigens recognized by mAb clones CMRF-44 and CMRF-56. Interestingly, CD1c, commonly used to demarcate peripheral CD141neg cDC,8, 9 could not be used for this purpose with thymic tissue. As exemplified with HLDA10 clone L161, a bright CD1c signal was obtained on thymic CD141neg CD11b+ cDC, but also CD141+ cDC were positive, as was the majority of all other tested thymic cell populations.
Expression of the thymic stromal lymphopoietin receptor (TSLP-R), which (by binding TSLP) enables thymic DC to induce regulatory T cells,16 was surprisingly found only on small subpopulations (5–10%) of both CD141+ and CD11b+ cDC, and in addition on approximately 20% of pDC. Recently, Martinez et al.13 obtained with the same mAb clone a bright staining of TSLP-R on the majority of BDCA3high (=CD141+) thymic cDC. Differences in the isolation procedure of thymic DC may only partially explain this discrepancy. Although tissue digestion (used by us) may have destroyed epitopes for mAb binding, isolation without digestion (used by Martinez et al.) led to a strongly reduced frequency of all thymic DC subsets, and may have resulted in the relative absence of TSLP-Rneg DC in the preparation. Of note, TSLP-R expression was also not detectable on the corresponding human peripheral blood DC subsets isolated without any digestion step17 (HLDA9, same mAb clone).
The staining of another set of analyzed HLDA10 mAb can be regarded as restricted to thymic cDC and pDC, because all other thymic populations were negative. Among these were two receptor tyrosine kinases, FLT3 (CD135) and Axl. In the bone marrow, FLT3 is expressed on hematopoietic progenitor cells, and is described to be required for DC development.18 In the spleen, FLT3 is expressed only on DC at steady state.19 In the human thymus, we were able to detect FLT3 expression only on DC subsets, with nearly all CD141+ cDC and pDC being positive. As recently reported for peripheral blood DC and lung DC,10 also, thymic CD141+ cDC showed significantly higher FLT3 levels compared with CD141neg CD11b+ (or CD1c+) DC. Axl kinase has been implicated in the clearance of apoptotic cells and in the facilitation of antigen cross-presentation by DC.20, 21 Expression of Axl on human thymic DC was not homogenous in that only subpopulations of the various subsets stained positive (approximately 30% of CD141+, 60% of CD11b+ and around 5–10% of pDC).
Clec7A (Dectin-1, now CD369) appears to be exclusively expressed on total thymic cDC, and this is consistent with previous reports of preferential expression by mouse and human DC.22, 23, 24 We found CD85h (ILT1, a member of the Ig-like transcripts family) to be similarly restricted to thymic cDC, which is congruent with a previous analysis of peripheral blood DC.25 Expression of CD85h was clearly higher on CD11b+ cDC as compared with CD141+ cDC.
The HLDA10 mAb recognizing Clec4D, Clec5A, Clec13A, DC-SIGN like, FDF03, FPR1 and FPRL1 were only detected on subpopulations of thymic CD11b+ cDC, so these molecules may be either candidates for the definition of new subpopulations of CD11b+ cDC, or they simply mark different activation/differentiation states within the same DC population. To address this issue, further experiments analyzing the transcriptional profile or specific functions of separated subpopulations will be required.
Few of the HLDA10 mAb tested specifically recognized only one thymic DC subset. CD85g (ILT7) was detected exclusively on thymic pDC, as already described for peripheral blood and tonsils.26, 27, 28 MAb recognizing Clec5A, Clec13A, FDF03 and FPR1 all stained a large majority of CD11b+ thymic cDC (each 60–90%, with variation between donors) and no other thymic population. However, none of the HLDA10 mAb was able to exclusively stain the entire CD11b+ cDC subset. The only mAb in the HLDA10 panel specifically recognizing CD141+ cDC were the clones 8F9 and 9A11, directed to Clec9A (now CD370). Clec9A/DNGR1 was previously shown to be expressed on cross-presenting DC in the mouse29 and on the homologous CD141+ cDC in human peripheral blood and various other tissues.10, 30, 31 The third Clec9A-specific clone FAB6049P, however, also exhibited significant binding to CD11b+ DC, and this issue needs further clarification.
In the human, Clec9A thus is a clear candidate for a lineage marker of cross-presenting DC. Other molecules known to be specifically and exclusively expressed on human CD141+ cDC (and their rhesus maqacue and mouse homologs) are the chemokine receptor XCR1 and the cell adhesion molecule CADM1. These three molecules can today be regarded as the best markers for cross-presenting DC in various tissues across various species. 12, 14, 32, 33, 34, 35
Methods
Cell isolation
Thymic tissue was obtained from newborns and infants up to the age of 8 months undergoing cardiac surgery for congenital heart defects and having no additional health issues. The analysis of DC in thymic tissues was approved by the Charité Ethics Committee. Individual tissues were used only after informed consent of the parents. Thymic tissue was cut into small pieces and digested for 20 min with collagenase D (500 μg ml−1) and DNase I (20 μg ml−1, both Roche, Mannheim, Germany) in RPMI 1640 containing 2% fetal calf serum (low endotoxin, Biochrom, Berlin, Germany) supplemented with 10 mM EDTA for the final 5 min of incubation. DC were further enriched by Nycodenz-gradient centrifugation (NycoPrep, Axis-Shield, Oslo, Norway). Cells were frozen in complete medium and 10% dimethyl sulfoxide, and stored in liquid nitrogen until use.
Flow cytometry
Detailed information on the HLDA10 mAb can be found at http://www.hcdm.org. For staining of cells, test mAb were used at concentrations recommended by the HLDA10 workshop. To define the DC subsets, antibodies directed to HLA-DR, CD11c, CD123, CD141 and CD11b were used, the lineage markers CD7 (or CD3), CD19 and CD56 were used for gating out developing T lymphocytes, B cells and NK cells. Before staining, cells were pre-incubated with 1 mg ml−1 human IgG (Endobulin, Baxter, Heidelberg, Germany) to block unspecific binding of mAb via the Fc-portion. After incubation with a biotinylated, unlabeled IgM, PE- or fluorescein isothiocyanate-coupled test mAb, cells were washed and stained with an antibody mix, which included the secondary reagent (PE-conjugated Streptavidin (SAv, eBioscience, San Diego, CA, USA) for biotinylated mAb, or mAb Bet-2 (ATCC HB-88, ATCC, Manassas, VA, USA), for unconjugated IgM mAb (Table 2). Bound unlabeled IgG test mAb were first reacted with goat anti-mouse IgG-Cy5 (Fcγ-specific, Dianova, Hamburg, Germany), washed, incubated with mouse gamma-globulin (Dianova) to block all free valancies of the secondary reagent, and then stained with the mAb-mix (Table 2). Background staining of unlabeled mAb was determined using the isotype control mAb MOPC-21 (IgG1, Sigma, St Louis, MO, USA), S43–10 (IgG2a36) or D3–13F1 (IgG2b, gift from K. Rajewsky). Staining was performed on ice according to standard methods.
Table 2. Staining scheme for testing HLDA10 mAb on primary human thymic DC.
| Type of test mAb | FITC | PE | PE-Cy7 | APC/Cy5 | A700 | PacB / BV421 | BV650 |
|---|---|---|---|---|---|---|---|
| mAb-Bio | CD7, CD19, CD56 | + SAv-PE | CD11c | CD141 | CD11b | HLA-DR | CD123 |
| mouse IgM mAb purified | CD7, CD19, CD56 | +α- IgM PE | CD11c | CD141 | CD11b | HLA-DR | CD123 |
| mAb-PE | CD7, CD19, CD56 | (mAb-PE) | CD11c | CD141 | CD11b | HLA-DR | CD123 |
| mAb-FITC | (mAb-FITC) | CD11b | CD11c | CD141 | HLA-DR | CD3, CD19 | CD123 |
| mouse IgG mAb purified/ascites | CD7, CD19, CD56 | CD11b | CD11c | +α-IgG Cy5 | HLA-DR | CD141 | CD123 |
Abbreviations: APC, antigen-presenting cell; DC, dendritic cells; FITC, fluorescein isothiocyanate; HLDA10, 10th Human Leukocyte Differentiation Antigens workshop; mAb, monoclonal antibody; PE, phycoerythrin.
Fluorophore-conjugated mAb recognizing human CD7 (clone CD7-6B7), CD56 (HCD56), CD11c (Bu15), CD123 (6H6) and CD141 (M80) were from BioLegend (San Diego, CA, USA), CD141 (AD5-14H12) from Miltenyi Biotec, Bergisch Gladbach, Germany. MAb recognizing human CD3 (OKT3, ATCC CRL-8001), CD11b (OKM1, ATCC CRL-8026), CD19 (BU1237), HLA-DR (L234, ATCC HB-55) or mouse IgM (Bet-2) were purified from hybridoma supernatants and coupled to PE, Pacific Blue or Alexa Fluor 700 (Molecular Probes, Eugene, OR, USA) by standard procedures.
For flow cytometry, analysis gates were set on live cells defined by scatter characteristics and exclusion of propidium iodide-positive events. Doublets were excluded using forward scatter (FSC) and side scatter (SSC) height versus area characteristics. Data were acquired on a LSR-Fortessa flow cytometer (BD Biosciences, San Jose, CA, USA) and analyzed using FlowJo software (Tree Star Inc., Ashland, OR, USA).
Acknowledgments
This work was supported by DFG grant Kr827/18–1 to RAK.
The authors declare no conflict of interest.
References
- 1Hart DN. Dendritic cells: unique leukocyte populations which control the primary immune response. Blood 1997; 90: 3245–3287. [PubMed] [Google Scholar]
- 2Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature 1998; 392: 245–252. [DOI] [PubMed] [Google Scholar]
- 3Klein L, Hinterberger M, Wirnsberger G, Kyewski B. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat Rev Immunol 2009; 9: 833–844. [DOI] [PubMed] [Google Scholar]
- 4Hadeiba H, Butcher EC. Thymus-homing dendritic cells in central tolerance. Eur J Immunol 2013; 43: 1425–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Bendriss-Vermare N, Barthelemy C, Durand I, Bruand C, Dezutter-Dambuyant C, Moulian N et al. Human thymus contains IFN-alpha-producing CD11c(-), myeloid CD11c(+), and mature interdigitating dendritic cells. J Clin Invest 2001; 107: 835–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Schmitt N, Cumont MC, Nugeyre MT, Hurtrel B, Barre-Sinoussi F, Scott-Algara D et al. Ex vivo characterization of human thymic dendritic cell subsets. Immunobiology 2007; 212: 167–177. [DOI] [PubMed] [Google Scholar]
- 7Vandenabeele S, Hochrein H, Mavaddat N, Winkel K, Shortman K. Human thymus contains 2 distinct dendritic cell populations. Blood 2001; 97: 1733–1741. [DOI] [PubMed] [Google Scholar]
- 8Dzionek A, Fuchs A, Schmidt P, Cremer S, Zysk M, Miltenyi S et al. BDCA-2, BDCA-3, and BDCA-4: three markers for distinct subsets of dendritic cells in human peripheral blood. J Immunol 2000; 165: 6037–6046. [DOI] [PubMed] [Google Scholar]
- 9Mittag D, Proietto AI, Loudovaris T, Mannering SI, Vremec D, Shortman K et al. Human dendritic cell subsets from spleen and blood are similar in phenotype and function but modified by donor health status. J Immunol 2011; 186: 6207–6217. [DOI] [PubMed] [Google Scholar]
- 10Haniffa M, Shin A, Bigley V, McGovern N, Teo P, See P et al. Human tissues contain CD141hi cross-presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity 2012; 37: 60–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Becker M, Güttler S, Bachem A, Hartung E, Mora A, Jäkel A et al. Ontogenic, phenotypic, and functional characterization of XCR1+ dendritic cells leads to a consistent classification of intestinal dendritic cells based on the expression of XCR1 and SIRPα. Front Immunol 2014; 5: 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Gurka S, Hartung E, Becker M, Kroczek RA. Mouse conventional dendritic cells can be universally classified based on the mutually exclusive expression of XCR1 and SIRPα. Front Immunol 2015; 6: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Martinez VG, Canseco NM, Hidalgo L, Valencia J, Entrena A, Fernandez-Sevilla LM et al. A discrete population of IFN λ-expressing BDCA3hi dendritic cells is present in human thymus. Immunol Cell Biol 2015; 93: 673–678. [DOI] [PubMed] [Google Scholar]
- 14Bachem A, Guttler S, Hartung E, Ebstein F, Schaefer M, Tannert A et al. Superior antigen cross-presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J Exp Med 2010; 207: 1273–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Crozat K, Guiton R, Contreras V, Feuillet V, Dutertre CA, Ventre E et al. The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8α+ dendritic cells. J Exp Med 2010; 207: 1283–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Watanabe N, Wang YH, Lee HK, Ito T, Wang YH, Cao W et al. Hassall's corpuscles instruct dendritic cells to induce CD4+CD25+ regulatory T cells in human thymus. Nature 2005; 436: 1181–1185. [DOI] [PubMed] [Google Scholar]
- 17Ding Y, Ju X, Azlan M, Hart DN, Clark GJ. Screening of the HLDA9 panel on peripheral blood dendritic cell populations. Immunol Lett 2011; 134: 161–166. [DOI] [PubMed] [Google Scholar]
- 18Waskow C, Liu K, Darrasse-Jeze G, Guermonprez P, Ginhoux F, Merad M et al. The receptor tyrosine kinase Flt3 is required for dendritic cell development in peripheral lymphoid tissues. Nat Immunol 2008; 9: 676–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Karsunky H, Merad M, Cozzio A, Weissman IL, Manz MG. Flt3 ligand regulates dendritic cell development from Flt3+ lymphoid and myeloid-committed progenitors to Flt3+ dendritic cells in vivo. J Exp Med 2003; 198: 305–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Seitz HM, Camenisch TD, Lemke G, Earp HS, Matsushima GK. Macrophages and dendritic cells use different Axl/Mertk/Tyro3 receptors in clearance of apoptotic cells. J Immunol 2007; 178: 5635–5642. [DOI] [PubMed] [Google Scholar]
- 21Subramanian M, Hayes CD, Thome JJ, Thorp E, Matsushima GK, Herz J et al. An AXL/LRP-1/RANBP9 complex mediates DC efferocytosis and antigen cross-presentation in vivo. J Clin Invest 2014; 124: 1296–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Ariizumi K, Shen GL, Shikano S, Xu S, Ritter R 3rd, Kumamoto T et al. Identification of a novel, dendritic cell-associated molecule, dectin-1, by subtractive cDNA cloning. J Biol Chem 2000; 275: 20157–20167. [DOI] [PubMed] [Google Scholar]
- 23Yokota K, Takashima A, Bergstresser PR, Ariizumi K. Identification of a human homologue of the dendritic cell-associated C-type lectin-1, dectin-1. Gene 2001; 272: 51–60. [DOI] [PubMed] [Google Scholar]
- 24Hermanz-Falcon P, Arce I, Roda-Navarro P, Fernandez-Ruiz E. Cloning of human DECTIN-1, a novel C-type lectin-like receptor gene expressed on dendritic cells. Immunogenetics 2001; 53: 288–295. [DOI] [PubMed] [Google Scholar]
- 25Cella M, Jarrossay D, Facchetti F, Alebardi O, Nakajima H, Lanzavecchia A et al. Plasmacytoid monocytes migrate to inflamed lymph nodes and produce large amounts of type I interferon. Nat Med 1999; 5: 919–923. [DOI] [PubMed] [Google Scholar]
- 26Rissoan MC, Duhen T, Bridon JM, Bendriss-Vermare N, Peronne C, de Saint Vis B et al. Subtractive hybridization reveals the expression of immunoglobulin-like transcript 7, Eph-B1, granzyme B, and 3 novel transcripts in human plasmacytoid dendritic cells. Blood 2002; 100: 3295–3303. [DOI] [PubMed] [Google Scholar]
- 27Ju XS, Hacker C, Scherer B, Redecke V, Berger T, Schuler G et al. Immunoglobulin-like transcripts ILT2, ILT3 and ILT7 are expressed by human dendritic cells and down-regulated following activation. Gene 2004; 331: 159–164. [DOI] [PubMed] [Google Scholar]
- 28Cabezon R, Sintes J, Llinas L, Benitez-Ribas D. Analysis of HLDA9 mAbs on plasmacytoid dendritic cells. Immunol Lett 2011; 134: 167–173. [DOI] [PubMed] [Google Scholar]
- 29Sancho D, Mourao-Sa D, Joffre OP, Schulz O, Rogers NC, Pennington DJ et al. Tumor therapy in mice via antigen targeting to a novel, DC-restricted C-type lectin. J Clin Invest 2008; 118: 2098–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30Huysamen C, Willment JA, Dennehy KM, Brown GD. CLEC9A is a novel activation C-type lectin-like receptor expressed on BDCA3+ dendritic cells and a subset of monocytes. J Biol Chem 2008; 283: 16693–16701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31Jongbloed SL, Kassianos AJ, McDonald KJ, Clark GJ, Ju X, Angel CE et al. Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J Exp Med 2010; 207: 1247–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32Contreras V, Urien C, Guiton R, Alexandre Y, Vu Manh TP, Andrieu T et al. Existence of CD8α-like dendritic cells with a conserved functional specialization and a common molecular signature in distant mammalian species. J Immunol 2010; 185: 3313–3325. [DOI] [PubMed] [Google Scholar]
- 33Crozat K, Tamoutounour S, Vu Manh TP, Fossum E, Luche H, Ardouin L et al. Cutting edge: expression of XCR1 defines mouse lymphoid-tissue resident and migratory dendritic cells of the CD8α+ type. J Immunol 2011; 187: 4411–4415. [DOI] [PubMed] [Google Scholar]
- 34Bachem A, Hartung E, Güttler S, Mora A, Zhou X, Hegemann A et al. Expression of XCR1 characterizes the Batf3-dependent lineage of dendritic cells capable of antigen cross-presentation. Front Immunol 2012; 3: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35Dutertre CA, Jourdain JP, Rancez M, Amraoui S, Fossum E, Bogen B et al. TLR3-responsive, XCR1+, CD141(BDCA-3)+/CD8α+-equivalent dendritic cells uncovered in healthy and simian immunodeficiency virus-infected rhesus macaques. J Immunol 2014; 192: 4697–4708. [DOI] [PubMed] [Google Scholar]
- 36Reth M, Hämmerling GJ, Rajewsky K. Analysis of the repertoire of anti-NP antibodies in C57BL/6 mice by cell fusion. I. Characterization of antibody families in the primary and hyperimmune response. Eur J Immunol 1978; 8: 393–400. [DOI] [PubMed] [Google Scholar]
- 37Flavell DJ, Flavell SU, Boehm DA, Emery L, Noss A, Ling NR et al. Preclinical studies with the anti-CD19-saporin immunotoxin BU12-SAPORIN for the treatment of human-B-cell tumours. Br J Cancer 1995; 72: 1373–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]